Abstract
The RNA-editing enzyme ADAR1 is a double-stranded RNA (dsRNA) binding protein that modifies cellular and viral RNA sequences by adenosine deamination. ADAR1 has been demonstrated to play important roles in embryonic erythropoiesis, viral response, and RNA interference. In human hepatitis virus infection, ADAR1 has been shown to target viral RNA and to suppress viral replication through dsRNA editing. It is not clear whether this antiviral effect of ADAR1 is a common mechanism in response to viral infection. Here, we report a proviral effect of ADAR1 that enhances replication of vesicular stomatitis virus (VSV) through a mechanism independent of dsRNA editing. We demonstrate that ADAR1 interacts with dsRNA-activated protein kinase PKR, inhibits its kinase activity, and suppresses the α subunit of eukaryotic initiation factor 2 (eIF-2α) phosphorylation. Consistent with the inhibitory effect on PKR activation, ADAR1 increases VSV infection in PKR+/+ mouse embryonic fibroblasts; however, no significant effect was found in PKR−/− cells. This proviral effect of ADAR1 requires the N-terminal domains but does not require the deaminase domain. These findings reveal a novel mechanism of ADAR1 that increases host susceptibility to viral infection by inhibiting PKR activation.
ADAR1 modifies the coding and noncoding sequences of cellular and viral RNAs by deamination that converts adenosine to inosine. ADAR1 is proposed to play a role in host defense mechanisms because it is induced by interferon and viral infection (20, 21). Extensive adenosine-to-inosine modifications have been observed in viral RNA during the late stages of polyomavirus infection (13). Biased hypermutations were found in single-stranded RNA viral genomes during lytic and persistent infections (3). In measles infection, the biased hypermutation events are associated with a change in viral pathogenesis and with persistent infection (4). Recently, ADAR1 has been shown to inhibit human hepatitis virus replication and to compromise virus viability as a potential antiviral mechanism (9, 12, 28). This antiviral function is ascribed to the editing activity of ADAR1, which modifies viral RNA. Nevertheless, ADAR1 is also a typical double-stranded RNA (dsRNA) binding protein (DRBP). It comprises two putative Z-DNA binding domains (Z-DBD) and three dsRNA binding repeats. Notably, DRBPs are commonly known to stabilize homo- or heterotypic protein-protein interactions and often mediate regulatory functions through the dsRNA-activated protein kinase (PKR) during viral infections (11, 25). We have previously shown that ADAR1 interacts with nuclear factor 90 (NF90) through cellular dsRNA components and affects NF90-mediated gene expression (16). NF90 is also a DRBP that interacts with and is phosphorylated by PKR in vitro and in vivo (18, 19). As PKR is a signal transducer of both translation and transcription in antiviral and stress responses (31), we propose that ADAR1 participates in mediation of viral and stress responses by mediating PKR-mediated signaling, in addition to directly modifying viral RNA.
MATERIALS AND METHODS
ADAR1 constructs.
The constructs of ADAR1 variants were made by PCR. To facilitate cloning, the XhoI cleavage site was added to the 5′ primer and the BamHI site was added to the 3′ primer. The ADAR1 cDNA (GenBank accession no. AF291050) was used as a template to amplify the fragments of ADAR1 LF150 (1 to 3459), ADAR1 SF110 (748 to 3459), ADAR1 SF80 (1561 to 3459), and Dcat (1 to 2263). Each fragment was cleaved with BamHI and XhoI and subcloned into the pcDNA3.1-His-Myc vector (Invitrogen) or the pBABE-hygromycin vector (15) for transient or stable expression, respectively. All of the constructs were sequenced in the Yale Keck Facilities. Protein expression was confirmed in 293T or NIH 3T3 cells and detected by immunoblotting using the antibody against His or Myc epitopes.
Cell lines.
The cell lines used in this study include HEK 293T, NIH 3T3, GP+E86, gastric cancer SGC7901, and neuroblastoma (N18). PKR−/− and PKR+/+ mouse embryonic fibroblasts (MEFs) were from Bryan Williams' laboratory. Stable cells expressing the ADAR1 variants were established as follows. The pBABE-ADAR1 constructs were transfected into phoenix packaging cells. Twenty-four hours after transfection, the culture medium containing the recombinant viruses was directly used to infect NIH 3T3 cells and PKR+/+ and PKR−/− MEFs. Stable cells were harvested after selection with 2 μg/ml of hygromycin. Expression of the ADAR1 variants was confirmed by immunoblotting.
Immunoprecipitation.
The double-tagged ADAR1 variants were transiently expressed in HEK 293T or mouse N18 cells. Cells (1 × 107) were transfected with His-Myc-tagged LF150, SF110, SF80, or vector using Lipofectamine 2000 (Invitrogen) for 32 h. The cells were lysed in buffer containing 150 mM NaCl, 25 mM Tris-HCl, pH 7.45, 0.5% Nonidet P40, 0.1% sodium dodecyl sulfate (SDS), 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride (pH 7.5), 10 U/ml RNase A, and a protease inhibitor cocktail (Roche). Cell lysates were cleared with rabbit or mouse immunoglobulin G (IgG)-immobilized protein A-Sepharose for 1 h at 4°C and immunoprecipitated with anti-His, anti-PKR, anti-NF90, or anti-RNA helicase p68 (RHp68) antibody-coated protein A-Sepharose (Amersham) at 4°C overnight. The agarose beads were extensively washed with high-stringency washing buffer containing 0.1% SDS to thoroughly remove nonspecific proteins. The associated proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) or immunoblotting.
RNase V1 digestion.
The immunoprecipitated proteins on the agarose beads were washed twice with RNase V1 digestion buffer (Ambion) and resuspended in the same buffer containing 0 to 0.4 U/μl of RNase V1 (Ambion). The samples were incubated at 37°C for 25 min and quickly pulled down by centrifugation. The beads containing the still-bound proteins and the supernatant containing the released proteins were mixed with the loading buffer, heated to 95°C for 5 min, and resolved on SDS-PAGE. The blots were analyzed by immunoblotting.
Immunoblotting.
293T cells were transfected with pcDNA3.1-myc-His-ADAR1 variants using Lipofectamine 2000 (Invitrogen) for 48 h. Cell lysates were prepared as previously described. Eighty to 120 μg of proteins were mixed with 2× protein-loading buffer (0.5 M Tris · HCl, pH 6.8, 4% SDS, and 100 mM dithiothreitol), heated to 95°C for 5 min, resolved on 4 to 20% SDS-PAGE, and transferred onto nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk and incubated with the primary antibodies overnight at 4°C. After being washed with phosphate-buffered saline (PBS), the membrane was incubated with an appropriate horseradish peroxidase-conjugated secondary antibody (Jackson Immunoresearch) for 1.5 h at room temperature. Protein bands were detected using Western Lightning Chemiluminescence Reagent (Perkin-Elmer). Typically, the membrane was first used to detect the phosphorylated proteins. It was stripped and reused to detect other proteins. The films were scanned, and the intensities of signals were quantitated (Scion Image). The primary antibodies against the following proteins were used: PKR, phosphorylated PKR (PT451; BioSource International), the α subunit of eukaryotic initiation factor 2 (eIF-2α) (Santa Cruz Biotechnology), phosphorylated eIF-2α (p-eIF-2α) (Ser51) (Cell Signaling Technology), mouse ADAR1 (developed in our laboratory), human ADAR1 (a gift from Brenda Bass, Utah University), NF90 (a gift from Peter Kao, Stanford University), RHp68 (a gift from Ralf Janknecht, Mayo Clinic, Rochester, NY), His, Myc (Santa Cruz Biotechnology), and β-actin (Sigma). The secondary antibodies were donkey anti-rabbit or donkey anti-mouse horseradish peroxidase-linked IgG (Jackson Immunoresearch).
siRNA.
For transfection, two complementary oligoribonucleotides were chemically synthesized and annealed to form a small interfering RNA (siRNA) against human ADAR1 (from 426 to 406; GenBank accession no. NM-001111). Validated siRNA against a bacterial RNA was used as a negative control (Ambion). Cells were grown to ∼80% of confluence, distributed in six-well plates at 2.5 × 106 cells/well, and allowed to adhere overnight. Typically, siRNA was mixed with Lipofectamine to transfect 293T cells at a final concentration of 10 nM. The cells were collected 48 h after transfection. Western blotting was performed to confirm the efficiency of silencing.
VSV infection and quantitation.
NIH 3T3, GP+E86, PKR−/−, or PKR+/+ MEFs were seeded in 24-well plates for 8 h at 1.5 × 105 cells/well. Recombinant vesicular stomatitis virus (VSV)-EGFP1 viruses (22) were added to the media at a multiplicity of infection (MOI) of 10 PFU/cell; GPE+86 cells were infected at an MOI of 100. The viruses were allowed to adsorb for 30 to 60 min. The cells were washed with warm PBS twice to remove excess viruses and continuously cultured for 12 h in Dulbecco's modified Eagle's medium with 5% fetal bovine serum. Cells infected with VSV-EGFP1 were visualized under fluorescence microscopy. To quantitate the titer, culture media were collected and passed through a 0.2-μm filter. Tenfold-dilution series were made with PBS to infect semiconfluent NIH 3T3 cells. Green fluorescent protein-positive plaques were counted under fluorescence microscopy 12 h postinfection, and the viral titer was calculated accordingly.
RESULTS AND DISCUSSION
ADAR1 interacts with PKR.
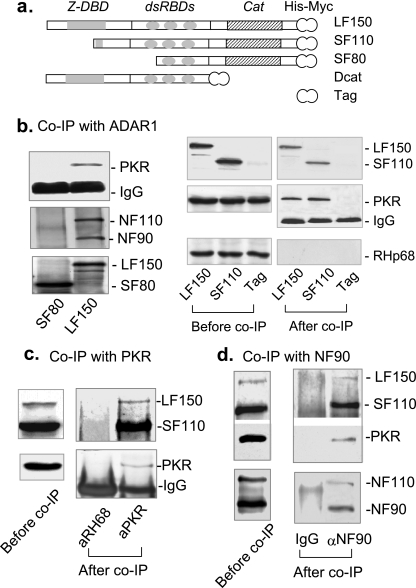
To test the postulated interaction with PKR, the long-form ADAR1 (LF150) and a short fragment (SF80) were tagged with the Myc-His epitope at their C termini and transiently expressed in human 293T cells (Fig. 1a). SF80 lacks the Z-DBD and the first dsRNA-binding domain (dsRBD). Cell lysates were immunoprecipitated with anti-His antibody, and the associated proteins were analyzed by immunoblotting using the antibodies against PKR. The NF90 proteins were used as positive controls (16). We found that PKR was efficiently immunoprecipitated by LF150 (Fig. 1b, left). SF80 did not interact with either the PKR or NF90 protein, which served as negative controls (16). To confirm the interaction, another short fragment of ADAR1 (SF110) and the Myc-His epitope were transiently expressed in 293T cells, immunoprecipitated, and compared. Similar to SF80, SF110 lacks the Z-DBD but still contains the intact dsRBD repeats. SF110 efficiently immunoprecipitated PKR, whereas the Myc-His tag did not (Fig. 1b, right). The specificity was supported by RHp68, another RNA binding protein (10), which was not coprecipitated. These results suggest that ADAR1 specifically interacts with PKR and that the dsRBD is required for the interaction. The same conclusion was obtained when mouse N18 cells were used (data not shown). Further experiments were performed to examine whether ADAR1 and PKR were reciprocally precipitated under endogenous conditions. Human SGC7901 cells were used for their higher levels of endogenous LF150. Whole-cell extracts were immunoprecipitated using the anti-PKR antibody. Endogenous ADAR1 LF150 and SF110 were efficiently coimmunoprecipitated by the PKR antibody (Fig. 1c). As a negative control, endogenous ADAR1 was not immunoprecipitated by the antibody against RHp68. Finally, we confirmed that NF90 coimmunoprecipitated both ADAR1 and PKR, suggesting that ADAR1, PKR, and NF90 are spontaneously associated in a complex (Fig. 1d). Thus, the specificity of the interaction between ADAR1 and PKR was supported by immunoprecipitation in three independent experiments.
FIG. 1.
Interaction between ADAR1 and PKR. (a) Diagram of naturally existing ADAR1 variants. LF150 is the full-length ADAR1 (1 to 3459). SF110, SF80, and Dcat (748 to 3459, 1561 to 3459, and 1 to 2263, respectively) are the truncated forms of ADAR1. Cat, catalytic domain; Tag, the His and Myc epitopes. (b) Immunoprecipitation with ADAR1. On the left, 293T cells were transfected with the His-Myc-tagged LF150 or SF80. Cell extracts were immunoprecipitated (IP) with anti-His antibody; the associated proteins were analyzed by immunoblotting using the antibodies against PKR and NF90. On the right, cells were transfected with LF150, SF110, or the vector (Tag; negative control). The immunoprecipitated proteins were analyzed using the antibody against PKR, RHp68 (negative control), or the Myc epitope (positive control). (c) Immunoprecipitation with PKR. Cell lysates were prepared from human SGC7901. Immunoprecipitation was performed using the immobilized antibody against PKR or RHp68. RHp68 was used as a negative control to exclude nonspecific interaction with other proteins. The associated proteins were detected by immunoblotting using the antibody against PKR (BioSource International) or the C-terminal end of the human ADAR1. Note: due to the poor efficiency of the anti-PKR antibody used for detection, these data might not correctly reflect the real molar ratio between ADAR1 and PKR. (d) Immunoprecipitation with NF90. Cell lysates were prepared from 293T cells without transfection. Coimmunoprecipitation was performed using the immobilized antibody against human immunoglobulin G (IgG) (negative control) or NF90. The associated proteins were analyzed by immunoblotting using the antibody against NF90 (positive control), PKR, or the C-terminal end of the human ADAR1.
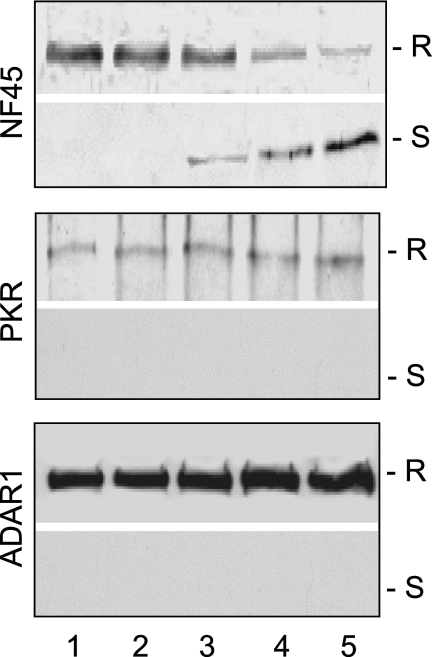
As ADAR1 interacts with NF90 and NF110 through cellular dsRNA components (16), it was essential to establish whether dsRNA was also involved in the interaction between ADAR1 and PKR. This question was addressed by RNase V1 digestion of the ADAR1 complex. RNase V1 specifically digests dsRNA and releases dsRNA-bridged proteins from the complex. We found that RNase V1 did not affect the interaction between PKR and ADAR1 (Fig. 2). In contrast, the NF45 proteins were quantitatively released from the complex in a concentration-dependent manner. These results suggest that ADAR1 closely interacts with PKR but is loosely connected to the NF90 proteins through cellular dsRNA. Because the dsRNA-binding domains usually facilitate protein-protein interactions, these data are consistent with the domains being important for the interactions between ADAR1 and PKR.
FIG. 2.
Analysis of dsRNA components in the ADAR complex. Proteins associated with ADAR1 p150 were digested with 0, 0.006, 0.025, 0.1, or 0.4 units/μl of RNase V1 (lanes 1 to 5). Protein complexes resistant (R) or sensitive (S) to RNase V1 digestion were analyzed by immunoblotting using antibodies against PKR, ADAR1, or NF45, respectively.
ADAR1 inhibits PKR activation.
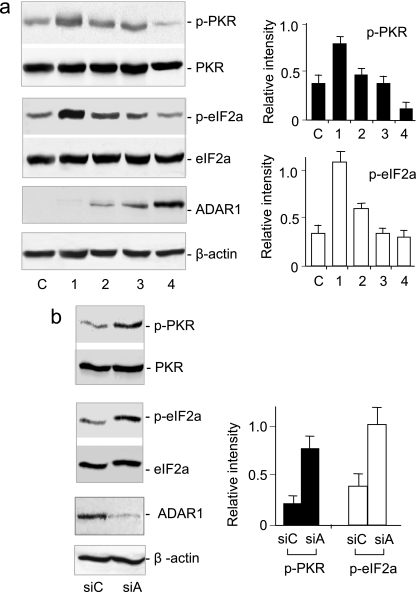
PKR, an important signal transducer, is activated by autophosphorylation in response to viral infection (31). Our findings suggest that ADAR1 might affect PKR phosphorylation. To test this speculation, we examined the effect of ADAR1 on the status of PKR phosphorylation in human 293T cells. Surprisingly, while the total PKR levels were unaffected by LF150, the phosphorylated PKR was downregulated as LF150 expression increased (Fig. 3a). Because the total PKR did not vary, immunoreactivity to the phosphorylated PKR represented the status of PKR activation. Thus, in contrast to the antiviral function by editing of viral RNA, these results suggest a potential proviral effect of ADAR1 that inhibits PKR phosphorylation and suppresses PKR activation. Next, we examined PKR phosphorylation in 293T cells by knockdown of endogenous ADAR1 using siRNA. A control siRNA against a bacterial gene was used as a reference to eliminate possible nonspecific siRNA effects (26). Although ADAR1 was only partially silenced, the phosphorylated PKR increased according to ADAR1 knockdown (Fig. 3b). Again, total PKR was not affected, indicating that PKR activation increased. These data consistently indicated that ADAR1 expression was inversely proportional to PKR activation.
FIG. 3.
ADAR1 inhibits the phosphorylation of PKR and eIF-2α. (a) 293T cells were transfected with increasing amounts of pcDNA3-ADAR1 LF150 plasmid by mixing them with the pcDNA3 vector in a ratio of 8:0, 6:2, 2:6, or 0:8 (μg:μg) (lanes 1 to 4). Lane c, 293T cells without transfection. Transiently expressed ADAR1 LF150 was detected by immunoblotting using the antibody against mouse ADAR1. (b) ADAR1 expression in human 293T cells was reduced with siRNA against ADAR1 (siA) or a control bacterial gene (siC). The phosphorylated PKR and total PKR, p-eIF-2α and total eIF-2α, and ADAR1 LF150 and β-actin were compared by immunoblotting using proper antibodies against human proteins (left). The intensity was digitalized with Scion Image software and normalized to total PKR, eIF-2α, or β-actin, respectively (right). The error bars represent standard deviations.
To confirm the inhibitory effect, we examined whether ADAR1 inhibits the downstream signaling of PKR activation. The eIF-2α protein is a well-studied cellular substrate of PKR that is phosphorylated to shut down protein synthesis during viral infection (6, 29). 293T cells were transfected with LF150, and the phosphorylation of eIF-2α was measured using specific antibodies. ADAR1 progressively inhibited the phosphorylation of eIF-2α in a concentration-dependent manner (Fig. 3a). Again, the total amounts of eIF-2α were not affected. These data showed that ADAR1 suppressed PKR activation and consequently inhibited the downstream signaling. In support, the knockdown of endogenous ADAR1 correspondingly increased eIF-2α phosphorylation (Fig. 3b).
ADAR1 increases viral replication.
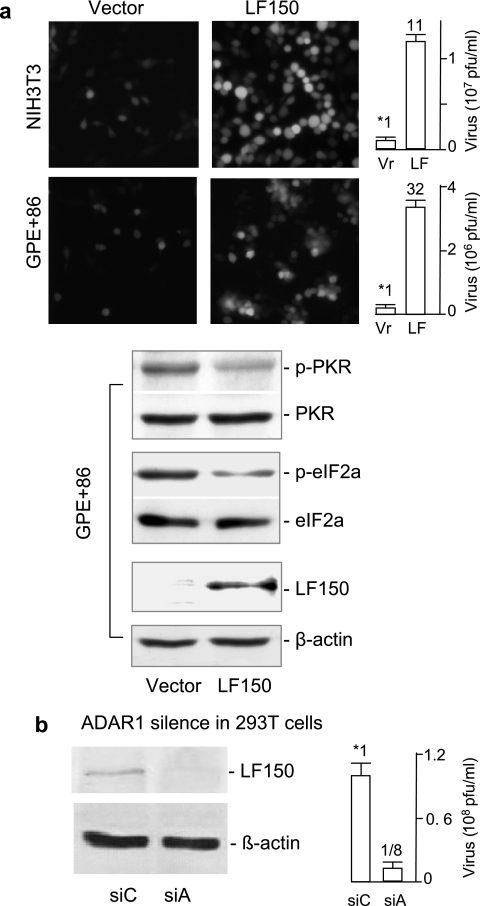
The key role of PKR activation in the innate response is to suppress virus replication in hosts ranging from insects to humans (14). Our findings consistently suggest a potential proviral effect of ADAR1 that might promote viral replication. We used VSV to test our speculation because its infection is very sensitive to PKR activation (1). Mice lacking PKR are predisposed to lethal intranasal infection by the usually innocuous VSV, and fibroblasts derived from PKR−/− mice are more permissive for VSV infection than wild-type fibroblasts (2, 8, 27). LF150 and a vector control were introduced into NIH 3T3 cells by a replication-deficient retroviral vehicle. Stable cells expressing LF150 or the control were infected with the recombinant VSV containing the green fluorescent protein marker (22). We found that LF150 expression significantly sensitized host cells for VSV infection as monitored under fluorescence microscopy (Fig. 4a). Quantitation by plaque-forming assay showed that the virus titer increased 11-fold in NIH 3T3 cells and 32-fold in GP+E86 fibroblast packaging cells when LF150 was compared with the control. To eliminate possible nonspecific effects, we further examined VSV infection by silencing endogenous ADAR1. Similarly, the level of ADAR1 was significantly reduced in 293T cells by siRNA (Fig. 3b and 4b). When these cells were infected with the recombinant VSV, the virus titer in ADAR1-silenced cells was reduced eightfold (Fig. 4b, right). Thus, the knockdown of ADAR1 made these cells more resistant to VSV infection. We conclude that ADAR1 is also a proviral mediator and that its expression is proportional to host susceptibility to viral infection.
FIG. 4.
ADAR1 mediates host susceptibility to VSV infection. (a) ADAR1 expression. NIH 3T3 (top) or GP+E86 (bottom) cells stably expressing LF150 or the vector were infected with the recombinant VSV-EGFP1 (5). NIH 3T3 cells were infected at an MOI of 10 PFU/cell for 20 min and GP+E86 cells at an MOI of 100 PFU/cell for 1 h. After 10 to 12 h of infection, the infected cells were monitored under fluorescence microscopy (left). The virus titer in the culture medium was quantitatively determined by the PFU (right; n = 4). The number over the bar indicates relative susceptibility to VSV infection. Levels of LF150, total PKR, and eIF-2α, and the phosphorylated PKR and eIF-2α in the testing cells, were examined by immunoblotting, and the results are shown below. (b) ADAR1 knockdown. 293T cells were transfected with siRNA against ADAR1 (siA) or a bacterial gene (siC) for 48 h and infected with VSV for 12 h. Endogenous ADAR1 was analyzed by immunoblotting using the antibody against human ADAR1. The virus titers in media were analyzed as described above. The error bars represent standard deviations.
The proviral effect of ADAR1 requires the involvement of PKR.
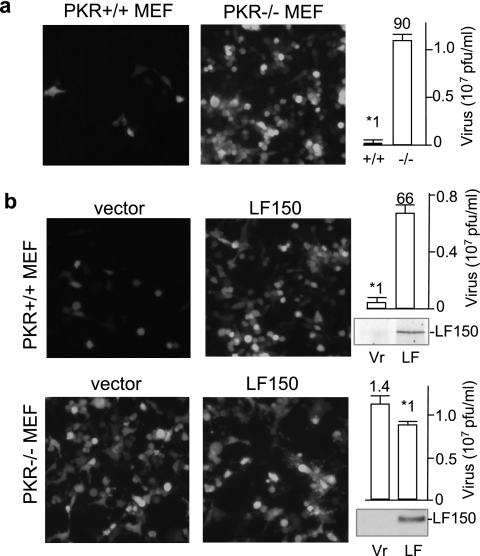
To examine whether the proviral effect of ADAR1 required the involvement of PKR, we measured the susceptibility of PKR−/− and PKR+/+ MEFs (32) to VSV infection. As previously reported (2, 27), the wild-type MEFs were normally resistant to VSV infection (Fig. 5a). Under our conditions, knockout of PKR increased VSV infection 90-fold. In comparison, stably expressing LF150 in the wild-type MEFs increased VSV infection 66-fold (Fig. 5b, top). This effect of LF150 was not significant in PKR−/− MEFs (Fig. 5b, bottom). Thus, PKR was required for the observed proviral effect. It revealed a novel mechanism by which ADAR1 inhibits PKR activation and consequently increases host susceptibility to viral infection.
FIG. 5.
PKR is required for the proviral effect of ADAR1. ADAR1 LF150 and a vector control were introduced into PKR+/+ and PKR−/− MEFs. The recombinant VSV-EGFP1 was used to infect (a) PKR+/+ and PKR−/− MEFs and (b) stable PKR+/+ (top) or PKR−/− (bottom) cells that expressed LF150 or the control (Vr), respectively. The photographs were taken 12 h postinfection under a fluorescence microscope. Virus titers in the media were quantitated as described above, and the results are shown on the right. Relative susceptibilities are indicated over the bars (n = 4). Transiently expressed LF150 was examined by immunoblotting, and the results are shown on the lower left. The error bars represent standard deviations.
The proviral effect of ADAR1 is independent of RNA editing.
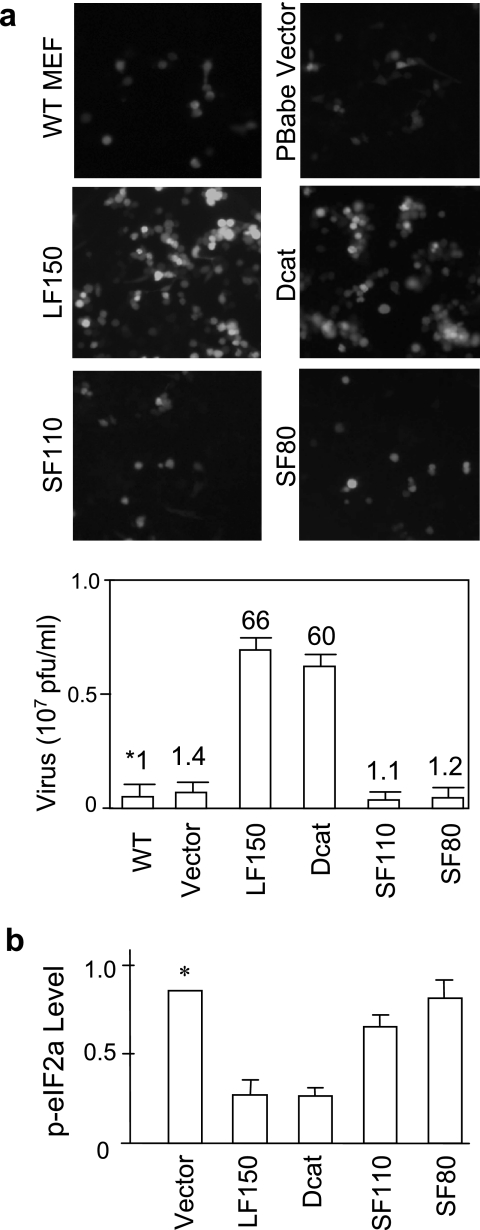
To further confirm the proviral function, we examined the effect of a few naturally existing ADAR1 variants on susceptibility to VSV infection. Notably, SF110, SF80, and Dcat are three ADAR1 variants that were previously detected in different mouse tissues (17). Among them, SF110 and SF80 were still active for dsRNA editing; however, the editing activity of Dcat was completely abolished (17). These variants were introduced into wild-type MEFs, and susceptibilities to VSV infection were compared. No increase of VSV infection was observed for SF110 and SF80 (Fig. 6a), suggesting that including both the dsRBD and the Z-DBD was very important for the function. Surprisingly, the C-terminally truncated fragment (Dcat) still efficiently stimulated VSV infection 60-fold, indicating that the deaminase domain was not essential for the function. We conclude that the proviral effect is independent of the RNA-editing activity of ADAR1. This result was further compared with PKR activation of the ADAR1 variants. Phosphorylation of eIF-2α was examined in the same cells and showed that LF150 and Dcat inhibited PKR activation; however, SF110 and SF80 did not (Fig. 6b). Thus, these data were consistent in showing that the dsRNA binding domains alone were not sufficient for the inhibitory function. An as-yet-unidentified motif covering the Z-DNA binding domain was also required.
FIG. 6.
Proviral effects of naturally existing ADAR1 variants. (a) Different ADAR1 variants, including LF150, Dcat, SF110, SF80, and the vector control, were introduced into wild-type (WT) MEFs. Stable cells were infected with the recombinant VSV-EGFP1 (MOI = 10), and infections were compared for different ADAR1 variants as described above. Note: as the basal level of VSV infection was high in PKR−/− MEFs, the differences for these ADAR1 variants were not as significant as in WT MEFs (data not shown). (b) Comparison of the naturally existing ADAR1 variants on PKR activation. 293T cells were transfected with the same set of ADAR1 constructs; p-eIF-2α and total eIF-2α in cell lysates were determined by immunoblotting and quantitated with Scion Image software. The ratios between p-eIF-2α and eIF-2α were calculated for each construct and normalized to the vector (*). All data are shown as mean plus standard deviation (n = 3).
PKR inhibitors are often encoded by viruses as a countermeasure to evade the antiviral function of PKR (14). VSV does not seem to encode such an inhibitor, and its infection is extremely sensitive to host PKR activation and the interferon antiviral response (1). We report that ADAR1 is a host-encoded PKR inhibitor that inhibits PKR-mediated antiviral response. Our data consistently show that ADAR1 interacts with PKR, inhibits PKR activation, and consequently increases the susceptibility of host cells to VSV infection. In contrast to the antiviral effect of ADAR1, which directly edits viral RNA (9, 12, 28), this proviral function is apparently independent of dsRNA editing and is ascribed to the dsRNA and Z-DNA binding motifs. It agrees with our hypothesis that the N-terminal motifs of ADAR1 are involved in the interaction with PKR and that this interaction is important for the proviral function. A question has been raised as to why the two apparently opposing effects are encoded in ADAR1. First, editing of viral RNA does not always seem to lead to an antiviral effect. When the editing sites are not critical for viral replication, hypermutations in viral RNA may be observed (4). Thus, RNA editing could also lead to a proviral effect to evade the adaptive antiviral responses by generation of hypermutations in viral RNA. RNA editing by ADAR1 could have either an antiviral or a proviral effect, depending on the editing sites. Second, the anti- and proviral effects may not spontaneously occur in the same host. The antiviral effect may occur only when viruses are colocalized with ADAR1 and accessible to dsRNA editing and the editing sites are essential for viral replication. The proviral effect may be more important when PKR activation is sensitive for viral infection. In addition, further investigation is needed to address why ADAR1 negatively regulates PKR activation. It is well known that PKR is important for cells to respond adequately to different stresses in addition to viral infection and to mediate different forms of stress-induced apoptosis (7, 31). Therefore, our data agree with the antiapoptotic role of ADAR1 in stresses (30). They suggest the potential mechanism of ADAR1 that negatively regulates the proapoptotic effect of PKR. VSV may have taken advantage of this mechanism to establish its infection in a host. Finally, recombinant VSV is known as an effective intranasal vaccine vector for gene therapy (23, 24). Our findings suggest a possible strategy to optimize recombinant-virus production and to develop new targets for antiviral therapeutics.
Acknowledgments
We thank Bryan Williams for providing the PKR-null MEFs; Ralf Janknecht, Brenda Bass, and Peter Kao for providing antibodies; Li Ding for technique assistance; and Alfred Bothwell, Ronald Salem, and Qian Gao for helpful discussions and critically reading the manuscript.
This work was supported by NIH grants GM-60426 and AI060701 to J.-H. Yang.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Balachandran, S., and G. N. Barber. 2000. Vesicular stomatitis virus (VSV) therapy of tumors. IUBMB Life 50:135-138. [DOI] [PubMed] [Google Scholar]
- 2.Balachandran, S., P. C. Roberts, L. E. Brown, H. Truong, A. K. Pattnaik, D. R. Archer, and G. N. Barber. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129-141. [DOI] [PubMed] [Google Scholar]
- 3.Cattaneo, R. 1994. Biased (A 3 I) hypermutation of animal RNA virus genomes. Curr. Opin. Genet. Dev. 4:895-900. [DOI] [PubMed] [Google Scholar]
- 4.Cattaneo, R., A. Schmid, D. Eschle, K. Baczko, V. ter Meulen, and M. A. Billeter. 1988. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55:255-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalton, K. P., and J. K. Rose. 2001. Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology 279:414-421. [DOI] [PubMed] [Google Scholar]
- 6.de Haro, C., R. Mendez, and J. Santoyo. 1996. The eIF-2α kinases and the control of protein synthesis. FASEB J. 10:1378-1387. [DOI] [PubMed] [Google Scholar]
- 7.Der, S. D., Y. L. Yang, C. Weissmann, and B. R. Williams. 1997. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc. Natl. Acad. Sci. USA 94:3279-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durbin, R. K., S. E. Mertz, A. E. Koromilas, and J. E. Durbin. 2002. PKR protection against intranasal vesicular stomatitis virus infection is mouse strain dependent. Viral Immunol. 15:41-51. [DOI] [PubMed] [Google Scholar]
- 9.Hartwig, D., C. Schutte, J. Warnecke, I. Dorn, H. Hennig, H. Kirchner, and P. Schlenke. 2006. The large form of ADAR 1 is responsible for enhanced hepatitis delta virus RNA editing in interferon-alpha-stimulated host cells. J. Viral Hepat. 13:150-157. [DOI] [PubMed] [Google Scholar]
- 10.Huang, Y., and Z. R. Liu. 2002. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J. Biol. Chem. 277:12810-12815. [DOI] [PubMed] [Google Scholar]
- 11.Jagus, R., and M. M. Gray. 1994. Proteins that interact with PKR. Biochimie 76:779-791. [DOI] [PubMed] [Google Scholar]
- 12.Jayan, G. C., and J. L. Casey. 2002. Increased RNA editing and inhibition of hepatitis delta virus replication by high-level expression of ADAR1 and ADAR2. J. Virol. 76:3819-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar, M., and G. G. Carmichael. 1997. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. USA 94:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langland, J. O., J. M. Cameron, M. C. Heck, J. K. Jancovich, and B. L. Jacobs. 2006. Inhibition of PKR by RNA and DNA viruses. Virus Res. 119:100-110. [DOI] [PubMed] [Google Scholar]
- 15.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nie, Y., L. Ding, P. N. Kao, R. Braun, and J. H. Yang. 2005. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol. Cell. Biol. 25:6956-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nie, Y., Q. Zhao, Y. Su, and J. H. Yang. 2004. Subcellular distribution of ADAR1 isoforms is synergistically determined by three nuclear discrimination signals and a regulatory motif. J. Biol. Chem. 279:13249-13255. [DOI] [PubMed] [Google Scholar]
- 18.Parker, L. M., I. Fierro-Monti, and M. B. Mathews. 2001. Nuclear factor 90 is a substrate and regulator of the eukaryotic initiation factor 2 kinase double-stranded RNA-activated protein kinase. J. Biol. Chem. 276:32522-32530. [DOI] [PubMed] [Google Scholar]
- 19.Patel, R. C., D. J. Vestal, Z. Xu, S. Bandyopadhyay, W. Guo, S. M. Erme, B. R. Williams, and G. C. Sen. 1999. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J. Biol. Chem. 274:20432-20437. [DOI] [PubMed] [Google Scholar]
- 20.Patterson, J. B., D. C. Thomis, S. L. Hans, and C. E. Samuel. 1995. Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology 210:508-511. [DOI] [PubMed] [Google Scholar]
- 21.Rabinovici, R., K. Kabir, M. Chen, Y. Su, D. Zhang, X. Luo, and J. H. Yang. 2001. ADAR1 is involved in the development of microvascular lung injury. Circ. Res. 88:1066-1071. [DOI] [PubMed] [Google Scholar]
- 22.Ramsburg, E., J. Publicover, L. Buonocore, A. Poholek, M. Robek, A. Palin, and J. K. Rose. 2005. A vesicular stomatitis virus recombinant expressing granulocyte-macrophage colony-stimulating factor induces enhanced T-cell responses and is highly attenuated for replication in animals. J. Virol. 79:15043-15053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts, A., E. Kretzschmar, A. S. Perkins, J. Forman, R. Price, L. Buonocore, Y. Kawaoka, and J. K. Rose. 1998. Vaccination with a recombinant vesicular stomatitis virus expressing an influenza virus hemagglutinin provides complete protection from influenza virus challenge. J. Virol. 72:4704-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnell, M. J., L. Buonocore, E. Kretzschmar, E. Johnson, and J. K. Rose. 1996. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc. Natl. Acad. Sci. USA 93:11359-11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp, T. V., F. Moonan, A. Romashko, B. Joshi, G. N. Barber, and R. Jagus. 1998. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology 250:302-315. [DOI] [PubMed] [Google Scholar]
- 26.Sledz, C. A., M. Holko, M. J. de Veer, R. H. Silverman, and B. R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834-839. [DOI] [PubMed] [Google Scholar]
- 27.Stojdl, D. F., N. Abraham, S. Knowles, R. Marius, A. Brasey, B. D. Lichty, E. G. Brown, N. Sonenberg, and J. C. Bell. 2000. The murine double-stranded RNA-dependent protein kinase PKR is required for resistance to vesicular stomatitis virus. J. Virol. 74:9580-9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor, D. R., M. Puig, M. E. Darnell, K. Mihalik, and S. M. Feinstone. 2005. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J. Virol. 79:6291-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomis, D. C., and C. E. Samuel. 1992. Mechanism of interferon action: autoregulation of RNA-dependent P1/eIF-2 alpha protein kinase (PKR) expression in transfected mammalian cells. Proc. Natl. Acad. Sci. USA 89:10837-10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Q., M. Miyakoda, W. Yang, J. Khillan, D. L. Stachura, M. J. Weiss, and K. Nishikura. 2004. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 279:4952-4961. [DOI] [PubMed] [Google Scholar]
- 31.Williams, B. R. 2001. Signal integration via PKR. Sci. STKE 2001:RE2. [DOI] [PubMed] [Google Scholar]
- 32.Zamanian-Daryoush, M., T. H. Mogensen, J. A. DiDonato, and B. R. Williams. 2000. NF-κB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-κB-inducing kinase and IκB kinase. Mol. Cell. Biol. 20:1278-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]