Abstract
Infection of cells with poliovirus induces a massive intracellular membrane reorganization to form vesicle-like structures where viral RNA replication occurs. The mechanism of membrane remodeling remains unknown, although some observations have implicated components of the cellular secretory and/or autophagy pathways. Recently, we showed that some members of the Arf family of small GTPases, which control secretory trafficking, became membrane-bound after the synthesis of poliovirus proteins in vitro and associated with newly formed membranous RNA replication complexes in infected cells. The recruitment of Arfs to specific target membranes is mediated by a group of guanine nucleotide exchange factors (GEFs) that recycle Arf from its inactive, GDP-bound state to an active GTP-bound form. Here we show that two different viral proteins independently recruit different Arf GEFs (GBF1 and BIG1/2) to the new structures that support virus replication. Intracellular Arf-GTP levels increase ∼4-fold during poliovirus infection. The requirement for these GEFs explains the sensitivity of virus growth to brefeldin A, which can be rescued by the overexpression of GBF1. The recruitment of Arf to membranes via specific GEFs by poliovirus proteins provides an important clue toward identifying cellular pathways utilized by the virus to form its membranous replication complex.
All known positive-strand RNA viruses replicate their genomes in association with membranous structures that are formed after the synthesis of viral proteins in infected cells by remodeling membranes from existing intracellular organelles. Different viruses target different organelle membranes (e.g., endoplasmic reticulum [ER], Golgi, endosomes, and mitochondria) and generate different morphological structures on which the replication complexes assemble (37).
Among the Picornavirus family members, RNA replication complexes from poliovirus-infected HeLa cells have been the best studied. Heterogeneously sized, vesicle-like structures that cluster in the perinuclear space in infected cells were observed by electron microscopy and described more than 40 years ago. Viral RNA replication occurs on the cytosolic surfaces of the vesicles, which aggregate into clusters (7-9). Little is known about how these structures are formed, although some recent observations have suggested that components of the cellular secretory and/or autophagy pathways are involved (24, 39, 41).
The process of membrane traffic begins at the ER, where polio proteins appear to be synthesized. Rust et al. (36) have visualized, by three-dimensional reconstruction of serial confocal microscope sections, poliovirus protein 2B sequences, and presumably other viral proteins, budding from multiple sites on the ER and showed extensive colocalization of the 2B sequences with COPII coatamer protein. Although it has not been determined which polio protein sequences are directly responsible for initiating the COPII-dependent budding process, it is generally believed that the P2 proteins 2C, 2BC, or precursors thereof, are required (2, 9, 12, 41). Normally, the COPII machinery establishes a membrane flow from the ER to the Golgi complex. However, poliovirus replication vesicles do not fuse with the Golgi complex; indeed, the Golgi complex appears to disassemble in poliovirus-infected cells, presumably because retrograde membrane traffic from the Golgi continues while anterograde traffic is rerouted to form the viral replication complexes.
A complicating observation is that polio RNA replication is inhibited by the fungal antimetabolite, brefeldin A (BFA) (18, 22, 27), whereas neither COPII-dependent traffic nor autophagous vesicle formation is known to be sensitive to this drug. Instead, BFA is known to inhibit the activation and function of some members of the small GTPase family, Arf, by interacting with specific guanine nucleotide exchange factors (GEFs) that recycle Arf from its inactive GDP-bound form to its active, GTP-bound form (30, 34). When activated, Arf-GTP recruits effectors such as coat complexes and lipid-modifying enzymes to specific membrane sites, creating domains competent for cargo transport (29, 31). Thus, the Arf GTPases play a central role in regulating membrane dynamics and protein transport. The recruitment of Arfs to their target membranes is mediated by a diverse group of GEFs, all of which share a Sec7 domain necessary for guanine nucleotide exchange (23). All Arf GEFs are peripherally associated membrane proteins either tethered to membranes through a pleckstrin homology domain that interacts with specific phosphoinositides or through interactions with specific membrane-bound proteins that are not well characterized. The latter group includes the high-molecular-weight GEFs GBF1, BIG1, and BIG2, which are sensitive to BFA (14). The binding of specific GEFs to various membrane sites confers specificity to the recruitment and activation of specific Arfs, serving to regulate different steps in the membrane trafficking pathway (4).
The sensitivity of poliovirus RNA replication to BFA suggested that, in addition (or subsequent) to the COPII-dependent budding of vesicles from the ER, an Arf-dependent membrane trafficking step may be required for polio replication complex formation. For example, Gazina et al. (21) reported that β-COP, a component of the coatamer COPI, whose recruitment to membranes is regulated by Arf1, localizes to the membranous replication complexes in cells infected by echovirus 11, a member of the Enterovirus genus whose growth, like that of poliovirus, is sensitive to BFA. Arf1 was not present on vesicles associated with the replication complexes of picornaviruses such as encephalomyocarditis virus, whose growth is resistant to BFA.
Previously, we demonstrated that Arf1 proteins colocalize with the newly formed poliovirus replication complexes in infected cells and that some members of the Arf family are recruited to cellular membranes by poliovirus proteins synthesized in HeLa cell extracts that support the complete translation and/or replication of viral RNA in vitro (5). These extracts have been used extensively to investigate many aspects of viral protein synthesis and processing, RNA replication, and assembly of infectious virions (3, 28), and we have shown previously that viral RNA synthesis, but not translation, is dependent upon the presence of intact membranes in the extracts (19). Recruitment of Arf to the membranes was induced independently by two viral proteins from the P3, but not the P2, region: 3A and 3CD.
We show here that the two viral proteins, 3A and 3CD, independently recruit different Arf GEFs (GBF1 and BIG1/2, respectively) to membranes. These GEFs then are responsible for Arf activation and translocation to these membranes. The requirement for these specific BFA-inhibited GEFs explains the sensitivity of virus growth to BFA, which can be rescued by overexpression of GBF1.
MATERIALS AND METHODS
Cells and viruses.
Monolayer cultures of HeLa or Vero cells, grown either in multiwell chambers for time-lapse microscopy or on 18-mm coverslips in 12-well plates for immunofluorescence, were infected with the Mahoney strain of poliovirus type 1 in Dulbecco modified Eagle medium containing ∼5 × 107 PFU of the virus.
Plasmids.
pXpA-SH contains full-length cDNA of poliovirus type 1 under control of the T7 promoter, with the hammerhead ribozyme coding sequence at the 5′end of the poliovirus sequence and additional unique restriction sites within the viral cDNA (5). pXpA-RenR plasmid encodes a poliovirus replicon with the Renilla luciferase gene from phRL-CMV (Promega) substituting for the capsid coding sequence in pXpA-SH. Plasmids used for transcription of RNAs coding for individual poliovirus proteins have been previously described (5). pArf1-GFP for Arf-GFP fusion expression, pHA-GBF1-Yc for expression of GBF1, and pHA-BIG2 have been previously described (32, 40, 43).
Transfection.
DNA transfections were performed with Fugene 6 reagent (Roche), and RNA transfections utilized the Trans-It mRNA transfection kit (Mirus) according to the manufacturer's instructions.
Cell-free translation assays.
In vitro transcription and translation of viral RNAs and Western blotting of the membrane fractions were performed essentially as described previously (5). Briefly, HeLa cell S10 extracts were programmed with poliovirus-specific RNAs and incubated for 3.5 h at 34°C, and the membrane material was collected by centrifugation at 15,700 × g for 20 min at 4°C prior to analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Antibodies.
Antibodies to BIG1 and BIG2 were provided by M. Vaughan and J. Moss (NHLBI). Anti-GTPase activating protein (GAP) and anti-GBF1 antibodies were from P. Randazzo (National Cancer Institute) and J. Bonifacino (National Institute of Child Health and Human Development). Monoclonal anti-poliovirus antibodies to 2C and 3A were a gift from K. Bienz, University of Basel, Basel, Switzerland. Secondary antibody conjugates used in immunofluorescence were from Molecular Probes; those used in Western blots were from Amersham.
Microscopy.
Live cell microscopy was performed on a Zeiss LSM510 line scanning confocal microscope. HeLa cells were grown in coverglass-bottom chambers (Lab-Tek). The cells were maintained at 37°C on the microscope stage for the duration of the experiments. High-spatial-resolution images were obtained with a ×63/1.4 numerical aperture plan Apochromat oil immersion objective lens with a small (1.2 Airy unit) pinhole aperture, whereas images for purposes of quantitation (e.g., fluorescence recovery after photobleaching [FRAP]) were acquired with a ×40/1.3 N.A. oil immersion objective lens with a fully open pinhole setting (∼14 Airy units). All images were acquired on 12-bit photomultiplier tubes. No saturated images were analyzed for quantitation.
For FRAP experiments, a region of interest (ROI) was drawn around the Arf1-GFP-labeled structures, and this ROI was photobleached for 5 s with a high-intensity 488-nm laser beam. Images of the samples were then taken with low-intensity light for 3 min. Input from lateral diffusion effects was excluded by photobleaching areas that did not have apparent contacts with other intracellular structures associated with Arf1-GFP. The FRAP data were analyzed with image analysis software which allowed us to measure the mean fluorescence per pixel in the defined ROIs. To obtain the total fluorescence associated with Arf1-GFP either at the Golgi apparatus or at cytoplasmic foci in an infected cell, we multiplied the background-subtracted mean fluorescence per pixel in the ROI with the area of that ROI. The total fluorescence at each time point was corrected for photobleaching (which typically occurs after a time series) by dividing it by the total fluorescence of the cell at that time.
For immunofluorescence, cells were grown on coverslips, fixed with 4% paraformaldehyde-phosphate-buffered saline (PBS) for 20 min, and permeabilized with 0.2% Triton X-100 in PBS for 5 min. The cells were then incubated in 3% nonfat dry milk solution for 1 h to block nonspecific binding sites. This solution also was used for dilution of primary and secondary antibodies in which cells were sequentially incubated for 1 h. Images were taken with Leica DMIRE microscope. Digital images were processed with Adobe Photoshop software.
Arf-GEF assay.
Detection of Arf GEF activity was performed as described previously (35). Briefly, cytoplasmic extract from ∼107 HeLa cells was incubated with recombinant myristoylated Arf1 in the presence of liposomes and [γ-35S]GTP. At various times aliquots of the reaction mix were put into stop solution and kept on ice. When all of the samples were collected, they were precipitated with 10% trichloroacetic acid on filter paper and washed twice with 5% trichloroacetic acid, and the precipitated radioactivity was measured by scintillation spectroscopy.
Arf-GTP pull-down assay.
HeLa cells were grown overnight on 10-cm petri dishes. The cells were then infected with 10 PFU of poliovirus/cell. At various times the cells were lysed on ice for 15 min in 1 ml of lysis buffer (50 mM Tris-Cl [pH 7.5], 100 mM NaCl, 2 mM MgCl2, 0.5% sodium deoxycholate, 1% Triton X-100, 10% glycerol) containing 100 μM phenylmethylsulfonyl fluoride and a 1/100 of volume of protease inhibitor cocktail (Sigma-Aldrich). Lysates were clarified by centrifugation at maximum speed for 20 min in an Eppendorf minifuge at 4°C. The supernatants were collected and stored at −80°C. The Arf-GTP pull-down assay was performed as described in reference 25. Briefly, lysates were precleared with 50 μl of a 50% slurry of glutathione-Sepharose 4B beads (Amersham-Biosciences) for 1 h and centrifuged. Supernatants were incubated with 50 μl of beads that contained 40 μg of prebound GST-GGA3-GAT fusion protein (expression construct kindly provided by Julie Donaldson, NHLBI, NIH). Beads were collected by centrifugation and washed three times with 250 μl (10 times the bed volume of beads) washing buffer. Proteins were eluted from beads by adding 1× SDS-PAGE sample buffer and heating the samples at 70°C for 15 min. All manipulations with lysates were carried out at 4°C.
Viral RNA replication assays.
HeLa cells grown in 96-well plates were transfected with specified plasmids and the next day were transfected with poliovirus replicon RNA (0.8 ng/well) transcribed from pXpA-RenR. After 1 h, normal growth medium containing 60 μM EnduRen Renilla luciferase substrate (Promega) and 1 μg of BFA/ml or solvent dimethyl sulfoxide was provided. Light from Renilla luciferase activity was measured at hourly intervals with a Molecular Devices MV microplate reader. After the 7-h time point, cells were lysed with DualGlo (Promega) firefly luciferase assay buffer, and the firefly luciferase activity was measured to assess DNA transfection efficiency.
RESULTS
Dynamics of Arf1-EGFP redistribution upon poliovirus infection.
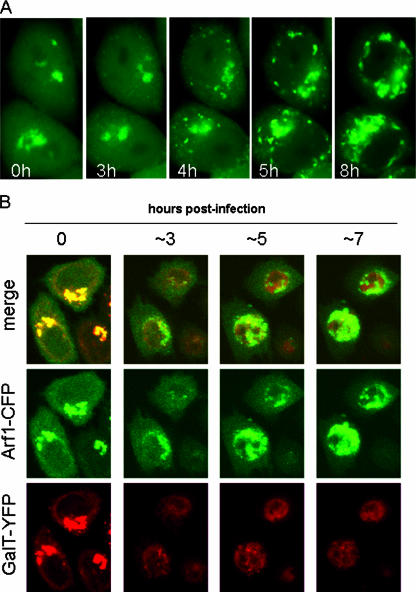
The Arf GTPases regulate vesicular traffic and organelle structure by recruiting coat proteins and regulating phospholipid metabolism (4). The sensitivity of poliovirus replication to BFA, which is known to inhibit the activation and function of Arf, previously prompted us to determine whether Arf was associated with the viral RNA replication complexes that formed in poliovirus-infected cells (5). The results showed a clear redistribution of Arf1-EGFP from its predominant concentration at the Golgi complex in uninfected cells to perinuclear structures that colocalized with viral replication proteins used to mark the replication complexes in infected cells. In order to monitor the kinetics and pathway of Arf translocation during the course of infection, HeLa cells transiently expressing Arf1-GFP fusion protein were infected with poliovirus and imaged by using time-lapse confocal microscopy at 6-min intervals throughout the course of infection (Fig. 1). The conditions were optimized to maintain cell viability during the entire virus replication cycle, allowing the molecular relocalization of Arf1-GFP to be monitored in individual live, infected cells. At the start of the infection, Arf1-GFP was primarily concentrated on the surface of Golgi membranes, with a soluble pool distributed diffusely throughout the cytoplasm. In most cells Arf1-GFP began to dissociate from the Golgi region between 2 and 3 h postinfection and, simultaneously, multiple new foci of fluorescence emerged in the cytoplasm (Fig. 1A). The foci began to coalesce and concentrate in the perinuclear region within approximately 30 min after they began to form. This pattern has been shown previously to correspond to the pattern of poliovirus replication complexes (9), which expands and remains perinuclear until cell death (Fig. 1; the imaging sequence can be viewed in movie form [http://www3.niaid.nih.gov/labs/aboutlabs/lid/picornavirusReplicationSection/ehrenfeld.htm]). In some cells all of the Golgi-associated Arf1-GFP distributed to the cytoplasm before relocalizing to the new structures, whereas in others some residual fluorescence remained in the original location.
FIG. 1.
Relocalization of Arf1-GFP in infected cells. (A) HeLa cells transfected with pArf1-GFP for 18 h were infected with poliovirus and monitored every 6 min for 16 h. The figure shows selected images taken at the indicated times postinfection. (For the complete sequence, see the movie [and corresponding Web site] referred to in Results.) (B) HeLa cells were cotransfected with pGalT-YFP and pArf-CFP. After 18 h of incubation, the cells were infected with poliovirus and monitored for fluorescence of both of the expressed proteins.
The loss of Arf1 from the Golgi region in poliovirus-infected cells coincided with the disassembly of the Golgi apparatus based on experiments in which Arf1-CFP was coexpressed with the Golgi enzyme marker, galactosyltransferase tagged with YFP (GalT-YFP). In these cells the two proteins initially completely colocalized in the Golgi area (Fig. 1B, 0 h postinfection). As infection proceeded, they both lost their juxtanuclear, Golgi localization and redistributed in different patterns. Based on these results, we concluded that Arf1 relocates from Golgi to non-Golgi membrane foci between 2 and 3 h postinfection.
To determine whether Arf relocalization occurred as a general response of infected cells to ER stress or induction of apoptosis, known to occur early in poliovirus infection (42), HeLa cells were treated with tunicamycin, which induces ER stress by interference with protein glycosylation, or with tumor necrosis factor, a potent inducer of receptor-mediated apoptosis. No significant change in the fluorescence pattern of Arf1-GFP was observed, and cells treated with these substances underwent apoptotic death with Arf1-GFP still associated with Golgi structures (not shown). Infection of cells in the presence of a caspase inhibitor, zVAD-fmk, previously shown to prevent apoptosis induction by poliovirus (1), also did not affect the translocation of Arf fluorescence. Thus, Arf1 redistribution in infected cells does not represent a response to general stress conditions but is specifically induced by poliovirus. Infection of Vero cells expressing Arf1-GFP resulted in a similar redistribution of Arf1-GFP as observed in HeLa cells, indicating that the cellular response to poliovirus infection is not host cell specific (not shown).
FRAP analysis of Arf1 on poliovirus replication complexes.
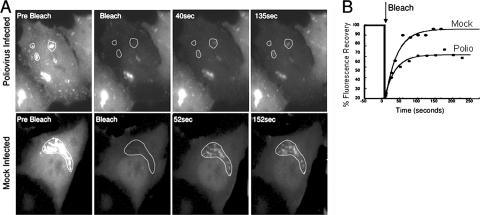
Normal function of Arf involves rapid exchange between membrane-associated and cytoplasmic Arf pools. To compare the rate of Arf cycling on and off membranes in polio-infected and mock-infected cells we performed FRAP experiments in cells expressing Arf1-GFP fusion protein. The FRAP experiments were performed in cells approximately 3 to 4 h postinfection, when Arf1-GFP containing foci were abundant in the cytoplasm. Regions of membrane-associated Arf1-EGFP fusion were selectively photobleached with laser irradiation, and the recovery of fluorescence, which occurs if there is an exchange of Arf between the membranes and the cytoplasmic pool, was monitored. Input from lateral diffusion was excluded by photobleaching areas that did not have apparent contacts with other intracellular membrane structures with associated Arf1-GFP. The rates of recovery of fluorescence in poliovirus- and mock-infected cells were similar (t1/2 = 22.2 ± 5.2 s in mock-infected cells and 19.5 ± 3.1 s in infected cells; however, infected cells contained a higher proportion [35% ± 13%] of noncycling Arf compared to 10% ± 7% in mock-infected cells [Fig. 2 ]). These results suggest that Arf1 undergoes less exchange between membrane and cytoplasmic pools in infected cells compared to uninfected cells. Alternatively, the cytoplasmic Arf1-GFP may be prevented from diffusing into and out of the photobleached regions due to the high concentration of membranes in this area.
FIG. 2.
Arf1-GFP dynamics in poliovirus-infected HeLa cells. (A) Regions of interest (dotted lines) were selected for photobleaching in cells ∼3 h postinfection. In mock-infected cells the Arf1-GFP-labeled Golgi apparatus was selected. A laser pulse was directed at the selected regions to bleach the fluorescence. Recovery of fluorescence was monitored with low intensity laser light every 5 s. (B) The recovery of fluorescence into the selected regions was quantified and plotted as described in Materials and Methods. Photobleaching was performed on 9 virus-infected and 12 mock-infected cells in two separate experiments. The mean rate of recovery (τ) for control cells was 32 ± 7.5 s−1, and for virus-infected cells it was 28 ± 4 s−1. The mean immobile fraction for control cells was 10% ± 7%, whereas for poliovirus-infected cells it was 35% ± 13%. Representative recovery curves from infected and mock-infected cells are plotted.
Poliovirus infection stimulates production of Arf-GTP in vivo.
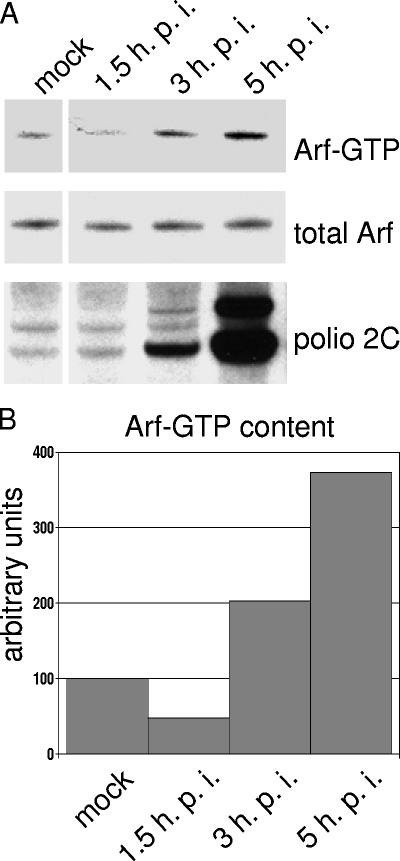
Inactive, GDP-bound Arf is located in the cytosol, and active GTP-bound Arf is associated with membranes. Arf activation then leads to recruitment of coat proteins and other factors that in turn regulate the protein sorting and membrane deformation events that direct intracellular trafficking. To confirm that the Arf associated with polio replication complexes was in an active, GTP-bound form and to determine whether the amount of activated Arf increased during the course of infection, we performed a pull-down assay using a GST-GGA3 fusion protein to monitor the amount of Arf-GTP protein in lysates of cells during the course of infection (Fig. 3) (25, 32, 38). GGA3 is an effector of Arf, and binds with high affinity specifically to Arf-GTP (10). Infected cells showed a three- to fourfold increase in Arf-GTP that accumulated steadily at least up to 5 h postinfection. Similar experiments were performed with antibodies specific for different members of the Arf family as probes. GTP-bound Arfs 1 and 3 (class I) showed the same pattern of increase during infection as the total Arf-GTP shown in Fig. 3; Arf5-GTP increased less dramatically, whereas Arf6-GTP showed no change during the virus growth cycle (data not shown).
FIG. 3.
GTP-bound Arf in poliovirus-infected cells. HeLa cells were mock infected or infected with poliovirus. (A) At the indicated times postinfection, cells were lysed, and the Arf-GTP pull-down assay was performed as described in Materials and Methods. The upper panel shows the amount of GTP-bound Arf recovered from each cell lysates, and the middle panel shows the total amount of Arf in each lysates. Anti-polio-2C staining (bottom panel) was performed on the same membrane used for Arf-GTP blot after stripping it with Chemicon Re-Blot Plus Mild solution. (B) Quantitation was performed by using Total Lab 1D gel image analysis software.
Arf GAPs do not accumulate on poliovirus replication complexes.
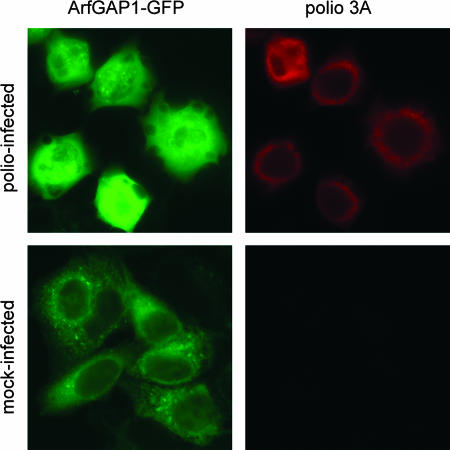
The hydrolysis of GTP bound to Arf proteins requires the activity of GAPs to enhance the weak intrinsic GTPase activity of Arf. The levels of cellular Arf-GTP result from a balance between GAP activity and GEF activity, which catalyzes the formation of Arf-GTP by exchanging GDP bound to Arf with GTP. To determine whether poliovirus protein-induced translocation of Arf to membranes was accompanied by changes in GAPs on the membranes, we translated full-length poliovirus RNA and RNAs coding for individual viral proteins in HeLa cell extracts, routinely used to study poliovirus translation-replication in vitro (3, 28). After the translation of poliovirus-specific RNAs we collected the membranes, with their associated proteins, from the extract by centrifugation and examined this material by immunoblotting with GAP-specific antibodies. None of the GAPs tested (ArfGAP1, ARAP1, ARAP2, ASAP1, ASAP2, AGAP1, AGAP2, and GIT1) manifested significant alterations in membrane association (not shown). These data suggest that the larger fraction of Arf1-GFP that is stably associated with membranes in poliovirus-infected cells (Fig. 2B) is due to limited GAP activity at new recruitment sites. This could explain the reduced exchange rate of Arf1-GFP on and off membranes as well as the accumulation of Arf-GTP during the course of infection. We therefore examined the distribution of ArfGAP1 tagged with GFP (ArfGAP1-GFP) (26) in poliovirus-infected cells. In agreement with the in vitro data, no accumulation of ArfGAP1-GFP on viral replication complexes was observed during infection (Fig. 4). In some fields, the fluorescence in virus-infected cells appeared to be more intense than in control cells, most likely because the shape and morphology of the cells change upon infection and the components appear more concentrated. In mock-infected cells, ArfGAP1-GFP was associated with ER, as previously described (26). After poliovirus infection, the ArfGAP1-GFP became more diffusely distributed but showed no colocalization with the viral replication complexes defined by the perinuclear localization of viral protein 3A (Fig. 4, bottom panels).
FIG. 4.
ArfGAP1 in poliovirus-infected cells. HeLa cells transfected with an ArfGAP1-EGFP expression plasmid for 18 h were infected with poliovirus. Cells were fixed at 5 h postinfection and stained with anti-poliovirus 3A antibodies to visualize the replication complexes.
Viral proteins show no GEF activity.
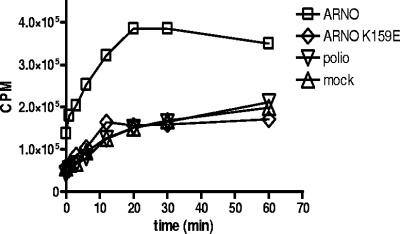
Cyclic regeneration of Arf from the GDP-bound to the active GTP-bound form requires the activity of a GEF. This activity could be provided by either preexisting cellular GEFs or by some viral protein(s). We therefore sought to identify which GEF(s) may be responsible for recruiting Arf. Initial experiments were performed to test whether any poliovirus proteins contributed a unique GEF activity for Arf activation during infection. Lysate from infected cells was incubated with recombinant Arf1 and the nonhydrolyzable GTP analog [γ-35S]GTP, and the production of radiolabeled Arf1 was measured in a standard GEF assay (35). As positive and negative controls we used lysates from HeLa cells transiently expressing the GEF, ARNO, or its mutated inactive version ARNO E156K (6). Although lysate from cells overexpressing ARNO demonstrated high levels of GEF activity, lysate from infected cells did not show any increase in GTP binding compared to the negative control (Fig. 5). Thus, we found no indication of significant GEF activity provided by any viral proteins in this assay.
FIG. 5.
GEF activity in poliovirus-infected cells. Cytoplasmic extracts from poliovirus-infected cells were assayed for guanine nucleotide exchange activity on purified recombinant Arf1 protein in vitro as described in Materials and Methods. GEF activity in infected cells was compared to that in mock-infected cells or cells expressing ARNO or an inactive, mutant protein, ARNO E156K.
Arf GEFs GBF1 and BIG1/2 are recruited to membranes by viral proteins 3A and 3CD.
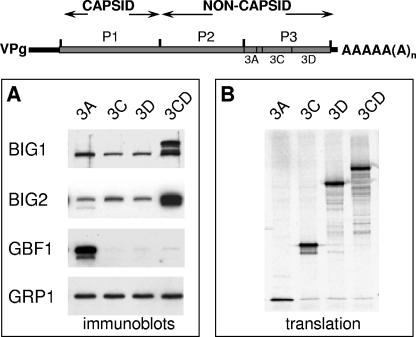
We examined the possible involvement of specific cellular GEFs in virus-induced membrane reorganization associated with Arf recruitment. RNAs coding for individual poliovirus proteins were translated in HeLa cell extracts (Fig. 6B), and the membranous pellets were collected by centrifugation and assayed by immunoblotting with GEF-specific antibodies. The synthesis of two viral proteins that were previously shown to induce translocation of Arf to the membranes (5) also demonstrated an increased association of specific GEFs with membranes. BIG1 and BIG2 were greatly enriched in the membrane fraction after the translation of RNA coding for protein 3CD (Fig. 6A). Translation of 3A RNA, on the other hand, did not recruit BIG1/2 but resulted in significant translocation to membranes of another GEF, GBF1 (Fig. 6A). Other GEFs tested (cytohesine 1, ARNO, and GRP1) showed no response to the synthesis of poliovirus proteins (see, for example, GRP1 in Fig. 6A, which serves also as a loading control).
FIG. 6.
Recruitment of GEFs to membranes by poliovirus proteins in vitro. (A) Poliovirus proteins were synthesized in HeLa cell extracts, and the membrane fractions were collected and assayed by immunoblotting with antibodies to BIG1, BIG2, GBF1, and GRP1. (B) Translation of each poliovirus-specific RNA was monitored by labeling an aliquot with [35S]methionine and analyzing the translation products by SDS-PAGE.
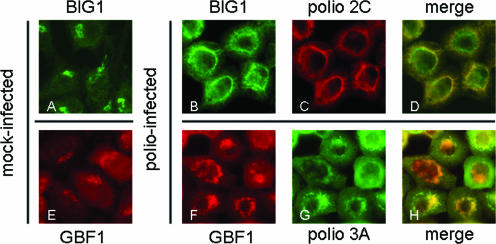
To confirm that the recruitment of specific GEFs to membranes after the synthesis of poliovirus proteins in vitro reflected processes that also occurred during poliovirus infection in vivo, we monitored the translocation of GEFs in poliovirus-infected cells. Staining of mock-infected cells with anti-BIG1 antibodies clearly showed the protein localized to the Golgi area (Fig. 7A), whereas in infected cells it was redistributed to the virus-induced perinuclear ring of vesicles, where the protein showed extensive colocalization with poliovirus replication protein 2C (Fig. 7B to D). Staining of GBF1 similarly demonstrated its relocalization to the perinuclear ring of poliovirus replication complexes (Fig. 7E to H). This pattern is in clear contrast to the nonspecific distribution of ArfGAP1-GFP in infected cells (cf. Figure 4). BIG2 antibodies suitable for immunofluorescence analysis were not available. The data in Fig. 6 and 7 together show a highly specific translocation of two Arf GEFs to the viral replication complex membranes and suggest that two different viral proteins may independently induce recruitment of GBF1 and BIG1/2.
FIG. 7.
Relocalization of BIG1 and GBF1 in HeLa cells upon poliovirus infection. (A) Mock-infected HeLa cells stained with anti-BIG1 antibodies. (B and C) Poliovirus-infected cells stained 3 h postinfection with antibodies against BIG1 and poliovirus nonstructural protein 2C, respectively. (D) Merged image of panels B and C. (E) Mock-infected cells stained with anti-GBF1 antibodies. (F and G) Poliovirus-infected cells stained 3 h postinfection with antibodies to GBF1 and poliovirus nonstructural protein 3A, respectively. (H) Merged image of panels F and G. Poliovirus proteins 2C and 3A both served as markers of viral RNA replication complexes.
GBF1 rescues poliovirus sensitivity to BFA.
The specificity of GEF recruitment by poliovirus proteins likely explains the previously reported sensitivity of poliovirus replication to BFA (15, 18, 27). BIG1, BIG2, and GBF1 belong to the family of high-molecular-weight GEFs whose members are sensitive to BFA (14). Specific amino acid residues within the Sec7 domain of these proteins bind BFA and trap a GEF/Arf-GDP complex in an inactive state, thus blocking nucleotide exchange. Recruitment of Arf to the membranes of the increasing number of RNA replication complexes that continue to form during the viral growth cycle is indicated by the sensitivity of virus production to BFA even when the drug is added in the middle or at later times in the growth cycle (27) (our unpublished observations).
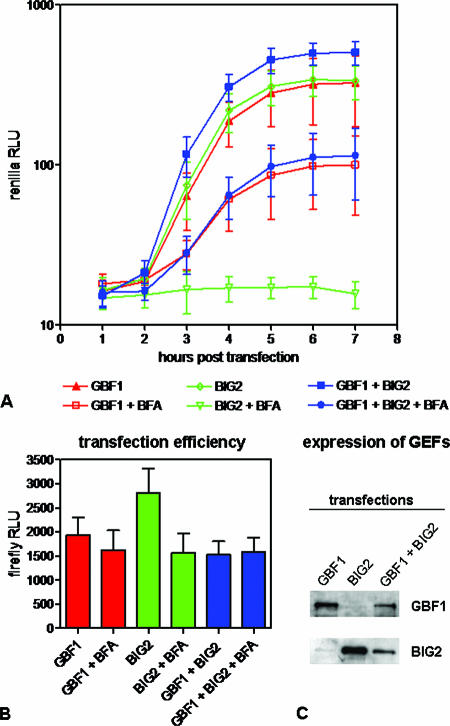
To test the functional significance of GEF involvement in poliovirus replication complex formation, we studied viral RNA replication in cells ectopically expressing GBF1 and BIG2. (Unfortunately, there is currently no reliable expression system for BIG1.) Initial attempts to lower endogenous levels of these GEFs with siRNAs resulted in a significant cytotoxic effect, making it difficult to reliably attribute a reduction in virus replication to the downregulation of a particular protein (not shown). Overexpression of GBF1 and BIG2 has been shown previously to rescue certain steps of intracellular membrane traffic from inhibition by BFA (25, 40). We tested whether those proteins can rescue BFA-induced inhibition of poliovirus replication. HeLa cells were transfected with (i) a plasmid encoding GBF1 fused to the C-terminal portion of YFP (HA-GBF1-Yc) that was shown previously to manifest the GEF activity of intact GBF1 (32); (ii) a BIG2 expression plasmid, pHA-BIG2 (40); or (iii) both plasmids together. To monitor the transfection efficiencies, 1% of a firefly luciferase expression plasmid pGL4.13 (Promega) was included in the transfection mix (Fig. 8B). The expression of GBF1 and BIG2 was confirmed by Western blotting (Fig. 8C). Reduced expression of both proteins in the sample transfected with both plasmids was likely due to promoter competition. After 18 h, the cells were transfected with a poliovirus replicon RNA containing the Renilla luciferase gene in place of the structural protein-coding region. Renilla luciferase activity was monitored in intact cells as a measure of viral RNA replication. Figure 8A shows similar replicon amplification in cells expressing GBF1, BIG2, or both GEFs together, and this level of RNA replication was generally the same as in control cells that were transfected with an empty vector (not shown). In the presence of BFA, however, GBF1 partially rescued RNA replication. It should be noted that rescue could only occur in the subpopulation of cells that had been transfected with GBF1-expressing plasmids, which we estimate to be 30 to 50% of total; thus, the actual rescue is significantly greater than that represented by the measured increase in replicon replication in the total cell population. No replication occurred in the presence of BFA in cells expressing BIG2 alone. This signal was indistinguishable from that observed in cells transfected with an empty vector or from a blank sample that was not transfected with the viral replicon (not shown). The rescue of replication by GBF1 was unaffected by simultaneous expression of BIG2. Overexpression of another GEF, ARNO, which is not BFA sensitive, had no effect on poliovirus replication in the presence of BFA (not shown), confirming the specificity of GBF1 action. The rescue of poliovirus replication in the presence of BFA by ectopic expression of GBF1 was not the result of decreasing the effective concentration of inhibitor because of binding BFA by extra GBF1 molecules, since BIG2 binds BFA and showed no such rescue. We also performed an additional experiment in cells expressing a GBF1 mutant (M832L) believed to be a fully functional GEF but unable to bind BFA (32). (Expression of yeast GBF1 homolog Gea1p with the corresponding M699L mutation made them resistant to BFA [34].) This mutant rescued viral RNA replication as well as the wild-type GBF1 did (not shown), indicating that the specific effect of GBF1 was not caused merely by the binding of BFA. Thus, it appears likely that at least GBF1 activity is required for formation and function of viral replication complexes and that this requirement is the cause of inhibition of virus growth by BFA.
FIG. 8.
Rescue of poliovirus RNA replication in the presence of BFA by GBF1. (A) Poliovirus replicon RNA containing the Renilla luciferase gene was introduced into HeLa cells previously transfected with expression plasmids for GBF1, BIG2, or both together. A total of 1 μg of BFA/ml was added where indicated. Each point is an average value from 16 wells of a 96-well plate. (B) Firefly luciferase activity was measured in the cells shown in panel A to evaluate transfection efficiencies. (C) Immunoblot showing expression of GBF1 and BIG2 in the transfected cells.
DISCUSSION
Although it is generally accepted that poliovirus replication requires the participation of cellular factors in various steps of its growth cycle, very few specific proteins or their functions have been identified. Exceptions are the cellular receptor, CD155, for virus entry, and several proteins that stimulate internal ribosome entry site utilization for initiation of translation. Viral RNA synthesis appears to require binding of poly(rC) binding protein to the 5′-terminal cloverleaf structure (20, 44) and perhaps hnRNP C binding to the 3′ untranslated region (11); however, the biochemical functions provided by these proteins are not known. The results described in the present study define a new class of cellular proteins (GEFs) that are “hijacked” for the purposes of remodeling intracellular membranes to generate a membranous replication complex scaffold and that are responsible for the sensitivity of virus growth to BFA. Our data suggest that viral protein 3A recruits GBF1, which in turn recruits Arf1 to induce changes in membrane architecture and function, and that protein 3CD, via BIG1/2/Arf recruitment, contributes to the formation of the characteristic perinuclear replication complexes.
Arf proteins regulate membrane transport by interacting with effectors that induce membrane deformation and budding and may recruit other cellular factors required for viral RNA replication. Poliovirus protein 3A is a transmembrane protein (16) that blocks ER to Golgi traffic, resulting in inhibition of cellular secretion during poliovirus infection (17, 18). While this work was in progress, Wessels et al. (45) reported that the 3A protein of coxsackievirus B3, a member of the enterovirus genus closely related to poliovirus, specifically binds to GBF1, trapping it on the membrane and preventing Arf activation and Arf1-dependent recruitment of COPI, a key player in maintenance of the ER-Golgi cycle of secretory pathway traffic. They concluded that when 3A was expressed in cells by itself, coxsackievirus 3A-GBF1 interaction inactivates GBF1, preventing it from recruiting Arf1, whereas our data show increased levels of activated Arf1 and recruitment of both GBF1 and Arf1 to replication complexes in infected cells. This discrepancy could be attributed to a difference in 3A behavior when expressed in cells as a separate protein versus expressed as part of the virus polyprotein, when other viral polypeptides can modulate its interactions with cellular counterparts (see for example, the dramatic change in epitope presentation of polio 3A expressed alone and in infected cells [13]). However, synthesis of the 3A protein alone in vitro caused Arf binding to membranes (5), and this apparent contradiction with the results of Wessels et al. is not understood.
The interference by 3A with GBF1/Arf metabolism results in the inhibition of protein transport in cells expressing 3A protein alone or in infected cells. Although this inhibition of protein transport may play a role in the virus's evasion of the host's immune responses, it is not absolutely required for replication in cultured cells, since mutations that fail to inhibit protein secretion are not lethal to virus replication (18). The rescue of poliovirus replication in the presence of BFA by overexpression of GBF1, however, suggests that there is a requirement for some activity of GBF1 for a successful infection.
Viral protein 3CD is a proteinase that generates individual capsid proteins from a precursor polyprotein, and also serves as an RNA-binding protein for at least two different steps in viral RNA synthesis (33). It has no inherent membrane-binding properties, but it interacts with another polio membrane-binding protein, 3AB (46). The mechanism by which 3CD induces association of BIG1 and BIG2 with membranes in the absence of other poliovirus proteins is not obvious and requires further investigation. Although overexpression of GBF1 rescued replication of poliovirus from inhibition by BFA, overexpression of BIG2 did not show this effect. The contribution made by each of these GEFs in the formation of poliovirus replication complexes is not yet understood. It is possible that these GEFs share some common functions and thus may substitute to a certain extent for one another or that GBF1 activated by 3A is the major player in supporting virus replication, whereas BIG1 and BIG2, brought to membranes by 3CD, may participate in other, nonessential processes. These interactions of virus and cellular proteins and their role in virus replication require further investigation.
The extensive remodeling of intracellular membranes that occurs after poliovirus and other plus-strand RNA virus infections of cultured cells was observed by electron microscopy many years ago. Despite considerable recent progress in understanding the biochemistry and cell biology of intracellular membrane trafficking and structural transformations, very little has been learned about the biochemical processes and morphological events induced by viruses to generate the functional and structural scaffolds that support viral RNA synthesis. Initial studies designed to characterize the events following poliovirus infection suggested that some products of the P2 region of the poliovirus genome (protein 2BC and/or its cleavage products) might initiate a COPII-dependent trafficking of viral proteins and cellular membranes to begin the remodeling required for the formation of viral RNA replication complexes (36). Our data suggest that cellular GEFs recruited by proteins from the P3 region of the poliovirus genome to the sites of poliovirus replication, GBF1 by 3A and BIG1/BIG2 by 3CD, in turn recruit and activate Arf1 (and likely other Arf species). This diverts these proteins from their normal activity of transforming the COPII-initiated membrane protrusions into early Golgi-like compartments and further transporting material through the secretory pathway. Instead, they participate in poliovirus-induced membrane remodeling, resulting in the clusters of vesicle-like replication complexes. The recruitment of GEFs to replication sites was not accompanied by increased binding of GAPs, thus allowing the accumulation of Arf-GTP at the replication complexes. Another series of studies provided both morphological and biochemical evidence that autophagy mechanisms may contribute to the formation of membranous vesicles that support poliovirus replication (24, 39, 41). The secretory pathway conducts traffic of cargo that is destined for insertion into the plasma membrane or secretion from the cell; autophagy is the major self-degradative process in eukaryotic cells, carrying material targeted for destruction in vesicles to the lysosomes. However, since all cellular membrane is generally thought to derive from the ER and since the points of divergence from cellular pathways to the new structures that form the viral replication complexes are not known, it may be that both of these pathways are initiated and ultimately utilized to support virus propagation.
The remodeling of cellular membrane structures to sites of viral RNA replication is complex and still far from understood. Multiple viral gene products and, no doubt, multiple cellular factors contribute to the various steps in this pathway. The data presented here suggest that viral proteins 3A and 3CD recruit Arf via the BFA-sensitive GEFs, GBF1 and BIG1/2, respectively, to participate in poliovirus-induced membrane remodeling. These activities, either sequentially or synergistically, contribute to the formation of the characteristic viral replication complexes. Thus, it seems that several nonstructural proteins from different regions of the poliovirus genome can induce interactions with components of different steps of the cellular secretory pathway in order to transform intracellular membranes into replication structures.
Acknowledgments
We are grateful to P. Randazzo for supplying purified recombinant Arf and for help with the assays for GEF activity.
This study was supported in part by the intramural research programs of the National Institute of Allergy and Infectious Disease and National Institute of Child Health and Human Development, National Institutes of Health.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Agol, V. I., G. A. Belov, K. Bienz, D. Egger, M. S. Kolesnikova, N. T. Raikhlin, L. I. Romanova, E. A. Smirnova, and E. A. Tolskaya. 1998. Two types of death of poliovirus-infected cells: caspase involvement in the apoptosis but not cytopathic effect. Virology 252:343-353. [DOI] [PubMed] [Google Scholar]
- 2.Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. [DOI] [PubMed] [Google Scholar]
- 3.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behnia, R., and S. Munro. 2005. Organelle identity and the signposts for membrane traffic. Nature 438:597-604. [DOI] [PubMed] [Google Scholar]
- 5.Belov, G. A., M. H. Fogg, and E. Ehrenfeld. 2005. Poliovirus proteins induce membrane association of GTPase ADP-ribosylation factor. J. Virol. 79:7207-7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beraud-Dufour, S., S. Robineau, P. Chardin, S. Paris, M. Chabre, J. Cherfils, and B. Antonny. 1998. A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the β-phosphate to destabilize GDP on ARF1. EMBO J. 17:3651-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 8.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1983. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology 131:39-48. [DOI] [PubMed] [Google Scholar]
- 10.Boman, A. L. 2001. GGA proteins: new players in the sorting game. J. Cell Sci. 114:3413-3418. [DOI] [PubMed] [Google Scholar]
- 11.Brunner, J. E., J. H. Nguyen, H. H. Roehl, T. V. Ho, K. M. Swiderek, and B. L. Semler. 2005. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 79:3254-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 13.Choe, S. S., and K. Kirkegaard. 2004. Intracellular topology and epitope shielding of poliovirus 3A protein. J. Virol. 78:5973-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox, R., R. J. Mason-Gamer, C. L. Jackson, and N. Segev. 2004. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell 15:1487-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuconati, A., A. Molla, and E. Wimmer. 1998. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J. Virol. 72:6456-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta, U., and A. Dasgupta. 1994. Expression and subcellular localization of poliovirus VPg-precursor protein 3AB in eukaryotic cells: evidence for glycosylation in vitro. J. Virol. 68:4468-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodd, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 75:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doedens, J., L. A. Maynell, M. W. Klymkowsky, and K. Kirkegaard. 1994. Secretory pathway function, but not cytoskeletal integrity, is required in poliovirus infection. Arch. Virol.(Suppl. 9):159-172. [DOI] [PubMed] [Google Scholar]
- 19.Fogg, M. H., N. L. Teterina, and E. Ehrenfeld. 2003. Membrane requirements for uridylylation of the poliovirus VPg protein and viral RNA synthesis in vitro. J. Virol. 77:11408-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gazina, E. V., J. M. Mackenzie, R. J. Gorrell, and D. A. Anderson. 2002. Differential requirements for COPI coats in formation of replication complexes among three genera of Picornaviridae. J. Virol. 76:11113-11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irurzun, A., L. Perez, and L. Carrasco. 1992. Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology 191:166-175. [DOI] [PubMed] [Google Scholar]
- 23.Jackson, C. L., and J. E. Casanova. 2000. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 10:60-67. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, W. T., T. H. Giddings, Jr., M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamoto, K., Y. Yoshida, H. Tamaki, S. Torii, C. Shinotsuka, S. Yamashina, and K. Nakayama. 2002. GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic 3:483-495. [DOI] [PubMed] [Google Scholar]
- 26.Liu, W., R. Duden, R. D. Phair, and J. Lippincott-Schwartz. 2005. ArfGAP1 dynamics and its role in COPI coat assembly on Golgi membranes of living cells. J. Cell Biol. 168:1053-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maynell, L. A., K. Kirkegaard, and M. W. Klymkowsky. 1992. Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 66:1985-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 29.Moss, J., and M. Vaughan. 1998. Molecules in the ARF orbit. J. Biol. Chem. 273:21431-21434. [DOI] [PubMed] [Google Scholar]
- 30.Mossessova, E., R. A. Corpina, and J. Goldberg. 2003. Crystal structure of ARF1*Sec7 complexed with brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol. Cell 12:1403-1411. [DOI] [PubMed] [Google Scholar]
- 31.Nie, Z., D. S. Hirsch, and P. A. Randazzo. 2003. Arf and its many interactors. Curr. Opin. Cell Biol. 15:396-404. [DOI] [PubMed] [Google Scholar]
- 32.Niu, T. K., A. C. Pfeifer, J. Lippincott-Schwartz, and C. L. Jackson. 2005. Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol. Biol. Cell 16:1213-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul, A. V. 2002. Possible unifying mechanism of picornavirus genome replication, p. 227-246. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 34.Peyroche, A., B. Antonny, S. Robineau, J. Acker, J. Cherfils, and C. L. Jackson. 1999. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3:275-285. [DOI] [PubMed] [Google Scholar]
- 35.Randazzo, P. A., and H. M. Fales. 2002. Preparation of myristoylated Arf1 and Arf6 proteins. Methods Mol. Biol. 189:169-179. [DOI] [PubMed] [Google Scholar]
- 36.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santy, L. C., and J. E. Casanova. 2001. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 154:599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinotsuka, C., Y. Yoshida, K. Kawamoto, H. Takatsu, and K. Nakayama. 2002. Overexpression of an ADP-ribosylation factor-guanine nucleotide exchange factor, BIG2, uncouples brefeldin A-induced adaptor protein-1 coat dissociation and membrane tubulation. J. Biol. Chem. 277:9468-9473. [DOI] [PubMed] [Google Scholar]
- 41.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolskaya, E. A., L. I. Romanova, M. S. Kolesnikova, T. A. Ivannikova, E. A. Smirnova, N. T. Raikhlin, and V. I. Agol. 1995. Apoptosis-inducing and apoptosis-preventing functions of poliovirus. J. Virol. 69:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasudevan, C., W. Han, Y. Tan, Y. Nie, D. Li, K. Shome, S. C. Watkins, E. S. Levitan, and G. Romero. 1998. The distribution and translocation of the G protein ADP-ribosylation factor 1 in live cells is determined by its GTPase activity. J. Cell Sci. 111(Pt. 9):1277-1285. [DOI] [PubMed] [Google Scholar]
- 44.Walter, B. L., T. B. Parsley, E. Ehrenfeld, and B. L. Semler. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008-12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wessels, E., D. Duijsings, K. H. Lanke, S. H. van Dooren, C. L. Jackson, W. J. Melchers, and F. J. van Kuppeveld. 2006. Effects of picornavirus 3A proteins on protein transport and GBF1-dependent COP-I recruitment. J. Virol. [DOI] [PMC free article] [PubMed]
- 46.Xiang, W., K. S. Harris, L. Alexander, and E. Wimmer. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]