Abstract
CXCR4-using (X4) human immunodeficiency virus type 1 (HIV-1) variants evolve from CCR5-using (R5) variants relatively late in the natural course of infection in 50% of HIV-1 subtype B-infected individuals and subsequently coexist with R5 HIV-1 variants. This relatively late appearance of X4 HIV-1 variants is poorly understood. Here we tested the neutralization sensitivity for soluble CD4 (sCD4) and the broadly neutralizing antibodies IgG1b12, 2F5, 4E10, and 2G12 of multiple coexisting clonal R5 and (R5)X4 (combined term for monotropic X4 and dualtropic R5X4 viruses) HIV-1 variants that were obtained at two time points after the first appearance of X4 variants in five participants of the Amsterdam Cohort Studies on HIV-1 infection and AIDS. Recently emerged (R5)X4 viruses were significantly more sensitive to neutralization by the CD4-binding-site-directed agents sCD4 and IgG1b12 than their coexisting R5 viruses. This difference was less pronounced at the later time point. Early (R5)X4 variants from two out of four patients were also highly sensitive to neutralization by autologous serum (50% inhibition at serum dilutions of >200). Late (R5)X4 viruses from these two patients were neutralized at lower serum dilutions, which suggested escape of X4 variants from humoral immunity. Autologous neutralization of coexisting R5 and (R5)X4 variants was not observed in the other patients. In conclusion, the increased neutralization sensitivity of HIV-1 variants during the transition from CCR5 usage to CXCR4 usage may imply an inhibitory role for humoral immunity in HIV-1 phenotype evolution in some patients, thus potentially contributing to the late emergence of X4 variants.
Entry of human immunodeficiency virus type 1 (HIV-1) into a host cell is mediated by binding of the viral envelope glycoprotein 120 (gp120) to CD4 and a coreceptor, of which CCR5 and CXCR4 are the most important on primary cells (7, 8, 9). Primary HIV-1 infections are generally established by R5 viruses, which remain present throughout the course of infection (27). In approximately 50% of therapy-naive individuals infected with subtype B HIV-1, X4 viruses evolve from R5 variants, preceding an increased CD4+ T-cell decline and accelerated progression to AIDS (4, 24, 27, 30).
The relatively late appearance of X4 HIV-1 variants is poorly understood. R5 and X4 subtype B HIV-1 variants can genetically be distinguished by the absence or presence of a positively charged amino acid on positions 11 and/or 25 in the third variable loop (V3) of the gp120 envelope (10). In vitro experiments revealed that these mutations in V3 (6), as well as other single or double mutations in V3 and other domains of gp120 (2, 12), are sufficient to change coreceptor usage. However, in spite of the high mutation rate of HIV-1, X4 viruses do not evolve rapidly in vivo and not in all infected patients. Moreover, the earliest detectable X4 variants in vivo show more than only one or two amino acid substitutions compared to coexisting R5 variants (16), suggestive of compensatory mutations. These observations point towards the existence of selective pressure against X4 HIV-1 evolution, the exact nature of which remains to be established.
During the conversion of CCR5 to CXCR4 usage in vitro and in vivo, HIV-1 has to traverse a phase with reduced replicative capacity and less-efficient coreceptor usage (16, 22, 34), indicating that HIV has to overcome a significant genetic hurdle to evolve from an R5 to an X4 phenotype. In addition, HIV-1 phenotype conversion may be suppressed by host immunity. In a previous study, we demonstrated that X4 variants emerged only after CD4 counts had dropped below 400 cells/μl blood (15). X4 variants may thus be considered opportunistic viruses that emerge as a result of immune failure and subsequently give rise to an accelerated loss of CD4+ T cells. Since the interaction between the viral envelope proteins and the host cell receptors may be prevented by neutralizing antibodies, humoral immunity could play a role in the evolution of HIV-1 coreceptor usage by differentially neutralizing R5 and X4 variants.
In our present study, we compared the neutralization sensitivities of clonal R5 and (R5)X4 (this term is used throughout the paper for both monotropic X4 and dualtropic R5X4) HIV-1 variants that coexisted in vivo and that were isolated from five individuals around the moment of the first appearance of X4 variants and at a later time point during symptomatic disease. We show that R5 and (R5)X4 variants have different susceptibilities to CD4-binding-site (CD4bs)-directed agents. For two out of four patients, the (R5)X4 variants isolated early after the appearance of X4 were potently neutralized by autologous serum. We postulate that the difference in neutralization by host humoral immunity may influence virus phenotype evolution in vivo in these patients. However, the absence of a detectable anti-X4 response in others argues against a major role for the humoral response in this process and indicates that other selective pressures are also involved.
MATERIALS AND METHODS
Patients and viruses.
Patients ACH0039, ACH0171, ACH1120, and ACH6052 were homosexual male participants of the Amsterdam Cohort studies on HIV-1 and AIDS who all developed X4 variants during a progressive disease course. Patient ACH9012 (female) was infected after a deliberate injection of a few ml of blood drawn from an AIDS patient (36). All patients were infected with subtype B HIV-1. None of the participants ever received multidrug antiretroviral therapy. Biological virus clones from time points early and late after the first appearance of X4 variants were obtained as previously described (27). For patient ACH9012, viruses of one time point, 3.5 months after seroconversion, were used. Phylogenetic analysis of coexisting R5 and X4 variants (data not shown) in combination with the equal contributions of R5 and X4 variants to total cellular viral load in the donor (35) implicate that the length of the period of R5 and X4 coexistence was such that the time point of virus isolation from ACH9012 equals the time point late after X4 emergence in the other patients. The moment of seroconversion was calculated as the midpoint between the last seronegative visit and the first seropositive visit. Similarly, the moment of the first appearance of X4 viruses was calculated as the midpoint between the last MT2-negative visit and the first MT2-positive visit.
For all of the HIV-1 variants studied here, the ability to replicate in the MT2 cell line was considered evidence of CXCR4 usage. In addition, CXCR4 usage for all clones was confirmed in peripheral blood mononuclear cells (PBMC) from a healthy donor homozygous for the 32-bp deletion in the CCR5 gene (CCR5Δ/Δ). For subjects ACH0039, ACH0171, ACH1120, and ACH6052, coreceptor usage was confirmed in U87 indicator cell lines expressing CD4 and either CCR5 or CXCR4. In previous studies, we determined that the replication of R5X4 and X4 virus variants of all patients in PBMC could be inhibited by AMD3100, indicating that these variants mainly use CXCR4 to enter these primary cells (13, 29). The sequences of the gp120 V3 domains of R5 and (R5)X4 variants from various time points have been determined, all showing the amino acid residues at positions 11 and/or 25 of the V3 domain that are commonly associated with an R5 or X4 virus phenotype (33; also data not shown). To prevent a change in sensitivity of the virus variants to neutralization during in vitro culture, a minimum number of passages of the viruses in PBMC was performed (1).
Primary cells.
PBMC were isolated from buffy coats obtained from healthy seronegative blood donors by Ficoll-Isopaque density gradient centrifugation. Cells (5 × 106/ml) were stimulated for 2 days in Iscove's modified Dulbecco medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and phytohemagglutinin (PHA) (5 μg/ml). Subsequently, cells (106/ml) were grown in the absence of PHA in medium supplemented with recombinant interleukin-2 (20 U/ml, Chiron Benelux, Amsterdam, The Netherlands).
Neutralization assay.
Viruses were tested for their relative neutralization sensitivities against recombinant soluble CD4 (sCD4) (Progenics, Tarrytown, NY) and the human monoclonal antibodies (MAbs) IGg1b12 (a kind gift from D. Burton), 2F5, 2G12, and 4E10 (Polymun Scientific, Vienna, Austria), autologous serum, and pooled serum from healthy uninfected individuals. From each virus isolate, a final inoculum of 10 50% tissue culture infective doses in a volume of 100 μl was incubated for 1 h at 37°C with increasing concentrations of the neutralizing agents. Subsequently, the mixtures of virus with sCD4 or antibodies were added to 105 PHA-stimulated PBMC derived from healthy blood donors. PBMC incubated with sera were washed after 4 h. On day 7, virus production in culture supernatants was analyzed by an in-house p24 antigen capture enzyme-linked immunosorbent assay (ELISA) (31). Experiments were performed in triplicate. The percent neutralization was calculated by determining the reduction in p24 production in the presence of the agent compared to that in the cultures with virus only. In experiments measuring the neutralization by autologous serum, the percentage of inhibition by pooled HIV-1-negative serum was subtracted from the neutralization obtained using patient serum samples. When possible, 50% inhibitory concentration (IC50s) were determined by linear regression.
Statistical analysis.
For analysis of the neutralization by sCD4 and monoclonal antibodies, IC50s were evaluated per virus variant using the Mann-Whitney U test. Viruses with IC50s of <0.2 or >12.5 were assigned a value of 0.1 or 12.5, respectively, for calculations and statistical analyses. IC50s of the autologous serum neutralization were evaluated for each time point and viral phenotype using the Wilcoxon signed ranks test. For this test, the IC50s for the two variants used per time point and phenotype were linked per serum sample to the IC50s of two other virus isolates of another time point or phenotype. Viruses with IC50s of <40 or >1,280 were assigned a value of 20 or 1,280, respectively.
RESULTS
Patients and viruses.
Longitudinally obtained coexisting R5 and dualtropic R5X4 and X4 [(R5)X4] viruses from five participants of the Amsterdam Cohort Studies were available from previous studies (14, 33, 34, 36). From four patients infected via homosexual contact (ACH0039, ACH0171, ACH1120, and ACH6052), biological virus clones were isolated shortly after the first detection of X4 variants and at a later time point 2 years before (ACH0171) or during symptomatic disease (Table 1). Patient ACH9012 (female) was parenterally infected with a mixture of R5 and X4 viruses that were first isolated 3.5 months after transmission (Table 1). Since X4 variants had already developed in the donor, the exact time since X4 appearance was not known, and these viruses were considered late post-X4 emergent (35) (see Materials and Methods).
TABLE 1.
Characteristics of R5 and (R5)X4 HIV-1 virus variants
| Patient no. (sexa; seropositivity at entry) | Viruses early post-X4
|
Viruses late post-X4
|
||||
|---|---|---|---|---|---|---|
| Time since SC or entry (mo)b | Time since X4 variant detected (mo) | Coreceptor usage (n) | Time since SC or entry (mo)b | Time since X4 variant detected (mo) | Coreceptor usage (n)c | |
| ACH0039 (M; no) | 11.2 | −4.9 | R5 (1) | 38.9 | 22.8 | R5 (2), X4 (1) |
| 18.2 | 2.1 | R5 (1), R5X4 (2) | 51.2 | 35.0 | R5 (1), X4 (3) | |
| 21.1 | 5.0 | R5 (2), R5X4 (1) | ||||
| ACH0171 (M; no) | 61.0 | −1.5 | R5 (1), R5X4 (1)c | 89.5 | 27.0 | R5 (4), R5X4 (4) |
| 67.0 | 4.5 | R5 (2), R5X4 (2) | ||||
| 73.0 | 10.5 | R5 (1), R5X4 (1) | ||||
| ACH1120 (M; no) | 53.3 | 6.0 | R5 (4), R5X4 (2), X4 (2) | 64.6 | 17.2 | R5 (4) |
| 66.4 | 19.1 | R5X4 (4) | ||||
| ACH6052 (M; yes) | 0d | Unknownd | R5 (3), X4 (5) | 32.0 | Unknownd | R5 (3), X4 (5) |
| ACH9012 (F; no) | NTe | NT | NT | 3.5 | 3.5 | R5 (3), X4 (3) |
M, male; F, female.
Time since seroconversion (SC) or entry of patient into HIV+ cohort.
As determined with transfected U87 indicator cell lines. n, no. of clones. For each time point, the presence of CXCR4-using variants in a bulk virus culture was determined in the MT2 assay, in which CXCR4 usage of single virus isolates can be missed.
ACH6052 was seropositive and carried X4 variants at the time of entry into the cohort.
NT, not tested.
Sensitivities of coexisting R5 and (R5)X4 variants to neutralization by CD4-binding-site-directed agents.
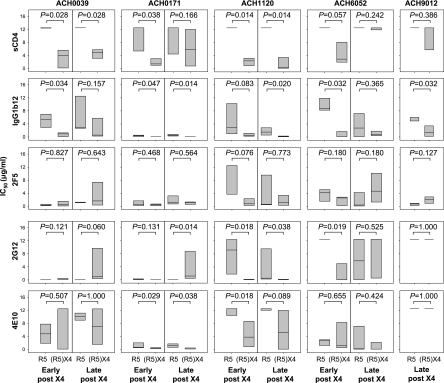
The sensitivities of clonal coexisting R5 and (R5)X4 viruses to neutralization by sCD4 and the CD4-binding-site-directed MAb IgG1b12 were analyzed. R5 viruses obtained early after X4 emergence from all patients were resistant to sCD4 neutralization (IC50s >12.5 μg/ml) (Fig. 1), while their coexisting (R5)X4 viruses were relatively susceptible (median IC50s ranging between 1.40 and 4.02 μg/ml) (Fig. 1), although this approached significance only in patient ACH6052. In line with this observation, (R5)X4 variants obtained early after X4 emergence showed an increased sensitivity to IgG1b12 compared to their coexisting R5 variants, although this was not statistically significant for viruses from patient ACH1120 (Fig. 1).
FIG. 1.
Sensitivities for sCD4-, IgG1b12-, 2F5-, 2G12-, and 4E10-mediated neutralization of coexisting R5 and (R5)X4 HIV-1 obtained early or late after X4 emergence. Neutralization was determined by a 7-day culture of clonal virus isolates on PBMC in the presence or absence of serial dilutions of neutralizing agent, followed by analysis of p24 production by ELISA. Distribution of IC50s for individual virus clones as determined by linear regression is shown. Bottom, middle, and top horizontal lines represent the 25th, 50th, and 75th percentiles, respectively. Per time point, IC50s of R5 and (R5)X4 viruses were evaluated using the Mann-Whitney U test. Data shown are from one representative experiment out of two to three performed.
For three out of five patients (ACH0039, ACH0171, and ACH1120), (R5)X4 viruses obtained late after X4 emergence still showed significantly higher sensitivities to neutralization by sCD4 and/or IgG1b12 than their coexisting R5 variants. For viruses isolated from the other patients, differential neutralization sensitivities were lost due to an increased susceptibility of the R5 viruses to IgG1b12 (although this reached statistical significance only for viruses from patient ACH6052 and not for those from patient ACH0039) and/or an increased resistance to sCD4 of the (R5)X4 variants (statistically significant for viruses from ACH6052 but not for viruses from ACH0171). X4 viruses from patient ACH9012 showed higher neutralization sensitivities to IgG1b12 and equal resistance to sCD4 compared to those of the coexisting R5 variants.
Equal sensitivities of coexisting R5 and (R5)X4 variants to zidovudine excluded that the differences in neutralization sensitivity for sCD4 and IgG1b12 observed here were the result of differences in the replication rate between the various virus variants (data not shown).
Sensitivities of coexisting R5 and (R5)X4 HIV-1 variants to neutralization by non-CD4bs-directed agents.
We next analyzed whether coexisting R5 and X4 variants also had differential susceptibilities to neutralization via epitopes outside the CD4-binding site. To this end we determined the sensitivities of the coexisting R5 and X4 viruses to neutralization by MAb 2G12 (recognizing a carbohydrate epitope on gp120) and monoclonal antibodies 2F5 and 4E10 (both binding to the membrane-proximal external region of gp41). For 2G12, increased neutralization sensitivity compared to that of their coexisting R5 variants was observed for early and late (R5)X4 viruses in patient ACH1120 and for the early (R5)X4 viruses in patient ACH6052 (Fig. 1). In contrast, late (R5)X4 viruses from patient ACH0171 were significantly more resistant to 2G12 neutralization than their coexisting R5 variants. For 4E10, early (R5)X4 viruses from patient ACH0171 and early and late (R5)X4 viruses from patient ACH1120 were more sensitive to neutralization than their coexisting R5 viruses (Fig. 1). No differences were observed for 2F5 (Fig. 1). Overall, some differences in neutralization susceptibilities were observed between R5 and (R5)X4 viruses for non-CD4bs-directed agents but not to the same extent as seen for sCD4 and IgG1b12.
Sensitivities of coexisting R5 and (R5)X4 HIV-1 variants to neutralization by autologous serum.
The differential neutralization sensitivities of coexisting R5 and (R5)X4 variants might suggest that X4 variants can emerge only in the absence of neutralizing humoral immunity. To examine whether the humoral immune response indeed deteriorated around the time point of the appearance of X4, we analyzed the neutralization sensitivities of coexisting R5 and (R5)X4 viruses to autologous serum samples obtained before and after the first appearance of X4 variants. Since ACH6052 and ACH9012 carried X4 variants at the time of entry into the cohort, only serum samples obtained after X4 emergence were available from these patients. Unfortunately, due to limited amounts of serum, it was not possible to analyze all virus variants. We therefore randomly chose two virus variants per time point and phenotype for determination of neutralization by autologous serum. Early (R5)X4 variants from patients ACH0171 and ACH6052 were highly sensitive to neutralization by sera from all time points (IC50 titers, >200) (Tables 2 and 3), which was significantly different from neutralization of the early R5 variants (for ACH0171, P = 0.018; for ACH6052, P = 0.027), as well as the late X4 variants (for ACH0171, P = 0.018; for ACH6052, P = 0.026). Virus variants from patients ACH0039, ACH1120, and ACH9012, irrespective of phenotype or time point of isolation, either were not neutralized or were neutralized at relatively low serum dilutions (Table 4, Table 5 and Table 6). No differences between virus variants from different time points or in viral tropism were observed for these patients (P > 0.050).
TABLE 2.
Autologous serum neutralization titers of early and late post-X4 virus clones of ACH0171
| Clone | Time since X4 (mo)a | Phenotype | Neutralization titer (IC50) at mo since X4a,b
|
|||
|---|---|---|---|---|---|---|
| −16.2 | −7.5 | 10.5 | 27.0 | |||
| 171.31.ROF6 | 4.5 | R5 | 125 | 79 | 69 | 87 |
| 171.33.ROA1 | 10.5 | R5 | 87 | 84 | 81 | 114 |
| 171.41.ROE2 | 27.0 | R5 | <40 | <40 | <40 | <40 |
| 171.41.ROC5 | 27.0 | R5 | <40 | <40 | <40 | <40 |
| 171.31.ROB9 | 4.5 | R5X4 | >1,280 | >1,280 | >1,280 | >1,280 |
| 171.33.ROA2 | 10.5 | R5X4 | 151 | 768 | 343 | NT |
| 171.41.D12 | 27.0 | R5X4 | 84 | <40 | 82 | <40 |
| 171.41.RAF5 | 27.0 | R5X4 | 51 | <40 | <40 | 80 |
Months since X4 variant was detected.
Neutralization was determined by 7-day culture of clonal virus isolates on PBMC in the presence or absence of serial dilutions of serum, followed by analysis of p24 production by ELISA. IC50s were determined by linear regression after subtracting the inhibition obtained using human pooled HIV-1-negative serum.
TABLE 3.
Autologous serum neutralization titers of early and late post-X4 virus clones of ACH6052
| Clone | Time since entry (mo)a | Phenotype | Neutralization titer (IC50) at mo since entrya,b
|
||
|---|---|---|---|---|---|
| 0 | 6.8 | 32.1 | |||
| 6052.1G2 | 0 | R5 | <40 | <40 | 50 |
| 6052.1E2 | 0 | R5 | <40 | <40 | <40 |
| 6052.4C4 | 32.1 | R5 | <40 | <40 | <40 |
| 6052.4C3 | 32.1 | R5 | <40 | <40 | <40 |
| 6052.1H4 | 0 | X4 | >1,280 | 339 | >1,280 |
| 6052.1G3 | 0 | X4 | >1,280 | 396 | 288 |
| 6052.4A10 | 32.1 | X4 | <40 | <40 | <40 |
| 6052.4B11 | 32.1 | X4 | <40 | <40 | <40 |
Months since entry of patient into HIV+ cohort.
Neutralization was determined by 7-day culture of clonal virus isolates on PBMC in the presence or absence of serial dilutions of serum, followed by analysis of p24 production by ELISA. IC50s were determined by linear regression after subtracting the inhibition obtained using human pooled HIV-1-negative serum.
TABLE 4.
Autologous serum neutralization titers of early and late post-X4 virus clones of ACH1120
| Clone | Time since X4 (mo)a | Phenotype | Neutralization titer (IC50) at mo since X4a,b
|
|||
|---|---|---|---|---|---|---|
| (−18.0) | (−8.7) | (6.0) | (17.3) | |||
| 1120.53.1B6 | 6.0 | R5 | <40 | <40 | <40 | <40 |
| 1120.53.1E4 | 6.0 | R5 | <40 | <40 | <40 | <40 |
| 1120.57.3E7 | 17.3 | R5 | <40 | <40 | <40 | 52 |
| 1120.57.3C4 | 17.3 | R5 | <40 | <40 | <40 | 85 |
| 1120.254 | 6.0 | X4 | 42 | <40 | <40 | 44 |
| 1120.255 | 6.0 | X4 | <40 | <40 | <40 | 41 |
| 1120.267 | 19.1 | R5X4 | <40 | <40 | <40 | <40 |
| 1120.268 | 19.1 | R5X4 | <40 | <40 | <40 | <40 |
Months since X4 variant was detected.
Neutralization was determined by 7-day culture of clonal virus isolates on PBMC in the presence or absence of serial dilutions of serum, followed by analysis of p24 production by ELISA. IC50s were determined by linear regression after subtracting the inhibition obtained using human pooled HIV-1-negative serum.
TABLE 5.
Autologous serum neutralization titers of early and late post-X4 virus clones of ACH0039
| Clone | Time since X4 (mo)a | Phenotype | Neutralization titer (IC50) at mo since X4a,b
|
|||
|---|---|---|---|---|---|---|
| −9.4 | −2.8 | 7.1 | 37.1 | |||
| 39.18.E11 | −4.9 | R5 | <40 | <40 | <40 | <40 |
| 39.21.D5 | 5.0 | R5 | <40 | 173 | 62 | 55 |
| 39.20.1E10 | 2.1 | R5X4 | 82 | 169 | 197 | 277 |
| 39.21.H10 | 5.0 | R5X4 | <40 | 177 | <40 | 60 |
| 39.28.A9 | 22.8 | X4 | <40 | <40 | <40 | <40 |
| 39.*1.C4 | 35.0 | X4 | <40 | 93 | 222 | <40 |
Months since X4 variant was detected.
Neutralization was determined by 7-day culture of clonal virus isolates on PBMC in the presence or absence of serial dilutions of serum, followed by analysis of p24 production by ELISA. IC50s were determined by linear regression after subtracting the inhibition obtained using human pooled HIV-1-negative serum.
TABLE 6.
Autologous serum neutralization titers of early and late post-X4 virus clones of ACH9012
| Clone | Time since entry (mo)a | Phenotype | Neutralization titer (IC50) at 3.5 mo since entrya,b |
|---|---|---|---|
| 9012.A10 | 3.5 | R5 | <40 |
| 9012.A2 | 3.5 | R5 | <40 |
| 9012.A7 | 3.5 | R5 | <40 |
| 9012.E6 | 3.5 | X4 | <40 |
| 9012.E10 | 3.5 | X4 | <40 |
| 9012.F3 | 3.5 | X4 | <40 |
Months since entry of patient into HIV+ cohort.
Neutralization was determined by 7-day culture of clonal virus isolates on PBMC in the presence or absence of serial dilutions of serum, followed by analysis of p24 production by ELISA. IC50s were determined by linear regression after subtracting the inhibition obtained using human pooled HIV-1-negative serum.
DISCUSSION
We compared the neutralization sensitivities of coexisting R5 and (R5)X4 viruses isolated early and late after X4 emergence. In a previous study, we (11) and others (3, 17, 20, 32) reported no difference in neutralization sensitivities between R5, X4, or dualtropic R5X4 HIV-1 from different patients. However, even if differences among the various viruses had been observed, these most likely would have reflected interpatient variability of the neutralization sensitivities of unrelated HIV-1 variants rather than differences determined by coreceptor usage. In our present study, we therefore compared R5 and (R5)X4 virus clones that had been isolated from the same patient at the same time points.
Our data show that (R5)X4 viruses obtained early after X4 conversion are more sensitive to CD4-binding-site-directed agents than their coexisting R5 variants, whereas this differential sensitivity was less pronounced at a later time point. Since the differential sensitivity for non-CD4-binding-site-directed antibodies was not a general phenomenon among the five patients included in this study, mutations in the (co)receptor binding site associated with evolution from CCR5 to CXCR4 usage most likely change the neutralization phenotype in this region but do not change the general envelope conformation. In addition, coexisting dualtropic R5X4 and X4 viruses early after X4 emergence, although available from only one patient, had comparable neutralization sensitivities (data not shown). We therefore propose that the acquisition of CXCR4 usage, rather than the loss of CCR5 usage, coincides with changes in gp120 that increase neutralization sensitivity. In line with this is an observation by Lusso et al. (19), who showed that X4 viruses constitutively expose the epitope for monoclonal antibody D19, whereas R5 viruses are neutralized by this antibody only after pretreatment with sCD4, which would indeed imply a difference in conformation between R5 and X4 variants.
The neutralization-sensitive conformation of the viral envelope during the evolution from an R5 phenotype to an X4 phenotype would suggest that transition is possible only when an efficient neutralizing-antibody response is absent or lost due to immune deterioration associated with disease progression. However, for two out of four patients, the early (R5)X4 variants were efficiently neutralized by autologous serum, suggestive for the presence of neutralizing antibodies in vivo. Late (R5)X4 variants from these patients were not neutralized, indicating that (R5)X4 viruses had evolved towards increased neutralization resistance. This is most clearly demonstrated with patient ACH0171, from whom the earliest (R5)X4 virus, isolated 4.5 months after X4 appearance, was neutralized very efficiently (IC50 titers of >1,280). A virus isolated 10.5 months after X4 appearance was partially adapted to the autologous antibody response (IC50 titers between 151 and 768), whereas the late viruses, isolated 27.0 months after X4 appearance, were almost completely resistant to autologous serum neutralization. We observed differential autologous neutralization only between recently emerged (R5)X4 viruses and their coexisting R5 variants. The use of X4 and R5 viruses from later time points in infection may explain the lack of differential neutralization by serum between R5 and X4 viruses, as observed in a previous study by others (28).
Neutralizing antibodies present in serum obtained at a certain time point in infection generally neutralize viruses found during earlier time points (25, 37). Continuous viral escape makes the neutralization of HIV-1 by contemporaneous serum a rare event (25, 37). It is therefore not surprising that the R5 and (R5)X4 virus variants that were isolated late after X4 emergence were not neutralized by serum obtained at the earlier time points. However, we had not expected that R5 viruses isolated early after X4 emergence would resist neutralization by serum obtained later in infection. This may point to a decreasing neutralizing humoral immune response late in infection.
We were also surprised that the (R5)X4 viruses isolated shortly after the first appearance of X4 variants in patient ACH0171 were potently neutralized by serum samples obtained at earlier time points, even before the appearance of X4 viruses. Possibly, the conformational requirements of an early X4 envelope led to the exposure of certain neutralization epitopes that were initially exposed on R5 trimeric envelopes before the R5 viruses escaped from the antibodies directed against these epitopes. Alternatively, early X4 viruses might expose epitopes that are vulnerable to antibodies directed against other envelope structures, such as monomeric gp120, and initially have an oligomeric envelope conformation that resembles the oligomeric gp120 of neutralization-sensitive T-cell-line-adapted HIV-1. Indeed, a large proportion of the antibody repertoire against HIV-1 envelope protein is directed against nonneutralizing epitopes on shed, misfolded, or otherwise nonfunctional forms of gp120 (21, 26), which could explain why such antibodies are already present before X4 viruses have emerged. Another possibility for the presence of antibodies directed against epitopes present on early X4 variants is that the virus has evolved towards CXCR4 usage earlier during infection but was previously inhibited by a potent humoral immune response. As a result of a decrease in selection pressure, the X4 variants may have been selected from the PBMC archive.
For three out of five patients, we did not observe effective neutralization of the early X4 viruses by serum samples from any time point. It has previously been shown that some patients fail to develop a humoral immune response against HIV-1 (25), which may explain the lack of neutralization observed in our study. Indeed, serum of patient ACH1120, which did not show neutralization of autologous virus, also did not contain broadly neutralizing antibodies (data not shown). In contrast, serum from ACH0171 displayed potent autologous neutralization as well as broadly neutralizing activity (data not shown). Patient ACH0039 rapidly progressed to AIDS (in 3.2 years after seroconversion), and may not have developed an effective immune response against HIV-1. As described above, patient ACH9012 became infected with a mixture of R5 and X4 variants, which prevented study of the role of autologous neutralization sensitivity in R5-to-X4 evolution. Interestingly, the proportion of X4 variants in serum in this patient increased preseroconversion but sharply decreased postseroconversion (5), suggestive for selective suppression of X4 variants by humoral immunity, although serum from ACH9012 did not show autologous neutralization. However, HIV-1 variants and the only serum from this patient were both obtained 3.5 months after seroconversion, again underscoring that neutralization of virus by contemporaneous serum is not commonly observed.
The observation that the emergence of X4 viruses is not prevented by the presence of neutralizing antibodies indicates that these viruses most likely appear in body compartments with lower antibody pressure than that in plasma. On the other hand, the early X4 variants used in this study have been isolated from PBMC, indicating that neutralization-sensitive X4 viruses were able to replicate in an environment containing neutralizing antibodies. The fact that X4 viruses are not hampered by the presence of neutralizing antibodies might suggest that these variants spread via direct cell-to-cell transmission, which limits the possibility of antibody neutralization.
In conclusion, we have shown here that early X4 variants are more susceptible to antibody neutralization than their coexisting R5 variants and that some patients mount a potent humoral immune response against these CXCR4-using variants. Although the humoral immune response in these patients was not sufficient to prevent the appearance of X4 variants, strong humoral immunity in certain patients could thus contribute to the inability of the virus to evolve to X4 usage. In this light, the much lower X4 conversion rate of subtype C HIV-1 (23) could be causally related to the stronger autologous neutralizing antibody titer for subtype C HIV-infected individuals (18). However, the observation that an anti-X4 response is absent for the other patients included in this study indicates that evolution of the virus phenotype may also be influenced by factors other than autologous humoral immunity.
Acknowledgments
The Amsterdam Cohort Studies on HIV infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Center of the University of Amsterdam, the Sanquin Blood Supply Foundation, and the University Medical Center Utrecht, are part of The Netherlands HIV Monitoring Foundation and are financially supported by The Netherlands National Institute for Public Health and the Environment. This study was financially supported by the Dutch AIDS fund (grants 2004064 and 7009).
We thank Dennis Burton for his generous supply of IgG1b12.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Beaumont, T., E. Quakkelaar, A. van Nuenen, R. Pantophlet, and H. Schuitemaker. 2004. Increased sensitivity to CD4 binding site-directed neutralization following in vitro propagation on primary lymphocytes of a neutralization-resistant human immunodeficiency virus IIIB strain isolated from an accidentally infected laboratory worker. J. Virol. 78:5651-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, M. T., G. R. Simpson, A. J. Cann, M. A. Johnson, and R. A. Weiss. 1993. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J. Virol. 67:3649-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelissen, M., G. A. Mulder-Kampinga, J. Veenstra, F. Zorgdrager, C. L. Kuiken, S. Hartman, J. Dekker, L. Van der Hoek, C. Sol, R. A. Coutinho, and J. Goudsmit. 1995. Syncytium-inducing (SI) phenotype suppression at seroconversion after intramuscular inoculation of a non-syncytium-inducing/SI phenotypically mixed human immunodeficiency virus population. J. Virol. 69:1810-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, H. K., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Suttons, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of the major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 8.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 9.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 10.Fouchier, R. A. M., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groenink, M., J. P. Moore, S. Broersen, and H. Schuitemaker. 1995. Equal levels of gp120 retention and neutralization resistance of phenotypically distinct primary human immunodeficiency virus type 1 variants upon soluble CD4 treatment. J. Virol. 69:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrowe, G., and C. Cheng-Mayer. 1995. Amino acid changes in the V3 loop are responsible for adaptation to growth in transformed T-cell lines of a primary human immunodeficiency virus type 1. Virology 210:490-494. [DOI] [PubMed] [Google Scholar]
- 13.Koning, F. A., D. Schols, and H. Schuitemaker. 2001. No selection for CCR5 coreceptor usage during parenteral transmission of macrophage-tropic syncytium-inducing human immunodeficiency virus type 1. J. Virol. 75:8848-8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koot, M., A. B. Van 't Wout, N. A. Kootstra, R. E. Y. De Goede, M. Tersmette, and H. Schuitemaker. 1996. Relation between changes in cellular load, evolution of viral phenotype, and the clonal composition of virus populations in the course of human immunodeficiency virus type 1 infection. J. Infect. Dis. 173:349-354. [DOI] [PubMed] [Google Scholar]
- 15.Koot, M., R. Van Leeuwen, R. E. Y. De Goede, S. A. Danner, J. K. M. Eeftink Schattenkerk, J. M. A. Lange, M. Tersmette, P. Reiss, and H. Schuitemaker. 1999. Conversion rate towards a syncytium inducing (SI) phenotype during different stages of HIV-1 infection and prognostic value of SI phenotype for survival after AIDS diagnosis. J. Infect. Dis. 179:254-258. [DOI] [PubMed] [Google Scholar]
- 16.Kuiken, C. L., J.-J. De Jong, E. Baan, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J. Virol. 66:4622-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaCasse, R. A., K. E. Follis, T. Moudgil, M. Trahey, J. M. Binley, V. Planelles, S. Zolla-Pazner, and J. H. Nunberg. 1998. Coreceptor utilization by human immunodeficiency virus type 1 is not a primary determinant of neutralization sensitivity. J. Virol. 72:2491-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, B., J. M. Decker, R. W. Johnson, F. Bibollet-Ruche, X. Wei, J. Mulenga, S. Allen, E. Hunter, B. H. Hahn, G. M. Shaw, J. L. Blackwell, and C. A. Derdeyn. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 80:5211-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lusso, P., P. L. Earl, F. Sironi, F. Santoro, C. Ripamonti, G. Scarlatti, R. Longhi, E. A. Berger, and S. E. Burastero. 2005. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J. Virol. 79:6957-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montefiori, D. C., R. G. Collman, T. R. Fouts, J. Y. Zhou, M. Bilska, J. A. Hoxie, J. P. Moore, and D. P. Bolognesi. 1998. Evidence that antibody-mediated neutralization of human immunodeficiency virus type 1 by sera from infected individuals is independent of coreceptor usage. J. Virol. 72:1886-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parren, P. W., M. C. Gauduin, R. A. Koup, P. Poignard, P. Fisicaro, D. R. Burton, and Q. J. Sattentau. 1997. Relevance of the antibody response against human immunodeficiency virus type 1 envelope to vaccine design. Immunol. Lett. 57:105-112. [DOI] [PubMed] [Google Scholar]
- 22.Pastore, C., A. Ramos, and D. E. Mosier. 2004. Intrinsic obstacles to human immunodeficiency virus type 1 coreceptor switching. J. Virol. 78:7565-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ping, L. H., J. A. Nelson, I. F. Hoffman, J. Schock, S. L. Lamers, M. Goodman, P. Vernazza, P. Kazembe, M. Maida, D. Zimba, M. M. Goodenow, J. J. Eron, Jr., S. A. Fiscus, M. S. Cohen, and R. Swanstrom. 1999. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 73:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richman, D. D., and S. A. Bozzette. 1994. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J. Infect. Dis. 169:968-974. [DOI] [PubMed] [Google Scholar]
- 25.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. Y. De Goede, R. P. Van Steenwijk, J. M. A. Lange, J. K. M. Eeftink Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skrabal, K., S. Saragosti, J. L. Labernardiere, F. Barin, F. Clavel, and F. Mammano. 2005. Human immunodeficiency virus type 1 variants isolated from single plasma samples display a wide spectrum of neutralization sensitivity. J. Virol. 79:11848-11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stalmeijer, E. H., R. P. van Rij, B. Boeser-Nunnink, J. A. Visser, M. A. Naarding, D. Schols, and H. Schuitemaker. 2004. In vivo evolution of X4 human immunodeficiency virus type 1 variants in the natural course of infection coincides with decreasing sensitivity to CXCR4 antagonists. J. Virol. 78:2722-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tersmette, M., J. M. A. Lange, R. E. Y. De Goede, F. De Wolf, J. K. M. Eeftink Schattenkerk, P. Th. A. Schellekens, R. A. Coutinho, J. G. Huisman, J. Goudsmit, and F. Miedema. 1989. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet i:983-985. [DOI] [PubMed] [Google Scholar]
- 31.Tersmette, M., I. N. Winkel, M. Groenink, R. A. Gruters, P. Spence, E. Saman, G. van der Groen, F. Miedema, and J. G. Huisman. 1989. Detection and subtyping of HIV-1 isolates with a panel of characterized monoclonal antibodies to HIV-p24 gag. Virology 171:149-155. [DOI] [PubMed] [Google Scholar]
- 32.Trkola, A., T. Ketas, V. N. KewalRamani, F. Endorf, J. M. Binley, H. Katinger, J. Robinson, D. R. Littman, and J. P. Moore. 1998. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J. Virol. 72:1876-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Rij, R. P., H. Blaak, J. A. Visser, M. Brouwer, R. Rientsma, S. Broersen, A. M. De Roda Husman, and H. Schuitemaker. 2000. Differential coreceptor expression allows for independent evolution of non-syncytium-inducing and syncytium-inducing HIV-1. J. Clin. Investig. 106:1039-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van 't Wout, A. B., H. Blaak, L. J. Ran, M. Brouwer, C. Kuiken, and H. Schuitemaker. 1998. Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J. Virol. 72:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van 't Wout, A. B., N. A. Kootstra, G. A. Mulder-Kampinga, N. Albrecht-van Lent, H. J. Scherpbier, J. Veenstra, K. Boer, R. A. Coutinho, F. Miedema, and H. Schuitemaker. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral and vertical transmission. J. Clin. Investig. 94:2060-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veenstra, J., R. Schuurman, M. Cornelissen, A. B. Van 't Wout, C. A. B. Boucher, H. Schuitemaker, J. Goudsmit, and R. A. Coutinho. 1995. Transmission of zidovudine-resistant human immunodeficiency virus type 1 variants following deliberate injection with blood of a patient with AIDS: characteristics and natural history of the virus. Clin. Infect. Dis. 21:556-560. [DOI] [PubMed] [Google Scholar]
- 37.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]