Abstract
Mouse cells do not support human immunodeficiency virus type 1 (HIV-1) replication because of host range barriers at steps including virus entry, transcription, RNA splicing, polyprotein processing, assembly, and release. The exact mechanisms for the suppression, however, are not completely understood. To elucidate further the barriers against HIV-1 replication in mouse cells, we analyzed the replication of the virus in lymphocytes from human CD4/CXCR4 transgenic mice. Although primary splenocytes and thymocytes allowed the entry and reverse transcription of HIV-1, the integration efficiency of the viral DNA was greatly reduced in these cells relative to human peripheral blood mononuclear cells, suggesting an additional block(s) before or at the point of host chromosome integration of the viral DNA. Preintegration processes were further analyzed using HIV-1 pseudotyped viruses. The reverse transcription step of HIV-1 pseudotyped with the envelope of murine leukemia virus or vesicular stomatitis virus glycoprotein was efficiently supported in both human and mouse cells, but nuclear import of the preintegration complex (PIC) of HIV-1 was blocked in mouse cells. We found that green fluorescent protein (GFP)-labeled HIV-1 integrase, which is known to be important in the nuclear localization of the PIC, could not be imported into the nucleus of mouse cells, in contrast to human cells. On the other hand, GFP-Vpr localized exclusively to the nuclei of both mouse and human cells. These observations suggest that, due to the dysfunction of integrase, the nuclear localization of PIC is suppressed in mouse cells.
A small animal model for AIDS would provide a valuable tool for the study of its pathogenesis and the evaluation of vaccine candidates and antiviral drugs. However, attempts to produce small animal models have been hampered thus far by species-specific host range barriers to infection by human immunodeficiency virus type 1 (HIV-1). CD4, the cellular receptor for HIV-1 (41, 49), was first identified as a host range barrier because mouse CD4 (muCD4) does not bind HIV-1 Env (46). Human CD4 (huCD4) transgenic (Tg) mice, however, were not susceptible to HIV-1 infection, suggesting the presence of additional barriers (47). Chemokine receptors were later identified as entry coreceptors (9, 22), but primary lymphocytes from mice transgenic for huCD4 and either huCXCR4 (70) or huCCR5 (13) exhibited little to no signs of productive infection.
Cyclin T1 (CycT1) is responsible for a transcriptional level barrier (3, 4, 26, 30, 58). CycT1 protein is a component of the TAK/pTEFb transcription factor complex (51, 78), and huCycT1 binds Tat and activates transcription from the promoter in the long terminal repeat (LTR). However, muCycT1 cannot bind Tat. Nevertheless, introduction of the huCycT1 protein to rodent cells together with a mixture of human receptors was insufficient to induce productive viral infection (11, 52).
Additional barriers have been reported in the late steps of the viral life cycle (11, 27, 40, 42, 43, 53). These late-stage defects can be rescued by fusing HIV-1-infected rodent cells to uninfected human cells (11, 52), indicating that the defects are due to the lack of necessary factors in rodent cells rather than the presence of dominant inhibitors of HIV-1 replication. CRM1, a nuclear export factor that functions in association with Rev, and p32, a splicing inhibitor and Rev-binding protein, are suggested to be necessary late-phase factors (67, 83).
We previously produced Tg mice carrying the HIV-1 proviral genome in which the pol gene is deleted (HIV-Tg) (36). Although transgene expression in lymphoid tissues is barely detectable under normal physiological conditions, relatively high levels of p24 Gag protein were detected in the serum (up to 400 pg/ml) after injection of bacterial lipopolysaccharide (74). All mRNA species, including unspliced, singly spliced, and multiply spliced mRNAs were produced normally. Thus, once the viral genome is integrated into the host chromosome, viral genes are expressed at a reasonable efficiency even in mouse cells, suggesting that the major host range barriers are present in the early stage of infection (prior to viral DNA integration) rather than in the late stage. However, it is not yet known whether there are any additional host range barriers in the early steps.
In this report, we investigated additional barrier steps of HIV-1 replication in mouse cells and found that the efficiency of viral genome integration into the host chromosome was low in huCD4/CXCR4 Tg mice. As this result suggested an additional barrier in the early steps of viral infection, we examined nuclear transport of the viral genome and demonstrated that integrase (IN)-dependent nuclear import of the preintegration complex (PIC) is blocked in mouse cells.
MATERIALS AND METHODS
Transgene construction.
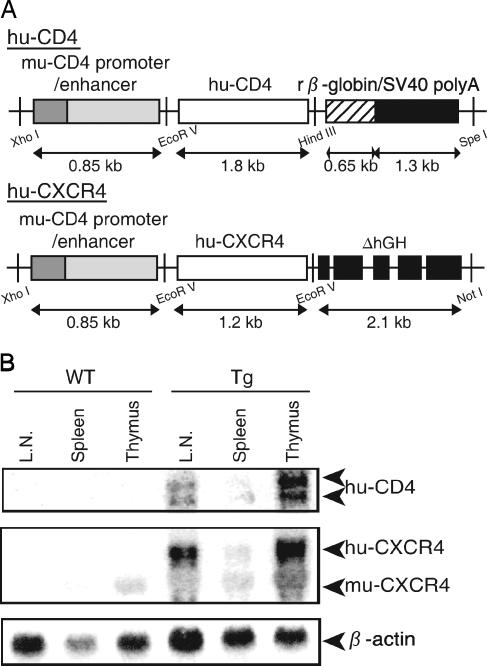
The huCD4 Tg vector (pCT4) was constructed as follows. The 0.85-kb XhoI-EcoRV fragment containing the muCD4 enhancer/promoter was ligated to a 1.8-kb EcoRV-HindIII fragment containing the huCD4 open reading frame, and then a 1.95-kb HindIII-SpeI fragment containing a rabbit β-globin intron sequence and a simian virus 40 (SV40) polyadenylation [poly(A)] signal was inserted into the HindIII-SpeI site downstream of the huCD4 gene (29) (Fig. 1A). To construct the huCXCR4 Tg vector, an XhoI-NotI fragment containing the entire coding region of huCXCR4 was isolated from pBCMGSNeo/HM89 (61), and the huCXCR4 fragment was blunted by T4 DNA polymerase, followed by insertion into the EcoRV site of pCDGH. pCDGH consisted of the muCD4 enhancer/promoter and a human growth hormone gene with its poly(A) signal but devoid of its initiation codon (pCFG) (80) (Fig. 1A). The XhoI-SpeI fragment from pCT4 and the XhoI-NotI fragment from pCFG were then purified and coinjected into the male pronuclei of fertilized mouse eggs (C3H/HeN) (80). The transgenes were detected by dot blot hybridization using DNA prepared from mouse tails (34). Mice were kept under specific-pathogen-free conditions in an environmentally controlled clean room at the Center for Experimental Medicine, the Institute of Medical Science, the University of Tokyo. All equipment and supplies were sterilized, including the cages, water bottles, wood chips, and food pellets. All experiments were conducted according to the institutional ethical and safety guidelines for animal experiments and safety guidelines for gene manipulation experiments.
FIG. 1.
(A) Transgene constructs. huCD4 or CXCR4 cDNA was placed downstream of the muCD4 enhancer/promoter and ligated to the SV40 poly(A) signal (huCD4) or to a defective human growth hormone gene containing the poly(A) signal (huCXCR4). (B) Transgene expression in lymphatic organs. Northern blot hybridization analysis was performed using 10 μg of total RNA prepared from the thymus, spleen, or lymph nodes of WT or Tg mice. The positions expected for huCD4, huCXCR4, muCXCR4, and β-actin mRNA are indicated on the right.
Northern blot hybridization.
Northern blot analyses were carried out as previously described (80). The EcoRV-SpeI fragment of huCD4 and the XhoI-NotI fragment of huCXCR4 used for transgene construction were used as templates to make probes detecting huCD4 and huCXCR4 mRNA, respectively. The autoradiograms were developed, and the radioactivity of each band was quantified with a BAS 2000 Bio-Image analyzer (Fuji Film, Tokyo, Japan).
Plasmids.
The HIV-1 pNL4-3 (X4-tropic, accession no. M19921) vector was obtained from A. Adachi (1). The HIV-1 pNL43lucΔenv vector, in which the env gene is defective and the nef gene is replaced by the firefly luciferase (Luc) gene, the pNL4-3 vector containing a mutation at the IN catalytic site (D116G), and an amphotropic Moloney murine leukemia virus (MuLV) envelope expression vector (pJD-1) were kindly provided by T. Masuda (54, 65, 76). A vesicular stomatitis virus G (VSV-G)-expressing plasmid (pMD-G) was obtained from L. Naldini (5, 62). The pGEM/NL-2-LTR plasmid was kindly provided by Y. Koyanagi (73).
The HIV-1 pNL43lucΔenv vector carrying a IN protein tagged with the SV40 nuclear localization signal (NLS) was constructed by using overlap extension PCR (33). First, two different PCRs were performed using HIV-1 pNL43lucΔenv vector as the template: one with the AflII-sense primer, 5′-CATCTTAAGACAGCAGTACAAATGGCAGTA-3′, and NLS-antisense primer, 5′-GGCCTTTCTCTTCTTTTTTGGATCCTCATCCTGTCTACTTGCC-3′, and the other with the NLS-sense primer, 5′-CCAAAAAAGAAGAGAAAGGCCTAACACATGGAAAAGATTAGT-3′, and PflMI-antisense primer, 5′-CTCTTTTTCCTCCATTCTATGGAGACTCCCTG-3′. These two PCR amplicons were then combined and used as the template for the third PCR with outer primers AflII-sense and PflMI-antisense. The final PCR product was digested with AflII and PflMI and ligated to the SpeI/AflII and SpeI/PflMI vector fragments of HIV-1 pNL43lucΔenv. The nucleotide sequence of the construct was confirmed by sequencing.
To prepare the expression vector for HIV-1 IN N-terminal fusion to enhanced green fluorescence protein (EGFP) (GFP-IN), the entire coding region of HIV-1 IN was amplified by PCR and inserted into the pEGFP-C2 expression vector (Clontech Laboratories, Palo Alto, CA) at its EcoRI and ApaI sites. The primers used to amplify the HIV IN were GFP-IN-sense, 5′-CCGGAATTCCGGGCCATAGCGGCCTTTTTAGATGGAATAGAT-3′, and GFP-IN-antisense, 5′-TCCGGGCCCGGATTAATCCTCATCCTGTCTACT-3′. To generate the expression vector for the HIV-1 Vpr N or C terminus fused to EGFP (GFP-Vpr, Vpr-GFP), the entire coding region of HIV-1 Vpr was amplified by PCR and inserted into the pEGFP-C2 or pEGFP-N1 expression vector at the HindIII and ApaI or HindIII and BamHI sites, respectively. The primers used to amplify HIV-1 Vpr were GFP-Vpr-sense, 5′-CCCAAGCTTGGGGGACGCCATGGAACAAGCCCCAGAA-3′, and GFP-Vpr-antisense, 5′-TCCGGGCCCGGACTAGGATCTACTGGCTCCATT-3′, or Vpr-GFP-sense, 5′-CCCAAGCTTGGGGACATGGAACAAGCCCCAGAA-3′, and Vpr-GFP-antisense, 5′-CGCGGATCCGCGGAGGATCTACTGGCTCCATT-3′, respectively. The amplified regions and the cloning junctions were confirmed by DNA sequencing.
Cell culture and isolation of human PBMCs and mouse splenocytes and thymocytes.
Human fibroblast-like cell lines (293T and HeLa), a mouse embryo fibroblast-like cell line (NIH 3T3) derived from an NIH Swiss mouse (Fv-1n), and a mouse rectum carcinoma cell line (Colon-26) from a BALB/c mouse (Fv-1b) were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS). The Colon-26 cell line was obtained provided from the Cell Resource Center for Biomedical Research Institute of Development, Aging, and Cancer, Tohoku University, Japan (75). Human T-cell lymphoma cell lines (MT-4 and Jurkat) and mouse T-cell lymphoma cell lines (EL4, YAC-1, and BW5147) were cultured in RPMI 1640 (SIGMA, Tokyo, Japan) containing 10% FBS. Human peripheral blood mononuclear cells (PBMCs) were obtained from peripheral blood. Briefly, buffy coats from the peripheral blood of healthy HIV-seronegative blood donors were separated over a Ficoll-Hypaque gradient (Ficoll-Paque PLUS; GE Healthcare Bio-Sciences, Tokyo, Japan). C3H/HeN mice (Charles River, Tokyo, Japan) were sacrificed at 8 weeks, and the splenocytes and thymocytes were isolated by passage through nylon mesh. Human PBMC suspensions and mouse splenocytes and thymocytes were stimulated with 1% phytohemagglutinin (SIGMA, Tokyo, Japan). These cells were grown in RPMI 1640 medium containing 4 ng/ml of recombinant human interleukin 2 or mouse interleukin 2 (Peprotech EC Ltd, London, United Kingdom) per ml and 10% FBS. After 1 week, human PBMCs and mouse splenocytes and thymocytes were >96% T cells and >40% activated cells, as judged by fluorescence-activated cell sorter analysis using anti-CD3 or anti-CD69 monoclonal antibodies (BD Biosciences, Tokyo, Japan), respectively (data not shown).
Virus preparation and infection assays.
HIV-1 strain NL4-3 was propagated in MT-4 cells, and the supernatants were filtered and stored at −80°C until use. For single-round infection assays, pseudotyped viruses were generated by cotransfection of 293T cells with pNL43lucΔenv vector and an amphotropic Moloney MuLV envelope expression vector (pJD-1) or a VSV-G envelope expression vector (pMD-G) using Lipofectamine PLUS (Invitrogen) (76). The pNL43lucΔenv vector containing a mutation at the IN catalytic site (D116G) was used as a control (54). The culture supernatants of the transfected 293T cells were harvested at 48 h posttransfection, filtered through 0.45-μm filters, and used as the virus preparations. Each virus preparation was treated with DNase I (40 μg/ml; Worthington Biochemical Co., Lakewood, NJ) in the presence of 10 mM MgCl2 at 37°C for 1 h to avoid DNA contamination. An aliquot of each virus preparation was incubated at 65°C for 1 h and used as a heat-inactivated control. To monitor viral gene expression from the pNL43lucΔenv vector carrying a IN protein tagged with the SV40 NLS, luciferase activity in transfected 293T cells was measured on a Lumat LB9507 luminometer (BERTHOLD, Technologies, Bad Wildbad, Germany). At 48 h posttransfection, 293T cells were lysed with 1 ml of luciferase lysis buffer (Promega). One microliter of each cell lysate was subjected to the luciferase assay. Human PBMCs (5 × 106) or HeLa (5 × 105), MT-4 (5 × 106), Jurkat (5 × 106), mouse splenocytes (5 × 106), mouse thymocytes (5 × 106), NIH 3T3 (5 × 105), BW5147 (5 × 106), EL4 (5 × 106), or YAC-1 (5 × 106) cells were infected with an aliquot (2 ml; containing approximately 500 ng [NL4-3], 200 ng [HIV-1/MuLV], or 50 ng [HIV-1/VSV-G] of p24) of DNase-treated virus. The infection proceeded in the presence of Polybrene (SIGMA, Tokyo, Japan) (10 μg/ml) at 37°C. After 6 h, the viruses were removed, and the cells were overlaid with fresh media and incubated at 37°C. For p24 CA analysis, the infected cell supernatants were removed on the indicated days following infection. The levels of HIV-1 p24 antigen were determined by an enzyme immunoassay system (RETRO-TEK; ZeptoMetrix Corp., Buffalo, NY). For luciferase analysis, infected cells were harvested 4 days after infection, and the total cell pellets from each well were washed twice with phosphate-buffered saline (PBS) and lysed in luciferase lysis buffer (Promega). Luciferase activity (measured in a relative light units [RLU]) was measured on a Lumat LB9507 luminometer (BERTHOLD, Technologies, Bad Wildbad, Germany).
Analysis of HIV-1 DNA synthesis and formation of 2-LTR circles.
Cells were harvested 24 h after infection. After washing with PBS, nucleic acids were extracted as described previously (81). Briefly, cells were disrupted in urea lysis buffer (4.7 M urea, 1.3%, sodium dodecyl sulfate [SDS], 0.23 M NaCl, 0.67 mM EDTA, and 6.7 mM Tris-HCl [pH 8.0]), phenol-chloroform extracted, and ethanol precipitated. The DNA pellet was resuspended in distilled H2O, and an aliquot of each sample was analyzed by PCR. For ex vivo infection of primary lymphocytes from huCD4/CXCR4 Tg mice, partial reverse transcripts of the viral DNA were quantified by semiquantitative PCR. The primers used were as follows (37, 45, 81): R-U5, R, 5′-GCCTCAATAAAGCTTGCCTTG-3′ (sense, positions 522 to 542); U5, 5′-CCACTGCTAGAGATTTTCCAC-3′ (antisense, positions 616 to 638); Gag forward, 5′-TGGGGGGACATCAAGCAGCCATGCA-3′ (sense, positions 1360 to 1385); Gag reverse, 5′-CTATGTCACTTCCCCTTGGTTCTCT-3′ (antisense, positions 1474 to 1498). The PCR program was 30 cycles at 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min in the presence of [32P]dCTP. The PCR products were electrophoresed on an 8% polyacrylamide-Tris-borate-EDTA gel. The autoradiograms were developed, and the radioactivity of each band was quantified by a BAS 2000 Bio-Image analyzer. For single-round infections, the DNA was measured by quantitative PCR using an ABI PRISM 7900HT qPCR machine (Applied Biosystems, Tokyo, Japan). The PCR primer pairs were as follows: R-U5 (M667/AA55), R-gag (M667/M661) (76), and the 2-LTR junction's sequence (2-LTR-S/2-LTR-AS) (73). The cycling conditions included a hot start (50°C for 2 min, 95°C for 10 min), followed by 40 cycles of denaturation (95°C for 15 s) and extension (60°C for 1 min). To compensate for varying DNA sample recovery, the data are presented as ratios of HIV-1 DNA to β-actin DNA.
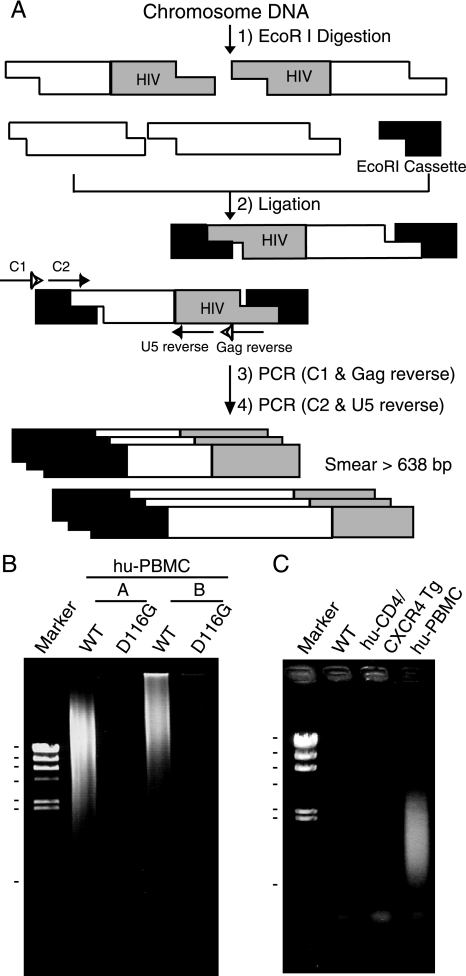
Cassette ligation-mediated PCR and integration analysis.
For the detection of the HIV-1 integration form, we designed a cassette ligation-mediated PCR system using an in vitro LA cloning kit (TaKaRa BIO, Shiga, Japan) (35) (see Fig. 3A). Briefly, 5 μg of DNA was digested with EcoRI and ligated to double-stranded DNA cassettes with compatible ends. The cassette-ligated restriction fragments were then subjected to two rounds of PCR using the cassette- and HIV-specific primers C1 (5′-GTACATATTGTCGTTAGAACGCGTAATACGACTCA-3′) and Gag reverse (described above) for the cassette, gag, and its upstream region and C2 (5′-CGTTAGAACGCGTAATACGACTCACTATAGGGAGA-3′) and U5 reverse (described above) for the cassette sequence downstream of C1 and the LTR region. PCR was performed according to the manufacturer's instructions. The amplification conditions were 30 cycles of 1 min at 94°C, 2 min at 54°C, 2 min at 72°C, and a final extension of 1 min at 72°C. The amplified products were resolved on 2% agarose gels and stained with SYBR green (FMC Bioproduct, Rockland, ME).
FIG. 3.
Suppression of NL4-3 virus DNA integration in the mouse chromosome. Human PBMCs and murine splenocytes were infected with equivalent doses of DNase-treated NL4-3 virus. After 6 h, the virus was removed and the cells were washed with growth medium. At 1 day postinfection, the cells were harvested and used to extract total DNA. The DNA (5 μg) was digested with EcoRI and ligated to double-stranded DNA cassettes with compatible ends. The cassette-ligated DNA fragments were used as templates for nested PCR using cassette- and HIV-specific primers. (A) Schematic representation of cassette ligation-mediated PCR and the primers used to detect HIV integration into the host chromosome. (B and C) Detection of the chromosome-integrated forms of viral DNA. (B) Human PBMC preparations from two donors (A and B) were infected with NL4-3-WT or integrase mutant (D116G), and the DNAs were subjected to PCR analysis. Markers: 23.1, 9.4, 6.6, 4.3, 2.3, 2.0, and 0.564 kb (λ/HindIII). (C) DNA was isolated from NL4-3-infected splenocytes or thymocytes from WT or Tg mice or human PBMCs following infection and subjected to PCR analysis. Note the smearing bands in virus-infected human PBMCs but not in huCD4/CXCR4 Tg mice. Marker: λ/HindIII.
Fluorescence microscopy.
HeLa (4 × 104), NIH 3T3 (3 × 104), and Colon-26 (3 × 104) cells were seeded onto 8-well culture slides (Nalge Nunc International, Rochester, NY) and transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen). At 24 h posttransfection, the cells were washed once in PBS and fixed with acetone for 5 min. After washing with PBS, the cells were mounted in 90% glycerol-50 mM NaHCO3-Na2CO3 and covered by a coverslip. Confocal microscopy was performed with a Nikon Optiphot-2 fluorescence microscope with a Bio-Rad MRC 1024 laser confocal imaging system, and the digital images were prepared using Adobe Photoshop software.
Particle preparation and Western blot analysis.
The culture supernatant (5 ml) of HIV-1 producing plasmid-transfected 293T cells was collected at 48 h postinfection. It was centrifuged thorough 20% (wt/vol) sucrose-PBS in an SW55 rotor (Beckman Coulter) at 4°C at 147,000 × g for 2 h (56), and the pellet was resuspended in PBS. The viral pellets were hearted at 90°C for 10 min in the presence of sample buffer (62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, 0.005% bromophenol blue). Then viral proteins were electrophoresed on a 12% SDS-polyacrylamide gel containing 0.2% SDS. Following blotting of proteins onto a polyvinylidene difluoride membrane, the membrane was first incubated with an antiserum from an AIDS patient (provided by Y. Inagaki, Tokyo Medical and Dental University, Tokyo, Japan, and Y. Koyanagi, Kyoto University, Kyoto, Japan), followed by horseradish peroxidase-conjugated anti-human immunoglobulin. HIV-1 proteins were visualized using an enhanced chemiluminescence detection system (GE Healthcare Bio-Science, Tokyo, Japan).
Statistical analysis.
Data were analyzed using Excel and Student's t test. A P value of <0.05 was considered statistically significant, and all results are presented as means ± standard errors of the means (SEM).
RESULTS
Lymphocytes from huCD4/CXCR4 Tg mice do not fully support HIV-1 infection. To elucidate the host range barriers of HIV-1 replication in mice, we analyzed the early processes of HIV-1 infection in huCD4/CXCR4 Tg mouse splenocytes. Transgenic mice were generated by introducing both huCD4 and huCXCR4 cDNA along with the muCD4 enhancer/promoter into fertilized C3H mouse eggs (Fig. 1A). As shown in Fig. 1B, the huCD4 and huCXCR4 mRNAs were detected mostly in the thymus but also in the lymph nodes and spleen. Two huCD4 mRNA species were detected due to alternative splicing of the SV40 gene that was ligated to the huCD4 gene (Fig. 1B). fluorescence-activated cell sorter analyses showed that Tg splenocytes and thymocytes both expressed huCD4 and huCXCR4 on their cell surfaces (data not shown).
To examine the susceptibility of these Tg mice to HIV infection, splenocytes and thymocytes were isolated from the mice and infected with T-tropic HIV-1 (NL4-3) or M-tropic HIV-1 (JR-CSF). However, we could not detect any p24 antigen in the culture supernatant of these Tg mouse-derived cells, although significant levels of p24 (up to 80 pg/ml) were produced in human PBMC culture supernatant 12 days after infection.
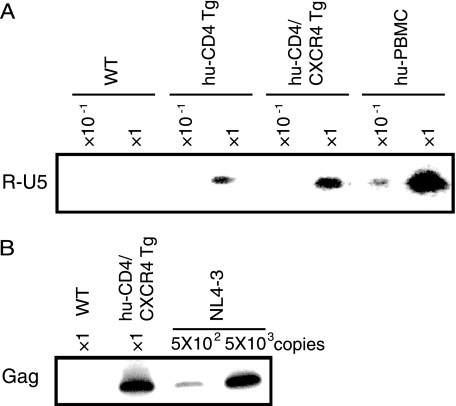
To determine the process by which the viral replication is blocked, we analyzed the early infection steps by examining the viral genomic structure. Twenty-four hours after HIV-1 infection of the huCD4/CXCR4 Tg splenocytes, cells were harvested and total DNA was extracted. Early (R-U5) and late-infectious intermediate products (R-gag) were determined using semiquantitative PCR with specific primers. As shown in Fig. 2A and B, both infectious intermediates were detected specifically in the DNA isolated from the splenocytes of huCD4/CXCR4 Tg mice exposed to HIV-1, indicating that viral entry and reverse transcription had proceeded normally in mouse cells provided that human viral receptors were supplied. Similar results were also obtained in huCD4/CCR5 Tg mouse splenocytes infected with HIV-1 JR-CSF (data not shown). In contrast, the early infectious intermediate was not detected in wild-type mouse splenocytes, indicating a block upon viral entry. Taken together, these results suggest that HIV-1 replication in huCD4/CXCR4 Tg splenocytes was blocked later than the reverse transcription step(s).
FIG. 2.
HIV infection of splenocytes and thymocytes of WT and huCD4/CXCR4 Tg mice. Splenocytes or thymocytes from WT mice, Tg mice, or human PBMCs were isolated and infected with equivalent amounts of DNase-treated NL4-3 virus at 37°C for 6 h. After 6 h, the viruses were removed and the cells were treated with trypsin (500 μg/ml) at 37°C for 10 min and then washed in growth medium. (A and B) Splenocytes were harvested at 24 h postinfection. Total DNA was extracted from the cells and subjected to PCR analysis with a primer pair for R/U5 (A) or R/gag (B). The reaction was carried out using the DNA preparation from 1 × 105 cells (×1) or 1 × 104 cells (×10−1). In panel B, the lanes marked 5 × 102 and 5 × 103 represent PCR products using 5 × 102 or 5 × 103 copies of the pNL4-3 plasmid as the template.
HIV-1 infection of mouse cells is blocked at steps preceding integration into the host chromosome.
We next examined the integration of the HIV-1 genome into the host chromosome. The pNL4-3 vector containing a mutation at the IN catalytic site (D116G) was used as a control (54). DNA from the infected cells was digested with EcoRI, which cut proviral DNA at only one site (nucleotide number 5743 of NL4-3, accession no. M19921). The DNA was then ligated to an EcoRI-specific cassette and subjected to the first round of PCR using primers specific for the cassette and the gag region, followed by the second round of PCR using primers for the cassette and the LTR region (Fig. 3A). As a result, the integrated viral DNA was visualized as smearing bands greater than 638 bp, which is the original length between C2 and U5 reverse primers without insertion. Smearing bands were clearly detected when the DNA from HIV-1 wild type (WT)-infected human PBMCs was analyzed. In contrast, no smearing bands were detected with the DNA from HIV-1-D116G infected human PBMC, HIV-1-WT infected splenocytes from WT, and huCD4/CXCR4 transgenic mice (Fig. 3B and C). These results suggest that HIV-1 replication is also blocked in mouse cells at steps between the entry and viral DNA integration steps or at the viral integration step in addition to the adhesion/entry step.
The infection of mouse cells with both HIV-1/pJD-1 and HIV-1/VSV-G pseudotyped virus are blocked at a postentry step.
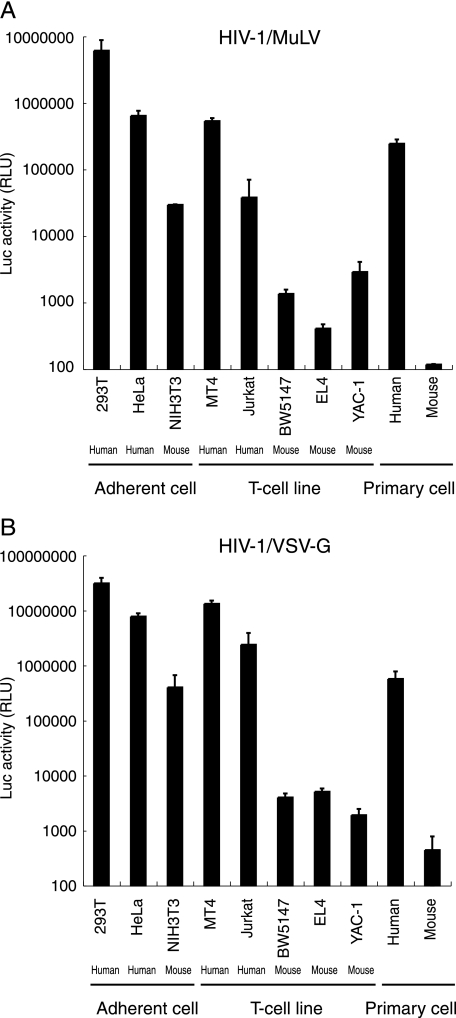
To examine the possibility that HIV-1 replication in mouse cells is blocked at steps between the viral entry and DNA integration steps, we analyzed the early steps of viral infection using HIV-1 pseudotyped viruses in which the Env is replaced by an amphotropic MuLV Env (HIV-1/pJD-1) or by the G protein of VSV (HIV-1/VSV-G) and the nef gene is replaced by the firefly luciferase gene (76). The MuLV envelope pseudotype uses a ubiquitously expressed phosphate transporter as the receptor (55), and the VSV-G envelope pseudotype is capable of infecting cells through a carbohydrate receptor and the endocytic pathway (2). By using these pseudotyped viruses, we overcame the barriers at the adhesion and entry steps.
Among the adherent cells tested, 293T and HeLa cells showed high luciferase activity upon infection with both types of pseudotyped viruses, whereas NIH 3T3 cells yielded 10- to 100-fold-lower signals (Fig. 4). Similarly, the mouse T-cell lines BW5147, EL4, and YAC-1 displayed 100- to 1,000-fold-lower signals than did the human T-cell lines MT4 and Jurkat. Furthermore, luciferase expression efficiency was lower, by more than 1,000-fold, in mouse primary splenocytes than in human PBMCs. The relative sensitivity to infection of these cells was similar between the two pseudotyped viruses, although the efficiency of infection was approximately 10-fold higher in the HIV-1/VSV-G infection. Thus, these results again support a species-specific block in mouse cells subsequent to the entry step.
FIG. 4.
Analysis of HIV-1 pseudotype virus replication in mouse cells. Human and murine cells were infected with equivalent doses of pNL43lucΔenv that was pseudotyped with either MuLV (A) or VSV-G (B) at 37°C for 6 h. After removal of the virus, cells were washed in growth medium. Luciferase activity was measured at 4 days postinfection, and normalized activities relative to the total protein quantity are shown. The data represent the means ± SEM of results from three wells. The data were reproduced in three independent experiments.
Reverse transcription of HIV-1 proceeds normally in mouse cells.
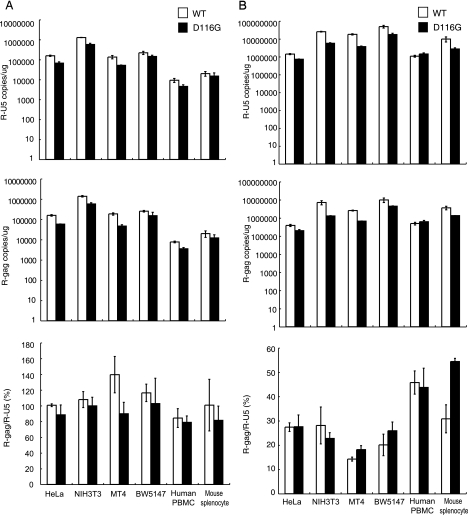
We next evaluated the ability of mouse cells to support HIV-1 DNA synthesis. One day after infection, total DNA was harvested from various infected cells and subjected to quantitative PCR analyses. Both early (R-U5) and late (R-gag) reverse transcription products were specifically detected in DNA isolated from human and mouse cells exposed to both of the pseudotyped viruses, HIV-1/pJD-1 (Fig. 5A) and HIV-1/VSV-G (Fig. 5B). No R-U5 or R-gag products were detected when heat-inactivated (65°C, 1 h) viruses were infected, indicating that these products were not derived from the transfected HIV-1 DNA carryover. The copy number of R-U5, which reflects the efficiency of viral entry, was higher in mouse cells (NIH 3T3, BW5147, and splenocytes) than in human cells (HeLa, MT4, and PBMCs). The ratio of R-gag/R-U5 was calculated to evaluate the efficiency of the reverse transcription because R-U5 reflects the efficiency of entry (Fig. 5A and B, lower panels). No significant difference in the efficiency of reverse transcription was observed between mouse cells and human cells exposed to HIV-1/pJD-1 (Fig. 5A) and HIV-1/VSV-G (Fig. 5B). These results indicated that mouse cells supported reverse transcription at an efficiency similar to that of human cells.
FIG. 5.
The efficiency of the reverse transcription of HIV-1 in mouse cells. Human and murine cells were infected with equivalent doses of DNase-treated WT or integrase mutant (D116G) MuLV pseudotyped virus (A) or VSV-G pseudotyped virus (B). At 1 day postinfection, the cells were harvested, and the total DNA was extracted and subjected to quantitative real-time PCR analysis using primer pairs for R/U5 (upper panels) or R/gag (middle panels). The copy numbers of HIV-1 DNA per 1 μg β-actin are shown. The reverse transcription (RT) efficiency is calculated by dividing the late RT product (R-gag) by the early RT product (R-U5) (lower panel). The data represent the means ± SEM of results from three wells. The data were reproduced in three independent experiments.
Nuclear import of the PIC is blocked in mouse cells.
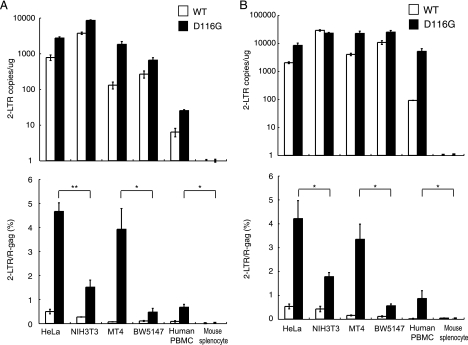
After completion of reverse transcription, the PIC crosses the nuclear membrane and enters the nucleus. Ligases within the nucleus then circularize the proviral DNA (2-LTR containing circular DNA) before its integration into the host chromosome (17, 57, 82). Although these 2-LTR circles are nonfunctional, they can serve as a measure of viral nuclear entry. To assess the efficiency of PIC transport into the nuclei of mouse cells, we estimated de novo-synthesized 2-LTR circular-form DNA by PCR using primer pairs that amplify sequences unique to this DNA form. The fragment corresponding to the 2-LTR circular junction was clearly detected at 1 day postinfection in the DNA samples from HeLa, NIH 3T3, MT-4, BW5147, and human PBMCs infected with HIV-1/pJD-1 pseudotyped virus (Fig. 6A, upper panel). However, only a small amount of 2-LTR circle was detected in the mouse splenocytes. A similar tendency was observed in cells infected with the HIV-1/VSV-G pseudotyped virus (Fig. 6B, upper panel). The 2-LTR DNA in mouse cells was also measured at 2 and 4 days postinfection using both pseudotyped viruses and provided similar results (data not shown).
FIG. 6.
Suppression of the 2-LTR circular form of DNA in mouse cells. Human and mouse cells were infected with equivalent doses of DNase-treated WT or integrase-mutant (D116G) MuLV pseudotype virus (A) or VSV-G pseudotype virus (B). The DNA from the infected cells was subjected to quantitative real-time PCR analysis using a primer pair specific for the 2-LTR circular form of the DNA (upper panels). The nuclear import efficiency was calculated by dividing the 2-LTR products by the late reverse transcription products (lower panels). The data represent the means ± SEM of results from three wells, and the data were reproduced in three independent experiments. *, P < 0.05; **, P < 0.01 (determined by Student's t test).
The ratio of 2-LTR/R-gag was calculated to evaluate the efficiency of nuclear import because the copy number of 2-LTR in the nucleus should be dependent on the amount of cytoplasmic R-gag, which represents the precursor of 2-LTR. The efficiency was very low and not significantly different between human and mouse when wild-type HIV-1 pseudovirus was infected (Fig. 6, lower panel). Because the nuclear concentration of 2-LTR is determined by the balance between accumulation of PIC by nuclear import and loss of PIC from the nucleoplasm by chromosome integration, we next used an integration-defective mutant, D116G, to examine only the efficiency of nuclear import. As shown in Fig. 6, the ratio was significantly lower in NIH 3T3 cells than in HeLa cells (33% or 43% of HeLa cells) and in BW5147 cells than in MT4 cells (12% or 17% of MT4 cells) when they were infected with HIV-1/pJD-1 or HIV-1/VSV-G, respectively (Fig. 6A and B, lower panels). We were unable to compare the efficiency of PIC import in human PBMCs and mouse splenocytes because the 2-LTR circle was not detected in mouse splenocytes. These results suggested that the nuclear import of the PIC is blocked in mouse cells, especially in splenocytes.
A block in the nuclear localization of the PIC is caused by a defect in IN nuclear localization.
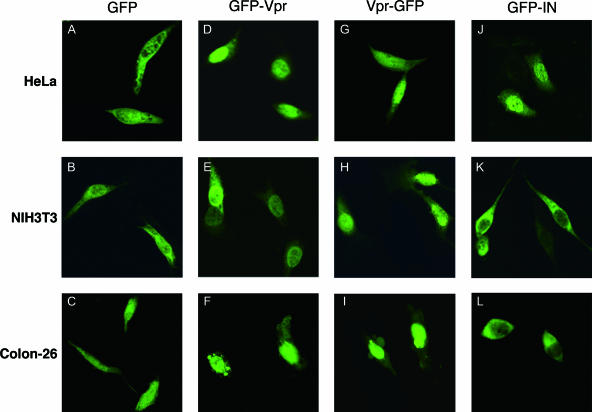
As Vpr and IN play important roles in importing the PIC into the nucleus, we hypothesized that Vpr and/or IN is nonfunctional in mouse cells due to the inability to utilize the cellular factors necessary for trafficking to the nucleus. To directly examine the karyophilic properties of HIV-1 IN in mouse cells, we generated an expression vector with HIV-1 IN in which the N terminus was fused to EGFP (GFP-IN). Since it was reported that β-galactosidase fusion to the C terminus of IN could not be located in the nucleus (44), we did not examine the C terminus fusion construct (IN-GFP). We also generated HIV-1 Vpr expression vectors in which the N or C terminus was fused to EGFP (GFP-Vpr and Vpr-GFP, respectively). At 24 h postinfection, HeLa, NIH 3T3, and Colon-26 cells were transfected with the GFP fusion vectors, and the subcellular localization of IN or Vpr was examined with a confocal microscope. We used Colon-26 cells (Fv-1n) in addition to NIH 3T3 cells (Fv-1b) because Fv-1 may exert its antiretroviral effect at a postentry step, after reverse transcription and prior to integration (38, 79). Colon-26 cells and NIH 3T3 cells showed similar levels of luciferase activity upon infection with both types of pseudotyped viruses. Control GFP without IN or Vpr was distributed uniformly throughout both the cytoplasm and the nuclei in all cells examined (Fig. 7A to C). GFP-Vpr, on the other hand, accumulated almost exclusively in the nuclei of HeLa (Fig. 7D), NIH 3T3 (Fig. 7E), and Colon-26 (Fig. 7F) cells, although low levels of nuclear membrane association were also observed in NIH 3T3 cells (Fig. 7E). Vpr-GFP also accumulated almost exclusively in the nuclei of HeLa (Fig. 7G), NIH 3T3 (Fig. 7H), and Colon-26 (Fig. 7I) cells. These results indicated that HIV-1 Vpr has strong karyophilic properties and that, even in mouse cells, it can be transported across the nuclear membrane. In contrast, although GFP-IN accumulated almost exclusively in the nuclei of HeLa cells (Fig. 7J), GFP-IN was localized only in the cytoplasm of NIH 3T3 (Fig. 7K) and Colon-26 (Fig. 7L) cells. Thus, our results demonstrate that IN-mediated nuclear transport of HIV-1 PIC is impaired in mouse cells of both Fv-1 genotypes.
FIG. 7.
Inhibition of IN-dependent GFP nuclear import in mouse cells. HeLa (A, D, G, and J), NIH 3T3 (B, E, H, and K), and Colon-26 cells (C, F, I, and L) were transfected with plasmids expressing GFP only (A, B, and C), GFP fused to the HIV-1 Vpr N terminus (GFP-Vpr) (D, E, and F), GFP fused to the HIV-1 Vpr C terminus (Vpr-GFP) (G, H, and I), and GFP fused to HIV-1 IN (J, K, and L) using Lipofectamine 2000. At 24 h posttransfection, the cells were fixed and visualized by confocal fluorescence microscopy. Note that the nuclear localization of GFP-IN is inhibited in mouse cells (K and L).
Addition of the SV40 NLS to the C terminus of HIV-1 integrase enhances viral infectivity in mouse cells.
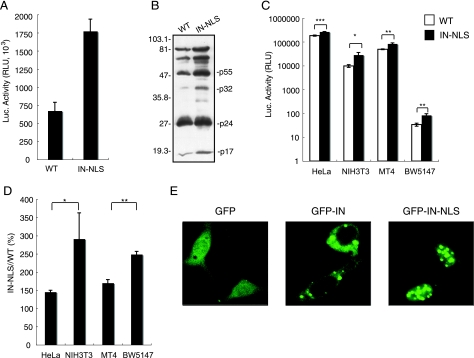
To analyze the role of IN in nuclear localization of HIV-1, we constructed an HIV-1 pNL43lucΔenv vector with the SV40 NLS at the C terminus of IN (IN-NLS), and pseudotyped virus was generated by cotransfection of 293T cells with the pNL43lucΔenv wild type or IN-NLS vector and VSV-G expression vector. Luciferase activity in the cell lysate of 293T cells transfected with IN-NLS was increased 2.5-fold compared to that transfected with wild-type virus (Fig. 8A). To verify that the gag-pol polyprotein processing was completed in IN-NLS virus particles, we performed Western blot analysis using an AIDS patient serum. No difference of the viral components was observed between parental WT and IN-NLS viruses (Fig. 8B). The content of p24 protein of the IN-NLS was also shown to be normal using a specific monoclonal antibody (data not shown). These results showed that addition of NLS to IN significantly activates viral replication.
FIG. 8.
Enhancement of viral infectivity to mouse cells by the addition of SV40 NLS to the C terminus of IN. (A) Luciferase activity in an HIV-1 pNL43lucΔenv vector (WT) or pNL43lucΔenv vector carrying an SV40 NLS-ligated IN (IN-NLS)-transfected 293T cells were measured at 2 days posttransfection. The data were reproduced in 10 independent experiments. (B) Virus particles were collected at 48 h posttransfection and resuspended in PBS. The viral pellets were hearted at 90°C for 10 min in the presence of sample buffer (62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, 0.005% bromophenol blue). Then viral proteins were electrophoresed on a 12% SDS-polyacrylamide gel. Viral proteins were detected with AIDS patient serum. Positions of the major viral proteins are indicated together with molecular weight markers. (C) Human and mouse cells were infected with equivalent amounts of WT (□) or IN-NLS (▪) virus at 37°C for 6 h, and then the virus was removed. The luciferase activity was measured 4 days after infection, and the activity was normalized relative to the total amount of protein. Means ± SEM are shown. (D) Relative Luc activities of IN-NLS virus-infected cells compared to WT virus-infected cells were calculated using the data shown in panel C. Means ± SEM are shown. (E) NIH 3T3 cells transfected with a plasmid expressing GFP, GFP-IN, or GFP-IN-NLS fusion protein were analyzed by confocal microscopy. At 24 h posttransfection, cells were fixed and GFP was detected by a confocal fluorescent microscope. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by Student's t test).
Then we tested the susceptibility of HeLa, NIH 3T3, BW5147, and MT4 cells to IN-NLS infection. Addition of the SV40 NLS to the C terminus of IN significantly enhanced viral infectivity in all cell lines used (Fig. 8C). As the NLS fusion at the C terminus of IN disrupted the vif gene, we did not analyze the infectivity to human PBMC, which was nonpermissive to vif-deficient virus (24, 69). Interestingly, the effect of the NLS on viral infection was significantly stronger in mouse cells than in human cells (Fig. 8D).
We also analyzed the karyophilic property of GFP-IN-NLS in which EGFP was fused to the N terminus and the SV40 NLS at the C terminus of IN. GFP-IN-NLS accumulated almost exclusively in the nucleus of NIH 3T3 cells, in contrast to GFP-IN (Fig. 8E). These results indicate that the addition of functional NLS to IN compensates the functional defects of IN in mouse cells.
DISCUSSION
HIV-1 replication in rodent cells is blocked at multiple steps, including viral entry, transcription, nuclear export of the mRNA, assembly, and budding (77). In this study, we demonstrated that an additional host range barrier is present in mouse cells at the PIC nuclear transport step. We suggest that this restriction is caused by the dysfunction of the HIV-1 IN-dependent PIC import system.
We found that the ratio of the 2-LTR circular product to the late reverse transcription product (R-gag), which are produced in the nucleus and cytoplasm, respectively, was decreased in mouse splenocytes relative to human PBMCs after infection with both HIV-1/MuLV and HIV-1/VSV-G pseudotyped viruses (Fig. 6, lower panels). This indicates a reduction in nuclear import of the PIC. The efficiency of nuclear transfer of the viral PIC was recently reported to be reduced in mouse T cells relative to human T cell lines, but not in NIH 3T3 cells (7). These results are consistent with ours, although we cannot explain their lack of reduction in the NIH 3T3 cells. Our system using the IN mutant virus is probably more sensitive in detecting the defect.
We showed that the infection efficiency, as determined by luciferase activity, was 10 to 100, 100 to 1,000, and >1.000 times lower than in human cells in mouse adherent cells, T cell lines, and primary cells, respectively (Fig. 4). As the PIC nuclear import reduction in mouse cells was at most 60 to 90% compared to human cells (Fig. 6), it is clear that other barriers are also involved in the HIV-1 replication restriction in mouse cells. In this regard, we showed that the 2-LTR/R-gag ratio following infection with WT HIV-1 was similar between HeLa and NIH 3T3 cells and between MT4 and BW5147 cells, in contrast to our observations with the IN-deficient mutant virus (Fig. 6, lower panels). These results are explainable if the R-gag products integrate rapidly into the host chromosome in human cells compared to mouse cells, assuming that a constant proportion of free R-gag products is converted into the 2-LTR form. Thus, integration of the R-gag products into the host chromosome may also be inhibited in mouse cells. In accordance with this idea, we demonstrated that the integration frequency of HIV-1 was greatly reduced upon infection with WT HIV-1 in mouse cells transgenic for the huCD4/CXCR4 genes.
There are two formal explanations for the inability of the mouse cells to support HIV-1 PIC nuclear import. One is the absence of a required human-specific factor, and the other is the presence of an inhibitory factor(s) in mouse cells. However, because mouse-human cell fusions allow viral replication, mouse cells most likely do not have such an inhibitory factor and are rather devoid of a critical factor for the import of the HIV-1 PIC (52).
Although the mechanisms of PIC nuclear import have not been elucidated completely, NLSs are present in three viral proteins (MA, Vpr, and IN) as well as in a central DNA flap produced during reverse transcription that also contributes to the successful nuclear targeting of the PIC (59, 64, 71). Although their respective contributions remain controversial and unclear, it has been clearly shown that both Vpr and IN are karyophilic and rapidly accumulate in the nuclei of infected cells (6, 16, 19, 20, 25, 32, 48, 63, 66, 68). Meanwhile, localization of MA to the nucleus is not well established (14, 23). In this report, we demonstrated that GFP-fused IN remained in the cytoplasm of NIH 3T3 cells, in contrast to its accumulation in the nuclei of HeLa cells. On the other hand, when GFP-fused Vprs were examined, they localized to the nuclei of both NIH 3T3 and HeLa cells. The nuclear distribution of GFP-IN, but not GFP-fused Vpr, was also inhibited in the Colon-26 mouse cell line. These observations indicate that Vpr can be imported into the nucleus using separate pathways. IN, on the other hand, cannot be imported efficiently into the nuclei of mouse cells, and this is probably due to the inability of IN to interact with mouse nuclear import system.
In support for this notion, we showed that nuclear import of IN was much more enhanced in mouse cells than in human cells when authentic SV40 NLS was added to IN (Fig. 8). Furthermore, the addition of this NLS to IN significantly enhanced the infectivity of HIV-1 pseudovirus to mouse cells. These results indicate that endogenous nuclear localization signals of IN are not fully functional in mouse cells.
In this context, it is known that the NLSs within HIV-1 IN are composed of basic amino acid-rich sequences that interact with importin β through the adapter importin (25). We previously showed, however, that mutational disruption of the suggested NLSs could not abolish the nuclear localization of a GFP-IN fusion protein (76). Recent studies have shown that nonclassical NLSs are necessary and sufficient to locate the viral PIC into the nuclei (12). Depienne et al. suggested that the in vitro nuclear import of IN does not require known cytosolic transport factors, including karyopherin β family proteins (18). Two proteins have recently been reported to mediate PIC import. The first is lens epithelium-derived growth factor (LEDGF/p75), a protein implicated in the regulation of gene expression and cellular stress responses. LEDGF interacts with HIV-1 IN in vitro and in living cells (50) and colocalizes with HIV-IN in the nuclei of human cells (15). The second is importin 7, a mildly hydrophobic protein belonging to the importin β superfamily. This protein is suggested to interact with basic proteins, such as IN, that bind viral nucleic acids (21). It is currently unclear which protein(s) is important in these processes and defective in mouse cells. Clearly, further work is necessary to identify the host cell factors that are associated with IN in human cells and defective in mouse cells.
Thus far, several host restriction factors are known to be involved in the suppression of HIV-1 replication in the early phase of its life cycle in mouse cells. Friend virus susceptibility factor-1 (Fv-1) is involved in the restriction of specific mouse cell genotypes to MuLV (10, 28). The Fv-1 targets the MuLV capsid and stops the nuclear import of the PIC (8, 39). However, recent reports have noted that there is no correlation between HIV-1 susceptibility and cellular Fv-1 genotype (7, 31). Tripartite motif 5 α (TRIM5α), encoded by the gene Lv-1, is another restriction factor (72). TRIM5α inhibits viral replication in rhesus macaque cells at a step after entry but before the reverse transcription of HIV-1 by targeting the viral capsid protein (60). Thus, both Fv-1 and TRIM5α function in processes other than the transport of PIC into the nucleus.
In conclusion, we have demonstrated that PIC nuclear import is blocked in mouse cells and that dysfunctional IN is at least partially responsible for the barrier. Further characterization and identification of factors that are involved in PIC nuclear import should provide new insight into the molecular mechanisms of the PIC import step and clues to the development of new therapeutics. Furthermore, identification of the factors responsible for this step will assist in our generation of transgenic small animal models that are permissive to HIV-1 infection.
Acknowledgments
We thank Takao Masuda (Tokyo Dental and Medical University) for providing pNL43lucΔenv and an amphotropic Moloney MuLV envelope expression vector (pJD-1) as well as for critical discussions. We also thank Yoshio Koyanagi (Kyoto University) for providing the 2-LTR plasmid and the AIDS patient serum and for important technical advice. We also thank Yoshio Inagaki (Tokyo Medical and Dental University) for providing the AIDS patient serum. We are grateful to Luigi Naldini (San Raffaele Telethon Institute for Gene Therapy) and Kenzaburo Tani (Kyushu University) for providing the VSV-G envelope-expressing plasmid (pMD-G).
This work was supported by Grants-in-Aid from the Japan Human Sciences Foundation.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken, C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71:5871-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, A., T. P. Cujec, and B. M. Peterlin. 1994. Effects of human chromosome 12 on interactions between Tat and TAR of human immunodeficiency virus type 1. J. Virol. 68:6505-6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso, A., D. Derse, and B. M. Peterlin. 1992. Human chromosome 12 is required for optimal interactions between Tat and TAR of human immunodeficiency virus type 1 in rodent cells. J. Virol. 66:4617-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai, Y., Y. Soda, K. Izawa, T. Tanabe, X. Kang, A. Tojo, H. Hoshino, H. Miyoshi, S. Asano, and K. Tani. 2003. Effective transduction and stable transgene expression in human blood cells by a third-generation lentiviral vector. Gene Ther. 10:1446-1457. [DOI] [PubMed] [Google Scholar]
- 6.Balliet, J. W., D. L. Kolson, G. Eiger, F. M. Kim, K. A. McGann, A. Srinivasan, and R. Collman. 1994. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology 200:623-631. [DOI] [PubMed] [Google Scholar]
- 7.Baumann, J. G., D. Unutmaz, M. D. Miller, S. K. Breun, S. M. Grill, J. Mirro, D. R. Littman, A. Rein, and V. N. KewalRamani. 2004. Murine T cells potently restrict human immunodeficiency virus infection. J. Virol. 78:12537-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benit, L., N. De Parseval, J. F. Casella, I. Callebaut, A. Cordonnier, and T. Heidmann. 1997. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 71:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berson, J. F., D. Long, B. J. Doranz, J. Rucker, F. R. Jirik, and R. W. Doms. 1996. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J. Virol. 70:6288-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 11.Bieniasz, P. D., and B. R. Cullen. 2000. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 74:9868-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 13.Browning, J., J. W. Horner, M. Pettoello-Mantovani, C. Raker, S. Yurasov, R. A. DePinho, and H. Goldstein. 1997. Mice transgenic for human CD4 and CCR5 are susceptible to HIV infection. Proc. Natl. Acad. Sci. USA 94:14637-14641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukrinsky, M., K. Manogue, and A. Cerami. 1995. HIV results in the frame. Other approaches. Nature 375:195-196. (Author's reply, 375:198.) [DOI] [PubMed] [Google Scholar]
- 15.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 16.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 17.Cullen, B. R. 2001. Journey to the center of the cell. Cell 105:697-700. [DOI] [PubMed] [Google Scholar]
- 18.Depienne, C., A. Mousnier, H. Leh, E. Le Rouzic, D. Dormont, S. Benichou, and C. Dargemont. 2001. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 276:18102-18107. [DOI] [PubMed] [Google Scholar]
- 19.Depienne, C., P. Roques, C. Creminon, L. Fritsch, R. Casseron, D. Dormont, C. Dargemont, and S. Benichou. 2000. Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp. Cell Res. 260:387-395. [DOI] [PubMed] [Google Scholar]
- 20.Farnet, C. M., B. Wang, J. R. Lipford, and F. D. Bushman. 1996. Differential inhibition of HIV-1 preintegration complexes and purified integrase protein by small molecules. Proc. Natl. Acad. Sci. USA 93:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J. M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 23.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabuzda, D. H., H. Li, K. Lawrence, B. S. Vasir, K. Crawford, and E. Langhoff. 1994. Essential role of vif in establishing productive HIV-1 infection in peripheral blood T lymphocytes and monocyte/macrophages. J. Acquir. Immune Defic. Syndr. 7:908-915. [PubMed] [Google Scholar]
- 25.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garber, M. E., and K. A. Jones. 1999. HIV-1 Tat: coping with negative elongation factors. Curr. Opin. Immunol. 11:460-465. [DOI] [PubMed] [Google Scholar]
- 27.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goff, S. P. 1996. Operating under a Gag order: a block against incoming virus by the Fv1 gene. Cell 86:691-693. [DOI] [PubMed] [Google Scholar]
- 29.Habu, K., J. Nakayama-Yamada, M. Asano, S. Saijo, K. Itagaki, R. Horai, H. Yamamoto, T. Sekiguchi, T. Nosaka, M. Hatanaka, and Y. Iwakura. 1999. The human T cell leukemia virus type I-tax gene is responsible for the development of both inflammatory polyarthropathy resembling rheumatoid arthritis and noninflammatory ankylotic arthropathy in transgenic mice. J. Immunol. 162:2956-2963. [PubMed] [Google Scholar]
- 30.Hart, C. E., C. Y. Ou, J. C. Galphin, J. Moore, L. T. Bacheler, J. J. Wasmuth, S. R. Petteway, Jr., and G. Schochetman. 1989. Human chromosome 12 is required for elevated HIV-1 expression in human-hamster hybrid cells. Science 246:488-491. [DOI] [PubMed] [Google Scholar]
- 31.Hatziioannou, T., S. Cowan, and P. D. Bieniasz. 2004. Capsid-dependent and -independent postentry restriction of primate lentivirus tropism in rodent cells. J. Virol. 78:1006-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 34.Hogan, B., E. Constantini, and E. Lacey. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Isegawa, Y., J. Sheng, Y. Sokawa, K. Yamanishi, O. Nakagomi, and S. Ueda. 1992. Selective amplification of cDNA sequence from total RNA by cassette-ligation mediated polymerase chain reaction (PCR): application to sequencing 6.5 kb genome segment of hantavirus strain B-1. Mol. Cell Probes 6:467-475. [DOI] [PubMed] [Google Scholar]
- 36.Iwakura, Y., T. Shioda, M. Tosu, E. Yoshida, M. Hayashi, T. Nagata, and H. Shibuta. 1992. The induction of cataracts by HIV-1 in transgenic mice. AIDS 6:1069-1075. [DOI] [PubMed] [Google Scholar]
- 37.Jackson, J. B., K. L. MacDonald, J. Cadwell, C. Sullivan, W. E. Kline, M. Hanson, K. J. Sannerud, S. L. Stramer, N. J. Fildes, S. Y. Kwok, et al. 1990. Absence of HIV infection in blood donors with indeterminate western blot tests for antibody to HIV-1. N. Engl. J. Med. 322:217-222. [DOI] [PubMed] [Google Scholar]
- 38.Jolicoeur, P., and D. Baltimore. 1976. Effect of Fv-1 gene product on proviral DNA formation and integration in cells infected with murine leukemia viruses. Proc. Natl. Acad. Sci. USA 73:2236-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jolicoeur, P., and E. Rassart. 1980. Effect of Fv-1 gene product on synthesis of linear and supercoiled viral DNA in cells infected with murine leukemia virus. J. Virol. 33:183-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keppler, O. T., W. Yonemoto, F. J. Welte, K. S. Patton, D. Iacovides, R. E. Atchison, T. Ngo, D. L. Hirschberg, R. F. Speck, and M. A. Goldsmith. 2001. Susceptibility of rat-derived cells to replication by human immunodeficiency virus type 1. J. Virol. 75:8063-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klatzmann, D., E. Champagne, S. Chamaret, J. Gruest, D. Guetard, T. Hercend, J. C. Gluckman, and L. Montagnier. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767-768. [DOI] [PubMed] [Google Scholar]
- 42.Koito, A., Y. Kameyama, C. Cheng-Mayer, and S. Matsushita. 2003. Susceptibility of mink (Mustera vision)-derived cells to replication by human immunodeficiency virus type 1. J. Virol. 77:5109-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koito, A., H. Shigekane, and S. Matsushita. 2003. Ability of small animal cells to support the postintegration phase of human immunodeficiency virus type-1 replication. Virology 305:181-191. [DOI] [PubMed] [Google Scholar]
- 44.Kukolj, G., K. S. Jones, and A. M. Skalka. 1997. Subcellular localization of avian sarcoma virus and human immunodeficiency virus type 1 integrases. J. Virol. 71:843-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwok, S., D. E. Kellogg, N. McKinney, D. Spasic, L. Goda, C. Levenson, and J. J. Sninsky. 1990. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 18:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landau, N. R., M. Warton, and D. R. Littman. 1988. The envelope glycoprotein of the human immunodeficiency virus binds to the immunoglobulin-like domain of CD4. Nature 334:159-162. [DOI] [PubMed] [Google Scholar]
- 47.Lores, P., V. Boucher, C. Mackay, M. Pla, H. Von Boehmer, J. Jami, F. Barre-Sinoussi, and J. C. Weill. 1992. Expression of human CD4 in transgenic mice does not confer sensitivity to human immunodeficiency virus infection. AIDS Res. Hum. Retrovir. 8:2063-2071. [DOI] [PubMed] [Google Scholar]
- 48.Lu, Y. L., P. Spearman, and L. Ratner. 1993. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J. Virol. 67:6542-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maddon, P. J., A. G. Dalgleish, J. S. McDougal, P. R. Clapham, R. A. Weiss, and R. Axel. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47:333-348. [DOI] [PubMed] [Google Scholar]
- 50.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528-33539. [DOI] [PubMed] [Google Scholar]
- 51.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mariani, R., B. A. Rasala, G. Rutter, K. Wiegers, S. M. Brandt, H. G. Krausslich, and N. R. Landau. 2001. Mouse-human heterokaryons support efficient human immunodeficiency virus type 1 assembly. J. Virol. 75:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mariani, R., G. Rutter, M. E. Harris, T. J. Hope, H. G. Krausslich, and N. R. Landau. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 74:3859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masuda, T., V. Planelles, P. Krogstad, and I. S. Chen. 1995. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 69:6687-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller, D. G., and A. D. Miller. 1994. A family of retroviruses that utilize related phosphate transporters for cell entry. J. Virol. 68:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morikawa, Y., S. Hinata, H. Tomoda, T. Goto, M. Nakai, C. Aizawa, H. Tanaka, and S. Omura. 1996. Complete inhibition of human immunodeficiency virus Gag myristoylation is necessary for inhibition of particle budding. J. Biol. Chem. 271:2868-2873. [DOI] [PubMed] [Google Scholar]
- 57.Neil, S., F. Martin, Y. Ikeda, and M. Collins. 2001. Postentry restriction to human immunodeficiency virus-based vector transduction in human monocytes. J. Virol. 75:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newstein, M., E. J. Stanbridge, G. Casey, and P. R. Shank. 1990. Human chromosome 12 encodes a species-specific factor which increases human immunodeficiency virus type 1 tat-mediated trans activation in rodent cells. J. Virol. 64:4565-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nisole, S., and A. Saib. 2004. Early steps of retrovirus replicative cycle. Retrovirology 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nisole, S., J. P. Stoye, and A. Saib. 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3:799-808. [DOI] [PubMed] [Google Scholar]
- 61.Nomura, H., B. W. Nielsen, and K. Matsushima. 1993. Molecular cloning of cDNAs encoding a LD78 receptor and putative leukocyte chemotactic peptide receptors. Int. Immunol. 5:1239-1249. [DOI] [PubMed] [Google Scholar]
- 62.Ory, D. S., B. A. Neugeboren, and R. C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 93:11400-11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petit, C., O. Schwartz, and F. Mammano. 2000. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J. Virol. 74:7119-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Piller, S. C., L. Caly, and D. A. Jans. 2003. Nuclear import of the pre-integration complex (PIC): the Achilles heel of HIV? Curr. Drug Targets 4:409-429. [DOI] [PubMed] [Google Scholar]
- 65.Planelles, V., A. Haislip, E. S. Withers-Ward, S. A. Stewart, Y. Xie, N. P. Shah, and I. S. Chen. 1995. A new reporter system for detection of retroviral infection. Gene Ther. 2:369-376. [PubMed] [Google Scholar]
- 66.Pluymers, W., P. Cherepanov, D. Schols, E. De Clercq, and Z. Debyser. 1999. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258:327-332. [DOI] [PubMed] [Google Scholar]
- 67.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 68.Popov, S., M. Rexach, G. Zybarth, N. Reiling, M. A. Lee, L. Ratner, C. M. Lane, M. S. Moore, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 17:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakai, H., R. Shibata, J. Sakuragi, S. Sakuragi, M. Kawamura, and A. Adachi. 1993. Cell-dependent requirement of human immunodeficiency virus type 1 Vif protein for maturation of virus particles. J. Virol. 67:1663-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sawada, S., K. Gowrishankar, R. Kitamura, M. Suzuki, G. Suzuki, S. Tahara, and A. Koito. 1998. Disturbed CD4+ T cell homeostasis and in vitro HIV-1 susceptibility in transgenic mice expressing T cell line-tropic HIV-1 receptors. J. Exp. Med. 187:1439-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sherman, M. P., and W. C. Greene. 2002. Slipping through the door: HIV entry into the nucleus. Microbes Infect. 4:67-73. [DOI] [PubMed] [Google Scholar]
- 72.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 73.Suzuki, Y., N. Misawa, C. Sato, H. Ebina, T. Masuda, N. Yamamoto, and Y. Koyanagi. 2003. Quantitative analysis of human immunodeficiency virus type 1 DNA dynamics by real-time PCR: integration efficiency in stimulated and unstimulated peripheral blood mononuclear cells. Virus Genes 27:177-188. [DOI] [PubMed] [Google Scholar]
- 74.Tanaka, J., H. Ozaki, J. Yasuda, R. Horai, Y. Tagawa, M. Asano, S. Saijo, M. Imai, K. Sekikawa, M. Kopf, and Y. Iwakura. 2000. Lipopolysaccharide-induced HIV-1 expression in transgenic mice is mediated by tumor necrosis factor-alpha and interleukin-1, but not by interferon-gamma nor interleukin-6. AIDS 14:1299-1307. [DOI] [PubMed] [Google Scholar]
- 75.Tsuruo, T., T. Yamori, K. Naganuma, S. Tsukagoshi, and Y. Sakurai. 1983. Characterization of metastatic clones derived from a metastatic variant of mouse colon adenocarcinoma 26. Cancer Res. 43:5437-5442. [PubMed] [Google Scholar]
- 76.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4795-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Maanen, M., and R. E. Sutton. 2003. Rodent models for HIV-1 infection and disease. Curr. HIV Res. 1:121-130. [DOI] [PubMed] [Google Scholar]
- 78.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 79.Yang, W. K., J. O. Kiggans, D. M. Yang, C. Y. Ou, R. W. Tennant, A. Brown, and R. H. Bassin. 1980. Synthesis and circularization of N- and B-tropic retroviral DNA Fv-1 permissive and restrictive mouse cells. Proc. Natl. Acad. Sci. USA 77:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yasuda, J., T. Miyao, M. Kamata, Y. Aida, and Y. Iwakura. 2001. T cell apoptosis causes peripheral T cell depletion in mice transgenic for the HIV-1 vpr gene. Virology 285:181-192. [DOI] [PubMed] [Google Scholar]
- 81.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 82.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]
- 83.Zheng, Y. H., H. F. Yu, and B. M. Peterlin. 2003. Human p32 protein relieves a post-transcriptional block to HIV replication in murine cells. Nat. Cell Biol. 5:611-618. [DOI] [PubMed] [Google Scholar]