Abstract
It is widely accepted that nucleocapsids of herpesviruses bud through the inner nuclear membrane (INM), but few studies have been undertaken to characterize the composition of these nascent virions. Such knowledge would shed light on the budding reaction at the INM and subsequent steps in the egress pathway. The present study focuses on glycoprotein M (gM), a type III integral membrane protein of herpes simplex virus 1 (HSV-1) that likely contains eight transmembrane domains. The results indicated that gM localized primarily at the perinuclear region, with especially bright staining near the nuclear membrane (NM). Immunogold electron microscopic analysis indicated that, like gB and gD (M. R. Torrisi et al., J. Virol. 66:554-561, 1992), gM localized within both leaflets of the NM, the envelopes of nascent virions that accumulate in the perinuclear space, and the envelopes of cytoplasmic and mature extracellular virus particles. Indirect immunofluorescence studies revealed that gM colocalized almost completely with a marker of the Golgi apparatus and partially with a marker of the trans-Golgi network (TGN), whether or not these markers were displaced to the perinuclear region during infection. gM was also located in punctate extensions and invaginations of the NM induced by the absence of a viral kinase encoded by HSV-1 US3 and within virions located in these extensions. Our findings therefore support the proposition that gM, like gB and gD, becomes incorporated into the virion envelope upon budding through the INM. The localization of viral glycoproteins and Golgi and TGN markers to a perinuclear region may represent a mechanism to facilitate the production of infectious nascent virions, thereby increasing the amount of infectivity released upon cellular lysis.
Herpes simplex virions are somewhat pleomorphic enveloped particles of at least 200 nm diameter. The particles contain a lipid envelope surrounding a proteinaceous tegument layer that lies between the internal surface of the envelope and the external surface of the nucleocapsid. Virally encoded membrane proteins are integrated into the lipid envelope, and 11 of these are glycosylated (46).
Considerable effort by a number of laboratories has been expended to understand the events leading to virion assembly. It is generally agreed that an important step in the production of infectious particles involves the envelopment of DNA-containing nucleocapsids at electron-dense patches within the inner nuclear membrane (INM) of infected cells. After this step, enveloped particles can be observed between the INM and outer nuclear membrane (ONM) (42). This compartment is termed the perinuclear space and is continuous with the lumen of the endoplasmic reticulum (ER).
Although the composition of perinuclear virions should greatly affect subsequent steps of virion egress, few studies have been undertaken to molecularly characterize these particles. Immunoelectron microscopy has revealed that herpes simplex virus (HSV) glycoproteins B and D (gB and gD, respectively), the HSV tegument protein encoded by UL11, and complexes of the HSV and pseudorabies virus (PRV) UL34 and UL31 proteins localize at the INM and in virions within the perinuclear space (1, 14, 41, 48). HSV-1 glycoprotein C and HSV-1 VP16 fused to green fluorescent protein (GFP) have also been noted in perinuclear virions (17, 36). These observations support the proposition that at least some integral membrane and tegument proteins become incorporated into virions upon budding through the INM.
To our knowledge, whether glycoproteins other than gB, gC, or gD localize in the nuclear membrane (NM) has not been investigated. The present study focuses on HSV-1 glycoprotein M (gM), encoded by UL10 (3, 31). The UL10 open reading frame predicts that gM is a hydrophobic integral membrane protein containing eight transmembrane domains, with both the N and the C termini predicted to lie within the cytosol (32, 49). Although the primary sequence of gM is variable, the hydropathy plots of gM homologs of other herpesviruses are virtually superimposable with that of HSV-1 gM, suggesting that the topology of the protein within membranes is conserved (J. Baines, unpublished observations).
The gM of HSV-1 is a virion component that is also associated with the plasma membrane of unfixed cells (3). As in other herpesviruses, HSV-1 gM forms a complex with another protein, encoded by UL49.5 in HSV (19, 25, 28-30, 43, 51). In viruses where HSV-1 UL49.5 protein orthologs are glycosylated (e.g., pseudorabies virus, human herpesvirus 8, human cytomegalovirus, and Epstein-Barr virus [EBV]), the gM interacting protein is designated gN (19, 25, 27, 29). Deletion of HSV-1 gM reduces infectious titers approximately 10-fold below those of wild-type viruses in Vero and BHK cells (3, 31). Similar defects in replication have been noted upon mutation of the gM homologs of PRV, equine herpesvirus, bovine herpesvirus, and laryngotracheitis virus (10, 15, 24, 37). The open reading frames encoding gM homologs of Mareks disease virus, human cytomegalovirus, and EBV are essential for normal replication (16, 27, 47). In the case of EBV, a gN-null mutant also lacks detectable gM and exhibits severe defects in viral egress and viral penetration into cells (27).
Whereas transient expression of glycoproteins B, D, H, and L are sufficient to cause cell-cell fusion, coexpression of gM with these proteins precludes cell fusion (23, 25). Interestingly, gM was also able to preclude fusion mediated by the bovine syncytial virus F protein, indicating a general rather than a specific effect (23, 24). Moreover, coexpression of gM decreased the surface expression of not only the Human respiratory syncytial virus F protein but also HSV-1 glycoproteins gD and gH/gL, influenza virus HA protein, and herpesvirus entry mediator, while Nectin 2 alpha or CD8 surface expression were not affected (9). Taken together with the observation that PRV gM and HSV-1 gM/UL49.5 colocalize with the trans-Golgi network (TGN) marker TGN46 when expressed in the absence of other viral proteins (9), these data suggest that gM functions to relocalize glycoproteins from the plasma membrane to putative sites of viral assembly, such as the TGN.
Also relevant to this report are studies of the US3 protein of HSV-1, a serine threonine kinase that phosphorylates a number of viral and cellular substrates and is present in nascent perinuclear virions (39, 41). US3 is antiapoptotic, and its kinase activity is also necessary for the efficient egress of virions from the perinuclear space (22, 35, 41, 44). In the absence of US3, virions accumulate in the perinuclear space, often within punctuate evaginations and/or extensions of the NM (22, 41). The virions within these pockets of NM contain a complex of UL31 and UL34, and both of these proteins are substrates of the US3 kinase (20, 38, 40, 41). Whether other proteins associate with these perinuclear virions has not been investigated.
The ultrastructural localization of gM in cells infected with HSV-1 and whether HSV-1 gM localizes within the TGN or Golgi network of infected cells have not been determined. To determine the site(s) at which gM becomes incorporated into virions, we characterized the localization of gM by light and electron microscopy in HSV-infected cells. As a control, cells infected with a deletion mutant lacking the capacity to synthesize gM were analyzed in parallel. It might be expected that, if the sole function of gM was to internalize surface proteins or participate in nucleocapsid envelopment at the TGN, then gM would localize preferentially in the TGN and plasma membrane in infected cells, similar to what is seen when gM and UL49.5 are coexpressed in uninfected cells (9). In contrast to the predictions of this hypothesis, however, gM localized primarily to the NM and also within perinuclear virions. Thus, HSV functions other than those mediated by UL49.5 play important roles in gM targeting to the NM where gM becomes incorporated into nascent virions. The data indicate that gM is incorporated into virions earlier in the egress pathway than previously appreciated.
MATERIALS AND METHODS
Indirect immunofluorescence.
HEp-2 cells were seeded on glass coverslips in six-well plates at 20% confluence and grown overnight at 37°C. Cells were infected with 3 PFU of either HSV-1(F), Us3-null mutant R7039, or gM-null mutant per cell (2, 11, 39). At various times after infection, the cells were washed three times with cold phosphate-buffered saline (PBS) and then fixed with ice-cold methanol for 15 min. Cells were washed again three times with PBS and then incubated 15 min at room temperature with 50 mM NH4Cl in PBS to quench autofluorescence. After another rinse, cells were permeabilized with 0.1% Triton X-100-PBS for 2 min and were washed as before. Cells were reacted with 10% goat serum and 10% filtered human serum in PBS for 15 min to decrease nonspecific binding and were then incubated with rabbit anti-UL10 antisera for 30 min at room temperature. In some experiments the permeabilized cells were reacted with a 1:100 dilution of antibody (Abcam, catalog no. ab6284) directed against Golgi 58K, a peripheral Golgi protein originally shown to be associated with microtubules (5), or 1:100 dilution of antibody directed against TGN38 (BD Transduction Labs, catalog no. 610898), a marker of the TGN and homolog of human TGN46 (26). After extensive rinsing in PBS, the cells were incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit secondary antibody at a 1:100 dilution for 30 min at room temperature. Samples were rinsed with PBS after the secondary incubation and then rinsed briefly with distilled water just before mounting with Vectashield mounting medium. (Vector Labs, United Kingdom). Glass slides were examined with an Olympus confocal laser scanning microscope equipped with krypton and argon lasers under ×63 oil immersion objective and ×10 ocular objective lenses. To assess overlap of emitted fluorescence, each channel was recorded separately with only one laser powered on. Digital images were acquired with Fluoview software (version 2.1.39).
Ultrastructural analysis of gM localization in infected cells.
To determine whether gM becomes incorporated into virions during budding at the INM, immunoelectron microscopy was performed. Cells were mock infected or were infected with HSV-1(F) or the gM deletion mutant virus, R7216. At 14 to 18 h after infection, cells were fixed in 4% paraformaldehyde and 0.25% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4) for 30 min at room termperature and then for 90 min at 4°C. The fixed cells were washed three times in 0.1 M sodium phosphate buffer (pH 7.4) at 4°C and dehydrated through a series of ethanol concentrations at 4°C and −20°C. Infiltration and embedding was done stepwise at −20°C with LRWhite (Electron Microscopy Sciences, Fort Washington, PA). The samples were then polymerized under UV light at −35°C overnight. Thin sections (60 to 90 nm thick) were collected on 300 mesh nickel grids (Ted Pella, Inc., Redding, CA) and floated on drops for the following procedures.
For electron microscopic immunostaining, grids were blocked with 10% normal goat sera and 10% human serum in PBS-0.05% Tween (PBST) and 1% fish gelatin for 15 min. at room temperature and were incubated on drops of gM-specific rabbit antibody diluted 1:250 in PBST plus 1% fish gelatin overnight in a humidity chamber at 4°C. After incubation, grids were washed by brief passage over a series of 3 drops in a high-salt buffer (5× PBS, 0.05% Tween and 1% fish gelatin) and then 5 drops of 1× PBST and fish gelatin. The secondary antibody, goat anti-rabbit immunoglobulin conjugated with 12-nm colloidal gold, was diluted 1:100 in PBST-1% fish gelatin and reacted for 1 h in a humidity chamber at room temperature. The grids were then washed as before on 6 successive drops of PBST-1% fish gelatin and then rinsed in a beaker of 200 ml of filtered water. Grids were air dried at room temperature prior to staining with 2% aqueous uranyl acetate for 20 min and then Reynolds lead citrate for 7 min. Stained grids were viewed in a Philips 201 transmission electron microscope. Conventionally rendered negatives of electron microscopic images were scanned by using a Microtek scanmaker 5 and Scanwizard Pro PPC 1.02 software. Positive images were rendered from digitized negatives with Adobe Photoshop software.
RESULTS
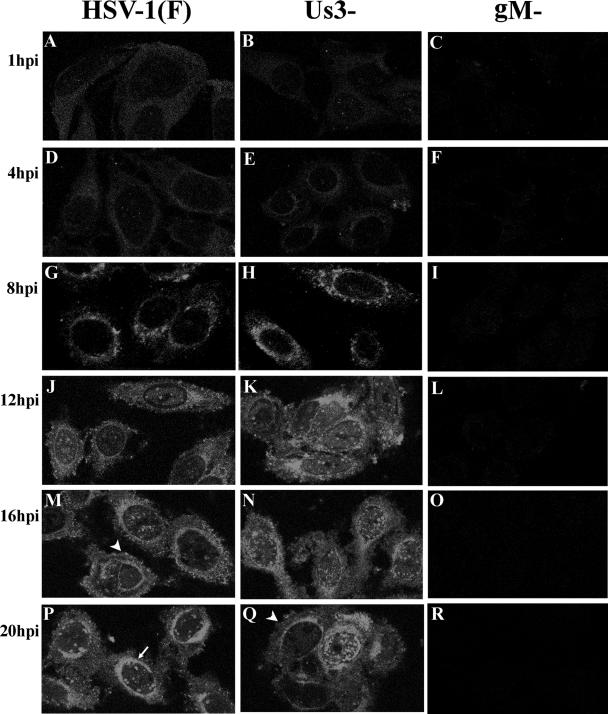
Cells were infected with HSV-1(F) and a deletion virus lacking the UL10 gene which encodes gM (2). HEp-2 cells were fixed at 1, 4, 8, 12, 16, and 20 h after infection and were immunostained with gM specific antibody as detailed in Materials and Methods. As seen in Fig. 1, significant immunostaining was not observed in cells infected with the gM-null mutant at any time postinfection. In contrast, gM-specific immunostaining was observed starting at 8 h after infection in cells infected with wild-type HSV-1(F). The staining appeared both smooth and as small foci distributed throughout the cytoplasm, around the nucleus (Fig. 1P, arrow), and at the plasma membrane (Fig. 1M, arrowhead). The latter finding is consistent with previous results showing that gM associates with the plasma membrane of unfixed cells (3). Although these regions stained consistently with gM after 8 h postinfection, the intensity of immunostaining in different regions changed over time. Specifically, staining was more evenly distributed throughout the cytoplasm at 8 h after infection, whereas more intense immunostaining was observed adjacent to the nucleus at later times (compare Fig. 1G and P).
FIG. 1.
Confocal indirect immunofluorescence localization of gM in HEp-2 cells infected with wild-type, gM-null mutant, or US3-null mutant viruses at various times after infection. Infected cells were fixed in methanol at the indicated times and reacted with gM-specific rabbit polyclonal antibody. Bound antibody was revealed by reaction with FITC-conjugated goat anti-rabbit immunoglobulin, followed by laser scanning confocal microscopy. Single optical sections are shown. The arrow in panel P indicates areas of nuclear fluorescence. The arrowheads in panels M and Q indicate fluorescence associated with the plasma membrane.
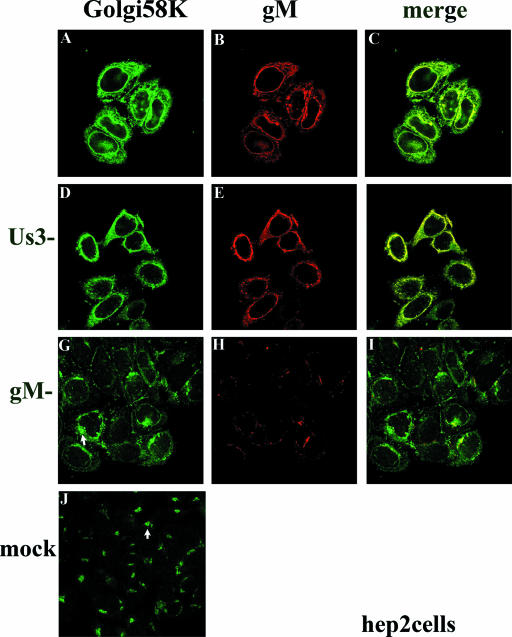
The patterns of gM-specific staining described above were reminiscent of the location of redistributed Golgi and TGN in infected HEp-2 cells (7). To determine whether gM was located in these organelles or their remnants, HEp-2 cells were infected with HSV-1(F) or the gM-null virus R7216 and were fixed at 16 h after infection. The fixed cells were then reacted with antibodies directed to gM and the Golgi marker 58K. As seen in Fig. 2, in uninfected HEp-2 cells Golgi 58K appeared predominantly in a single discrete region located near the nucleus but within the cytoplasm (Fig. 2J, arrow). In contrast, Golgi 58K-specific immunostaining localized around the perimeter of the nuclei of cells infected with HSV-1(F) (Fig. 2A). In addition, some staining in a punctate pattern was located elsewhere in the cytoplasm. Importantly, much of the gM-specific immunostaining localized in regions that stained brightly with Golgi 58K-specific antibody, and this colocalization was especially pronounced near and adjacent to the nucleus.
FIG. 2.
Indirect immunofluorescence of gM and Golgi localization in HEp-2 cells infected with wild type (top row), gM-null, or US3-null viruses at 16 h postinfection. HEp-2 cells were infected with the indicated viruses and were reacted with antibody to gM and mouse monoclonal antibody to Golgi 58K, a resident protein of the Golgi apparatus. Bound immunoglobulins were revealed by reaction with FITC conjugated goat anti-rabbit or Texas Red-conjugated anti-mouse immunoglobulin G followed by laser scanning confocal microscopy. Single optical sections are shown. Both the red channel and the green channel were scanned independently with only the fluorescence-stimulating laser powered on. The rightmost column shows scanned images of the left columns merged by using Adobe Photoshop software; a yellow color indicates regions where the two antibodies coincide. The arrows in panels G and J indicate different staining patterns of the Golgi marker in cells that were infected with the gM-null virus and mock infected, respectively.
The pattern of Golgi 58K immunostaining in HEp-2 cells infected with the gM-null mutant differed from that of cells infected with HSV-1(F) and mock-infected cells. Specifically, overall immunostaining was slightly reduced compared to infection with wild-type HSV-1, and the Golgi staining was not smoothly distributed around the nucleus. Rather, the immunostaining appeared in perinuclear patches, with some areas staining brightly with Golgi 58K antibody, and other regions staining faintly or not at all (e.g., Fig. 2G, arrow). These patches were less discrete and more widely distributed around the perinuclear region than was the case in mock-infected cells (compare regions indicated by arrows in Fig. 2G and J). We therefore conclude that gM is not required for the redistribution of the Golgi apparatus toward the perinuclear region but is required for the even redistribution of Golgi markers around the nucleus, as seen in cells infected with wild-type HSV-1(F).
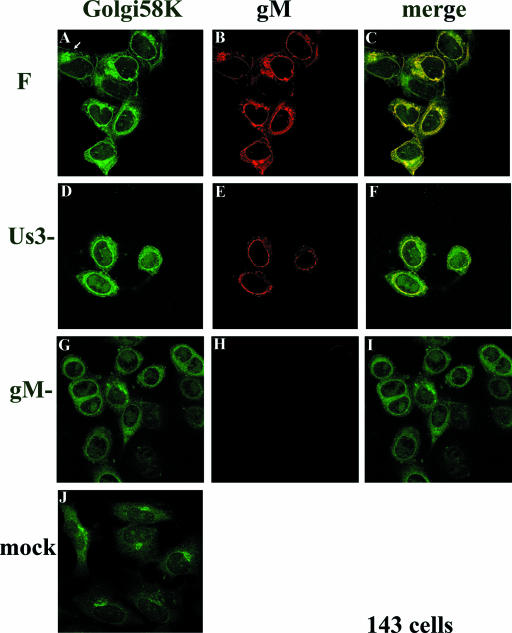
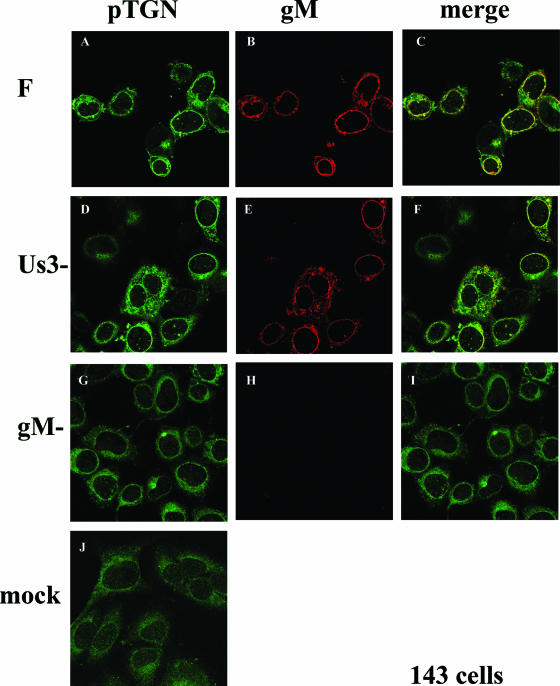
It has been shown that 143 cells are more resistant to HSV-mediated Golgi redistribution than are HEp-2 cells, although at late times after infection, the Golgi is redistributed in a portion of 143 cells (7). To determine whether the effects of gM on Golgi redistribution also occurred in 143 cells, we fixed HSV-1(F)-infected 143 cells at 16 h postinfection, when a portion of the cells maintain the Golgi such that it is not redistributed toward the nucleus. At 16 h postinfection with HSV-1(F) an estimated 25% of the 143 cells contained the Golgi 58K marker in a discrete perinuclear region (e.g., Fig. 3A, arrow, and data not shown), and this localization was reminiscent of the appearance of uninfected cells stained with the Golgi 58K specific antibody (Fig. 3J). In other cells in the same experiment (Fig. 3A), the pattern of Golgi 58K staining was similar to that of HEp-2 cells infected with HSV-1(F), i.e., surrounding the nucleus. Regardless of the pattern of immunostaining in various 143 cells, gM and Golgi 58K immunostaining colocalized almost completely. In contrast to observations in HEp-2 cells (Fig. 2), the pattern of Golgi 58K immunostaining in 143 cells infected with the gM-null virus was similar to that of cells infected with HSV-1(F) (compare Fig. 3A and G) and was arranged in a smooth pattern surrounding the nucleus. The overall intensity of Golgi 58K staining was reduced relative to that of HSV-1(F)-infected cells immunostained in parallel (compare Fig. 3A and G). Although less immunostaining appeared to concentrate at the nuclear rim in cells infected with the gM-null mutant compared to those infected with HSV-1(F), it is not clear whether this apparent difference was simply a consequence of the overall decreased intensity of staining. In a control experiment, little immunostaining was apparent in 143 cells infected with the gM-null virus reacted with the gM specific antibody (Figure 3H). These data therefore indicate that gM colocalizes with both redistributed and unredistributed Golgi 58K in infected 143 cells. The data also indicate that gM has an effect on the appearance or distribution of Golgi marker immunostaining in different infected cell lines.
FIG. 3.
Indirect immunofluorescence of infected and mock-infected 143 cells stained with gM or a Golgi-specific marker. 143 cells were infected with the indicated viruses. At 16 h postinfection, the cells were fixed and immunostained as described in the legend to Fig. 2. All images are of single optical sections collected using identical laser and photomultiplier tube (PMT) settings. The arrow in panel A indicates a cell with a Golgi 58K staining pattern that is reminiscent of that of mock-infected cells and different from the staining pattern of other infected cells in the same panel. F, HSV-1(F).
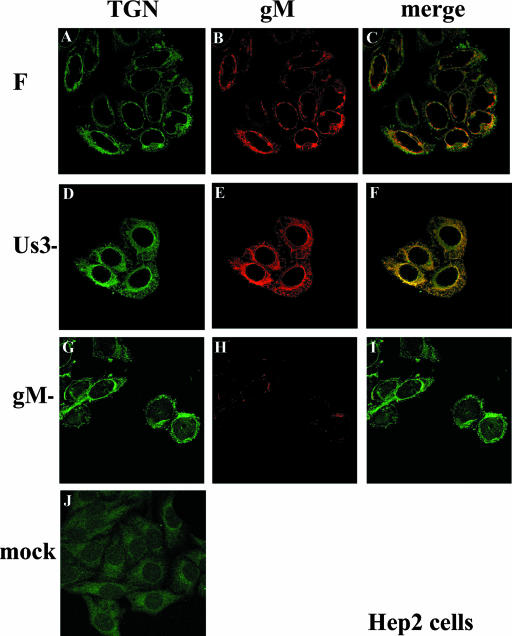
To determine the fate of the TGN in infected cells and whether gM was located there as suggested by experiments with transiently expressed gM and gN (9), HEp-2 cells were infected with HSV-1(F) and at 16 h postinfection were fixed and immunostained with a mouse monoclonal antibody directed against TGN38 and the rabbit polyclonal antibody directed against gM. As a control, cells were also infected with the gM-null virus. In uninfected cells (Fig. 4J), TGN38-specific immunostaining was distributed diffusely in the cytoplasm, with slightly greater intensity within a single location in the cytoplasm near the nuclear rim. In infected cells however, the pattern of the TGN38 immunostaining circled the nuclei of cells infected with HSV-1(F) and the gM-null virus but was also contained in the cytoplasm in a punctate pattern and occasionally on the plasma membrane (Fig. 4A and G). gM staining colocalized most extensively with TGN-specific staining located near the nucleus, with only occasional colocalization within the punctate staining seen in the cytoplasm and plasma membrane (Fig. 4C). There was very little nonspecific immunostaining in cells infected with the gM-null mutant reacted with the gM-specific antibody (Fig. 4H). Taken with the data shown in Fig. 2, these results indicate that gM colocalized more extensively with the Golgi apparatus of infected cells than the TGN.
FIG. 4.
Indirect immunofluorescence of infected and mock-infected HEp-2 cells stained with TGN- or gM-specific antibodies. HEp-2 cells infected 16 h previously were fixed and reacted with gM specific antibody or a mouse monoclonal antibody to TGN38, a marker of the TGN. Confocal microscopy, image processing, and analyses were performed as described in the legend to Fig. 2. All images are of single optical sections taken at identical laser intensities and PMT settings. For illustrative purposes, the brightness of the lower left image (mock) was increased by 30% using Adobe Photoshop software. F, HSV-1(F).
In a separate experiment, TGN38 immunostaining was performed in uninfected 143 cells or in 143 cells infected with HSV-1(F) or the gM-null mutant. The results, shown in Fig. 5, indicated that gM extensively colocalized with TGN38 at the nuclear rim and less with TGN38 that was not located adjacent to the nucleus (Fig. 5C). This localization was similar to that of gM in HEp-2 cells (Fig. 4). On the other hand, the absence of gM altered the appearance of TGN38 immunostaining in 143 cells, inasmuch as the distribution of TGN38 was less intense at the nuclear rim and localized in a smooth pattern in cells infected with the gM-null mutant rather than the intense mostly punctate pattern seen at the nuclear rim of cells infected with wild-type HSV-1(F) (compare Fig. 5A and G).
FIG. 5.
Indirect immunofluorescence of infected and mock-infected 143 cells stained with TGN or gM-specific antibodies. 143 cells were mock infected or were infected with the indicated viruses and were fixed at 16 h after infection. The cells were immunostained with antibodies to TGN38 and gM, and bound immunoglobulin was revealed by reaction with appropriately conjugated antisera. Images of cells were taken at identical laser and PMT settings. Image collection, processing, and analysis were performed as in the legend to Fig. 2. F, HSV-1(F).
Effects of US3 on localization of gM and markers of Golgi and TGN.
We and others have previously noted that the absence of U33 or its associated kinase activity alters the conformation of the NM of HSV-infected cells (41, 44). Specifically, aberrantly large numbers of enveloped virions localize within the perinuclear space and within punctate extensions or evaginations of this compartment. These punctate extensions contain substantial amounts of the UL34 and UL31 proteins, whereas the UL31 and UL34 proteins are normally distributed around the nuclear rim in a smooth even pattern (41). In order to determine whether normal distribution of gM around the nuclear rim was also dependent on US3, HEp-2 and 143 cells were infected with a US3 deletion mutant previously described (39) and at various times after infection were examined by indirect immunofluorescence confocal microscopy.
As shown in Fig. 1, punctate gM-specific staining associated with the nuclear rim was more apparent in some cells infected with the US3 mutant late in infection (Fig. 1N and Q) compared to the appearance of cells infected with wild-type virus. Detection of this punctate staining depended on the plane of the optical section, with more extensive punctate structures visible at the top and bottom of the nucleus. Thus, we conclude that normal smooth distribution of gM near the nuclear rim is partially dependent on US3. On the other hand, gM staining around the nucleus also accumulated in a smooth staining pattern in most cells.
In light of evidence that US3 is necessary for efficient virion egress from the perinuclear space, it was of interest to determine whether part of this function was associated with putative effects of US3 on Golgi or TGN localization in infected cells. To examine this possibility, HEp-2 and 143 cells were infected with wild-type virus and the US3 deletion mutant and were immunostained at 16 h after infection with antibodies directed against TGN38 and the Golgi marker 58K. As shown in Fig. 2 to 5, the Golgi and TGN markers were redistributed to the perinuclear region whether or not US3 was expressed in infected cells. We therefore conclude that US3 is largely dispensable for the HSV-mediated relocalization of TGN and Golgi markers toward the perinuclear region.
Ultrastructural localization of gM.
Inasmuch as localization near or at the nuclear rim does not identify whether a given protein localizes in the INM, ONM, or ER, efforts were expended to characterize the gM-specific immunostaining at the ultrastructural level. Cells were infected with HSV-1(F) or the gM-null virus, fixed in paraformaldehyde, embedded, and subjected to immunoelectron microscopy using the gM-specific antibody as detailed in Materials and Methods.
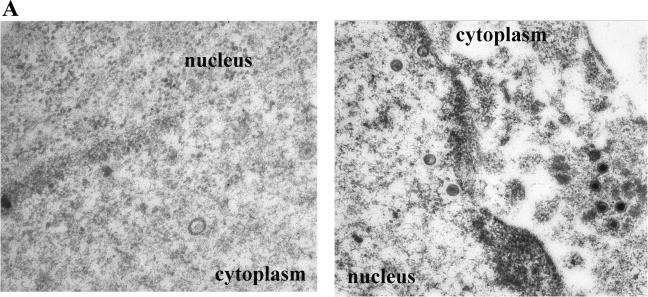
Thin sections of cells infected with the gM deletion virus and reacted with the gM-specific antisera contained very few colloidal gold beads, and rare gold beads (one to three per view at magnification 45,000) were distributed equally throughout the nucleoplasm, cytoplasm, and extracellular space (Fig. 6A). Moreover, the gold beads were not associated with any particular structure. Despite the fact that many structures apparently delimited by membranes were present in the cytoplasm, none of these contained large numbers of gold beads. Similar low background levels were obtained upon reaction of thin sections of mock-infected HEp-2 cells with gM-specific antiserum (not shown).
FIG.6.
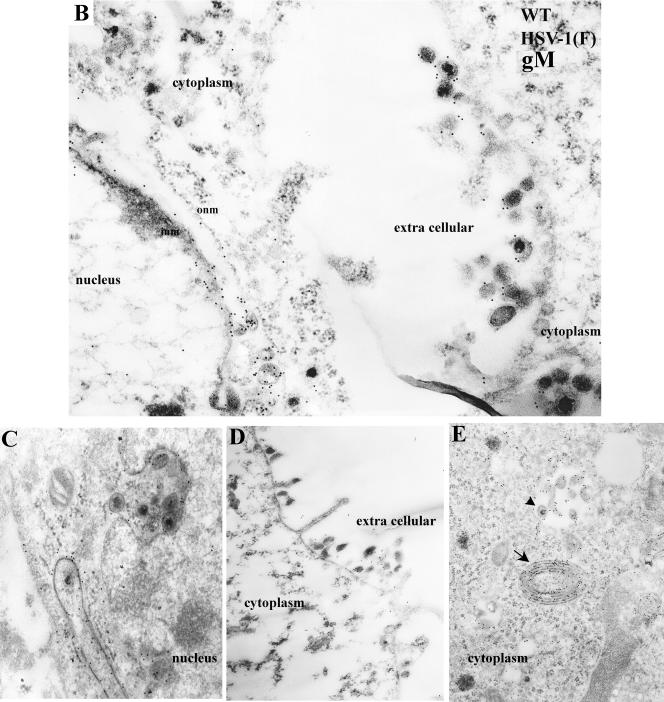
(A) Immune transmission electron microscopy of HEp-2 cells infected with the gM-null mutant and reacted with gM specific antibody. HEp-2 cells were infected with the gM-null virus R7216, fixed at 14 h after infection, and reacted with anti-gM rabbit polyclonal serum. Bound antibody was visualized by reaction with the F(ab)2 fragments of a goat anti-rabbit antibody conjugated to 12-nm colloidal gold particles. In this and subsequent electron photomicrographs, capsids of approximately 125 nm serve as a size standard. Magnifications: left panel, ×45,000; right panel, ×30,000. (B to E) Electron photomicrograph of a thin section of HEp-2 cells infected with HSV-1 (F) and stained with antibody to gM. HEp-2 cells were infected at 5.0 PFU per cell, harvested at 14 h after infection, fixed, embedded, sectioned, and reacted with rabbit gM-specific antibody and, subsequently, goat anti-rabbit IgG conjugated to 12 nm gold beads as described in Materials and Methods. In panel D, an arrow indicates heavily immunolabeled reduplicated membrane in cytoplasm. Arrowhead indicates a group of immunolabeled virions located within a cytoplasmic vacuole. Magnifications: C, D, and E, ×30,000; B, ×45,000.
In contrast, sections of cells infected with HSV-1(F) and reacted with gM-specific antibody exhibited extensive immunogold labeling. Representative results are shown in Fig. 6B to E and can be summarized as follows. (i) Figure 6B and C show a portion of the nuclei and perinuclear regions of HEp-2 cells infected with HSV-1(F) and reacted with the gM-specific antiserum. Gold beads were associated with the NM and the INM in particular. Viral particles in the perinuclear space were heavily labeled with gold beads. (ii) Whereas many colloidal gold beads representing the localization of gM associated with the ribbon-like electron-dense NM, beads were virtually absent from the nucleoplasm, intranuclear capsids, and other intranuclear structures (Fig. 6B and C). (iii) Cytoplasmic immunoreactivity was largely restricted to electron-dense viral particles or membranous structures, some of which exhibited extensive duplication (Fig. 6E, arrow). Immunoreactivity was detected in cytoplasmic virions and within vacuoles contained throughout the cytoplasm (Fig. 6E, arrowhead). (iv) gM specific immunoreactivity was also detected in association with extracellular virions (Fig. 6B and D).
These results indicate that gM localizes at the INM and ONM of infected cells and within nascent virions that accumulate between these membranes. gM also associates with cytoplasmic vesicular membranes and cytoplasmic and extracellular virions.
To determine whether gM accumulates within the NM extensions seen in the absence of US3, cells were infected with HSV-1(F) or the US3 deletion mutant, and immunoelectron microscopy using the gM-specific antibody was performed as described above.
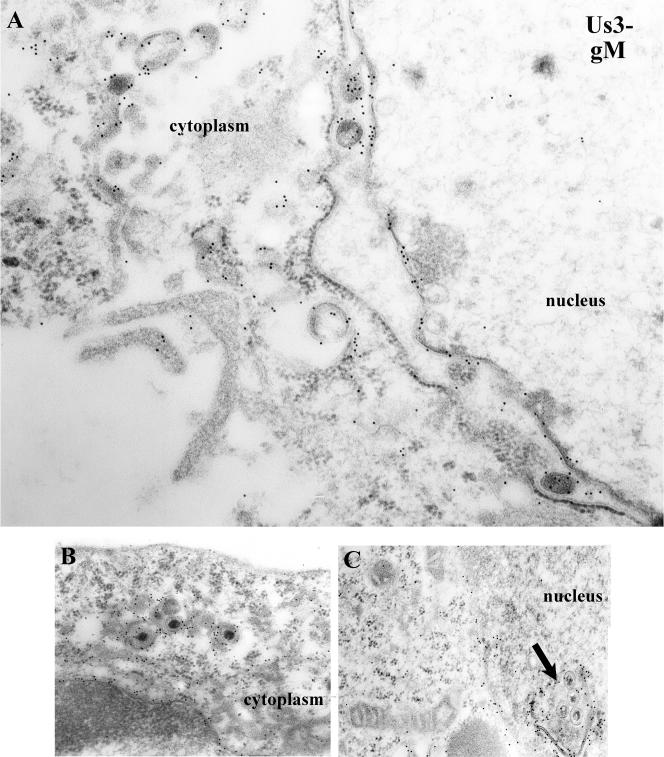
As demonstrated in the electron photomicrographs shown in Fig. 6 and Fig. 7, gM specific immunolabeling in cells infected with the US3 mutant was similar to that seen in cells infected with HSV-1(F) except that gM localized in both the NM and within virions within the punctate nuclear extensions or evaginations peculiar to cells infected with the US3 mutant. These data indicate that US3 is dispensable for incorporation of gM into perinuclear virions and association of gM with the INM and ONM. On the other hand, the even distribution of gM throughout the NM is dependent on US3, probably as a consequence of US3's effects on NM morphology.
FIG. 7.
Electron photomicrograph of thin sections of HEp-2 cells infected with a US3 mutant of HSV-1(F) and stained with gM-specific antibody. Cells were infected with R7039, a viral mutant lacking US3. At 14 h postinfection, the cells were fixed and embedded. Thin sections were reacted with gM-specific antibody, followed by reaction with 12-nm gold-conjugated anti-rabbit IgG, and examined in a Phillips 201 electron microscope. In panel C, an arrow indicates a cross-section of a punctate extension or invagination of the nuclear membrane containing virions. This feature is peculiar to cells infected with the US3 deletion virus. Magnifications: A, ×45,000; B and C, ×30,000.
DISCUSSION
The composition of perinuclear virions has not been well studied but is essential to understanding any of the three models of herpesvirion egress that have been proposed (18, 45, 50). In two of these models, the initial virion envelope obtained at the INM and present in perinuclear virions is retained by extracellular virions (18, 50). This implies that the composition of the perinuclear virion should be very similar to that of the extracellular virion and elicits important questions concerning how such glycoproteins become targeted en masse to envelopment sites at the INM in order to interact with nucleocapsids. Relevant to this are recent studies suggesting that targeting to the INM requires a nuclear localization signal in the cytosolic domains of integral membrane proteins (21).
In the third model, which is perhaps the most widely supported, the initial virion envelope is lost by fusion of the ONM with the envelope of the perinuclear virion (6, 34, 45). The composition of perinuclear virions is also critical to understanding this model inasmuch as the membrane fusion event must presumably be mediated by proteins associated with the perinuclear virion envelope and ONM. Regulation of such a fusion event also seems likely. For example, virions accumulate aberrantly in the perinuclear spaces of cells infected with viral mutants bearing mutations that fuse a truncated UL20 gene with UL20.5 or preclude US3 kinase activity, suggesting that these mutations impair ONM or virion envelope fusion or other means of virion egress from the perinuclear space (4, 12, 22, 41).
The present study indicates that gM localizes in both wild-type and US3(−) perinuclear virions and is associated with the INM and ONM of infected cells. These observations suggest that gM is incorporated into virions during envelopment at the INM. Thus, gM joins a group of proteins shown to associate with nascent virions, including those encoded by US3, UL11, UL31, UL34, VP16/GFP, and glycoproteins B, C, and D (1, 17, 36, 40, 48). Although such a conclusion is consistent with any of the three models of virion egress, it is critical for models in which the initial virion envelope is retained. The localization of gM in the present study and gB, gC, and gD in previous studies is not consistent with models of egress of PRV virions, in which perinuclear virions are shown to lack glycoproteins (17, 33, 34, 48).
We have also noted that, like pUL31 and pUL34, gM localizes in distended NMs that result upon infection with viral mutants lacking the US3 kinase (41). Unlike pUL31 and pUL34, however, gM-specific immunostaining was also associated with regions of the NM other than the extended pouches of NM. Thus, gM molecules likely become incorporated into virions by association with the pUL31/pUL34-containing envelopment sites, but at late times after infection some gM molecules within the INM also remain separate from these sites.
The localization of gM at the nuclear rim of infected cells is different from that in uninfected cells, where gM colocalizes primarily with TGN markers in the cytoplasm (9). The Golgi is a highly dynamic organelle, and its structure is maintained through a balance between delivery of vesicles from the ER and exit of vesicles toward the TGN and plasma membrane. Similarly, the TGN is maintained, in part, by bidirectional vesicular trafficking to and from the plasma membrane. Because it has been well documented that Golgi components redistribute to the perinuclear space as a consequence of viral infection (7), it is logical to propose that localization of gM toward the NM in infected cells is consequential to HSV-mediated effects on the Golgi and TGN. In the present study, gM colocalized most closely with a Golgi 58K marker within and around the vicinity of the nuclear rim in infected cells. This marker is normally associated with the Golgi periphery, i.e., regions of the Golgi most distal from the nucleus and close to the TGN (5). The observation that the TGN38 marker also overlaps with gM in the vicinity of the nuclear rim suggests that some components of TGN and peripheral Golgi have been shunted toward the nuclear rim or ER, at least in HEp-2 cells and most 143 cells late in infection. In contrast, the TGN38 marker located more distally from the nucleus did not usually colocalize with gM, suggesting that some components of the TGN remain in the cytoplasm. Since the final virion contains gM, the absence of substantial amounts of gM from these peripheral regions argues against their utility as the major site of final virion assembly late in infection, although it does not exclude this possibility. In addition, if gM is involved in trafficking surface proteins from the plasma membrane internally into the TGN, as suggested in transient-expression experiments (9), the current data indicate that only a minor population of gM is responsible for this function, at least late in infection.
Whereas gM contributed modestly to HSV-mediated effects on the TGN in 143 cells, its presence more dramatically increased the extent of localization of Golgi 58K around the nuclear rim in HEp-2 cells. It is unclear whether these are the consequences of direct or indirect effects of gM on components of the secretory pathway. In any event, the gM-dependent relocalization of the secretory apparatus toward the NM in HEp-2 cells is functionally different from complete dissolution of the Golgi induced by overexpression of HSV-1 gK or treatment with brefeldin A, inasmuch as both virion and glycoprotein egress are precluded under the latter two conditions (8, 13).
The results presented here are consistent with but do not prove that the Golgi and some components of the TGN are incorporated into the ER or contiguous perinuclear space by virus-induced retrograde trafficking. This might occur if retrograde vesicular trafficking from the Golgi to the ER became dominant in HSV-infected cells. If so, it might work to the advantage of the virus because it would allow Golgi enzymes responsible for glycoprotein processing access to perinuclear virions soon after budding from the INM. Such a feature would potentially maximize the production of virion infectivity in vivo when nascent virions are released by immune mediated or apoptotic lysis of the infected cell. Whether or not future ultrastructural studies will reveal a merging of the Golgi and ER in HSV-infected cells, the localization of gM at the INM and within perinuclear virions suggest roles for gM earlier in the egress pathway than previously realized.
Acknowledgments
We thank Jarek Okulicz-Kozaryn for excellent technical assistance.
This study was supported by Public Health Service grant AI52341 from the National Institutes of Health.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The UL11 gene products of herpes simplex virus 1 are present in the nuclear and cytoplasmic membranes and intranuclear dense bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10 and UL16 are dispensable for the growth of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1993. The UL10 gene of herpes simplex virus 1 encodes a novel glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J. Virol. 67:1441-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom, G. S., and T. A. Brashear. 1989. A novel 58-kDa protein associates with the Golgi apparatus and microtubules. J. Biol. Chem. 264:16083-16092. [PubMed] [Google Scholar]
- 6.Browne, H., S. Bell, T. Minson, and D. W. Wilson. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 70:4311-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campadelli, G., R. Brandimarti, C. Di Lazzaro, P. L. Ward, B. Roizman, and M. R. Torrisi. 1993. Fragmentation and dispersal of Golgi proteins and redistribution of glycoproteins and glycolipids processed through the Golgi apparatus after infection with herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 90:2798-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, P., B. W. Banfield, and F. Tufaro. 1991. Brefeldin A arrests the maturation and egress of herpes simplex virus particles during infection. J. Virol. 65:1893-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crump, C. M., B. Bruun, S. Bell, L. E. Pomeranz, T. Minson, and H. M. Browne. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J. Gen. Virol. 85:3517-3527. [DOI] [PubMed] [Google Scholar]
- 10.Dijkstra, J. M., N. Visser, T. C. Mettenleiter, and B. G. Klupp. 1996. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J. Virol. 70:5684-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 12.Foster, T. P., J. M. Melancon, J. D. Baines, and K. G. Kousoulas. 2004. The herpes simplex virus type 1 UL20 protein modulates membrane fusion events during cytoplasmic virion morphogenesis and virus-induced cell fusion. J. Virol. 78:5347-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, T. P., G. V. Rybachuk, X. Alvarez, O. Borkhsenious, and K. G. Kousoulas. 2003. Overexpression of gK in gK-transformed cells collapses the Golgi apparatus into the endoplasmic reticulum inhibiting virion egress, glycoprotein transport, and virus-induced cell fusion. Virology 317:237-252. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, W., and T. C. Mettenleiter. 1999. DNA sequence of the UL6 to UL20 genes of infectious laryngotracheitis virus and characterization of the UL10 gene product as a nonglycosylated and nonessential virion protein. J. Gen. Virol. 80(Pt. 8):2173-2182. [DOI] [PubMed] [Google Scholar]
- 16.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, H. L., and B. Norrild. 1998. Herpes simplex virus type 1-infected human embryonic lung cells studied by optimized immunogold cryosection electron microscopy. J. Histochem. Cytochem. 46:487-496. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, D. C., and P. G. Spear. 1982. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 43:1101-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jöns, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, A., M. Yamamoto, T. Ohno, H. Kodaira, Y. Nishiyama, and Y. Kawaguchi. 2005. Identification of proteins phosphorylated directly by the Us3 protein kinase encoded by herpes simplex virus 1. J. Virol. 79:9325-9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King, M. C., C. Lusk, and G. Blobel. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442:1003-1007. [DOI] [PubMed] [Google Scholar]
- 22.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 23.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konig, P., K. Giesow, and G. M. Keil. 2002. Glycoprotein M of bovine herpesvirus 1 (BHV-1) is nonessential for replication in cell culture and is involved in inhibition of bovine respiratory syncytial virus F protein induced syncytium formation in recombinant BHV-1-infected cells. Vet. Microbiol. 86:37-49. [DOI] [PubMed] [Google Scholar]
- 25.Koyano, S., E. C. Mar, F. R. Stamey, and N. Inoue. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 84:1485-1491. [DOI] [PubMed] [Google Scholar]
- 26.Ladinsky, M. S., and K. E. Howell. 1992. The trans-Golgi network can be dissected structurally and functionally from the cisternae of the Golgi complex by brefeldin A. Eur. J. Cell Biol. 59:92-105. [PubMed] [Google Scholar]
- 27.Lake, C. M., and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 74:11162-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lake, C. M., S. J. Molesworth, and L. M. Hutt-Fletcher. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J. Virol. 72:5559-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mach, M., B. Kropff, P. Dal Monte, and W. Britt. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 74:11881-11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mach, M., B. Kropff, M. Kryzaniak, and W. Britt. 2005. Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J. Virol. 79:2160-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacLean, C. A., L. M. Robertson, and F. E. Jamieson. 1993. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigation of its role in vivo. J. Gen. Virol. 74:975-983. [DOI] [PubMed] [Google Scholar]
- 32.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 33.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9:423-429. [DOI] [PubMed] [Google Scholar]
- 35.Munger, J., A. V. Chee, and B. Roizman. 2001. The US3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 75:5491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naldinho-Souto, R., H. Browne, and T. Minson. 2006. Herpes simplex virus tegument protein VP16 is a component of primary enveloped virions. J. Virol. 80:2582-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osterrieder, N., A. Neubauer, C. Brandmuller, B. Braun, O.-K. Kaaden, and J. D. Baines. 1996. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell to cell spread of virions. J. Virol. 70:4110-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poon, A. P., and B. Roizman. 2005. Herpes simplex virus 1 ICP22 regulates the accumulation of a shorter mRNA and of a truncated US3 protein kinase that exhibits altered functions. J. Virol. 79:8470-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purves, F. C., R. M. Longnecker, D. P. Leader, and B. Roizman. 1987. The herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 61:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purves, F. C., D. Spector, and B. Roizman. 1992. UL34, the target gene of the herpes simplex virus US3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J. Virol. 66:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the HSV-1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roizman, B., and D. Furlong. 1974. The replication of herpesviruses, p. 229-403. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology. Plenum Press, Inc., New York, NY.
- 43.Rudolph, J., C. Seyboldt, H. Granzow, and N. Osterrieder. 2002. The gene 10 (UL49.5) product of equine herpesvirus 1 is necessary and sufficient for functional processing of glycoprotein M. J. Virol. 76:2952-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryckman, B. J., and R. J. Roller. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78:399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stackpole, C. W. 1969. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J. Virol. 4:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. L. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, NY.
- 47.Tischer, B. K., D. Schumacher, M. Messerle, M. Wagner, and N. Osterrieder. 2002. The products of the UL10 (gM) and the UL49.5 genes of Marek's disease virus serotype 1 are essential for virus growth in cultured cells. J. Gen. Virol. 83:997-1003. [DOI] [PubMed] [Google Scholar]
- 48.Torrisi, M. R., C. Di Lazzaro, A. Pavan, L. Pereira, and G. Campadelli-Fiume. 1992. Herpes simplex virus envelopment and maturation studies by fracture label. J. Virol. 66:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 50.Wild, P., M. Engels, C. Senn, K. Tobler, U. Ziegler, E. M. Schraner, E. Loepfe, M. Ackermann, M. Mueller, and P. Walther. 2005. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 79:1071-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, S. X., X. P. Zhu, and G. J. Letchworth. 1998. Bovine herpesvirus 1 glycoprotein M forms a disulfide-linked heterodimer with the U(L)49.5 protein. J. Virol. 72:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]