Abstract
Recently a cell culture model supporting the complete life cycle of the hepatitis C virus (HCV) was developed. Searching for host cell determinants involved in the HCV replication cycle, we evaluated the efficiency of virus propagation in different Huh-7-derived cell clones. We found that Huh-7.5 cells and Huh7-Lunet cells, two former replicon cell clones that had been generated by removal of an HCV replicon by inhibitor treatment, supported comparable levels of RNA replication and particle production, whereas virus spread was severely impaired in the latter cells. Analysis of cell surface expression of CD81 and scavenger receptor class B type I (SR-BI), two molecules previously implicated in HCV entry, revealed similar expression levels for SR-BI, while CD81 surface expression was much higher on Huh-7.5 cells than on Huh7-Lunet cells. Ectopic expression of CD81 in Huh7-Lunet cells conferred permissiveness for HCV infection to a level comparable to that for Huh-7.5 cells. Modulation of CD81 cell surface density in Huh-7.5 cells by RNA interference indicated that a certain amount of this molecule (∼7 × 104 molecules per cell) is required for productive infection with a low dose of HCV. Consistent with this, we show that susceptibility to HCV infection depends on a critical quantity of CD81 molecules. While infection is restricted in cells expressing very small amounts of CD81, susceptibility rapidly rises within a narrow range of CD81 levels, reaching a plateau where higher expression does not further increase the efficiency of infection. Together these data indicate that a high density of cell surface-exposed CD81 is a key determinant for productive HCV entry into host cells.
Hepatitis C virus (HCV) is a highly variable enveloped virus. Based on sequence comparison of patient isolates, the different strains are grouped into 6 genotypes and more than 100 subtypes within the genus Hepacivirus of the family Flaviviridae (27). HCV is transmitted parenterally, and infection results in persistent virus replication in 55% to 85% of cases (13). In the course of decades, such chronic infection often causes substantial morbidity, including progressive hepatic fibrosis, cirrhosis, and hepatocellular carcinoma (26). Therefore, and in light of currently 170 million persistently infected individuals, HCV represents a considerable global health problem.
HCV possesses a positive-strand RNA genome of approximately 9.6 kb that is composed of a 5′ and a 3′ nontranslated region and a large open reading frame that encodes a polyprotein. Co- and posttranslational cleavages liberate the structural proteins (core and envelope proteins 1 and 2) which constitute the viral particle, a small hydrophobic polypeptide designated p7, and six nonstructural proteins (NS2, -3, -4A, -4B, -5A, and -5B) from the polyprotein (for a recent review, see reference 2). Several reports suggest that ribosomal frame shifting and internal translation initiation yield a further so-called core + 1 protein, but its function is currently not known.
Investigation of the molecular mechanisms of virus replication, the functions of individual viral proteins, and their interplay with host determinants critically rely on the availability of suitable tissue culture models to propagate the virus. In the case of HCV, studies aimed at dissecting the complete viral replication cycle were long hampered by the poor replication of HCV in cultured cells. However, recently this limitation was overcome by the molecular cloning of the JFH1 genome from a Japanese patient suffering from fulminant hepatitis, as this virus isolate replicates efficiently in transfected Huh-7 cells and produces virus particles infectious both in vitro and in vivo (19, 33, 38).
The construction of hybrid genomes consisting essentially of the JFH1 replicase and the core-to-NS2 region of other HCV isolates from divergent subtypes or even genotypes, as well as the construction of a highly cell culture-adapted variant of the H77 isolate, substantially extended the scope of this cell culture system (18, 24, 35), although particle yields are very low with the latter. Clearly, viral determinants contribute to the efficiency of virus assembly and release (18, 24), but it is likely that cellular proteins also modulate the efficiency of HCV production. This is illustrated by the differential outcomes of passaging of naive Huh-7 cells or a subclone of Huh-7.5 cells, designated Huh-7.5.1, that had been transfected with JFH1 RNA: Wakita and colleagues reported that JFH1 copy numbers in transfected naive Huh-7 cells slowly decreased upon continuous passaging, and even 13 days posttransfection, only 50 to 60% of the cells were HCV antigen positive (33); in contrast, Zhong and colleagues observed a robust increase of JFH1 RNA at later passages, overall about 50-fold-higher virus titers, and eventually the spread of JFH1 to almost 100% of cells (see reference 38; also discussed in reference 3). These data suggest that Huh-7 cells differ substantially with respect to their permissiveness for HCV.
Knowledge of crucial determinants governing permissiveness for HCV in vitro not only may help to further improve the currently available model system but additionally should shed light on key virus-host interactions that may be a target for the development of future antiviral therapies. Therefore, in this study we determined the reasons underlying the different levels of permissiveness of naive Huh-7 cells and various Huh-7-derived cell clones. While Huh-7.5 cells supported the most efficient virus propagation, Huh7-Lunet cells, which represent another “cured” replicon cell clone supporting a high level of HCV RNA replication, sustained only limited virus spread. This defect was linked to a low expression of CD81 on the surface of these cells and was reconstituted by overexpression of CD81. Moreover, we found that copy numbers of cell surface-expressed CD81 molecules modulate host cell permissiveness and are a key determinant for productive infection by HCV.
MATERIALS AND METHODS
Cell culture and cell lines.
All cell lines were propagated in Dulbecco's modified Eagle medium (DMEM) (Invitrogen, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 10% fetal calf serum (DMEM complete). Huh7-Lunet cells and Huh-7.5 cells represent cell clones that were generated by “curing” Huh-7 replicon cells with a selective drug or alpha interferon, respectively (7, 12). Culture medium of Huh-7.5- or Huh7-Lunet-derived cell lines was supplemented with G418 or Zeocin as given below.
Generation of stable Huh7-Lunet- and Huh-7.5-derived cell lines.
The Lunet/V control cell line was produced by stable transfection of Huh7-Lunet cells with the vector pEF1/V5-HisA (Invitrogen, Karlsruhe, Germany) using Lipofectamine 2000 (Invitrogen) and subsequent selection using DMEM complete supplemented with G418 (750 μg/ml). Likewise, Lunet/CD81 cells were generated by Lipofectamine 2000 transfection with the pEF1/V5-HisA/CD81 expression construct (see below) and selection using the same growth medium. Stable transfectants were stained with a CD81-specific monoclonal antibody (1.3.3.22; Ancell, Bayport, MN) and sorted for high CD81 expression using a FACSAria sorter (Becton Dickinson Biosciences, Heidelberg, Germany). Subsequently, both cell lines were propagated in medium with 750 μg/ml G418.
To knock down CD81 expression in Huh-7.5 cells, we created two pBABE/H1/SV40/EGZ/ΔU3-based retroviral vectors (17) encoding CD81-specific small-hairpin RNA (shRNA) (pBABE-shCD81.1 and pBABE-shCD81.2). Moreover, a control vector was prepared carrying a CD13-specific shRNA (pBABE-shCD13). In each case, forward and reverse oligonucleotides encoding the shRNAs were annealed and inserted into pBABE/H1/SV40/EGZ/ΔU3 restricted with XhoI and BglII. While shCD81.1 was chosen based on a recently described small interfering RNA mediating potent knockdown of CD81 in hepatic stellate cells (22), all remaining shRNA sequences were selected based on results obtained with the Block-iT RNAi Designer software (Invitrogen web site). The following oligonucleotides were used: shCD81.1-forward, 5′-GATCCCCATCTGGAGCTGGGAGACAATTCAAGAGATTGTCTCCCAGCTCCAGATTTTTTGGAAAAC-3′; shCD81.1-reverse, 5′-TCGAGTTTTCCAAAAAATCTGGAGCTGGGAGACAATCTCTTGAATTGTCTCCCAGCTCCAGATGGG-3′; shCD81.2-forward, 5′-GATCCCCGCCCAACACCTTCTATGTATTCAAGAGATACATAGAAGGTGTTGGGCTTTTTGGAAAAC-3′; shCD81.2-reverse, 5′-TCGAGTTTTCCAAAAAGCCCAACACCTTCTATGTATCTCTTGAATACATAGAAGGTGTTGGGCGGG-3′; shCD13-forward, 5′-GATCCCCGGTGAAGGACAGCCAGTATTTCAAGAGAATACTGGCTGTCCTTCACCTTTTTGGAAAAC-3′; shCD13-reverse, 5′-TCGAGTTTTCCAAAAAGGTGAAGGACAGCCAGTATTCTCTTGAAATACTGGCTGTCCTTCACCGGG-3′. Retroviral particles transducing given shRNAs were created by calcium phosphate-mediated cotransfection of 293T cells (28) with equal amounts of pHIT60 (28), pcz-VSV-G (14), and the respective retroviral vector essentially as described previously (28). Briefly, 2 × 106 293T cells were seeded into 6-cm-diameter culture dishes 24 h prior to transfection with 5 μg DNA of each plasmid. Cell-free culture fluids were harvested 48 h posttransfection and directly used to inoculate Huh-7.5 cells. Afforded by the simian virus 40 promoter-dependent expression of an enhanced green fluorescent protein (EGFP)-zeocin resistance fusion protein (EGFP-Zeor) from the pBABE-vector backbone (17), shRNA-expressing cells were selected by using DMEM complete supplemented with 5 μg/ml zeocin. After five rounds of passaging in the presence of this antibiotic, fluorescence-activated cell sorting (FACS) analysis revealed expression of EGFP-Zeor in more than 90% of cells.
Plasmid construction.
The plasmid pFK-J6/C3 encodes a chimeric virus genome, designated “Jc1,” which consists of codons 1 to 846 derived from J6/CF (GenBank accession no. AF177036) and all remaining viral sequences from the JFH1 isolate (GenBank accession no. AB047639) (24). To allow detection and quantification of HCV infection by autofluorescence of a fluorescence protein, we created pFK-Venus-Jc1 by replacing the firefly luciferase coding region within the pFK-Luc-Jc1 reporter virus construct (16) with the coding sequence of Venus (a variant of yellow fluorescent protein) (23). Analogous to the configuration of the firefly luciferase coding region in the parental construct, the Venus gene is fused in frame via an AscI site to the first 16 codons of the JFH1 core coding region (Fig. 1A).
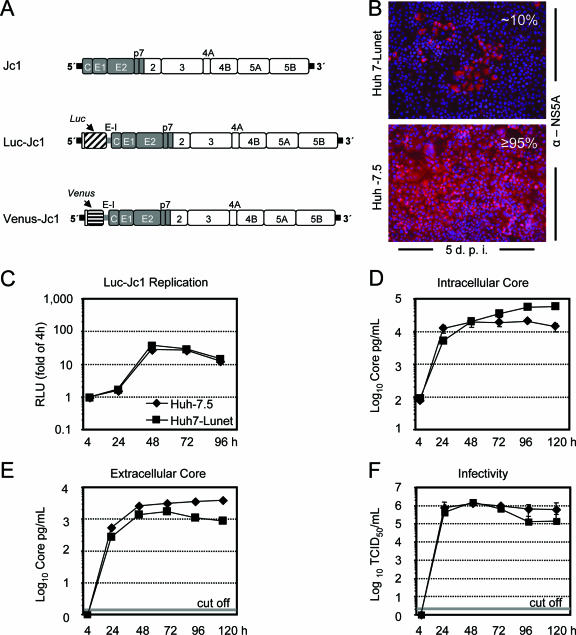
FIG. 1.
Susceptibility to infection and efficiency of HCV replication and production of infectious particles in Huh7-Lunet and Huh-7.5 cells. (A) Schematic representation of Jc1 and Jc1-based bicistronic reporter virus constructs Luc-Jc1 and Venus-Jc1. The chimeric HCV polyprotein is indicated by a large box; J6CF-derived regions are shown in gray, while JFH1 parts are depicted by open boxes. HCV 5′ and 3′ nontranslated regions are denoted by black bars, and the internal ribosome entry site of the encephalomyocarditis virus (E-I) is given as a gray bar. (B) Huh-7.5 or Huh7-Lunet cells were inoculated with a low dose of Jc1 (MOI of ca. 0.1 TCID50/cell). Five days postinoculation (d.p.i.), infected cells were visualized by using NS5A-specific indirect immunofluorescence (red staining). Nuclei were counterstained by using DAPI (blue). The percentage of HCV-infected cells is given in the upper right of each panel. (C) Luc-Jc1 RNA was transfected into Huh7-Lunet or Huh-7.5 cells (black squares and black diamonds, respectively), and replication was monitored by measuring luciferase activity at the given time points posttransfection. Mean values of duplicate measurements are given and are expressed relative to the reporter activity determined 4 h posttransfection, which was set to 1. (D) Intracellular core protein expression, (E) core release into the culture fluid, and (F) accumulation of infectivity in culture supernatant of Jc1 RNA-transfected Huh7-Lunet or Huh-7.5 cells as determined by a core-specific immunoassay (D and E) or a limiting-dilution assay using Huh-7.5 target cells (F). Mean values and standard errors of the means of two independent experiments are given. Where applicable, the cut-off value of the immunoassay or the limiting-dilution assay is indicated by a gray bar.
CD81 expression in Huh7-Lunet cells was reconstituted by using plasmid pEF1-CD81, which encodes the cDNA of human CD81 under the control of the EF1-α promoter. This construct was created by restricting pEF/V5-HisA (Invitrogen) with SpeI and EcoRI and inserting the NheI and EcoRI fragment derived from pcDNA3.1(+)CD81 (kindly provided by M. Ott, Gladstone Institute of Virology and Immunology, University of California, San Francisco). The CD81 protein encoded by pEF1-CD81 and the parental construct matches the primary sequence of human TAPA-1 (GenBank no. M33680) except for one amino acid exchange (K52 to M52).
In vitro transcription, electroporation, and transient HCV replication assays using luciferase reporter genomes.
Prior to in vitro transcription, plasmid DNA was restricted with MluI, extracted with phenol and chloroform, precipitated with ethanol, and dissolved in RNase-free water. In vitro transcriptions were performed in a total volume of 100 μl deionized water containing the following reagents: 80 mM HEPES (pH 7.5), 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), 3.125 mM of each nucleoside triphosphate, 100 U of RNasin (Promega, Mannheim, Germany), 10 μg of plasmid DNA, and 80 U of T7 RNA polymerase (Promega). After 2 h at 37°C, an additional 40 U of T7 RNA polymerase was added, and the reaction was incubated for another 2 h. Transcription was terminated by the addition of 7.5 U of RNase-free DNase (Promega) and a 30-min incubation at 37°C. RNA was purified by extraction with acidic phenol and chloroform, precipitated with isopropanol, and dissolved in RNase-free water. To generate virus stocks for infection assays and for transient replication assays, cells were transfected by electroporation. Cells were detached by trypsin treatment, washed with phosphate-buffered saline (PBS), counted, and resuspended at 107 cells per ml in Cytomix (32) containing 2 mM ATP and 5 mM glutathione. Five micrograms of in vitro-transcribed RNA was mixed with 400 μl of the cell suspension by pipetting, electroporated, and immediately transferred to 20 ml of complete DMEM. Subsequently, the cells were seeded at a density of 4.16 × 104 cells/cm2, which corresponds to 2 ml of the cell suspension per well of a six-well plate. Electroporation conditions were 960 μF and 270 V by using a Gene Pulser system (Bio-Rad, Munich, Germany) and a cuvette with a gap width of 0.4 cm (Bio-Rad). For assaying the luciferase activity, cells were washed once with PBS, lysed directly on the plate with 1 ml ice-cold lysis buffer (1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, and 1 mM DTT; pH 7.8) and frozen. Upon thawing, lysates were resuspended by pipetting, and 100 μl was mixed with 360 μl of assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP, 15 mM K2PO4; pH 7.8) and, after the addition of 200 μl of a luciferin solution (200 μM luciferin, 25 mM glycylglycine; pH 8.0), measured in a luminometer (Lumat LB9507; Berthold, Freiburg, Germany) for 20 s. All luciferase assays were done at least in duplicate measurements.
Immunohistochemical staining and virus titration.
HCV was titrated essentially as described recently (18). In brief, Huh-7.5 cells were seeded in 96-well plates at a density of 1 × 104 cells per well 24 h prior to inoculation with serial dilutions of filtered cell culture supernatant (at least six wells were used per dilution). Two to three days later, cells were washed with PBS, fixed for 20 min with methanol at −20°C, washed again three times with PBS, and then permeabilized and blocked for 1 h with PBS containing 0.5% saponin, 1% bovine serum albumin, 0.2% dried skim milk, and 0.02% sodium azide. Endogenous peroxidases were blocked by incubating cells for 5 min with PBS containing 0.3% hydrogen peroxide. Upon three washes with PBS and one with PBS containing 0.5% saponin (PBS-saponin), NS5A was detected by overnight incubation at 4°C with PBS-saponin supplemented with a 1:400 dilution of hybridoma supernatant 9E10 (18). Cells were washed as described above, and bound 9E10 was detected by 1 h of incubation with peroxidase-conjugated antimouse antibody (Sigma-Aldrich) diluted 1:200 in PBS-saponin and washed thereafter as specified above. Finally, peroxidase activity was detected by using the Vector NovaRED substrate kit (Linaris Biologische Produkte GmbH, Wertheim, Germany), and virus titers (50% tissue culture infective dose [TCID50]/ml) were calculated based on the statistical method described by Spearman and Kärber (15, 29).
Quantitative detection of HCV core protein.
The HCV core protein was quantified using the Trak C Core (Ortho Clinical Diagnostics, Neckargemund, Germany) enzyme-linked immunosorbent assay according to the instructions of the manufacturer and as described recently (16).
Indirect immunofluorescence and quantification of virus spread.
Cells were seeded on glass coverslips in 24-well plates at a density of 3 × 104 per well 24 h before infection, followed by inoculation with 250 μl of filtered cell culture supernatant. At different time points, cells were fixed with 3% paraformaldehyde in PBS and permeabilized with 0.5% Triton X-100 in PBS. Staining of NS5A was performed by using the 9E10 hybridoma supernatant (18) at a dilution of 1:250 in PBS-5% NGS. Bound primary antibodies were detected using goat antibodies conjugated to Alexa-Fluor 546 at a dilution of 1:1,000 in PBS-5% NGS. DNA was stained with 4′, 6′-diamidino-2-phenylindole (DAPI) (Molecular Probes, Karlsruhe, Germany).
To assess the kinetics of virus spread, cells seeded as described above were inoculated at a multiplicity of infection (MOI) of 0.1 TCID50/cell for 4 h. At different time points after inoculation, cells were fixed and the proportion of infected cells was monitored by NS5A-specific indirect immunofluorescence. For each time point at least 1,000 cells were evaluated, and mean values and the standard deviation of at least two independent experiments are given.
Flow cytometry and antibodies.
Cells were detached by using PBS supplemented with 0.2% (wt/vol) EDTA, washed twice with PBS, and passed through a 16-gauge needle. Approximately 1 × 106 cells per ml were stained for 1 h at 4°C with CD81-specific (1.3.3.22; Ancell, Bayport, MN) or scavenger receptor class B type I (SR-BI)-specific monoclonal antibody (anti-CLA1; Becton Dickinson Biosciences) diluted to 10 μg/ml in PBS containing 0.2% bovine serum albumin and 0.02% sodium azide (FACS sample buffer). Subsequently, cells were washed with PBS and bound antibodies were detected by incubation for 1 h at 4°C with mouse-specific secondary antibodies conjugated with phycoerythrin (PE) (eBioscience, San Diego, CA) at a dilution of 1:100 in FACS sample buffer. For of two-color analysis of CD81 surface expression and Venus-Jc1-derived autofluorescence, we employed mouse-specific secondary antibodies conjugated to allophycocyanin (Dianova, Hamburg, Germany) at a dilution of 1:100 in FACS sample buffer. Stained cells were washed with PBS, resuspended in 500 μl FACS sample buffer, and analyzed immediately using a FACSCalibur apparatus and the Cell Quest Pro software (both from Becton Dickinson Biosciences). Copy numbers of CD81 molecules on the cell surface were estimated by using the QuantiBRITE quantitation kit (Becton Dickinson Biosciences) according to the instructions of the manufacturer.
Quantitative analysis of relationship between CD81 receptor expression and HCV infection.
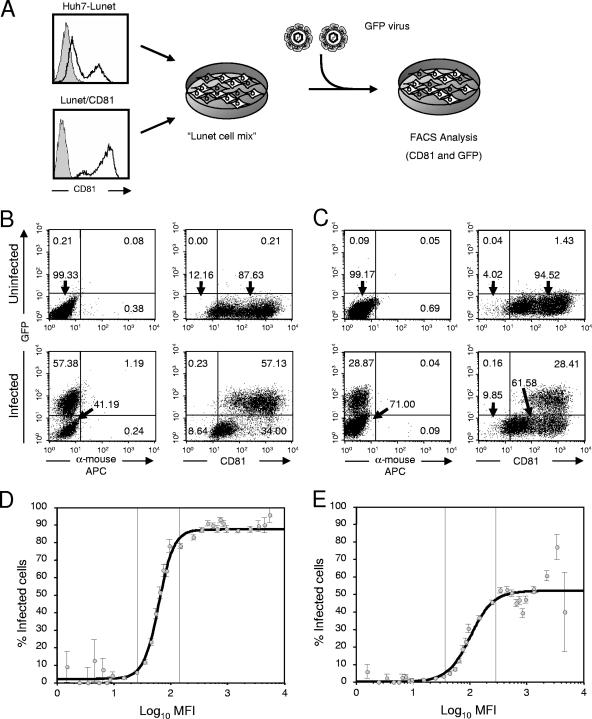
Huh-7.5 cells were transfected with 5 μg of Venus-Jc1 RNA as described above. Forty-eight hours posttransfection, culture fluid was harvested, cleared by passing through 0.45-μm filters, and concentrated using Amicon Ultrafiltration devices (Millipore, Schwalbach, Germany) according to the manufacturer's protocol. Concentrated virus preparations were used to inoculate a 1:1 mixture of Huh7-Lunet and Lunet/CD81 cells that had been seeded at a density of 1 × 105 cells per well of a six-well plate 24 h earlier. Seventy-two hours later, cells were analyzed by FACS as described above. Results were grouped into 32 discrete classes according to their mean fluorescence intensity (MFI) on the x axis, which represents divergent copy numbers of CD81 on the cell surface. For each class, the proportion of infected cells was determined by using quadrant statistics and employing a threshold level of 15 MFI on the y axis to distinguish infected from uninfected cells. The percentage of infected cells was plotted against the center of the respective MFI interval of CD81 expression. Mean values and the standard deviation of the means are given. The relationship between CD81 levels (with a logarithmic scale) and the proportion of infected cells was fitted with a maximum-likelihood approach using ordinary and rescaled probit or logit regression models.
RESULTS
Propagation of HCV in Huh-7 cell clones.
Since the first establishment of HCV subgenomic replicons in Huh-7 cells, several alternative cell lines were identified which support HCV RNA replication. These include human hepatoma cell lines, such as HepG2 (11) and Huh6 (34), but also human nonhepatoma cells lines (293 and HeLa [1, 39]) and even nonhuman cells (Hepa1-6 [39] and mouse embryonic fibroblasts [8]). While this host range indicates that key host factors for HCV RNA replication are broadly expressed, replication efficiency markedly differs and is by far the best in Huh-7 cells. Moreover, in particular, “cured” Huh-7 replicon cell clones, such as, for instance, Huh-7.5 (7) or Huh7-Lunet (12) and some high-passage naive Huh-7 cells (20), were found to be the most permissive for HCV RNA replication. Assuming that high-level RNA replication is only one prerequisite for efficient virus propagation, which requires in addition efficient assembly, virus release, and infection, we sought to identify cells supporting optimal propagation of infectious HCV by screening a panel of high-passage naive Huh-7 cells and cured Huh-7 replicon cell clones. We chose our most efficient virus chimera, called Jc1, which consists of J6CF- and JFH1-derived sequences (Fig. 1A) (24), and assessed the efficiency of virus propagation 5 days after inoculation with a low virus dose (MOI of ∼0.1 TCID50/cell). By using NS5A-specific immunofluorescence staining, we found that virus spread was most efficient in Huh-7.5 cells, leading to infection of almost all cells 5 days postinfection, whereas naive Huh-7 cells and other Huh-7-derived cell clones displayed various percentages of infected cells (Fig. 1B; also data not shown). This was also true for Huh7-Lunet cells, which are “cured” replicon cells supporting high-level HCV RNA replication. Only a fraction (ca. 10%) of cells was infected with Jc1 under these conditions, arguing that certain host cell factors important for virus propagation are differentially expressed in Huh-7.5 and Huh7-Lunet cells, thus limiting virus propagation in the latter.
To examine which step of the viral replication cycle was impaired in Huh7-Lunet cells, we compared the efficiency of RNA replication and virus production between transfected Huh-7.5 and Huh7-Lunet cells. Since luciferase-based reporter constructs permit measurement of HCV RNA replication with high accuracy, we chose the recently described Luc-Jc1 reporter virus (16), transfected Huh-7.5 cells and Huh7-Lunet cells with equal amounts of Luc-Jc1 RNA, and monitored replication by measuring luciferase activity at 4 to 96 h posttransfection. The results presented in Fig. 1C show that Luc-Jc1-expressed reporter activity was indistinguishable between Huh-7.5 and Huh7-Lunet cells, indicating that both cell lines support similar HCV RNA replication levels. To rule out any bias caused by different half-lives of firefly luciferase in the two cell clones, we transfected these cells with Jc1 RNA, monitored transfection efficiency by NS5A-specific immunofluorescence (data not shown), and assessed replication by measuring intracellular core protein levels, particle release by determining extracellular core protein amounts, and release of infectivity by a limiting-dilution assay using Huh-7.5 target cells (Fig. 1D, E, and F, respectively). In both cell lines, intracellular core protein levels increased about 100-fold between 4 and 24 h posttransfection, indicating vigorous Jc1 RNA replication (Fig. 1D). Intracellular core production did not further increase at subsequent time points in the case of Huh-7.5 but kept slowly rising in transfected Huh7-Lunet cells, eventually reaching peak levels approximately threefold over those present in Huh-7.5 cells at 120 h posttransfection. In contrast, accumulation of core protein and infectivity in the cell culture supernatant of Jc1-transfected Huh-7.5 was slightly more efficient and more sustained than those for Huh7-Lunet cells (Fig. 1E and F), which may be related to the limited ability of HCV to spread in the latter cells. In summary, these data demonstrate that Luc-Jc1 and Jc1 replicate with comparable efficiencies in transfected Huh-7.5 and Huh7-Lunet cells. Although reproducibly slightly more infectious particles were produced in Huh-7.5 cells upon transient transfection, we concluded that this marginal difference could not account for the dramatic difference of virus spread in the two cell clones.
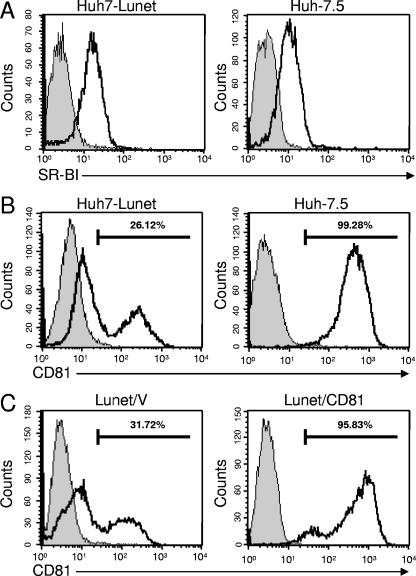
Expression of SR-BI and CD81 on the surfaces of Huh-7.5 and Huh7-Lunet cells.
Assuming that a block at the level of virus entry may impede virus spread in Huh7-Lunet cells, we investigated the cell surface expression level of SR-BI and CD81, two molecules known to be involved in productive HCV infection, by using FACS analyses. As shown in Fig. 2A and B, the surface expression of SR-BI was similar in the two cell lines, whereas CD81 expression clearly differed. While essentially all Huh-7.5 cells expressed uniformly high levels CD81 on their cell surfaces (Fig. 2B), the majority of Huh7-Lunet cells displayed low to almost undetectable CD81 surface expression, and only a minor fraction expressed well-detectable levels of CD81 on the cell surface. Since staining with an alternative CD81-specific antibody resulted in a similar profile (data not shown), we conclude that most cells do express CD81, albeit at rather different levels. Although the reason for this bimodal CD81 expression profile is currently not known, we speculated that the rather low level of CD81 expression on the surfaces of Huh7-Lunet cells restricts HCV propagation in these cells.
FIG. 2.
Expression of SR-BI and CD81 on the surfaces of Huh7-Lunet or Huh-7.5 cells. Cells were stained by using SR-BI-specific (A) or CD81-specific (B) mouse monoclonal antibody and secondary antibodies conjugated with PE. (C) CD81 expression on the surfaces of Huh7-Lunet cells transfected with an empty vector (Lunet/V, left panel) or a CD81 expression vector (Lunet/CD81, right panel). Gray profiles represent cells that were stained only with the secondary antibodies.
Augmenting CD81 surface expression in Huh7-Lunet cells enhances virus spread.
To confirm this hypothesis, we generated a Huh7-Lunet cell pool, called Lunet/CD81, expressing high levels of CD81 from a stably transfected CD81 transgene. An additional cell pool, named Lunet/V, was generated by transfection of Huh7-Lunet cells with the empty expression vector and served as a negative control. As demonstrated in Fig. 2C, transfection of the vector alone did not markedly influence CD81 cell surface expression, while stable transfection of the CD81 expression construct led to cell surface levels comparable with those of Huh-7.5 cells.
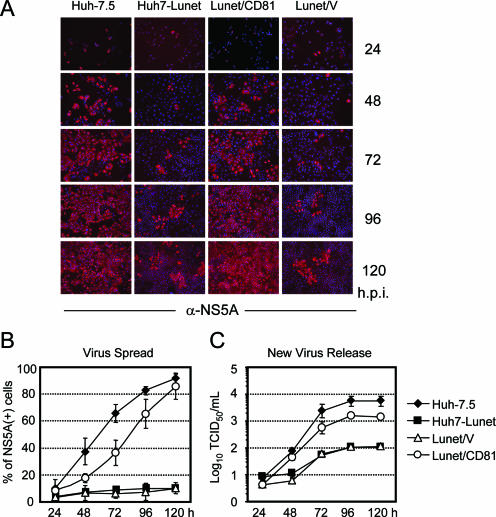
To assess the influence of ectopic CD81 expression on HCV propagation, we inoculated Huh7-Lunet, Lunet/V, Lunet/CD81, and as a reference, Huh-7.5 cells with a low dose of Jc1 virus (MOI of ∼0.1 TCID50/cell) and analyzed the percentages of HCV antigen-positive cells by using NS5A-specific immunofluorescence at different time points postinoculation (Fig. 3A and B). In the case of Huh-7.5, the number of NS5A-expressing cells rapidly increased to about 90% of the cells at 120 h postinoculation. In contrast, during the same time interval, most Huh7-Lunet cells were refractory to Jc1 infection. Similarly, only about 10% of Lunet/V cells expressed NS5A 5 days postinoculation, whereas 85% of Lunet/CD81 cells became infected. In parallel, we monitored the release of infectious Jc1 progeny from the inoculated cells over time. As depicted in Fig. 3C, the release of infectivity into the culture medium of Huh7-Lunet and Lunet/V cells was low and did not exceed 100 TCID50/ml, likely due to the small number of infected cells. In contrast, Lunet/CD81 cells produced progeny virus with kinetics very similar to that of Huh-7.5 cells and released a peak titer more than 10-fold higher than that with the parental Huh7-Lunet cells and only about 3- to 5-fold lower than that with Huh-7.5 cells.
FIG. 3.
Jc1 spread and production of virus progeny in various Huh-7-derived cell lines. (A) Cells denoted above each column were seeded into replicate wells and inoculated with a low dose of Jc1 (MOI of ca. 0.1 TCID50/cell). At the time points indicated at the right (hours postinfection [h.p.i.]), cells were fixed to analyze the quantity of infected cells by using NS5A-specific indirect immunofluorescence (red staining). Nuclei were counterstained by using DAPI (blue color). (B) Quantification of Jc1 spread in different Huh-7-derived cell lines. The fraction of NS5A-expressing cells was counted at the given time points and is expressed as a percentage of the total population. Mean values from three independent experiments and the standard errors of the means are given. (C) Supernatants of the inoculated cells were collected at the given time points, and the virus titer was determined by a limiting-dilution assay using Huh-7.5 target cells (one representative experiment of three independent repetitions is presented). Mean values and the respective standard deviations are given.
In conclusion, these results show that the low cell surface expression of CD81 in Huh7-Lunet cells limits the propagation of Jc1 in these cells and that this impediment can be overcome by augmenting CD81 cell surface expression.
High-level CD81 cell surface expression is required for efficient HCV infection.
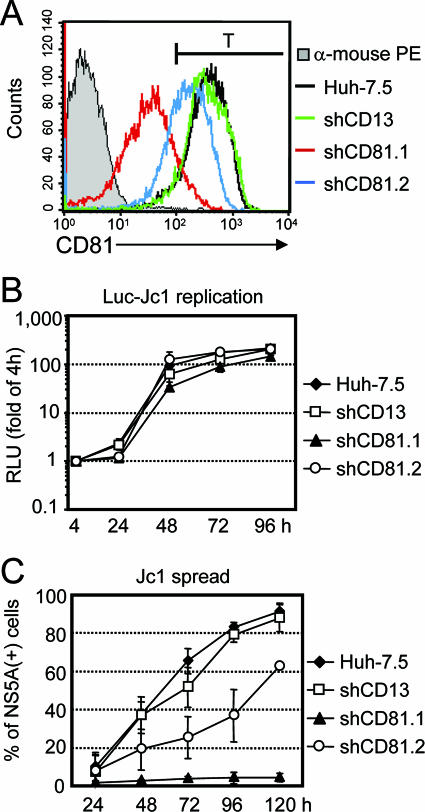
The data presented so far clearly indicated a correlation between the level of CD81 cell surface expression and susceptibility to HCV infection. To investigate this assumption in more detail, we modulated CD81 expression in Huh-7.5 cells by employing RNA interference. Huh-7.5 cells were transduced by retroviral vectors delivering either of two different shRNAs directed against CD81 (shCD81.1 and shCD81.2) or a third shRNA targeting CD13, a cell surface molecule not required for HCV infection (shCD13). Analysis of CD81 surface expression in the resulting cell pools revealed that the CD13-specific shRNA did not affect CD81 expression, whereas shCD81.2 moderately and shCD81.1 strongly down modulated CD81 cell surface levels (Fig. 4A). The expression of the different shRNAs did not significantly affect either SR-BI expression (as determined by FACS analysis; data not shown) or the efficiency of HCV RNA replication after transient transfection (Fig. 4B). In contrast, when cells were inoculated with a low dose of Jc1 (ca. 0.1 TCID50/cell), the efficiency of virus spread was clearly compromised in Huh-7.5 cells expressing lower copy numbers of CD81 due to RNA interference (Fig. 4C). More specifically, only about 60% of Huh-7.5-shCD81.2 cells and less than 5% of Huh-7.5-shCD81.1 cells were infected 5 days after inoculation with Jc1 (Fig. 4C). It is interesting to note that despite rather efficient silencing of CD81 by shCD81.1 in Huh-7.5 cells, more than 90% of the cells express well-detectable levels of CD81 on their surfaces (Fig. 4A). Nevertheless, under the experimental conditions chosen, more than 95% of the total cell population was not infected with Jc1 5 days postinoculation. Similarly, in the case of Huh-7.5-shCD81.2 cells, essentially all cells displayed high levels of CD81, but only about 60% of these were infected during the time course. Based on these findings, we defined a theoretical threshold of fluorescence intensity dividing the respective cell populations into a permissive fraction and a nonpermissive fraction (Fig. 4A and C) (threshold level [T] = 100 arbitrary fluorescence units). The percentages of the respective subpopulations which were defined by this marker correlated well with the actual percentages of infected cells obtained by the Jc1 infection assay depicted in Fig. 4A and C. To estimate the copy numbers of cell surface-resident CD81 molecules correlating with this threshold, we used quantitative flow cytometry and determined the number of CD81 molecules equivalent to this fluorescence intensity. These data are summarized in Table 1 and indicate that upon inoculation with Jc1 at a low MOI (0.1 TCID50/cell), only cells expressing more than about 7 × 104 copies of CD81 on the cell surface are susceptible to infection.
FIG. 4.
Jc1 spread in Huh-7.5-derived cell lines expressing CD81- and CD13-specific shRNAs. (A) Huh-7.5 cells were transduced with retroviral vectors encoding either of two CD81-specific shRNAs or a CD13-specific shRNA. Subsequently, cell surface expression in these cell lines was analyzed by FACS and compared to that in Huh-7.5 cells. The gray profile represents staining of the parental cell line with antimouse PE secondary antibody alone, while the black line denotes CD81 expression in these cells. The green, red, and blue lines represent CD81 expression on the surface of cells transduced with shCD13, shCD81.1 or shCD81.2, respectively. The black bar depicted above the FACS profiles defines a threshold level (T) of CD81 surface expression required for susceptibility to Jc1 infection under the experimental conditions employed (compare with Table 1). (B) Efficiency of HCV RNA replication in cell lines described for panel A as determined by transfection of Luc-Jc1 RNA and luciferase assays. Mean values of duplicate measurements are given and expressed relative to the reporter activity determined 4 h posttransfection, which was set to 1. (C) The efficiency of Jc1 spread in these cell lines was assessed by inoculation with a low virus dose (0.1 TCID50/cell) and quantification of the fraction of infected cells by using indirect immunofluorescence. Mean values from three independent experiments and the respective standard deviations are given.
TABLE 1.
CD81 quantification
| Huh-7 clone | CD81 thresholda (T ≥ 7 × 104/cell) | NS5Ab |
|---|---|---|
| Huh7-Lunet | 17 | 11.2 ± 3.9 |
| Lunet/V | 17 | 10.2 ± 0.7 |
| Lunet/CD81 | 86 | 85.8 ± 9.7 |
| Huh-7.5 | 93 | 91.5 ± 4.2 |
| Huh-7.5/shCD81.1 | 11 | 4.4 ± 2.1 |
| Huh-7.5/shCD81.2 | 68 | 62.4 ± 0.4 |
| Huh-7.5/shCD13 | 93 | 87.8 ± 7.2 |
Fraction of cells (%) among the total cell population expressing CD81 copy numbers equal to or higher than 7 × 104 molecules per cell as determined using the QuantiBRITE fluorescence quantification kit.
Percentage of cells expressing NS5A 5 days postinoculation with a low dose of Jc1 (MOI, ca. 0.1 TCID50/cell). Mean values from three independent experiments and the respective standard errors are given.
To further corroborate these findings, we utilized a novel bicistronic Jc1 reporter virus, called Venus-Jc1, which encodes a modified green fluorescent protein in lieu of luciferase, thus giving us the opportunity to detect HCV infection and CD81 receptor expression at a single-cell level. In order to characterize the relationship between CD81 receptor density and susceptibility to HCV infection in more detail, we prepared concentrated Venus-Jc1 stocks and inoculated target cell populations consisting of a mixture of Huh7-Lunet and Lunet/CD81 cells and thus comprising cells with a diverse spectrum of CD81 expression levels (Fig. 5). In two independent experiments performed with different doses of Venus-Jc1 (MOI of ∼5 or ∼1 TCID50/cell), a substantial proportion of target cells was successfully infected (ca. 57% and 29%, respectively; Fig. 5B and C). Importantly, when we correlated the percentage of infected cells with their CD81 receptor expression level, we detected a reproducible relationship between rising CD81 expression and the proportion of infected cells. This relationship was best approximated by a rescaled logistic function which was centered around 61.9 MFI of CD81 expression, rising from 2% to a maximum of 87.6% of infected cells upon inoculation with 5 TCID50/cell, or centered on 102.5 MFI, rising from 0% to 52% infected cells after inoculation with 1 TCID50/cell (Fig. 5D and E, respectively). Together these data indicate that cells carrying undetectable or very low copy numbers of CD81 (0 to ca. 20 MFI) on their surfaces are not infectible or are only poorly infectible, whereas beyond this threshold susceptibility continuously and rapidly increases within a narrow range of CD81 receptor densities (ca. 25 to 290 MFI). Beyond a CD81 receptor density equivalent to approximately 290 MFI, the proportion of infected cells did not further increase. This implies that the surface density of CD81 did not further enhance susceptibility and indicates that other factors, such as the MOI or availability of other components required for HCV entry, limited infection.
FIG. 5.
Dependence of HCV infection on CD81 receptor density. (A) Schematic representation of the experimental procedure. A mixed population of Huh7-Lunet and Lunet/CD81 cells was seeded and inoculated with concentrated virus stocks of Venus-Jc1. Seventy-two hours later, cells were harvested and analyzed for infection and CD81 surface expression by FACS analysis. FACS data obtained by inoculation of target cells with an MOI of ca. 5 or 1 TCID50/cell are depicted in B) and C, respectively. Uninfected cells are given in the upper panels, whereas infected cells are depicted below. Relative numbers of cells in the respective quadrants are given. (D and E) Relationship between rising CD81 levels with a logarithmic scale and the proportion of infected cells (indicating susceptibility to infection) was fitted with a maximum-likelihood approach using ordinary and rescaled probit or logit regression models for both experiments (MOI of 5 or 1 TCID50/cell, respectively). Cells were grouped into discrete classes according to their MFI on the x axis. The proportion of infected cells was determined for each class and is plotted against the center of the respective MFI interval of CD81 expression. Mean values and standard deviations are given. Note that the changes of susceptibility concentrate in a region between 26.5 and 144.5 MFI or 36.5 and 287.9 MFI, respectively, whereas absolute changes outside these regions are less than 5%.
DISCUSSION
The HCV infection system based on the JFH1 strain and JFH1-derived chimeras (18, 24, 33, 38) has opened the opportunity to study the complete viral replication cycle in cell culture and has thus paved the way for comprehensive analyses of virus-host interaction, including viral egress and entry pathways. In order to identify key host cell determinants required to support the viral life cycle, in this study we compared different Huh-7-derived host cells with respect to their permissiveness for HCV propagation.
We observed a pronounced variation in the efficiencies of virus spread between divergent Huh-7 cell clones. This discrepancy was particularly evident when comparing Huh-7.5 and Huh7-Lunet cells, which both formerly carried HCV replicons that had been removed by inhibitor treatment (7, 12), suggesting that host cell factors essential for HCV RNA replication and virus production, egress, or entry are differentially expressed in these cells. Further analyses demonstrated that the poor permissiveness of Huh7-Lunet cells was primarily due to insufficient cell surface expression of CD81.
The reason why virus spread is slightly more rapid in Huh-7.5 cells than in Lunet cells with comparable CD81 levels (Lunet/CD81) is currently not known. However, it has been shown that Huh-7.5 cells carry a defective allele of RIG-I(30), a molecule known to sense virus infection by recognizing double-stranded RNA, which is a key pathogen-associated molecular pattern produced during virus infection (36). As a consequence, the signaling cascade which in response to virus infection leads to the induction of the beta interferon promoter, release of beta interferon, and triggering of an antiviral state is ablated in these cells (30), which may in turn allow a more-rapid virus spread in these cells. In line with this interpretation, Loo and colleagues recently observed that HCV infection triggers RIG-I-dependent signaling that transiently limits virus replication and is subsequently overcome when viral proteins accumulate (21).
To further characterize the requirements for CD81-dependent HCV infection of Huh-7-derived cell clones, we utilized RNA interference and modulated the cell surface density of this molecule. We observed a direct correlation between permissiveness for virus propagation and relative expression level of CD81. These results are in line with a previous report by Zhang and colleagues, who noted that silencing of CD81 expression reduced infection of HCV pseudoparticles (37). More specifically, using quantitative FACS analyses, we defined a minimal threshold of CD81 surface expression (approximately 7 × 104 molecules/cell) required for susceptibility of Huh7-Lunet- or Huh-7.5-derived clones to infection with a low dose of Jc1. In addition, when we used a green fluorescent protein-expressing reporter virus, we noted a preferential infection of Huh7-Lunet cells expressing high levels of CD81. By using mathematical modeling to describe the relationship between CD81 levels and the proportion of infected cells expressing a given density of CD81, we obtained a symmetric sigmoid curve encompassing a narrow interval of CD81 receptor density across which the fraction of infected cells rapidly increased, indicating a rapid change of susceptibility. Interestingly, while the maximal proportion of infected cells at the highest receptor density differed depending on the virus dose applied for infection, the overall properties of the relationship did not. We therefore conclude that susceptibility to HCV infection is strongly linked to CD81 receptor density on the cell surface, which restricts HCV infection at low receptor density but supports HCV entry beyond copy numbers equivalent to or higher than ca. 290 arbitrary fluorescent units (at least in the case of Huh7-Lunet cells). This observation not only defines an important prerequisite for culturing HCV in vitro but also indicates that accessibility of CD81 or a protein complex containing this molecule may be limiting in certain tissues in vivo, contributing to HCV tropism.
As for other enveloped viruses, HCV entry appears to be a multistep process involving interactions of the virus particle with multiple cellular factors. These mediate in a coordinated manner a sequence of steps of initial docking and adhesion, subsequent wrapping, clathrin-mediated endocytosis, and finally a pH-dependent exit from the endocytic vesicle into the cytoplasm (6, 16, 31). Although at present several factors have been implicated with HCV entry (for recent reviews, see references 5 and 9), their individual contributions to HCV entry are not fully understood. While interactions of the virus particle with specific cell surface-resident proteoglycans carrying highly sulfated glycosaminoglycan moieties likely contribute to initial virus attachment (4, 16), interaction of HCV with CD81 is believed to be important for a step post-initial virus binding. This assumption is primarily based on the observation that CD81-specific antibodies neutralize infection with similar potencies irrespective of their application during or after virus binding (10, 16).
Considering that CD81 was initially identified as a putative HCV receptor based on its ability to interact with HCV E2 and that a soluble form of the large extracellular loop of CD81 is capable of capturing serum-derived HCV particles (25), it is somewhat surprising that this molecule does not appear to significantly contribute to the efficiency of virus adsorption. On the other hand, in vivo CD81 is broadly expressed in many tissues, including lymphoid and nonlymphoid cells, and therefore a high-affinity interaction of the virus particle with this molecule may trap the virus on the surfaces of nonpermissive cells, thus precluding preferential infection of hepatocytes, which are the primary host cells of the virus. In light of these considerations, it is possible that interaction of HCV with CD81 or CD81-containing complexes may, for instance, be required for routing the virus into endocytic vesicles. In this model, a high CD81 receptor density may be important for multivalent high-avidity interactions assisting particle wrapping and uptake into endocytic vesicles and providing an entry port for the uptake of surface-bound virions into host cells.
In summary, by comparing the efficiencies of HCV propagation in two Huh-7 cell-derived cell clones, we defined CD81 as a key host cell factor essential for culturing HCV in vitro. More specifically, we provide evidence that a critical threshold level of this molecule is indispensable for productive infection with HCV and that availability of CD81 is a rate-limiting step in the entry program. These findings shed further light on the molecular events during HCV invasion of host cells, contribute to our understanding of HCV pathogenesis, and help in devising strategies for interference with HCV infection.
Acknowledgments
We are grateful to Urlike Herian for excellent technical assistance, to Julia Lenz for help with cell sorting, to Melanie Ott and Miyawaki Atsushi for expression constructs, to Charles Rice and Tim Tellinghuisen for the Huh-7.5 cell line and 9E10 hybridoma supernatant, to Takaji Wakita for the gift of the JFH1 isolate, to Jens Bukh for the J6CF strain, to all members of the laboratory for helpful discussions, and to Eike Steinmann for critical reading of the manuscript.
This work was supported by an Emmy Noether fellowship from the Deutsche Forschungsgemeinschaft to T.P. (PI 734/1-1), a grant from the Ministry of Science, Research and the Arts of Baden-Württemberg (Az. 23-7532.24-22-21-12/1) to T.P. and R.B., and a grant from the Bristol-Myers-Squibb Foundation to R.B.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Ali, S., C. Pellerin, D. Lamarre, and G. Kukolj. 2004. Hepatitis C virus subgenomic replicons in the human embryonic kidney 293 cell line. J. Virol. 78:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., and T. Pietschmann. 2005. Efficient hepatitis C virus cell culture system: what a difference the host cell makes. Proc. Natl. Acad. Sci. USA 102:9739-9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. [DOI] [PubMed] [Google Scholar]
- 5.Bartosch, B., and F. L. Cosset. 2006. Cell entry of hepatitis C virus. Virology 348:1-12. [DOI] [PubMed] [Google Scholar]
- 6.Blanchard, E., S. Belouzard, L. Goueslain, T. Wakita, J. Dubuisson, C. Wychowski, and Y. Rouille. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, K. S., Z. Cai, C. Zhang, G. C. Sen, B. R. Williams, and G. Luo. 2006. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. J. Virol. 80:7364-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocquerel, L., C. Voisset, and J. Dubuisson. 2006. Hepatitis C virus entry: potential receptors and their biological functions. J. Gen. Virol. 87:1075-1084. [DOI] [PubMed] [Google Scholar]
- 10.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Date, T., T. Kato, M. Miyamoto, Z. Zhao, K. Yasui, M. Mizokami, and T. Wakita. 2004. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. J. Biol. Chem. 279:22371-22376. [DOI] [PubMed] [Google Scholar]
- 12.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21-S29. [DOI] [PubMed] [Google Scholar]
- 14.Kalajzic, I., M. L. Stover, P. Liu, Z. Kalajzic, D. W. Rowe, and A. C. Lichtler. 2001. Use of VSV-G pseudotyped retroviral vectors to target murine osteoprogenitor cells. Virology 284:37-45. [DOI] [PubMed] [Google Scholar]
- 15.Kärber, G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch. Exp. Pathol. Pharmakol. 162:480-487. [Google Scholar]
- 16.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kronke, J., R. Kittler, F. Buchholz, M. P. Windisch, T. Pietschmann, R. Bartenschlager, and M. Frese. 2004. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J. Virol. 78:3436-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 19.Lindenbach, B. D., P. Meuleman, A. Ploss, T. Vanwolleghem, A. J. Syder, J. A. McKeating, R. E. Lanford, S. M. Feinstone, M. E. Major, G. Leroux-Roels, and C. M. Rice. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. USA 103:3805-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 103:6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazzocca, A., S. C. Sciammetta, V. Carloni, L. Cosmi, F. Annunziato, T. Harada, S. Abrignani, and M. Pinzani. 2005. Binding of hepatitis C virus envelope protein E2 to CD81 up-regulates matrix metalloproteinase-2 in human hepatic stellate cells. J. Biol. Chem. 280:11329-11339. [DOI] [PubMed] [Google Scholar]
- 23.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87-90. [DOI] [PubMed] [Google Scholar]
- 24.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 26.Seeff, L. B. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35-S46. [DOI] [PubMed] [Google Scholar]
- 27.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 28.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spearman, C. 1908. The method of “right and wrong cases” (“constant stimuli”) without Gauss's formulae. Br. J. Psychol. 2:227-242. [Google Scholar]
- 30.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van den Hoff, M. J., V. M. Christoffels, W. T. Labruyere, A. F. Moorman, and W. H. Lamers. 1995. Electrotransfection with “intracellular” buffer. Methods Mol. Biol. 48:185-197. [DOI] [PubMed] [Google Scholar]
- 33.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Windisch, M. P., M. Frese, A. Kaul, M. Trippler, V. Lohmann, and R. Bartenschlager. 2005. Dissecting the interferon-induced inhibition of hepatitis C virus replication by using a novel host cell line. J. Virol. 79:13778-13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu, Q., J. T. Guo, and C. Seeger. 2003. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 77:9204-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]