Abstract
The RNA genome of Turnip mosaic virus is covalently linked at its 5′ end to a viral protein known as VPg. This protein binds to the translation eukaryotic initiation factor iso 4E [eIF(iso)4E]. This interaction has been shown to be important for virus infection, although its exact biological function(s) has not been elucidated. In this study, we investigated the subcellular site of the VPg-eIF(iso)4E interaction using bimolecular fluorescence complementation (BiFC). As a first step, eIF(iso)4E, 6K-VPg-Pro, and VPg-Pro were expressed as full-length green fluorescent protein (GFP) fusions in Nicotiana benthamiana, and their subcellular localizations were visualized by confocal microscopy. eIF(iso)4E was predominantly associated with the endoplasmic reticulum (ER), and VPg-Pro was observed in the nucleus and possibly the nucleolus, while 6K-VPg-Pro-GFP induced the formation of cytoplasmic vesicles budding from the ER. In BiFC experiments, reconstituted green fluorescence was observed throughout the nucleus, with a preferential accumulation in subnuclear structures when the GFP split fragments were fused to VPg-Pro and eIF(iso)4E. On the other hand, the interaction of 6K-VPg-Pro with eIF(iso)4E was observed in cytoplasmic vesicles embedded in the ER. These data suggest that the association of VPg with the translation factor might be needed for two different functions, depending of the VPg precursor involved in the interaction. VPg-Pro interaction with eIF(iso)4E may be involved in perturbing normal cellular functions, while 6K-VPg-Pro interaction with the translation factor may be needed for viral RNA translation and/or replication.
One of the initial steps in protein synthesis is the recruitment of mRNAs by the translation eukaryotic initiation factor 4F (eIF4F) complex (17). In plants, this complex is composed of two proteins, eIF4E and eIF4G (4). eIF4E is the cap-binding protein, while eIF4G is a large polypeptide containing binding sites for eIF4E, eIF4A, eIF3, and the poly(A) binding protein. eIF4F simultaneously interacts with the cap structure of mRNA (through eIF4E) and the ribosome-associated eIF3 (through eIF4G). Thus, eIF4F executes the pivotal function of bridging mRNAs with ribosomes. Another cap-binding complex, eIF(iso)4F, has been identified in plants (5) and is composed of a smaller eIF(iso)4E subunit and a larger eIF(iso)4G subunit relative to the eIF4F subunits. Those two complexes perform essentially the same task in translation but have different affinities for certain classes of mRNA substrates and may be involved in different cellular events (14).
Potyviruses have a positive-strand RNA genome of approximately 10 kb (53). The genome codes for a single long polyprotein that is processed by viral proteinases into 10 mature proteins. The viral RNA is polyadenylated at its 3′ end and has a covalently linked virus-encoded protein (VPg) instead of a cap structure at the 5′ end. Being derived from a polyprotein, VPg exists in various forms (i.e., VPg, VPg-Pro, 6K-VPg-Pro) that fulfill specific functions. For instance, VPg-Pro is one of the three proteinases responsible for maturation of the polyprotein (6). 6K-containing forms (e.g., 6K-VPg-Pro) induce the formation of cytoplasmic vesicles necessary for replication complex formation (48). VPg is uridylated by the viral RNA-dependent RNA polymerase (RdRP), which may then act as a primer for negative-strand RNA synthesis (38). VPg and its precursor forms thus play a central role in the viral replication cycle. Another function has recently emerged. A yeast two-hybrid screen has shown that eIF(iso)4E of Arabidopsis thaliana interacts with the VPg of Turnip mosaic virus (TuMV) (57). This interaction (31) has also been reported to take place in infected cells (32). Similar interactions are taking place between the VPg of other potyviruses and either eIF(iso)4E or eIF4E (20, 46). The cap analogue m7GDP and VPg bound to eIF4E or eIF(iso)4E at two distinct sites, although binding of one ligand to the translation factor reduced its affinity for the other ligand (31, 33, 34). This suggests that the potyviral VPg interferes with the formation of a translational initiation complex on cellular mRNAs by sequestering the translation factor. A similar interaction with eIF(iso)4E has also been shown for the proteinase of the nepovirus Tomato ringspot virus (30). The interaction of a VPg and eIF4E is not limited to plant viruses: direct interaction of murine, feline, and human calicivirus VPgs with eIF4E was recently demonstrated (18). Several lines of evidence suggest that eIF4E and/or eIF(iso)4E plays an important role in potyvirus replication. First, knockout A. thaliana plants for eIF(iso)4E were phenotypically indistinguishable from wild-type plants but were resistant to several potyviruses (29). In lettuce, mo11 and mo12 are recessive resistance genes toward common isolates of Lettuce mosaic virus and code for different alleles of eIF4E (36). It was also shown that resistance to potyviruses attributed to the alleles at the pvr2 locus in Capsicum annuum (44) and the pot-1 locus in Lycopersicon esculentum (45) resulted from mutations in eIF4E. Complementing those experiments was the demonstration that the virulence determinant toward these recessive resistances was VPg (35) and that failure of eIF4E alleles to bind VPg correlated with resistance in most cases (26).
Although, these experiments clearly indicate that the interaction between potyvirus VPg and either eIF4E or eIF(iso)4E is important for virus infection, the aim or consequence of the interaction has not yet been deciphered. Involvement in viral RNA translation is likely, but the interaction might also result in perturbation of some host functions or even be required for cell-to-cell trafficking (15). Although the interaction domain has been mapped to the VPg, it is not known if precursor forms of VPg are capable of interacting with the factor in plants and in what subcellular compartment the interaction is taking place. To further investigate the cellular localization of this interaction, eIF(iso)4E, 6K-VPg-Pro, and VPg-Pro were expressed as GFP fusions in Nicotiana benthamiana, and their localizations were visualized by confocal microscopy. Bimolecular fluorescence complementation (BiFC) experiments showed that the interaction of eIF(iso)4E with VPg-Pro took place in the nucleus, while the interaction with 6K-VPg-Pro took place in cytoplasmic vesicles. The subcellular interaction sites may then reflect the role of the interaction in the virus replication cycle.
MATERIALS AND METHODS
Plasmid constructions.
Plasmids for colocalization were constructed as follows. EGFP and DsRed2 (Clontech) genes were amplified by PCR with primers EGFP-F/EGFP-R and DsR-F/DsR-R (Table 1), respectively. Amplified fragments were inserted into the BamHI/EcoRI sites located in the multiple cloning site downstream of the Cauliflower mosaic virus 35S promoter in the pGreen 0029 vector (21), resulting in plasmids pGreenGFP and pGreenDsRed, respectively. The cDNA for eIF(iso)4E was obtained from an A. thaliana mRNA extract with the eIF(iso)4E-F and eIF(iso)4E-R primers (Table 1) and inserted into the XbaI/BamHI site of the same vector. The resulting plasmids were identified as pGreen/eIF(iso)4E-GFP. 6K-VPg-Pro and VPg-Pro were amplified with the 6K-F/Pro-R and VPg-F/Pro-R primers (Table 1), respectively, from the pET/6K2-NIa Quebec isolate of TuMV, which has been modified at the junction of 6K2 and VPg and at the active site of Pro to prevent Pro-mediated proteolysis (28). 6K-VPg-Pro and VPg-Pro cDNA fragments were inserted into HindIII/BamHI sites of the pGreenDsRed (for VPg-Pro only) and pGreenGFP vectors. The resulting plasmids were identified as pGreen/6K-VPg-Pro-GFP, pGreen/VPg-Pro-GFP, and pGreen/VPg-Pro-DsRed.
TABLE 1.
Primers used for vector construction
| Primer | Sequence (5′→3′) | Comments |
|---|---|---|
| EGFP-F | TCTAGAGGATCCCCCATGGTGAGCAAGGGCGAG | 5′ EGFP + BamHI + NcoI |
| EGFP-R | GAATTCGAGCTCCCCCTTGTACAGCTCGTCCAT | 3′ EGFP + EcoRI |
| DsRed-F | TCTAGAGGATCCCCCATGGCCTCCTCCGAGAAC | 5′ DsRed + BamHI |
| DsRed-R | TAATTAAAGGAATTCTTACAGGAACAGGTGGTGGCGGCCCA | 3′ DsRed + EcoRI |
| eIF(iso)4E-F | GCTTTCTAGAGGATCGAGTAATTTAGCTCAACT | 5′ eIF(iso)4E + XbaI |
| eIF(iso)4E-R | TCACCATGGGGGATCCGACAGTGAACCGGCTTCT | 3′ eIF(iso)4E + BamHI |
| 6K-F | ATGGAGGCAGTTAAGCTTATGAGCACCAACGAA | 5′ 6K-VPg-Pro + HindIII |
| Pro-R | AGGCCATGGGGGATCCTTGTGCGTAGACTGCCGT | 3′ 6K-VPg-Pro + BamHI |
| VPg-F | TCTGAACCCGTAAAGCTTATGGCAAAAGGCAAGA | 5′ VPg-Pro + HindIII |
| NT-F | CTATCATTATCCGGATCCAAATGAGTAAAGGAGAA | 5′ NTGFP + BamHI |
| NT-R | TTCAGCGTACCGGAATTCTTGCTTGTCGGCCATGAT | 3′ NTGFP + EcoRI |
| CT-F | ATTTGGAGAGGACAGCCCGGGATCCAAGAACGGCATCAAAGCC | 5′ CTGFP + BamHI |
| CT-R | TTTCAGCGTACCGGAATTCTATTTGTATAGTTCATCCAT | 3′ CTGFP + EcoRI |
| NTeIF(iso)4E-F | AACTCAACTGCGGAATTCATGGCGACCGAT | 5′ eIF(iso)4E + EcoRI |
| NTeIF(iso)4E-R | GTGATTTCAGCGGAATTCTCAGACAGTGAACCGGCT | 3′ eIF(iso)4E + EcoRI |
Plasmids for complementation were obtained as follows. N-terminal GFP (ntGFP) and C-terminal GFP (ctGFP) fragments were amplified from plasmid pBin-mGFP5-ER by the NT-F/NT-R and CT-F/CT-R primers (Table 1), respectively, and inserted into the BamHI/EcoRI sites of pGreen0029. pGreen/ntGFP-eIF(iso)4E was obtained by amplification of eIF(iso)4E from plasmid pGreen/eIF(iso)4E-GFP with the NTeIF(iso)4E-F and NTeIF(iso)4E-R primers (Table 1) and inserted into the EcoRI site in pGreen/ntGFP. pGreen/VPg-Pro-ctGFP was obtained by transferring the HindIII/BamHI VPg-Pro fragment from pGreen/VPg-Pro-GFP into the corresponding sites of pGreen/ctGFP. pGreen/6K-VPg-Pro-ctGFP was derived from pGreen/6K-VPg-Pro-GFP in a similar manner.
Protein expression in plants.
Vectors containing genes for fluorescent and fusion proteins were introduced into Agrobacterium tumefaciens AGL1 by electroporation. Transformed cells were selected on kanamycin-ampicillin plates. Cultures of the bacterium containing both plasmids were prepared overnight, the culture medium was removed, and cells were resuspended in water supplemented with 10 mM MgCl2 and 150 μM acetosyringone. The resulting preparation was used to agroinfiltrate leaves from 3-week-old N. benthamiana plants, along with Agrobacterium containing plasmids allowing the expression of suppressors of silencing P19 or HcPro prepared the same way. Plants were then kept for 4 days in a growth chamber before observation.
Confocal microscopy.
Sections from agroinfiltrated leaves were cut out and placed in immersion oil on a microscope coverslide. The coverslide was then inverted over a microscope slide, presenting a depression above which was placed the leaf section. Individual cells were observed with a 40× oil immersion objective on a Radiance 2000 confocal microscope from Bio-Rad. Fluorescent proteins were excited with an Argon-Krypton laser. The data for green and red channels were collected simultaneously. Images were collected with a charge-coupled-device camera and treated with Adobe Photoshop or Image J (http://rsb.info.nih.gov/ij/) software.
Cellular fractionation.
Subcellular fractionation from plants was carried out according to the method of Davies and Abe (10). Brassica perviridis plants (three-leaf stage) were infected with TuMV or mock inoculated with phosphate-buffered saline (PBS). At 12 days postinoculation, leaves that developed next to the inoculated leaves were harvested. Leaf tissue (0.5 g) was minced in 5 volumes (2.5 ml) of buffer A (200 mM Tris-HCl, pH 8.5-50 mM KCl-25 mM MgCl2). Nuclei, chloroplasts, cell wall, and debris were removed by centrifugation at 3,700 × g at 4°C for 10 min. The supernatant (S3) was centrifuged at 27,000 × g at 4°C for 10 min, resulting in soluble (S30) and crude membrane (P30) fractions. The pellet containing membranes was resuspended in the same volume as the supernatant in buffer A with 40 mM octyl-β-d-glucopyranoside (Bioshop). Twenty microliters of total, soluble, and membrane fractions were collected, diluted 1:5 in protein dissociation buffer, and subjected to immunoblot analysis following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Immunoreactions were detected with the enhanced chemiluminescence (ECL)-based secondary antibody system (Amersham).
Nucleus isolation.
Nucleus isolation from plants was carried out according to the method of Gaudino and Pikaard (16). Brassica perviridis plants (three-leaf stage) were infected with TuMV or mock inoculated with PBS. At 12 days postinoculation, leaves that developed next to the inoculated leaves were harvested. Leaf tissue (5 g) was cut in pieces with a blade and minced in 3 volumes (15 ml) of nucleus isolation buffer (NIB) (50 mM Tris-HCl, pH 7.2-5 mM KCl-5 mM MgCl2-1 M sucrose-10 mM β-mercaptoethanol-0.2 mM phenylmethylsulfonyl fluoride). The homogenate was filtered through nylon membranes (160 μm and 41 μm) and subjected to centrifugation at 14,000 × g for 15 min at 4°C. The pellet was resuspended in 1 volume (5 ml) of NIB and homogenized. Seven milliliters of nuclei in Percoll 5% was layered on a step gradient of Percoll with 5-ml layers of 15%, 30%, 45%, and 60% and subjected to centrifugation at 530 × g for 10 min and at 8,500 × g for 20 min at 4°C. Nuclei accumulated between the 60%-45% Percoll layers and the 30%-45% Percoll layers. Nuclei were diluted in 5 volumes of NIB, mixed gently by inversion, and subjected to centrifugation at 1,500 × g for 10 min. Nuclei were gently resuspended in 25 volumes of NIB and centrifuged again. Nuclei were resuspended in 100 μl of storage buffer (50 mM Tris-HCl, pH 8-0.3 mM sucrose-5 mM MgCl2-1.5 mM NaCl-0.1 mM CaCl2-5 μM β-mercaptoethanol). Proteins from S14 and nucleus preparations were diluted 1:5 in loading SDS buffer, separated by SDS-PAGE, and detected by immunoblot analysis with the ECL-based secondary antibody system (Amersham).
RESULTS AND DISCUSSION
Distribution of VPg precursors and eIF4E isomers among soluble and membrane-enriched fractions in TuMV-infected cells.
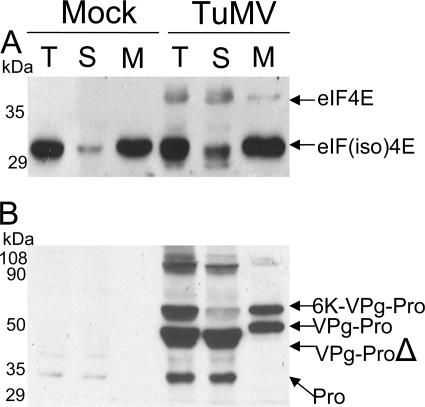
Brassica perviridis plants were infected with TuMV or mock inoculated, and leaves that developed next to the inoculated ones were harvested 12 days postinfection. Leaves were homogenized, and nuclei, chloroplasts, cell wall, and debris were removed by centrifugation at 3,700 × g. Soluble proteins were then separated from membrane-associated proteins by centrifugation at 27,000 × g. To allow a quantitative evaluation of the distribution of the proteins among the two fractions, the pellet containing the membrane-associated proteins was resuspended in the same volume as that of the supernatant. Total, soluble, and membrane-associated proteins were separated by SDS-PAGE and subjected to immunoblot analysis with a rabbit serum raised against a recombinant form of eIF(iso)4E from A. thaliana. This antibody also cross-reacts with eIF4E (32). eIF4E and eIF(iso)4E of A. thaliana have calculated sizes of 26.5 and 22.5 kDa, respectively (32). Although the exact sizes of the B. perviridis isomers are not known, they are expected to be very similar, since both plants belong to the same family. Figure 1A shows that in the total protein fraction of mock-inoculated plants, only eIF(iso)4E was detected, while both eIF4E and eIF(iso)4E were observed in TuMV-infected leaves, although the eIF(iso)4E signal was stronger than that of eIF4E. The anti-eIF(iso)4E serum recognized with equal intensity the recombinant forms of the two isomers (32), suggesting that the signal intensity reflected the amount of the protein in infected cells. After cellular fractionation, eIF(iso)4E was predominantly found in the membrane-enriched fraction for both mock- and TuMV-infected leaves. In contrast, eIF4E was generally found in the soluble fraction. Because eIF4E isomers are associated with ribosomal complexes (22), the results of the subcellular fractionation experiment suggest that eIF(iso)4E is mainly associated with the rough endoplasmic reticulum and that eIF4E is associated with free ribosomes in TuMV-infected cells. eIF4F functionally differs from eIF(iso)4F in promoting internal initiation, cap-independent translation, and translation of structured mRNAs (14); this differential activity may be reflected in the uneven distribution to free or membrane-bound ribosomes.
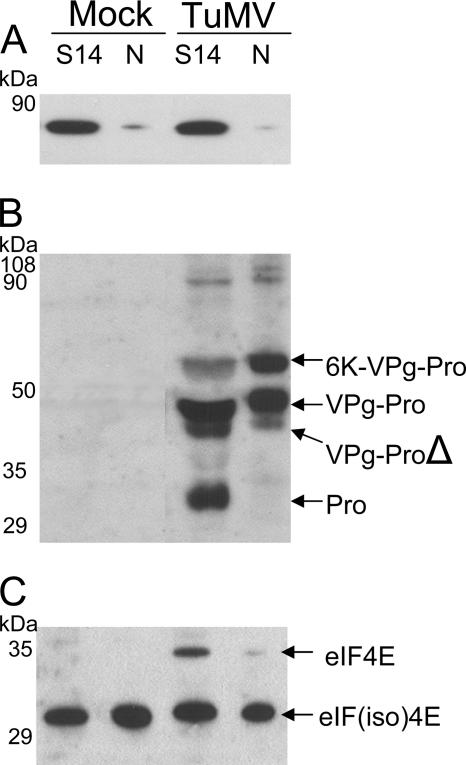
FIG. 1.
Immunoblot analysis of soluble and membrane-associated proteins from healthy or TuMV-infected plants. B. perviridis plants were mock inoculated or infected with TuMV. Twelve days later, total proteins (T) were extracted and soluble proteins (S) were separated from membrane-associated proteins (M) by centrifugation at 27,000 × g. Proteins were separated by SDS-PAGE and analyzed by Western blotting using a rabbit serum against eIF(iso)4E (A) or VPgPro (B). The text on the right shows the electrophoretic migration positions of the indicated proteins.
The subcellular fractions were also analyzed with a rabbit serum raised against the TuMV VPg-Pro (Fig. 1B). Bands corresponding to 6K-VPg-Pro, VPg-Pro, the C-terminally deleted form of VPg-Pro, and Pro were detected in the total protein fraction of infected leaves. In addition, several bands of approximately 100 kDa were detected. The exact nature of these bands is not known; they may correspond to large polyprotein precursors or to dimeric forms of 6K-VPg-Pro, VPg-Pro, and Pro. No polypeptide corresponding to the mature VPg was detected. A fraction of VPg-Pro, the C-terminally deleted form of VPg-Pro, and Pro were detected in the soluble fraction. On the other hand, 6K-VPg-Pro and the remaining population of VPg-Pro were found in the membrane fraction. The association of 6K-VPg-Pro with membranes is explained by the presence of the 6K domain, which contains a stretch of hydrophobic amino acids. This protein was previously shown to be an integral membrane protein (48). The presence of a substantial quantity of VPg-Pro in the membrane-associated fraction is likely the result of its interaction with 6K-VPg-Pro (27).
Cellular localizations of GFP fusions of eIF(iso)4E, VPg-Pro, and 6K-VPg-Pro.
To further refine their cellular localizations, eIF(iso)4E, VPg-Pro, and 6K-VPg-Pro were fused to the green fluorescent protein (GFP). In order to prevent proteolysis by Pro at the 6K-VPg and VPg-Pro junctions, the glutamic acid residue preceding the cleavage bond was modified to histidine. These modifications were shown to prevent processing at the cleavage sites (28). Transient expression of VPg precursors was performed by agroinfiltration in N. benthamiana, which is a host for TuMV. HCPro or P19 was used as a suppressor of gene silencing; no difference in VPg-Pro expression or localization (see below) was noticed with either suppressor. Expression of the GFP fusions was assessed by immunoblot analyses using a rabbit serum raised against VPg-Pro. In each case, a signal corresponding to the expected molecular mass of the analyzed protein was observed, indicating that no degradation or Pro-mediated processing of the proteins had taken place (Fig. 2).
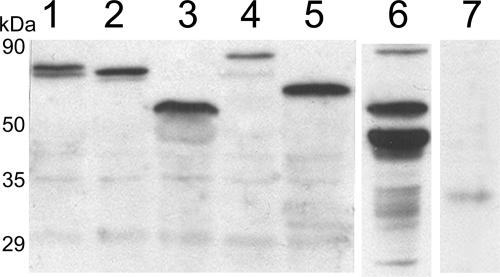
FIG. 2.
Expression of GFP fusions in N. benthamiana. Leaves were infiltrated with A. tumefaciens; 4 days later, total proteins were extracted, separated by SDS-PAGE, and analyzed by Western blotting using a rabbit serum against VPgPro. A. tumefaciens suspensions contained binary Ti plasmids encoding VPg-Pro-GFP (lane 1), VPg-Pro-DsRed2 (lane 2), VPg-Pro-ctGFP (lane 3), 6K-VPg-Pro-GFP (lane 4), 6K-VPg-Pro-ctGFP (lane 5), and GFP (lane 7). Proteins from TuMV-infected B. perviridis were loaded in lane 6.
GFP fluorescence was visualized between 2 and 4 days postinfiltration by confocal microscopy. No notable differences in cellular localization were observed during this time period. Fluorescence was generally observed in 30 to 50% of the cells in the infiltrated area. To facilitate the identification of membrane structures, the red fluorescent protein containing ER targeting and retention signals (ER-DsRed2) (58) was coexpressed along with the GFP fusions. In the case of ER-DsRed2, red fluorescence was not detected before 3 days postinfiltration. In order to ascertain that the green fluorescence that was observed was not emitted by the DsRed2 protein, control experiments were conducted with the GFP fusions only. In every case, the patterns of green fluorescence were similar, whether ER-DsReD2 was coexpressed or not.
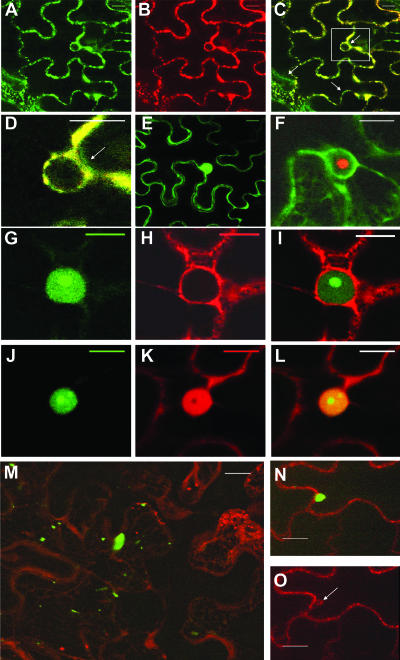
Fluorescence associated with eIF(iso)4E-GFP was concentrated around the nucleus or in the cytoplasm (Fig. 3A). A similar distribution was observed with the fluorescence emitted by ER-DsRed2 (Fig. 3B). Merging of the fluorescence observed for eIF(iso)4E-GFP and ER-DsRed2 showed that most of eIF(iso)4E was associated with the ER, although some green emission was observed in the merged fluorescence (Fig. 3C and D). A similar pattern was observed when eIF(iso)4E was fused to DsRed2 and coexpressed with GFP targeted to the ER (data not shown). Fluorescence associated with free GFP was distributed throughout the cytoplasm and the nucleus, to the exclusion of the nucleolus (Fig. 3E). This indicates that localization of eIF(iso)4E-GFP to the ER was due to the translation factor and not to the fluorescent protein. This result is in accordance with the cellular fractionation data, in which eIF(iso)4E was detected mostly in the membrane-enriched fraction.
FIG. 3.
Subcellular localizations of eIF(iso)4E and VPg precursors. N. benthamiana leaves were infiltrated with A. tumefaciens, and expression of fluorescent proteins was visualized by confocal microscopy 4 days later. A. tumefaciens suspensions contained binary Ti plasmids encoding eIF(iso)4E-GFP and ER-DsRed2 (A to D), GFP (E), VPg-Pro-DsRed2 and GFP-ER (F), VPg-Pro-GFP and ER-DsRed2 (G to I), VPg-Pro-GFP and DsRed2 (J to L), and 6K-VPg-Pro-GFP and ER-DsRed2 (M to O). Panel M is a “maximum-intensity Z projection” of 50 1-μm slices stacked on top of each other. Panels A, E, G, and J show fluorescence emitted by the green channel only; panels B, H, K, and O show fluorescence emitted by the red channel only. Panel D is a close-up view of the rectangle depicted in panel C. The arrow in panel O shows an outgrowth of ER. Bar, 15 μm.
Although some fluorescence was observed in the cytoplasm, VPg-Pro-GFP was observed principally in the nucleus (Fig. 3G), which was made evident by the accumulation of ER-DsRed2 in the perinuclear ER membranes (Fig. 3H and I). The same distribution with VPg-Pro-DsRed2 was observed (Fig. 3F), indicating that nuclear accumulation was not dependent on the fluorescent protein. Nuclear localization is not unexpected, as it was shown to be the case for the VPg-Pro of at least two other potyviruses (47). Nuclear distribution is possibly the consequence of an active process, since a putative nuclear localization signal similar to the one identified with the VPg-Pro of tobacco etch virus (TEV) (40) was found within the TuMV protein (data not shown). VPg-Pro not only localized to the nucleus, it accumulated preferentially to a subnuclear structure, possibly the nucleolus. To confirm that VPg-Pro is effectively distributed in the nucleolus, VPg-Pro-GFP was expressed (Fig. 3J) along with the soluble form of DsRed2 (Fig. 3K). Red fluorescence was distributed in the cytoplasm and the nucleoplasm, to the exclusion of the nucleolus. The presence of free dsRed2 (and free GFP) in the nucleus has been documented previously and is the result of passive diffusion through the nucleus pore (19). Fluorescence observed from VPg-Pro-GFP colocalized with that of DsRed in the nucleus, except for a brighter area exactly where no red fluorescence was observed (Fig. 3L). Similar data were obtained with the coexpression of VPg-Pro-DsRed2 with GFP (data not shown). VPg-Pro-DsRed2 was also coexpressed with fibrillarin 1 from A. thaliana fused to GFP, which is a nucleolar protein (2). Green and red fluorescence were found to colocalize (data not shown). These experiments then indicate that the TuMV VPg-Pro accumulated in the nucleus and preferentially in the nucleolus when synthesized independently of other viral proteins. Nucleolar localization was also noticed for the TEV homologue (47).
The fluorescence emitted by 6K-VPg-Pro-GFP was associated with cytoplasmic vesicles distributed throughout the cortical ER membrane system (Fig. 3M) and very often was also associated with the perinuclear ER (not shown). On average, there were one to three vesicles of 10 μm in diameter per cell, although several smaller fluorescing structures were also observed. These vesicles were clearly seen budding from ER membranes (Fig. 3N and O). This ER association of 6K-VPg-Pro agrees with our previous observation using sucrose gradient fractionation of plant cell extracts (32). The membranous vesicles are reminiscent of those induced by the TEV 6K protein (48). In the case of TEV, it was suggested that these vesicles correspond to the replication complexes.
Nuclear localization of the interaction of VPg-Pro with eIF(iso)4E.
BiFC (3, 54) was used to determine the subcellular localization of the interaction between VPg precursors and eIF(iso)4E. In this assay the GFP is split into two nonoverlapping fragments, ntGFP and ctGFP. The ntGFP fragment is fused to the N-terminal end of one binding protein, while the ctGFP fragment is fused to the C-terminal end of the corresponding partner. Reconstitution of a fluorescing GFP chromophore takes place upon interaction of the two partner proteins. Consequently, eIF(iso)4E was fused to the N-terminal portion whereas the VPg precursors were fused to the C-terminal portion of GFP. Transient expression was achieved by agroinfiltration in N. benthamiana leaves along with ER-DsRed2. Fluorescence was observed after 3 or 4 days postinfiltration by confocal microscopy. The 1-day delay in fluorescence observation may be explained by the fact that intact (full-size) GFP requires several hours to mature in the cell (51), and it is conceivable that the intermolecular reconstitution of a split fluorophore may take longer. Immunoblot analysis with the anti-VPg-Pro rabbit serum showed that no degradation or Pro-mediated proteolysis took place for the VPg precursors (Fig. 2).
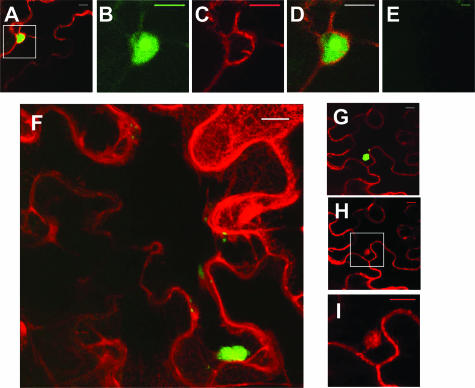
Green fluorescence was observed throughout the nucleus, with a preferential accumulation in subnuclear structures when VPg-Pro-ctGFP was coexpressed with ntGFP-eIF(iso)4E (Fig. 4A to D). The subnuclear structures were of the same size as those observed for the localization of VPg-Pro-GFP. Thus, the cellular localization of VPg-Pro coincides with that for the interaction with the translation factor. No fluorescence was observed for the following combinations: ntGFP-eIF(iso)4E with ctGFP (Fig. 4E) or VPg-Pro-ctGFP with ntGFP (data not shown). Nuclear import of mammalian eIF4E is an active process mediated by the eIF4E transporter (11). Since eIF(iso)4E-GFP was not observed in the nucleus when expressed alone, it maybe because the N. benthamiana transporter complex does not recognize the fusion protein or is not expressed when agroinoculation is applied. eIF(iso)4E nuclear interaction with VPg-Pro suggests, however, that the viral protein may act as a transporter protein. This was confirmed by the fusion of eIF(iso)4E with Ds-Red2 and of VPg-Pro with GFP. eIF(iso)4E-DsRed2 expressed alone was observed exclusively in the cytoplasm but was also observed in the nucleolus along with VPg-Pro-GFP (data not shown).
FIG. 4.
Subcellular localizations of the interaction between eIF(iso)4E and VPg precursors. N. benthamiana leaves were infiltrated with A. tumefaciens, and expression of fluorescent proteins was visualized by confocal microscopy 4 days later. A. tumefaciens suspensions contained binary Ti plasmids encoding VPg-Pro-ctGFP, ntGFP-eIF(iso)4E, and ER-DsRed2 (A to D); ctGFP and ntGFP-eIF(iso)4E (E); or 6K-VPg-Pro-ctGFP, ntGFP-eIF(iso)4E, and ER-DsRed2 (F to I). Panel F is a “maximum-intensity Z projection” of 13 1-μm slices stacked on top of each other. Panels B to D and I are close-up views of the rectangles depicted in panels A and H, respectively. Panel B shows fluorescence emitted by the green channel only; panels C, H, and I show fluorescence emitted by the red channel only. Bar, 15 μm.
Nuclear localization of the interaction between VPg-Pro and eIF(iso)4E was visualized in the absence of other viral proteins. In an attempt to confirm the presence of VPg-Pro and eIF(iso)4E in the nucleus of TuMV-infected cells, nuclei were purified from mock-inoculated and TuMV-infected leaves as described in Materials and Methods. Isolation of nuclei was confirmed by phase-contrast microscopy and 4′,6-diamidino-2-phenylindole staining (data not shown). The S14 fraction and the purified nuclei were analyzed by immunoblot assay using antibodies raised against BiP, which is a marker of the ER (7). For the postnuclear fraction, 1/1,000 of the preparation was loaded on the acrylamide gel, whereas one-fifth was loaded for the nuclear fraction. A strong signal for BiP was detected in the S14 fraction, while a faint band was observed in the nuclear preparation of mock- and TuMV-infected cells (Fig. 5A), suggesting that the purified nuclei were essentially free of ER membranes.
FIG. 5.
Immunoblot analysis of nuclear and postnuclear fraction proteins from healthy or TuMV-infected plants. B. perviridis plants were mock inoculated or infected with TuMV. Twelve days later, leaves were homogenized and centrifuged at 14,000 × g to separate the “soluble” fraction (S) from crude nuclei, which were further purified by Percoll gradient centrifugation (N). Proteins were separated by SDS-PAGE and analyzed by Western blotting using a rabbit serum against Bip (A), VPg-Pro (B), or eIF(iso)4E (C). The text on the right shows the electrophoretic migration positions of the indicated proteins.
These fractions were analyzed with a rabbit serum raised against VPg-Pro. Bands corresponding to 6K-VPg-Pro and VPg-Pro were found in both fractions (Fig. 5B). However, Pro was detected in the S14 fraction only. This suggests that the nuclei were free of cytoplasmic proteins, since the viral protein is a soluble cytoplasmic protein (Fig. 1) and does not contain a nuclear localization signal. Consequently, a fraction of VPg-Pro is localized to the nucleus of infected cells, which confirms the GFP fusion and BiFC experiments. The presence of 6K-VPg-Pro in the nuclear fraction is explained by its often perinuclear localization, as observed in the GFP fusion experiments. The purification of nuclei probably resulted in the copurification of some of the 6K-VPg-Pro-induced vesicles.
When the postnuclear and nuclear fractions were analyzed with a serum raised against eIF(iso)4E, the signal for the translation factor was detected in both fractions with similar intensities. In contrast, the BiP signal was observed at a much lower level in the nuclear than in the postnuclear fraction, indicating that the detection of eIF(iso)4E in purified nuclei was not the result of impurities in the preparation. The presence of VPg-Pro and eIF(iso)4E in nucleus-enriched fractions of infected cells provides additional support for the suggested interaction of these two proteins in the nucleus. Interestingly, eIF4E was found mainly in the S14 fraction. Again, this differential localization might reflect differential activity of the two isomers.
Although positive-sense RNA viruses replicate in the cytoplasm, there are several examples of proteins from such viruses that have been found in the nucleus, often in the nucleolus (for reviews, see references 23 and 24). The general hypothesis that has been presented is that the presence of these viral proteins is required for disruption of nuclear functions and/or inhibition of antiviral responses (56). However, the exact mode of action of these proteins in the nucleus remains to be fully described. Nuclear interaction with eIF(iso)4E may thus offer a novel perspective for the mode of action of VPg-Pro in this organelle. The traditional role of eIFs is to initiate protein synthesis in the cytoplasm, which requires the coordinated activities of a large number of eIFs (37). However, a wide variety of studies showed that a number of “classical” translation machinery components are located in the nucleus of mammalian cells, including eIF4E (for a review, see reference 49). eIF4E controls the expression of certain growth stimulatory proteins by regulating mRNA export from the nucleus (42, 43). Additionally, nuclear eIF4E might be involved in nuclear translation, possibly required for proofreading of transcripts before they are transported to the cytoplasm (25). Consequently, nuclear/nucleolar interaction of VPg-Pro with eIF(iso)4E may perturb the nuclear role of the factor by sequestration. Interestingly, VPg-Pro possesses DNase and RNase activities (1, 8). Association with eIF(iso)4E might thus target VPg-Pro to particularly sensitive areas for DNA and RNA degradation. This possibility may then explain the host gene shutoff that has been observed in Pea seedborne mosaic virus-infected cells (55).
Vesicular localization of 6K-VPg-Pro and eIF(iso)4E interaction.
Green fluorescence was observed in cytoplasmic vesicles when 6K-VPg-Pro-ctGFP was coexpressed with ntGFP-eIF(iso)4E. Figure 4F represents a “maximum-intensity Z projection” of 13 1-μm slices stacked on top of each other for one interaction event. One vesicle is clearly depicted: it was oblong, with a length of 15 μm, a height of 7 μm, and a depth of at least 13 μm. It was embedded on the surface of the ER, which was seen as a convoluted web-like reticular network of interconnected tubules. Smaller vesicles around ER membranes were also observed. Possibly, these structures were in transit for the formation of the larger vesicles. The ∼10-μm vesicles were an outgrowth of the ER membrane system (Fig. 4G to I). No fluorescence was observed when ntGFP-eIF(iso)4E was expressed with ctGFP or when ntGFP was expressed with 6K-VPg-Pro-ctGFP (data not shown). This result suggests that the vesicles induced by 6K-VPg-Pro are the site for interaction with eIF(iso)4E.
The interaction of eIF(iso)4E with 6K-VPg-Pro in cytoplasmic vesicles points to a direct involvement of the translation factor in virus replication. The very first events upon entry of a positive-strand RNA virus into a cell are uncoating of the virion, followed by translation of the genome. This leads to the production of viral proteins that induce the formation of cytoplasmic vesicles (48, 52). Upon maturation, these vesicles house replication complexes for virus RNA replication to take place (12). However, it is not clear yet whether translation and replication are physically separate events or whether they are tightly coupled processes. 6K-VPg-Pro is a likely component of the replication complex and interacts with the viral RdRP (9). The 6K domain of TEV has been shown to be an integral membrane protein (41) responsible for the formation of ER-derived cytoplasmic vesicles sheltering replication complexes (48). The vesicles induced by TuMV 6K-VPg-Pro (this study) are very similar in size and origin to those described for the TEV replication complex (48). The presence of eIF(iso)4E in these vesicles thus suggests a close physical relationship between translation and replication of the viral RNA. The presence of host translation factor in highly purified replication complexes has been described for other plant viruses. For instance, Tobacco mosaic virus and Brome mosaic virus replication complexes include a subunit of eIF3 (39, 50). Also, it has been shown that functional poliovirus replication complexes are formed in cis in a coupled process involving viral translation, membrane modification, vesicle budding, and viral RNA synthesis (13). Our results raise the possibility that 6K-VPg-Pro not only induces the formation of cytoplasmic vesicles that house replication complexes but may also be responsible for the coupling of viral RNA translation and replication through interaction with both eIF(iso)4E and the viral RdRP within a single vesicle.
This study thus provides a fine example that precursor proteins derived from a polyprotein can have quite different functions. In the case of TuMV, VPg-Pro interaction with eIF(iso)4E may be involved in perturbing normal cellular functions, while 6K-VPg-Pro interaction with the translation factor may be required for viral replication, which is a complex process involving translation and replication of the RNA genome.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council (NSERC) of Canada.
We thank Marcel Desrosiers for help with the confocal microscope, A. Vitale for the anti-BiP serum, M. Echeverría for AtFibrillarin1-GFP, and H. Sanfaçon for the generous gift of ER-dsRed2, as well as for critically reading the manuscript.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Anindya, R., and H. S. Savithri. 2004. Potyviral NIa proteinase, a proteinase with novel deoxyribonuclease activity. J. Biol. Chem. 279:32159-32169. [DOI] [PubMed] [Google Scholar]
- 2.Barneche, F., F. Steinmetz, and M. Echeverria. 2000. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275:27212-27220. [DOI] [PubMed] [Google Scholar]
- 3.Bracha-Drori, K., K. Shichrur, A. Katz, M. Oliva, R. Angelovici, S. Yalovsky, and N. Ohad. 2004. Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40:419-427. [DOI] [PubMed] [Google Scholar]
- 4.Browning, K. S. 1996. The plant translational apparatus. Plant Mol. Biol. 32:107-144. [DOI] [PubMed] [Google Scholar]
- 5.Browning, K. S., C. Webster, J. K. Roberts, and J. M. Ravel. 1992. Identification of an isozyme form of protein synthesis initiation factor 4F in plants. J. Biol. Chem. 267:10096-10100. [PubMed] [Google Scholar]
- 6.Carrington, J. C., R. Haldeman, V. V. Dolja, and M. A. Restrepo-Hartwig. 1993. Internal cleavage and trans-proteolytic activities of the VPg-proteinase (NIa) of tobacco etch potyvirus in vivo. J. Virol. 67:6995-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cascardo, J. C., R. S. Almeida, R. A. Buzeli, S. M. Carolino, W. C. Otoni, and E. P. Fontes. 2000. The phosphorylation state and expression of soybean BiP isoforms are differentially regulated following abiotic stresses. J. Biol. Chem. 275:14494-14500. [DOI] [PubMed] [Google Scholar]
- 8.Cotton, S., P. J. Dufresne, K. Thivierge, C. Ide, and M. G. Fortin. 2006. The VPgPro protein of Turnip mosaic virus: in vitro inhibition of translation from a ribonuclease activity. Virology 351:92-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daros, J. A., M. C. Schaad, and J. C. Carrington. 1999. Functional analysis of the interaction between VPg-proteinase (NIa) and RNA polymerase (NIb) of tobacco etch potyvirus, using conditional and suppressor mutants. J. Virol. 73:8732-8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, E., and S. Abe. 1995. Methods for isolation and analysis of polyribosomes. Methods Cell Biol. 50:209-222. [DOI] [PubMed] [Google Scholar]
- 11.Dostie, J., M. Ferraiuolo, A. Pause, S. A. Adam, and N. Sonenberg. 2000. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 19:3142-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egger, D., and K. Bienz. 2005. Intracellular location and translocation of silent and active poliovirus replication complexes. J. Gen. Virol. 86:707-718. [DOI] [PubMed] [Google Scholar]
- 13.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallie, D. R., and K. S. Browning. 2001. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J. Biol. Chem. 276:36951-36960. [DOI] [PubMed] [Google Scholar]
- 15.Gao, Z., E. Johansen, S. Eyers, C. L. Thomas, T. H. Noel Ellis, and A. J. Maule. 2004. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 40:376-385. [DOI] [PubMed] [Google Scholar]
- 16.Gaudino, R. J., and C. S. Pikaard. 1997. Cytokinin induction of RNA polymerase I transcription in Arabidopsis thaliana. J. Biol. Chem. 272:6799-6804. [DOI] [PubMed] [Google Scholar]
- 17.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 18.Goodfellow, I., Y. Chaudhry, I. Gioldasi, A. Gerondopoulos, A. Natoni, L. Labrie, J. F. Laliberte, and L. Roberts. 2005. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 6:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodin, M. M., R. G. Dietzgen, D. Schichnes, S. Ruzin, and A. O. Jackson. 2002. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31:375-383. [DOI] [PubMed] [Google Scholar]
- 20.Grzela, R., L. Strokovska, J. P. Andrieu, B. Dublet, W. Zagorski, and J. Chroboczek. 2006. Potyvirus terminal protein VPg, effector of host eukaryotic initiation factor eIF4E. Biochimie 88:887-896. [DOI] [PubMed] [Google Scholar]
- 21.Hellens, R., P. Mullineaux, and H. Klee. 2000. Technical focus: a guide to Agrobacterium binary Ti vectors. Trends Plant Sci. 5:446-451. [DOI] [PubMed] [Google Scholar]
- 22.Hiremath, L. S., S. T. Hiremath, W. Rychlik, S. Joshi, L. L. Domier, and R. E. Rhoads. 1989. In vitro synthesis, phosphorylation, and localization on 48 S initiation complexes of human protein synthesis initiation factor 4E. J. Biol. Chem. 264:1132-1138. [PubMed] [Google Scholar]
- 23.Hiscox, J. A. 2002. The nucleolus—a gateway to viral infection? Arch. Virol. 147:1077-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiscox, J. A. 2003. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 95:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iborra, F. J., D. A. Jackson, and P. R. Cook. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293:1139-1142. [DOI] [PubMed] [Google Scholar]
- 26.Kang, B. C., I. Yeam, J. D. Frantz, J. F. Murphy, and M. M. Jahn. 2005. The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 42:392-405. [DOI] [PubMed] [Google Scholar]
- 27.Kang, S. H., W. S. Lim, and K. H. Kim. 2004. A protein interaction map of soybean mosaic virus strain G7H based on the yeast two-hybrid system. Mol. Cell 18:122-126. [PubMed] [Google Scholar]
- 28.Laliberté, J. F., O. Nicolas, C. Chatel, C. Lazure, and R. Morosoli. 1992. Release of a 22-kDa protein derived from the amino-terminal domain of the 49-kDa NIa of turnip mosaic potyvirus in Escherichia coli. Virology 190:510-514. [DOI] [PubMed] [Google Scholar]
- 29.Lellis, A. D., K. D. Kasschau, S. A. Whitham, and J. C. Carrington. 2002. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12:1046-1051. [DOI] [PubMed] [Google Scholar]
- 30.Leonard, S., J. Chisholm, J. F. Laliberte, and H. Sanfacon. 2002. Interaction in vitro between the proteinase of Tomato ringspot virus (genus Nepovirus) and the eukaryotic translation initiation factor iso4E from Arabidopsis thaliana. J. Gen. Virol. 83:2085-2089. [DOI] [PubMed] [Google Scholar]
- 31.Leonard, S., D. Plante, S. Wittmann, N. Daigneault, M. G. Fortin, and J. F. Laliberte. 2000. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 74:7730-7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leonard, S., C. Viel, C. Beauchemin, N. Daigneault, M. G. Fortin, and J. F. Laliberte. 2004. Interaction of VPg-Pro of turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J. Gen. Virol. 85:1055-1063. [DOI] [PubMed] [Google Scholar]
- 33.Michon, T., Y. Estevez, J. Walter, S. German-Retana, and O. LeGall. 2006. The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J. 273:1312-1322. [DOI] [PubMed] [Google Scholar]
- 34.Miyoshi, H., N. Suehiro, K. Tomoo, S. Muto, T. Takahashi, T. Tsukamoto, T. Ohmoriand, and T. Natsuaki. 2006. Binding analyses for the interaction between plant virus genome-linked protein (VPg) and plant translational initiation factors. Biochimie 88:329-340. [DOI] [PubMed] [Google Scholar]
- 35.Moury, B., C. Morel, E. Johansen, L. Guilbaud, S. Souche, V. Ayme, C. Caranta, A. Palloix, and M. Jacquemond. 2004. Mutations in potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol. Plant-Microbe Interact. 17:322-329. [DOI] [PubMed] [Google Scholar]
- 36.Nicaise, V., S. German-Retana, R. Sanjuan, M. P. Dubrana, M. Mazier, B. Maisonneuve, T. Candresse, C. Caranta, and O. LeGall. 2003. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus lettuce mosaic virus. Plant Physiol. 132:1272-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestova, T. V., and C. U. Hellen. 2000. The structure and function of initiation factors in eukaryotic protein synthesis. Cell. Mol. Life Sci. 57:651-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puustinen, P., and K. M. Makinen. 2004. Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J. Biol. Chem. 79:38103-38111. [DOI] [PubMed] [Google Scholar]
- 39.Quadt, R., C. C. Kao, K. S. Browning, R. P. Hershberger, and P. Ahlquist. 1993. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 90:1498-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Restrepo, M. A., D. D. Freed, and J. C. Carrington. 1990. Nuclear transport of plant potyviral proteins. Plant Cell 2:987-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Restrepo-Hartwig, M. A., and J. C. Carrington. 1994. The tobacco etch potyvirus 6-kilodalton protein is membrane associated and involved in viral replication. J. Virol. 68:2388-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenwald, I. B., R. Kaspar, D. Rousseau, L. Gehrke, P. Leboulch, J. J. Chen, E. V. Schmidt, N. Sonenberg, and I. M. London. 1995. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J. Biol. Chem. 270:21176-21180. [DOI] [PubMed] [Google Scholar]
- 43.Rousseau, D., R. Kaspar, I. Rosenwald, L. Gehrke, and N. Sonenberg. 1996. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA 93:1065-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruffel, S., M. H. Dussault, A. Palloix, B. Moury, A. Bendahmane, C. Robaglia, and C. Caranta. 2002. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32:1067-1075. [DOI] [PubMed] [Google Scholar]
- 45.Ruffel, S., J. L. Gallois, M. L. Lesage, and C. Caranta. 2005. The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genomics 274:346-353. [DOI] [PubMed] [Google Scholar]
- 46.Schaad, M. C., R. J. Anderberg, and J. C. Carrington. 2000. Strain-specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology 273:300-306. [DOI] [PubMed] [Google Scholar]
- 47.Schaad, M. C., R. Haldeman-Cahill, S. Cronin, and J. C. Carrington. 1996. Analysis of the VPg-proteinase (NIa) encoded by tobacco etch potyvirus: effects of mutations on subcellular transport, proteolytic processing, and genome amplification. J. Virol. 70:7039-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strudwick, S., and K. L. Borden. 2002. The emerging roles of translation factor eIF4E in the nucleus. Differentiation 70:10-22. [DOI] [PubMed] [Google Scholar]
- 50.Taylor, D. N., and J. P. Carr. 2000. The GCD10 subunit of yeast eIF-3 binds the methyltransferase-like domain of the 126 and 183 kDa replicase proteins of tobacco mosaic virus in the yeast two-hybrid system. J. Gen. Virol. 81:1587-1591. [DOI] [PubMed] [Google Scholar]
- 51.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 52.Turner, K. A., T. L. Sit, A. S. Callaway, N. S. Allen, and S. A. Lommel. 2004. Red clover necrotic mosaic virus replication proteins accumulate at the endoplasmic reticulum. Virology 320:276-290. [DOI] [PubMed] [Google Scholar]
- 53.Urcuqui-Inchima, S., A.-L. Haenni, and F. Bernardi. 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74:157-175. [DOI] [PubMed] [Google Scholar]
- 54.Walter, M., C. Chaban, K. Schutze, O. Batistic, K. Weckermann, C. Nake, D. Blazevic, C. Grefen, K. Schumacher, C. Oecking, K. Harter, and J. Kudla. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40:428-438. [DOI] [PubMed] [Google Scholar]
- 55.Wang, D. W., and A. J. Maule. 1995. Inhibition of host gene-expression associated with plant-virus replication. Science 267:229-231. [DOI] [PubMed] [Google Scholar]
- 56.Weidman, M. K., R. Sharma, S. Raychaudhuri, P. Kundu, W. Tsai, and A. Dasgupta. 2003. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res. 95:75-85. [DOI] [PubMed] [Google Scholar]
- 57.Wittmann, S., H. Chatel, M. G. Fortin, and J. F. Laliberte. 1997. Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology 234:84-92. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, S. C., G. Zhang, L. Yang, J. Chisholm, and H. Sanfacon. 2005. Evidence that insertion of Tomato ringspot nepovirus NTB-VPg protein in endoplasmic reticulum membranes is directed by two domains: a C-terminal transmembrane helix and an N-terminal amphipathic helix. J. Virol. 79:11752-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]