Abstract
Human parechoviruses (HPeV), members of the Parechovirus genus of Picornaviridae, are frequent pathogens but have been comparatively poorly studied, and little is known of their diversity, evolution, and molecular biology. To increase the amount of information available, we have analyzed 7 HPeV strains isolated in California between 1973 and 1992. We found that, on the basis of VP1 sequences, these fall into two genetic groups, one of which has not been previously observed, bringing the number of known groups to five. While these correlate partly with the three known serotypes, two members of the HPeV2 serotype belong to different genetic groups. In view of the growing importance of molecular techniques in diagnosis, we suggest that genotype is an important criterion for identifying viruses, and we propose that the genetic groups we have defined should be termed human parechovirus types 1 to 5. Complete nucleotide sequence analysis of two of the Californian isolates, representing two types, confirmed the identification of a new genetic group and suggested a role for recombination in parechovirus evolution. It also allowed the identification of a putative HPeV1 cis-acting replication element, which is located in the VP0 coding region, as well as the refinement of previously predicted 5′ and 3′ untranslated region structures. Thus, the results have significantly improved our understanding of these common pathogens.
Human parechoviruses (HPeVs) are frequent pathogens (seroprevalence of over 90% has been reported), usually causing acute respiratory and gastrointestinal infections in young children (2, 7, 19, 45). These infections are normally mild, although more serious symptoms requiring hospitalization are sometimes observed (8, 19). The spectrum of symptoms is similar to that shown by enteroviruses, and HPeVs were first classified in the Enterovirus genus of the Picornaviridae, a family which currently consists of 9 genera containing around 300 virus serotypes (42). However, subsequent sequence analysis revealed that HPeVs are genetically distinct, and they are now classified in the Human parechovirus species of the Parechovirus genus, of which Ljungan virus, including viruses which infect rodents, is a second species (44, 46). Two HPeV serotypes (HPeV1 and HPeV2) have been known for several years, and a third (HPeV3), including strains currently circulating in Japan, North America, and Europe, has been recognized recently (1, 3, 16).
Five HPeV and 4 Ljungan virus complete nucleotide sequences are available and reveal that parechoviruses have a number of distinctive features (3, 9, 15, 16, 33). For instance, whereas the single-stranded positive-sense RNA genome of most picornaviruses is enclosed in a capsid made of 60 copies of 4 proteins VP1 to 4, parechoviruses have only 3 proteins because the cleavage of VP0 to VP4 and VP2 seen in other picornaviruses does not occur (43). In addition, in common with that of members of another picornavirus genus Kobuvirus, the 2A protein is of an unusual form, sharing motifs with a family of cellular proteins involved in control of cell proliferation (14). Studies on entry, translation, and replication have started to underscore the significance of molecular features revealed by sequence analysis, but the relative paucity of sequence information hinders a fuller understanding of HPeV molecular biology, evolution, and epidemiology (4, 20, 21, 22, 30, 31, 39). To address this issue, we have analyzed 7 HPeV strains isolated in California between 1973 and 1992. We found that, on the basis of VP1 sequences, these fall into two genetic groups, one of which has not been previously observed, bringing the number of known groups to five. In view of the growing importance of molecular techniques in diagnosis, we suggest that genotype is an important criterion for identifying viruses, and we propose that these groups should be termed human parechovirus types 1 to 5. Complete nucleotide sequence analysis of two of the Californian isolates, representing two types, confirmed the identification of a new genetic group and suggested for the first time the occurrence of recombination in parechovirus evolution. It also allowed the identification of a putative HPeV1 CRE (cis-acting replication element) as well as the refinement of previously predicted RNA structures.
MATERIALS AND METHODS
Virus strains and sequences.
Virus strains used were isolated in California between 1973 and 1992 (41). The strains, dates of isolation, and accession numbers for sequence data obtained are as follows: T75-4077, 1975, genome sequence accession no. AM235750; T92-15, 1992, genome sequence accession no. AM235749; T83-2051, 1983, VP1 sequence accession no. AM234724; T82-659, 1981, VP1 sequence accession no. AM234726; T82-203, 1981, VP1 sequence accession no. AM234727; T73-838, 1973, VP1 sequence accession no. AM234725; T82-0169, 1982, VP1 sequence accession no. AM234728. Accession numbers of other sequences used are as follows: complete sequences, HPeV1 Harris, S45208; HPeV2 Williamson, AJ005695; HPeV3 A308/99, AB084913; HPeV2 CT86-6760, AF055846; HPeV3 Can82853-01, AJ889918; P1 sequences, A1086-99, AB112485; A10987-00, AB112487; A942-99, AB112486; A354-99, AB112483; A317-99, AB112482; A628-99, AB112484; partial VP1 sequences (isolates having 6 digit numbers shown in Fig. 1a), DQ172416 to DQ172451 (2).
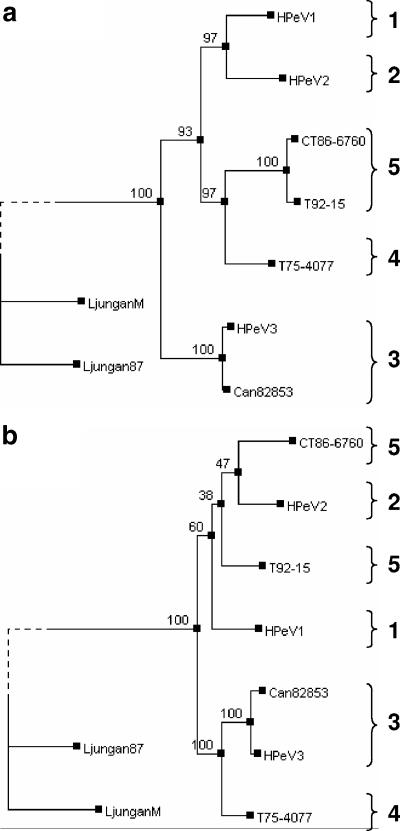
FIG. 1.
(a) Phylogenetic tree, based on the partial amino acid sequence of VP1, showing the relationship between the 54 available HPeV sequences for this region. Sequences were aligned with CLUSTALW, and the tree was constructed by the PHYML program, using the JTT model of amino acid substitution. The corresponding VP1sequences of two more distantly related parechovirues, Ljungan virus M1146 and 87-012, are included as out-groups, and the branches to the Ljungan virus nodes have been truncated for space reasons, as indicated by the dotted lines. Bootstrap values from 100 pseudoreplicates are shown for the major nodes. The HPeVs cluster into 5 genoypes, labeled types 1 to 5. (b) Alignment of the C-terminal region of VP1 showing the RGD motif, seen in 4 of the types, and well-conserved flanking residues in boldface type.
Oligonucleotides.
Oligonucleotides were designed to amplify cDNA from all HPeVs by aligning all of the known complete HPeV sequences and identifying conserved regions. The oligonucleotides used were as follows (positions of binding sites relative to the HPeV1 Harris sequence are indicated in parentheses): OL1053, 5′-GACGTTTGAAAGGGGTCTCCTAGA-3′ (1 to 20); OL1019, 5′-ACCTTGGCTTTTGGCCCCAG-3′ (455 to 437); OL1018, 5′-CTTATGCCCACACAGCCATC-3′ (306 to 325); OL990, 5′-CAAATTCATCAANACATGNGG-3′ (1354 to 1334); OL991, 5′-GTGGCTTTCATTTYCARGTNCA-3′(1203 to 1224); OL992, 5′-CAAGTGGGCCATCRTCYTGNGC-3′(2465 to 2445); OL993, 5′-TCATGGGGTTCNCARATGGA-3′ (2339 to 2358); OL994, 5′-CTTGGCAATGGTYTCRCARTT-3′ (3302 to 3282); OL1040, 5′-TGGAGATAATGTGTACCAATTG-3′(3115 to 3136); OL1041, 5′-CAAGTCATCTATAAGGTGGATGTCT-3′(4408 to 4384); OL995, 5′-CGGATGACATTTTYAARACNGC-3′(3141 to 3162); OL996, 5′-CTTGTTTGTGGTGGCNACNAC-3′ (4552 to 4532); OL997, 5′-GCTAGTGAATTYATGGAYGGNTA-3′(4358 to 4380); OL1004, 5′-TTCCCAGCAATRTGCATNCC-3′ (5785 to 5766); OL1046, 5′-ATTAAAACACGGGAGGGAACTGAG-3′(5648 to 5671); OL1005, 5′-CAAAGGAATGTGYGGNGGNCT-3′ (5707 to 5727); OL1057, 5′-GCTTAAATGTGCTGAAATTYTTCATCCACAT-3′(7085 to 7056); OL1146, 5′-CAGCTGGGTATTCATTTGTC-3′ (6181 to 6200); OL1002, 5′-GAGAGAACGCGT16[poly(A)].
Cell and virus propagation.
Monolayers of A549 (lung carcinoma) cells were maintained in minimal essential medium containing 10% heat-inactivated fetal calf serum, 1% nonessential amino acids (all from Invitrogen), and 100 μg/ml gentamicin (Sigma) in 25-cm2 tissue culture flasks (Nunc). HPeV isolates were adsorbed to the cells for 1 h at room temperature in 1 ml of growth medium, then 4 ml of medium was added, and the cells were incubated at 37°C for 48 h. Virus was liberated by freeze-thawing, and the supernatant was used directly for RNA isolation.
RNA isolation and reverse transcription (RT)-PCR.
To isolate viral RNA from cell cultures, a NucleoSpin RNA II kit (Macherey Nagel), allowing purification of up to 20 μg of highly pure total RNA, was used according to the manufacturer's instructions. Briefly, 100 μl of virus-containing cell culture supernatant and 350 μl of RA1 lysis buffer were mixed vigorously to lyse residual cells. The lysate was mixed once again with 350 μl of ethanol (70%) to adjust RNA binding conditions. The suspension was transferred to a NucleoSpin RNA II column and centrifuged (11,000 rpm, 30 s). The supernatant was passed through a silica membrane, then desalted by addition of 350 μl membrane desalting buffer, and centrifuged (14,000 rpm, 1 min) to dry the membrane. Genomic DNA was eliminated by applying 95 μl of DNase reaction mixture directly on the silica membrane. After a 15-min incubation at room temperature, the column was washed with 200 μl of buffer RA2 to inactivate DNase and centrifuged (11,000 rpm, 30 s). Then the column was washed with 600 μl and 250 μl of buffer RA3, respectively, to remove salts and other cellular components. The membrane was then completely dried by centrifugation for 2 min at 14,000 rpm. Total RNA was finally eluted with 40 μl RNase-free water and centrifuged at 14,000 rpm for 1 min.
Extracted RNA and two oligonucleotides complementary to the opposite ends of the region of interest were subjected to RT-PCR using a SuperScript one-step RT-PCR kit (Invitrogen) according to the manufacturer's instructions. For the RT-PCR, 5 μl of template RNA, 50 to 100 pmol of each primer, 1 μl of RT-Taq mix, and 25 μl of 2× reaction mix were mixed together in a total volume of 50 μl. Thermal cycling was carried out automatically using a thermal cycler (Perkin Elmer 2400 or Appligene Crocodile III). The specific amplicon was isolated by agarose gel electrophoresis and cloned into the pGEM-T Easy vector (Promega). The DNA was sequenced from both ends, and the sequence was completed using primers specific for the newly generated sequences. The sequences obtained were then used to design a second set of primers for RT-PCR. As these matched the viral RNA sequences exactly, they gave improved PCR products which could be sequenced directly. All of the sequences were determined directly in both orientations from these products.
Computer analysis.
Sequences were aligned using ClustalW and phylogenetic trees produced using the maximum likelihood approach implemented in the program PHYML (11, 48). Potential recombination was studied with SimPlot version 3.5.1, using the Kimura 2 parameter (25), and RNA folding was predicted using MFOLD (28, 51).
RESULTS
VP1 sequences.
The 7 HPeV strains analyzed in this study were isolated in California between 1973 and 1992 (41). Using RT-PCR, we amplified then sequenced cDNA encoding most (223/234 codons) of the VP1 region from the isolates. This region was chosen as it has been used extensively in studies on the epidemiology and evolution of another picornavirus genus, Enterovirus. We then used a 207-amino-acid section of the VP1 sequence, as this is present in the majority of partial parechovirus sequences in the database. Figure 1a shows a phylogenetic tree expressing the relationship between 54 available sequences. It can be seen that the HPeVs fall into five clear groups, and these partly correlate with the three established HPeV serotypes, the prototype strains of which are indicated HPeV1 to 3 in the tree. The recent isolates from Japan, North America, and Europe (1-3, 16) cluster tightly within two groups, either being loosely related to the HPeV1 prototype or closely related to the HPeV3 prototype. A surprising feature is that two isolates previously serotyped as HPeV2, the HPeV2 prototype Williamson and CT86-6760, fall into different genetic groups. The Californian strains are distributed among two genetic groups, one exemplified by CT86-6760 (isolates T82-0169, T83-2051, T92-15, and T82-659), and the second (isolates T82-203, T75-4077, and T-838) constituting a novel genetic group. The amino acid sequence identity between members of any of these groups does not exceed 75% in the region analyzed. This level of divergence is comparable to that which separates distinct types in enteroviruses (34, 35), and so we propose that the genetic groups are termed HPeV types 1 to 5. The numbering is based on the inclusion of a recognized prototype within the type, with the definition of two additional types: type 4, which includes the Californian isolates T82-203, T75-4077, and T-838, and type 5, which includes viruses related to CT86-6760.
All of the Californian isolates contain a C-terminal proximal RGD motif, shown to be essential for infectivity in HPeV1, as do all the HPeVs where this region has been sequenced, except those from HPeV type 3 (Fig. 1b). The HPeVs conform to the previously observed RGDM/L context seen in most other picornaviruses with a functional RGD motif (5, 6, 9), and the downstream amino acid, usually A, is also strongly conserved. There is weaker conservation of the +4 position than in other picornaviruses, where RGDXXXL is frequently seen.
Sequences of T75-4077 and T92-15.
The virtually complete sequences of T75-4077 (type 4) and T92-15 (type 5), representatives of each of the two genetic groups including the Californian isolates, were determined by RT-PCR and sequencing of overlapping cDNA fragments of the genome. The first 20 nucleotides of T75-4077 were not determined, as this region was used for priming the PCR. In common with previous analyses of HPeVs, it proved difficult to amplify the first few nucleotides of T92-15, and a downstream primer was used, meaning that the unique sequence starts at position 46. Both T75-4077 and T92-15 possess previously identified HPeV-specific sequence features, including the N-terminal extension to VP3, relative to any other picornavirus, which contains several basic amino acids and the conservation of 2A motifs previously shown to be shared with a family of cellular proteins involved in the control of cell proliferation (14, 43) (data not shown).
A phylogenetic tree showing the relationship between the P1 amino acid sequences of completely sequenced HPeVs confirms the existence of the same 5 types defined by the VP1 analysis (Fig. 2a). In contrast, when the 3Dregion nucleotide sequences were analyzed, the isolates grouped differently (Fig. 2b). Here, T75-4077 clusters with the two type 3 viruses, which were distinct from other HPeVs in the P1 region. T92-15 is more diverse from its fellow type 5 virus CT86-6760 than in P1. Apart from the HPeV3 prototype and Can82853, which are 96% identical, there is no clear grouping in the 5′ untranslated region (UTR) (data not shown).
FIG. 2.
Genetic relationships between completely sequenced HPeVs. The designation of each of the types, as defined in Fig. 1a, is indicated by the number (1 to 5) on the right hand side of each tree. (a) Tree based on amino acid sequences of the P1 region. Sequences were aligned with CLUSTALW, and the tree was constructed by the PHYML program, using the JTT model of amino acid substitution. The corresponding P1 sequences of two more distantly related parechoviruses, Ljungan virus M1146 (Ljungan M) and 87-012 (Ljungan 87), are included as out-groups, and the branches to the Ljungan virus nodes have been truncated for space reasons, as indicated by the dotted lines. Bootstrap values from 100 pseudoreplicates are shown for each node. (b) Tree based on nucleotide sequences of the 3D region. Sequences were aligned with CLUSTALW, and the tree was constructed by the PHYML program, using the HKY model of nucleotide substitution and a transition/transversion ratio of 4. The corresponding 3D sequences of two more distantly related parechoviruses, Ljungan virus M1146 (Ljungan M) and 87-012 (Ljungan 87), are included as outgroups, and the branches to the Ljungan virus nodes have been truncated for space reasons, as indicated by the dotted lines. Bootstrap values from 100 pseudoreplicates are shown for each node.
SimPlot analysis.
To further investigate the noncongruence of these trees, we performed SimPlot analysis on the seven complete genomic RNA sequences available (Fig. 3). As expected from the P1 tree, T92-15 shows a very high degree of nucleotide similarity to CT86-6760 across the P1 region, but 3′ of position 4500, CT86-6760 ceases to be the closest relative. There is also a loss of linkage to CT86-6760-like sequences in the 5′ UTR. The comparison of T92-15 with HPeV2 Williamson is particularly interesting. In the first part of the nonstructural (P2/3) region, these viruses are relatively dissimilar, but between 5250 and 6500, HPeV2 Williamson becomes the closest relative of T92-15. After position 6500, the closest relative of T92-15 is the HPeV1 prototype.
FIG. 3.
SimPlot analysis of the relationship between completely sequenced HPeVs, based on nucleotide similarities of other HPeVs to T92-15 (top panel) and T75-4077 (lower panel). The Kimura 2 correction was applied, with a Ts/Tv ratio of 3, and a window of 600 nucleotides and a step of 10 nucleotides were used.
Although T75-4077 is less similar to the HPeV3 strains than to any of the other viruses in P1, in accord with the 3D tree, there is a surprising degree of similarity in the P2/3-encoding region, particularly after position 5500 (Fig. 3). Direct sequence comparisons indicates that the second half of the 5′ UTR is also more similar to HPeV3 (93% identity) than to any of the other viruses (88 to 89%) (data not shown). The noncongruence of P1 and P2/P3 is suggestive of past recombination events, as demonstrated extensively in enterovirus evolution but not studied before in parechoviruses (23, 26, 36, 40).
Identification of a putative HPeV CRE.
The CRE has been shown to be critical for RNA replication in several other picornaviruses and to consist of an RNA stem-loop, generally but not exclusively located in the coding region (10, 24, 27, 38, 50). Alignment of the seven complete and six partial HPeV sequences revealed a section of the VP0-encoding gene with a high degree of nucleotide sequence identity (Fig. 4a). This sequence is not constrained by the occurrence of methionine or tryptophan codons or by a preponderance of twofold degenerate codons (data not shown). MFOLD analysis of the region shows that it can be predicted to fold into a stem-loop (Fig. 4b). Covariance in this stem provides evidence that the structure is significant. Several of the HPeV sequences differ from those of HPeV1 Harris at position 1392 and its predicted partner 1423, and there is also variation at the base pair predicted between positions 1395 and 1420, in each case, the stem structure being maintained by the pairs of differences. Within the predicted loop there is an AAA sequence motif common to other picornavirus CREs, and alignment of representative CREs with that predicted in HPeVs shows strong conservation of the larger motif CAAAC (Fig. 4c). The other Parechovirus species Ljungan virus (18) does not have a putative CRE at the corresponding location, but a similar analysis (data not shown) reveals a potential structure in the VPg-encoding region. Again a CAAAC motif is seen in the loop structure (Fig. 4c). Only one covariant position is seen, but the limited number of Ljungan virus sequences available probably reduces the possibility of observing covariance.
FIG. 4.
(a) Alignment of the putative CRE region of HPeV strains. < and > denote nucleotides participating in the predicted stem, and covariant residues differing from the HPeV1 sequence are shaded. U/C differences maintaining the structure are underlined. (b) MFOLD-derived structure of the HPeV1 CRE. (c) Alignment of the loop regions of CREs in picornaviruses, showing the strong conservation of the motif CAAAC.
5′ UTR structure.
A secondary structure for the HPeV1 5′ UTR has been proposed, based on comparisons between 2 HPeV sequences and similarities with more distantly related picornaviruses (9). Use of the two sequences presented here, together with the other available HPeV sequences, allows this structure to be tested. In general, covariance between the structures provided support for the structural features originally identified, stem-loop (SL) domains A, B, and D to L and a tertiary structure domain C, although in some cases, there is little sequence variation. An example is SL-F, where very extensive variation is seen but virtually all differences are covariant, and a common structure is seen in all HPeVs (data not shown). In contrast, SL-G shows differences in only one HPeV, although these are covariant.
3′ UTR structure.
The 3′ UTRs of other picornaviruses contain secondary and, in the case of enteroviruses, tertiary structure elements important in virus replication (29). Analysis of the HPeV sequences shows that in each case much of the 3′ UTR is made up of two highly conserved repeats of the sequence AUUAGAACACUAAUUUG arranged in tandem (Fig. 5). The 3′ UTR structure of HPeVs was analyzed using MFOLD, and in accord with a recent report (1), it is predicted to be composed of a single stem-loop with extensive base pairing between the poly(A) tail and the highly U-rich 5′ part of the 3′ UTR (predicted base paired regions shown in Fig. 5). The low degree of variability between the sequences means that there is little covariance support for the structure, but differences are largely concentrated in predicted loops, which is consistent with this being the structure of the region.
FIG. 5.
Alignment of the 3′ UTRs of HPeVs showing the tandem repeats (shaded). < and > denote nucleotides participating in the predicted stem-loop structure.
DISCUSSION
HPeVs are being increasingly recognized as significant human pathogens but have received relatively little attention in molecular terms compared to other human pathogens of the Enterovirus, Rhinovirus, and Hepatovirus genera of the Picornaviridae. We have shown here, on the basis of sequence relationships in VP1, that 7 HPeV strains isolated in California between 1973 and 1992 fall into two genetic groups (Fig. 1a). One of these has not been described before, and the other contains viruses related to a previously sequenced isolate, CT86-6760, which has been serotyped as HPeV2. Interestingly, this latter group is substantially different from the HPeV2 prototype Williamson in the P1 region. Our analysis therefore takes the number of HPeV genotypes to 5.
In enteroviruses, there is a good correlation between VP1 sequence and serotype, and this has led to several new enteroviruses being defined entirely on the basis of sequence identity (32, 34, 37). These are called types, rather than serotypes, as neutralization data are now frequently not obtained. The relationship between HPeV genotype and serotype appears to be less clear than in enteroviruses, since HPeV2 Williamson and HPeV2 CT86-6760 represent two different genotypes (Fig. 1a). Thus, we are not suggesting that the new HPeV genotype necessarily represents a new serotype, although it should be noted that it has been shown that viruses belonging to this genotype are not neutralized by antisera against HPeV1 and HPeV2 reference strains (41). As virus detection is now mainly being performed using PCR, molecular features become increasingly important in defining differences between viruses. We therefore propose that, by analogy with enteroviruses, the 5 genetic groups we have defined should be considered HPeV types. Each of these types contains viruses which cluster relatively tightly, although the recent type 1 viruses are relatively different from the HPeV1 prototype. At around 90% amino acid identity in the region of VP1 used, these type 1 isolates would be on the margin of defining separate types in enteroviruses, and there seems to be no compelling case to separate them into different HPeV types. The analysis of more type 1 HPeVs, isolated during the long period (nearly 50 years) between the prototype and the recent isolates, may help to resolve this issue.
Our results show that there were at least two HPeV types circulating in California between 1973 and 1992. There is no clear-cut correlation between date of isolation and type. The type 4 viruses were isolated in the 1970s and 1980s and the type 5 viruses in the 1980s and 1990s, but more isolates would need to be analyzed to clarify whether they were cocirculating or whether type 4 was superseded by type 5. Currently, the situation from Japanese, Canadian, and Dutch studies seems to be that HPeV types 1 and 3 are the predominant viruses, with no recent isolations of the other types among more than 50 viruses studied (1-3, 16).
In the P1 region, type 3 appears to be relatively diverse from the other types (Fig. 2a), and it is interesting that these viruses lack the RGD motif seen in VP1 in the other viruses compared here (Fig. 1b). This has been shown to be essential for replication of the HPeV type 1 prototype Harris, presumably being required for interaction with αvβ1 and αvβ3, the RGD-dependent ligands reported to be its receptors (4). In another picornavirus, coxsackievirus A9 (CAV9), an RGD-less mutant is viable and grows efficiently in some cells but shows altered pathogenicity in a mouse model (12, 13). There is some evidence that HPeV type 1 and 3 infections differ in age profile and symptoms, and receptor tropism could be a factor in this (2). It would be interesting to determine whether HPeV type 3 recognizes these molecules by an RGD-independent interaction, or whether another, unknown receptor is used. In all of the HPeV isolates studied here, the RGD motif is followed by the amino acid M or L (Fig. 1b), also well conserved in the other picornaviruses with VP1 RGD motifs (foot-and-mouth disease virus, CAV9, and echovirus 9) and presumably involved in RGD function (5, 6). Integrin αvβ6 appears to play a role in infection of both foot-and-mouth disease virus and CAV9, and the similarity in RGD context could imply that this integrin is also recognized by HPeV1 (17, 49). Interestingly, there is considerable conservation of the RGD region between the viruses. For instance, the final 11 VP1 amino acids of the HPeV type 1 prototype Harris and the type 5 viruses T92-15 and T82-659 are identical. This conservation is consistent with the observation that the RGD region is not a major determinant of antigenicity (21).
The diverse nature of HPeV type 3 in the P1 region makes surprising the observation that both fully sequenced type 3 viruses are comparatively closely related to the type 4 virus T75-4077 in the nonstructural protein region (Fig. 2b). This raises the possibility that recombination plays a role in the evolution of these viruses. SimPlot analysis (Fig. 3) shows that T75-4077 is much more closely related to type 3 than to other HPeVs throughout the final 3,000 nucleotides of the genome, particularly the final 1,500, although this similarity is not great enough to prove that it is due to recombination (Fig. 3). It is also interesting that the other strain sequenced, T92-15, is highly related to CT86-6760 in the capsid region, but the degree of similarity falls off rapidly after position 4500, suggesting acquisition of 3′ sequences with a different evolutionary history by recombination, rather than gradual drift. These observations could suggest that frequent recombination in HPeVs leads to the generation of mosaic viruses which do not necessarily have a clear linkage between the P1 region and P2/3, but more complete HPeV sequences, which may include those with genomic regions with more clearly identifiable origins, are required to investigate this aspect of parechovirus evolution. As recombination has been well documented in another genus of important human picornaviruses, Enterovirus, it is not surprising that it should occur in parechoviruses, given that these are common pathogens and so there is potentially a high incidence of dual infections.
The availability of significantly more HPeV sequence information gives insights into the molecular biology of these viruses. In addition to the conservation of the RGD motif in most HPeVs, alignment of the sequences shows the presence of the N-terminal, basic extension to VP3 relative to other picornaviruses and conservations of the 2A motifs shared with cellular proteins involved in the control of cell proliferation (data not shown) (14, 43-45). The new data are also consistent with the previously predicted cleavage sites. The sequences are particularly useful in defining RNA structural features which are likely to play a role in RNA replication and/or translation. Mapping of nucleotide sequence differences suggests that the previously predicted 5′ UTR structure is essentially correct (9). The derived structure is similar to those present in picornaviruses with a type 2 internal ribosome entry site, and HPeVs share functional similarity with these viruses (30, 47). We have also presented a predicted structure for the HPeV 3′ UTR (Fig. 5). An interesting feature of the 3′ UTR is an apparent tandem duplication of 17 nucleotides, which is highly conserved in all of the HPeVs studied. The 3′ UTR is also highly U-rich following the stop codon, a feature seen in a number of other picornaviruses. Much of the 3′ UTR has the potential to fold into a single, imperfect stem-loop. The prediction of the 3′ UTR gives a framework for the analysis of the role of this region in HPeV replication, which will be useful, as the picornavirus 3′ UTR is still relatively ill understood.
One of the most important aspects of the structural predictions is the probable identification of the HPeV CRE, an element which plays a critical role in positive-sense RNA synthesis in other picornaviruses through VPg uridylylation (38). The element was detected through the identification of a region of HPeV with high nucleotide sequence identity (Fig. 4a and b). This contains an AAA motif and can be folded to give a simple stem-loop (Fig. 4b). There is strong phylogenetic support for this stem-loop, as two different pairs of covariant nucleotide substitutions maintain the structure among all the HPeVs analyzed. Although the predicted loop is larger than that seen in the CREs of other picornaviruses, this structural support and presence of the AAA strongly suggests that this is the parechovirus CRE. The putative CRE is located in VP0, which is also the location of the CRE seen in cardioviruses, but the two locations are not analogous, as the cardiovirus CRE is in the βA1 strand, as opposed to the βG strand for HPeVs. Interestingly, the predicted CRE in Ljungan virus is largely in the VPg-encoding region. The identification of a CRE in different positions in two parechovirus species is not surprising, as the two rhinovirus species have CREs in VP1 (Human rhinovirus B) and 2A (Human rhinovirus A) (50). An analysis of the loop region of all known picornavirus CREs shows little overall conservation, except that the AAA motif, which is the site of VPg uridylylation, is part of a strongly conserved CAAAC motif (Fig. 4c).
Human parechoviruses, members of the Parechovirus genus of the Picornaviridae, are frequent pathogens but have been comparatively poorly studied. The analysis we have presented here greatly extends the information available on parechoviruses and gives important insights into their molecular biology as well as significantly improving our understanding of their diversity and evolution.
Acknowledgments
M.A.-S. thanks the Saudi Arabian Ministry of Culture for the generous provision of a studentship. This work was supported by the Wellcome Trust, grant number 060055.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Abed, Y., and G. Boivin. 2005. Molecular characterization of a Canadian human parechovirus (HPeV)-3 isolate and its relationship to other HPeVs. J. Med. Virol. 77:566-570. [DOI] [PubMed] [Google Scholar]
- 2.Benschop, K. S. M., J. Schinkel, R. P. Minnaar, D. Pajkrt, L. Spanjerberg, H. C. Kraakman, B. Berkhout, H. L. Zaaijer, M. G. H. M. Beld, and K. C. Wolthers. 2006. Human parechovirus infections in Dutch children and the association between serotype and disease severity. Clin. Infect. Dis. 42:204-210. [DOI] [PubMed] [Google Scholar]
- 3.Boivin, G., Y. Abed, and F. D. Boucher. 2005. Human parechovirus 3 and neonatal infections. Emerg. Infect. Dis. 11:103-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boonyakiat, Y., P. J. Hughes, F. Ghazi, and G. Stanway. 2001. An arginine-glycine-aspartic acid motif is a critical determinant for human parechovirus 1 entry. J. Virol. 75:10000-10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, K. H., P. Auvinen, T. Hyypiä, and G. Stanway. 1989. The nucleotide sequence of coxsackievirus A9; implications for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269-3280. [DOI] [PubMed] [Google Scholar]
- 6.Chang, K. H., C. Day, J. Walker, T. Hyypiä, and G. Stanway. 1992. The nucleotide sequences of wild-type coxsackievirus A9 strains imply that an RGD motif in VP1 protein is functionally significant. J. Gen. Virol. 73:621-626. [DOI] [PubMed] [Google Scholar]
- 7.Ehrnst, A., and M. Eriksson. 1993. Epidemiological features of type 22 echovirus infection. Scand. J. Infect. Dis. 25:275-281. [DOI] [PubMed] [Google Scholar]
- 8.Figueroa, J. P., D. Ashley, D. King, and B. Hull. 1989. An outbreak of acute flaccid paralysis in Jamaica associated with echovirus type 22. J. Med. Virol. 29:315-319. [DOI] [PubMed] [Google Scholar]
- 9.Ghazi, F., P. J. Hughes, T. Hyypiä, and G. Stanway. 1998. Molecular analysis of human parechovirus type 2 (formerly echovirus 23). J. Gen. Virol. 79:2641-2650. [DOI] [PubMed] [Google Scholar]
- 10.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 12.Harvala, H., H. Kalimo, G. Stanway, and T. Hyypiä. 2003. The role of the viral arginine-glycine-aspartic acid motif in the mouse pathogenesis of coxsackievirus A9. J. Gen. Virol. 84:2375-2379. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, P. J., C. Horsnell, T. Hyypiä, and G. Stanway. 1995. The coxsackievirus A9 RGD motif is not essential for virus infectivity. J. Virol. 69:8035-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, P. J., and G. Stanway. 2000. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev-107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 80:201-207. [DOI] [PubMed] [Google Scholar]
- 15.Hyypiä, T., C. Horsnell, M. Maaronen, M. Khan, N. Kalkkinen, P. Auvinen, L. Kinnunen, and G. Stanway. 1992. A distinct picornavirus group identified by sequence analysis. Proc. Natl. Acad. Sci. USA 89:8847-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, M., T. Yamashita, H. Tsuzuki, N. Takeda, and K. Sakae. 2004. Isolation and identification of a novel human parechovirus. J. Gen. Virol. 85:391-398. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, T., D. Sheppard, M. Denyer, W. Blakemore, and A. M. Q. King. 2000. The epithelial integrin alpha v beta 6 is a receptor for foot-and-mouth disease virus. J. Virol. 74:4949-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson, S., B. Niklasson, J. Maizel, A. E. Gorbalenya, and A. M. Lindberg. 2002. Molecular analysis of three Ljungan virus isolates reveals a new, close-to-root lineage of the Picornaviridae with a cluster of two unrelated 2A proteins. J. Virol. 76:8920-8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joki-Korpela, P., and T. Hyypiä. 2001. Parechoviruses, a novel group of human picornaviruses. Ann. Med. 33:466-471. [DOI] [PubMed] [Google Scholar]
- 20.Joki-Korpela, P., V. Marjomäki, C. Krogerus, J. Heino, and T. Hyypiä. 2001. Entry of human parechovirus 1. J. Virol. 75:1958-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joki-Korpela, P., M. Roivainen, H. Lankinen, T. Pöyry, and T. Hyypiä. 2000. Antigenic properties of human parechovirus 1. J. Gen. Virol. 81:1709-1718. [DOI] [PubMed] [Google Scholar]
- 22.Krogerus, C., D. Egger, O. Samuilova, T. Hyypiä, and K. Bienz. 2003. Replication complex of human parechovirus 1. J. Virol. 77:8512-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindberg, A. M., P. Andersson, C. Savolainen, M. N. Mulders, and T. Hovi. 2003. Evolution of the genome of human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J. Gen. Virol. 84:1223-1235. [DOI] [PubMed] [Google Scholar]
- 24.Lobert, P. E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukashev, A. N., V. A. Lashkevich, O. E. Ivanova, G. A. Koroleva, A. E. Hinkkanen, and J. Ilonen. 2003. Recombination in circulating enteroviruses. J. Virol. 77:10423-10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason, P. W., S. V. Bezborodova, and T. M. Henry. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 76:9686-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 29.Mirmomeni, M., P. J. Hughes, and G. Stanway. 1997. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J. Virol. 71:2363-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nateri, A. S., P. J. Hughes, and G. Stanway. 2000. In vivo and in vitro identification of structural and sequence elements of the human parechovirus 5′ untranslated region required for internal initiation. J. Virol. 74:6269-6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nateri, A. S., P. J. Hughes, and G. Stanway. 2002. Terminal replication elements in human parechovirus 1. J. Virol. 76:13116-13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norder, H., L. Bjerregaard, L. Magnius, B. Lina, M. Aymard, and J. J. Chomel. 2003. Sequencing of ‘untypable’ enteroviruses reveals two new types, EV-77 and EV-78, within human enterovirus type B and substitutions in the BC loop of the VP1 protein for known types. J. Gen. Virol. 84:827-836. [DOI] [PubMed] [Google Scholar]
- 33.Oberste, M. S., K. Maher, and M. A. Pallansch. 1998. Complete sequence of echovirus 23 and its relationship to echovirus 22 and other human enteroviruses. Virus Res. 56:217-223. [DOI] [PubMed] [Google Scholar]
- 34.Oberste, M. S., K. Maher, D. R. Kilpatrick, M. R. Flemister, B. A. Brown, and M. A. Pallansch. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oberste, M. S., K. Maher, and M. A. Pallansch. 2004. Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J. Virol. 78:855-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberste, M. S., S. M. Michele, K. Maher, D. Schnurr, D. Cisterna, N. Junttila, M. Uddin, J.-J. Chomel, C-S Lau, W. Ridha, S. al-Busaidy, H. Norder, L. O. Magnius, and M. A. Pallansch. 2004. Molecular identification and characterization of two proposed new enterovirus serotypes, EV74 and EV75. J. Gen. Virol. 85:3205-3212. [DOI] [PubMed] [Google Scholar]
- 38.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samuilova, O., C. Krogerus, I. Fabrichniy, and T. Hyypiä. 2006. ATP hydrolysis and AMP kinase activities of nonstructural protein 2C of human parechovirus 1. J. Virol. 80:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santti, J., T. Hyypiä, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnurr, D., M. Dondero, D. Holland, and J. Connor. 1996. Characterization of echovirus 22 variants. Arch. Virol. 141:1749-1758. [DOI] [PubMed] [Google Scholar]
- 42.Stanway, G., F. Brown, P. Christian, T. Hovi, T. Hyypiä, A. M. Q. King, N. J. Knowles, S. M. Lemon, P. D. Minor, M. A. Pallansch, A. C. Palmenberg, and T. Skern. 2004. Picornaviridae, p. 757-778. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy, VIIIth report of the ICTV. Elsevier/Academic Press, London, United Kingdom.
- 43.Stanway, G., N. Kalkkinen, M. Roivainen, F. Ghazi, M. Khan, M. Smyth, O. Meurman, and T. Hyypiä. 1994. Molecular and biological characteristics of echovirus 22, a representative of a new picornavirus group. J. Virol. 68:8232-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanway, G., and T. Hyypiä. 1999. Parechoviruses. J. Virol. 73:5249-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanway, G., P. Joki-Korpela, and T. Hyypiä. 2000. Human parechoviruses-biological and clinical significance. Rev. Med. Virol. 10:57-69. [DOI] [PubMed] [Google Scholar]
- 46.Stanway, G., T. Hovi, N. J. Knowles, and T. Hyypiä. 2002. Biological and molecular basis of picornavirus classification, p. 17-24. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 47.Stewart, S. R., and B. L. Semler. 1997. RNA determinants of picornavirus cap-independent translation initiation. Semin. Virol. 8:242-255. [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams, Ç. H., T. Kajander, T. Hyypiä, T. Jackson, D. Sheppard, and G. Stanway. 2004. Integrin αvβ6 is an RGD-dependent receptor for coxsackievirus A9. J. Virol. 78:6967-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, Y., R. Rijnbrand, K. L. McKnight, E. Wimmer, A. Paul, A. Martin, and S. M. Lemon. 2002. Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 14. J. Virol. 76:7485-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]