Abstract
The retinoic acid-inducible gene I product (RIG-I) has been identified as a cellular sensor of RNA virus infection resulting in beta interferon (IFN-β) induction. However, many viruses are known to encode viral products that inhibit IFN-β production. In the case of influenza A virus, the viral nonstructural protein 1 (NS1) prevents the induction of the IFN-β promoter by inhibiting the activation of transcription factors, including IRF-3, involved in IFN-β transcriptional activation. The inhibitory properties of NS1 appear to be due at least in part to its binding to double-stranded RNA (dsRNA), resulting in the sequestration of this viral mediator of RIG-I activation. However, the precise effects of NS1 on the RIG-I-mediated induction of IFN-β have not been characterized. We now report that the NS1 of influenza A virus interacts with RIG-I and inhibits the RIG-I-mediated induction of IFN-β. This inhibition was apparent even when a mutant RIG-I that is constitutively activated (in the absence of dsRNA) was used to trigger IFN-β production. Coexpression of RIG-I, its downstream signaling partner, IPS-1, and NS1 resulted in increased levels of RIG-I and NS1 within an IPS-1-rich, solubilization-resistant fraction after cell lysis. These results suggest that RIG-I, IPS-1, and NS1 become part of the same complex. Consistent with this idea, NS1 was also found to inhibit IFN-β promoter activation by IPS-1 overexpression. Our results indicate that, in addition to sequestering dsRNA, the NS1 of influenza A virus binds to RIG-I and inhibits downstream activation of IRF-3, preventing the transcriptional induction of IFN-β.
The type I interferon (alpha and beta interferon [IFN-α/β]) response constitutes one of the first lines of defense against virus infections (14). Most cells respond to viral infection by the production and secretion of IFN-β. This cytokine induces the activation of innate antiviral genes through the JAK/STAT pathway and participates in the stimulation of downstream immune events resulting in the activation of specific cells involved in innate and adaptive antiviral immunity (6, 47). Critical elements in the induction of IFN-α/β are the cellular components involved in sensing viral infection through the recognition of viral products. These sensor molecules initiate the molecular events leading to the transcriptional induction of IFN-α/β. Initially, it was recognized that toll-like receptors (TLR) participate in the detection of products derived from many pathogens, including viruses (1). These transmembrane proteins sample the extracytoplasmic environment and initiate a signaling cascade from their cytoplasmic tails resulting in activation of the IFN-α/β promoters (24). Particularly important for the detection of RNA viruses are TLR3, TLR7, and TLR8, which recognize double-stranded RNA (dsRNA) and single-stranded RNA, mainly in endosomal compartments (2, 10, 18, 33). However, the lack of major deficiencies associated with IFN-α/β induction by viruses in mice and cell lines lacking critical components of the TLR pathway indicated the existence of additional cellular sensors responsible for triggering IFN-α/β production after viral infection (12, 31, 34).
Recently, the retinoic acid-induced gene I product (RIG-I) was identified as a cytoplasmic sensor of dsRNA and of RNA virus infection (48, 55). This protein contains a DExD/H box helicase domain at its carboxy terminus and, upon binding to dsRNA, appears to expose an amino-terminal caspase-recruiting domain (CARD). The RIG-I CARD motif mediates interaction with a second CARD-containing protein, named IPS-1, MAVS, VISA, or CARDIF, and an activation signal is then transmitted to the kinases TBK1 (TANK-binding kinase 1) and IKKɛ (IκB kinase ɛ) (25, 28, 37, 44, 54). The activation of these kinases results in phosphorylation and activation of IRF-3 and IRF-7, two related transcription factors that are involved in activation of IFN-α/β expression (19). The CARD-containing DExD/H box helicase MDA-5, a protein highly related to RIG-I, also appears to be able to sense dsRNA in a similar fashion, resulting in IFN-α/β induction (3). The importance of RIG-I and MDA-5 in mediating IFN-α/β expression in response to RNA virus infections and in inducing antiviral responses has been recently demonstrated with murine knockout cells and animals (23). In particular, RIG-I appears to be essential for the production of IFN-α/β by several RNA viruses, including paramyxoviruses, flaviviruses, and influenza viruses (23).
In order to overcome the antiviral response induced by IFN-α/β, most viruses have evolved viral products that antagonize this response at multiple levels (for a recent review, see reference 17). In the case of influenza A virus, a segmented negative-strand RNA virus, the viral nonstructural protein 1 (NS1) has been shown to be essential for the inhibition of the IFN-α/β response to levels that allow efficient viral replication in vivo (15). The NS1 protein antagonizes both the induction of IFN-β (49) and the activity of PKR and OAS, IFN-induced proteins with antiviral activities (5, 27, 38). It has been shown that NS1 inhibits IFN-β expression by preventing the activation of IRF-3 and IRF-7 transcription factors during viral infection (45, 49). This might be attributed to the ability of NS1 to bind to dsRNA and to possibly sequester this molecule from recognition by cellular sensors (13, 51). However, we recently found that an NS1 mutant protein unable to bind to dsRNA still partly retains its IFN-inhibitory activity, raising the possibility that the NS1 protein counteracts the IFN response by a dsRNA-binding independent mechanism (11). In this report, we describe that the NS1 interacts with the RNA helicase RIG-I. We also show that RIG-I-mediated induction of IFN-β is prevented by expression of NS1 in the absence of dsRNA. Thus, targeting of the RIG-I pathway by NS1 results in inhibition of the host IFN-mediated antiviral response, and this is likely to have an important role in the virulence and pathogenesis of influenza A viruses.
MATERIALS AND METHODS
Cells and viruses.
293T and Vero cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Stocks of Newcastle disease virus (NDV) expressing green fluorescent protein (GFP) (41) or monomeric red fluorescence protein (mRFP), Sendai virus (SeV) Cantell, and influenza A/PR/8/34 (PR8) virus were grown in 10-day-old embryonated chicken eggs as previously described. delNS1, a recombinant PR8 virus lacking the NS1 gene, was grown in 7-day-old embryonated eggs (50).
rNDV-mRFP.
The monomeric red fluorescence protein has been described previously (9). Roger Tsien kindly provided the pRSETb-mRFP vector containing the mRFP open reading frame (ORF). To clone the mRFP gene into the full-length NDV cDNA, the mRFP gene was amplified with the following primers: mRFP/NDV/5′ (5′-CGCGTCTAGATTAGAAAAAATACGGGTAGAACCGCCACCATGGCCTCCTCCGAGGACGTCATC-3′), containing an XbaI site (underlined), NDV gene end (bold), NDV intergenic region (T), NDV gene start (italic), and Kozak sequence (CCGCCACC) before the mRFP start codon; and mRFP/NDV/3′ (5′-CGCGTCTAGATTAGGCGCCGGTGGAGTGGCGGCC), containing an XbaI site (underlined). The PCR product was digested with XbaI and subcloned into the same site of pNDV-F3A (42). Recombinant NDV expressing the mRFP (rNDV-mRFP) was rescued as described in reference 39. Virus expression of mRFP was confirmed after infection of chicken embryo fibroblast cells and the detection of mRFP expression by fluorescence microscopy twenty-four hours postinfection (data not shown).
Plasmids.
The pEGFP-IRF-3 and pCAGGS-NS1 plasmids have already been described (4). pCAGGS-Flag-RIG-I expression plasmids were obtained by cloning RIG-I-containing sequences into pCAGGS-Flag. The RIG-I cDNA was first obtained from RNA recovered from IFN-treated A549 cells and then cloned into pEF-Tak (a pEF-derived vector). DNA coding sequences corresponding to wild-type RIG-I, amino-terminal mutant RIG-IN, and carboxy-terminal mutant RIG-IC were amplified from pEF-Tak-RIG-I using primer sets 5RigNot (5′-ATAAGAATGCGGCCGCGACCACCGAGCAGCGACGCAGC-3′) and 3RigNhe (5′-CTAGCTAGCTTATCATTTGGACATTTCTGCTGG-3′), 5RigNot and 3′NheIRIG-I (5′-CTAGCTAGCTAGTTATCATTTTTTAAGATGATGTTCAC-3′), and 5RigCNot (5′-GATAAGGCGGCCGCGGAATGCCAGAATCTTAGTGAG-3′) and 3RigNhe. PCR fragments were digested with NotI and NheI and cloned into NotI- and NheI-digested pCAGGS-Flag. IPS-1 was amplified by PCR from the human cDNA clone pCMV-SPO T6 (catalog no. 5751684; ATCC) and subcloned into pCAGGS-HA-NH2 by using SacI and SmaI. pCAGGS-HA-NH2 is a derivative of pCAGGS-MCS (40) that contains the influenza virus hemagglutinin (HA)-derived epitope YPYDVPDYA and expresses NH2-terminal HA-tagged proteins. pCAGGS-NS1-GFP, encoding a NS1-GFP fusion, was generated by cloning the influenza PR8 virus NS1 ORF into the EcoRI and SmaI restriction sites of the pCAGGS-GFP expression vector. The pCAGGS-GFP plasmid is a pCAGGS-MCS variant containing two multiple-cloning sites that flank the GFP coding sequence and thus expresses amino- and/or carboxy-terminal GFP fusion proteins. Reporter plasmids pIFNβ-GFP/CAT and pHISG54-GFP/CAT each encode a chimeric GFP-chloramphenicol acetyltransferase (CAT) protein under the control of IFN-β and interferon-stimulated gene-responsive element (ISRE) promoters, respectively, and they were made by cloning the GFP ORF in frame with the CAT gene present in pIFNβ-CAT (52) and pHISG54-CAT (7) by using appropriate restriction endonucleases.
Bioassays.
To measure the amount of interferon produced by 293T cells following transfection with the indicated plasmids and/or by infection with the indicated viruses, bioassays were performed as described elsewhere (41) with the following modifications. Virus present in the culture media collected from infected 293T cells were inactivated by exposure to UV light for 10 min. After inactivation, Vero cells seeded in 96-well plates were treated with the 293T-cell media overnight. The cells were then infected with rNDV-mRFP at a multiplicity of infection (MOI) of 1. At 24 h postinfection, red fluorescence was read in a FLUOstar OPTIMA plate reader. To determine whether this new bioassay was based on IFN-β production, culture media of 293T cells infected with SeV were incubated in the presence or absence of a sheep polyclonal anti-IFN-β antibody (PBL Biochemical Laboratories, NJ) prior to addition to Vero cells. As controls, cells were pretreated with the indicated international units of IFN-β (catalog no. 407297; Calbiochem).
Indirect immunofluorescence assays.
293T cells that were transfected with the indicated plasmids and/or mock infected or infected with SeV were fixed with 2.5% paraformaldehyde and permeabilized with 0.5% NP-40. To detect SeV infection, cells were treated with 1 μg/ml of monoclonal antibodies 11F3 and 5F5 (Mount Sinai Hybridoma Center) and then with secondary fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin antibody (catalog no. F 0261; Dakocytomation).
Western blotting.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto nitrocellulose filters. After a blocking with 10% milk, the filter was incubated with monoclonal anti-Flag M2 antibody (Sigma) and monoclonal or polyclonal anti-NS1 antibodies that were kindly provided by Jonathan Yewdell. After being washed with phosphate-buffered saline (PBS) containing 0.5% Tween-20, the filters were incubated with the secondary horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin antibody (Amersham Biosciences). The horseradish peroxidase immunocomplexes were detected using an enhanced chemiluminescence kit from PerkinElmer (catalog no. NEL101). NS1-GFP protein expression was detected in Western blot analyses with an anti-GFP monoclonal antibody (catalog no. 632380; Clontech).
Reversible cross-linking immunoprecipitation.
Transfected and/or infected cells were washed, collected, and resuspended in PBS. Dithio-bis(succynimidyl propionate) was added into the suspension at a final concentration of 1 mM, and this was incubated on ice for 30 min. The cross-linking reaction was stopped by the addition of 100 mM glycine and further incubation on ice for 15 min. Cells were collected by centrifugation and lysed in radioimmunoprecipitation assay (RIPA) buffer (Tris-HCl [pH 7.4], 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA). The lysates were incubated with anti-Flag agarose (Sigma) at 4°C for several hours, and the beads were washed three times with RIPA buffer. The beads were resuspended in PBS containing 150 ng/ml 3 × Flag peptide (Sigma) and incubated at 4°C for 30 min. After centrifugation, the immunoprecipitated protein complexes were eluted from the beads and subjected to SDS-PAGE and Western blotting.
CAT and luciferase assays.
Cells were lysed in reporter lysis buffer (Promega) and cleared by centrifugation. For CAT assays, 50 μl of lysate was incubated in 250 mM Tris-HCl (pH 7.5), 875 μg/ml acetyl coenzyme A, and 10 nmol/ml 14C-chloramphenicol at 37°C for 2 h. After incubation, the reaction mixture was extracted with ethylacetate. The extract was evaporated under a spin vacuum and redissolved with 30 μl of ethylacetate. The samples were spotted on a silica gel thin-layer chromatography plate and developed in chloroform-methanol (95:5). Acetylated and nonacetylated 14C-chloramphenicol were detected by autoradiography and quantified in a Storm phosphorimager (Amersham Biosciences). For quantification, lysates were serially diluted 10-fold until the percentage of acetylated 14C-chloramphenicol was within the linear range. For luciferase assays, 10 μl of lysate from each sample was examined in the luciferase assay system used (Promega) by following the manufacturer's instructions. Luciferase activity was measured by a Lumat luminometer.
RNA interference.
The target sequences for small interfering RNAs (siRNA) are 5′-CCACAGATTCTTGTGAACAACCTTA-3′ (RIG-I) and 5′-CCATTAGGTTCAAGTCAACCACTTA-3′ (control). siRNAs were synthesized as Stealth_1199 (RIG-I specific) and Stealth_Control_1199 (Invitrogen), respectively. 293T cells were transfected with siRNA (40 fmol) using 3 μl of Lipofectamine 2000 (Invitrogen) and incubated at 37°C for 36 h.
RESULTS
The NS1 protein of influenza A virus interacts with RIG-I.
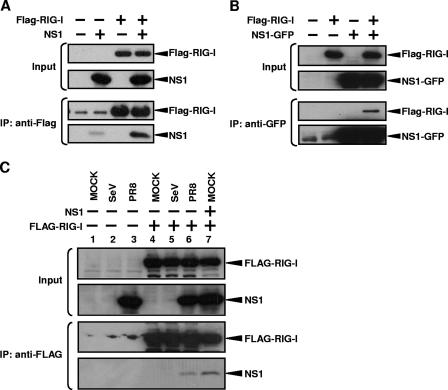
RIG-I has been described as a critical intracellular sensor molecule that, upon recognition of dsRNA and of RNA virus infection, results in transcriptional activation of IFN-β (55). Because the NS1 of influenza virus is known to inhibit transcriptional activation of IFN-β during influenza and Sendai virus infection or after dsRNA transfection (32, 49, 52), we sought to investigate whether RIG-I and NS1 interact. 293T cells were transfected with NS1 and Flag-tagged RIG-I expression plasmids, and cell lysates were prepared. The cell lysates were subjected to reversible cross-linking immunoprecipitation assays using anti-Flag antibody. Figure 1A shows that NS1 is coprecipitated with Flag-RIG-I. Similar results were obtained without cross-linking, but the use of a cross-linker before immunoprecipitation resulted in a slightly stronger NS1 signal (data not shown). In a second experiment, we tagged NS1 with GFP and performed an immunoprecipitation assay using GFP antibodies in 293T cells coexpressing NS1-GFP and Flag-tagged RIG-I. Figure 1B shows that RIG-I comes down when NS1-GFP is expressed. To confirm the interaction of NS1 with RIG-I in influenza virus-infected cells, 293T cells were transfected with Flag-RIG-I plasmid and then infected with influenza PR8 virus or SeV. NS1 was coprecipitated with Flag-RIG-I not only when it was expressed by plasmid transfection but also when expressed by influenza virus infection (Fig. 1C). These results indicate that, at least in 293T cells, NS1 interacts with RIG-I.
FIG. 1.
RIG-I and NS1 interact in coimmunoprecipitation experiments. IP, immunoprecipitation. (A) 293T cells were transfected with 3.5 μg of pCAGGS-Flag-RIG-I (third and fourth lanes) and/or 3.5 μg of pCAGGS-NS1 (second and fourth lanes) by use of Lipofectamine 2000. The total DNA amount was normalized with empty pCAGGS plasmid. At 48 h posttransfection, cells were subjected to reversible cross-linking immunoprecipitation assays using anti-Flag antibodies. RIG-I and NS1 were detected by Western blotting using monoclonal anti-Flag and polyclonal anti-NS1 antibodies. (B) 293T cells were transfected with 3.5 μg of pCAGGS-Flag-RIG-I (second and fourth lanes) and/or 3.5 μg of pCAGGS-NS1-GFP (third and fourth lanes) by use of Lipofectamine 2000. The total DNA amount was normalized with empty pCAGGS plasmid. At 48 h posttransfection, cells were subjected to the reversible cross-linking immunoprecipitation assays using monoclonal anti-GFP antibodies. RIG-I and NS1-GFP were detected by Western blotting using monoclonal anti-Flag and monoclonal anti-GFP antibodies. (C) 293T cells were transfected with 3.5 μg of pCAGGS-Flag-RIG-I (lanes 4 to 7) with (lane 7) or without 3.5 μg of pCAGGS-NS1 by use of Lipofectamine 2000. The total DNA amount was normalized with empty pCAGGS plasmid. At 48 h posttransfection, cells were mock infected (lanes 1, 4, and 7) or infected with SeV (lanes 2 and 5) or PR8 (lanes 3 and 6). Cells were collected at 9 h postinfection and subjected to the reversible cross-linking immunoprecipitation assays using anti-Flag antibodies. RIG-I and NS1 were detected by Western blotting using monoclonal anti-Flag and polyclonal anti-NS1 antibodies. Weak bands in the first and second lanes and lanes 1, 2, and 3, of the third blots in panels A and C, respectively, most likely correspond to anti-Flag antibody used in the IP and appearing at low levels after peptide elution of immunocomplexes from the beads.
Influenza A virus NS1 inhibits RIG-I-mediated activation of the IFN-β promoter.
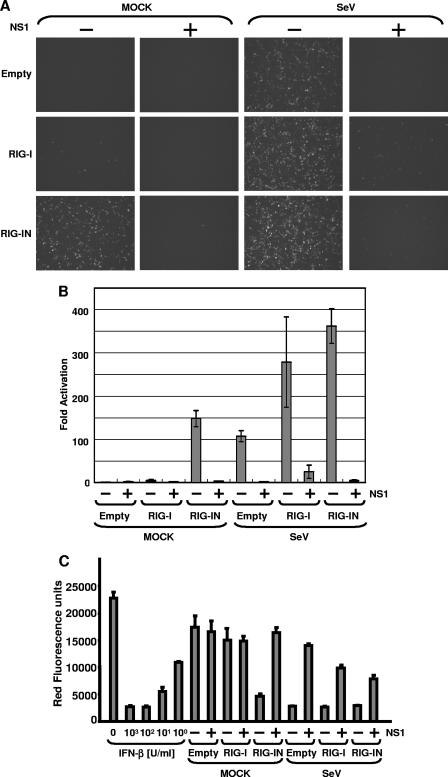
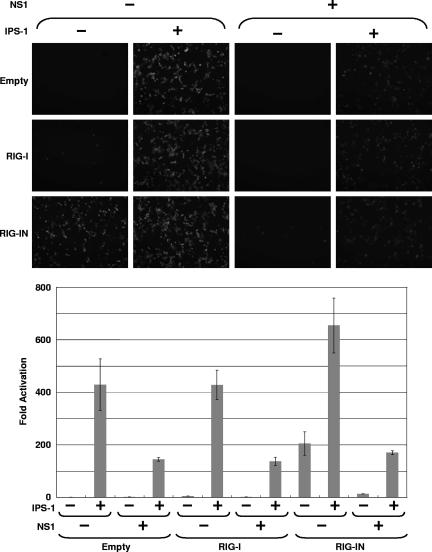
We next investigated whether expression of influenza A virus NS1 would inhibit RIG-I-mediated activation of the IFN-β promoter. Yoneyama et al. have reported that overexpression of RIG-I enhances the induction of the IFN-β by intracellular dsRNA and NDV (55). Moreover, expression of a carboxy-terminally truncated RIG-I (RIG-IN) containing the CARD and lacking the helicase domain of RIG-I activated the IFN-β promoter in the absence of any other stimuli. Flag-RIG-I or Flag-RIG-IN expression plasmids were transfected into 293T cells together with a GFP-CAT fusion reporter gene under the control of the IFN-β promoter (pIFNβ-GFP/CAT). In Fig. 2A and B, it is shown that RIG-IN constitutively activated the IFN-β promoter without virus infection as expected. The activation of the IFN-β promoter by RIG-IN was inhibited in the presence of NS1, as demonstrated by reductions in GFP expression (Fig. 2A) and in CAT activity (Fig. 2B). CAT assay data were consistent with the observed levels of fluorescence in the GFP-expressing cells. Overexpression of wild-type RIG-I resulted in a weak activation of the GFP-CAT reporter gene but potentiated the effects of SeV infection on IFN-β promoter activation. In all cases, NS1 expression inhibited reporter gene expression. Thus, NS1 inhibited activation of the IFN-β promoter by SeV, RIG-I, or RIG-IN.
FIG. 2.
Inhibition of RIG-I-mediated IFN-β production by NS1. (A) Fluorescent reporter assays. 293T cells were transfected with 0.5 μg of pIFN-β-GFP/CAT, 0.5 μg of pCAGGS-Luc, 250 ng of pCAGGS-Flag-RIG-I or pCAGGS-Flag-RIG-IN (as indicated at the left of each panel), and 2.5 μg of pCAGGS-NS1 (as indicated at the top of each panel) by using CaPO4. The total DNA amount was normalized with empty pCAGGS plasmid. At 24 h posttransfection, cells were mock infected or infected with SeV at an MOI of 10 (as indicated at the top). Twenty-four hours postinfection, the cells were observed by use of a fluorescence microscope. (B) Enzymatic reporter assays. After the fluorescence was observed, cells were collected and lysed in reporter lysis buffer (Promega). The lysates were cleared by centrifugation, and supernatants were subjected to CAT and luciferase assays. Relative CAT activity was normalized to the values obtained for luciferase activity. The y axis indicates activation (n-fold) of the IFN-β promoter by setting the reporter activity in the absence of RIG-I, RIG-IN, and NS1 at 1. The reporter assays were conducted in triplicate. Error bars indicate standard deviations. (C) Bioassays. Before the collection of cells, culture media were harvested and subjected to a bioassay for determination of IFN levels/production based on RFP expression in NDV-mRFP-infected Vero cells. The y axis indicates the red fluorescence produced by NDV-mRFP.
In order to confirm that results obtained with the IFN-β-GFP-CAT reporter gene mirrored the induction of endogenous IFN-β expression, we measured (in the same set of experiments) the amount of IFN-α/β secreted into supernatants by a bioassay that is based on the inhibition of replication of an IFN-sensitive virus expressing RFP, NDV-mRFP. Expression of RFP is used as a marker for NDV replication (41). Red fluorescence was measured in a microplate fluorescence reader. The level of NDV-mRFP replication inversely correlates with the levels of IFN-α/β from transfected and/or infected 293T cells. As seen in Fig. 2C, medium from RIG-IN-transfected cells strongly inhibited replication of NDV-mRFP in Vero cells. This inhibition was suppressed by the expression of NS1. SeV infection with or without overexpression of RIG-I induces production of IFN-α/β, and this induction is again inhibited by NS1 expression. These results indicate that NS1 inhibits RIG-I-mediated IFN-β production.
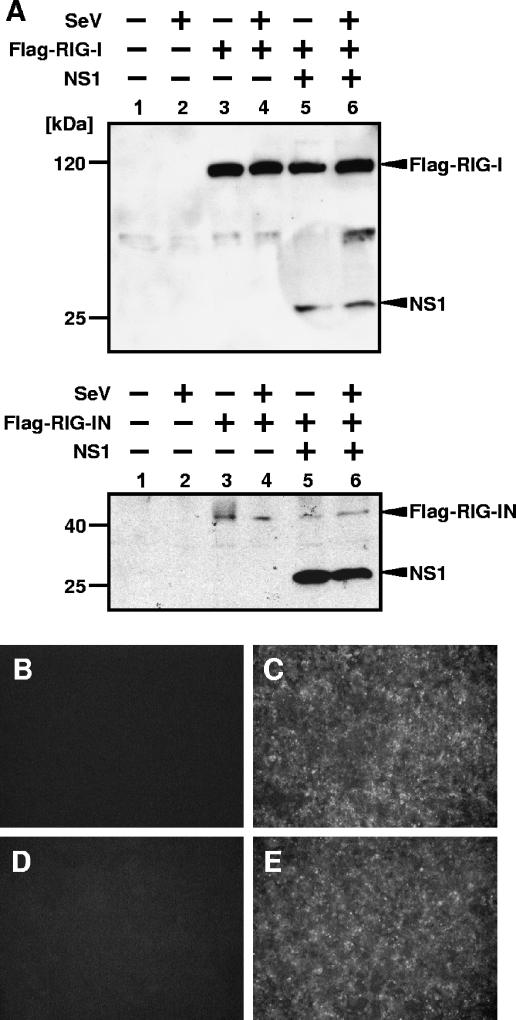
One consequence of the NS1 inhibiting RIG-I-mediated activation of the IFN-β promoter is decreased expression of IFN-β-responsive genes. To test this aspect, 293T cells were cotransfected with a reporter plasmid expressing GFP fused to CAT protein under an ISRE promoter (pHISG54-GFP/CAT), RIG-I or RIG-IN expression plasmids, and empty or NS1 expression plasmids. A subset of these transfected cells were subsequently infected with SeV at 24 h posttransfection. Cell lysates were prepared at 24 h postinfection and subjected to Western blotting with anti-FLAG and anti-NS1 antibodies (Fig. 3A). Activation of the ISRE promoter by RIG-IN transfection and by SeV infection was inhibited by NS1 (data not shown), while cells transfected with empty plasmid showed a clear activation of the promoter as demonstrated by the observed GFP fluorescence levels and enzymatic measurements of CAT activity (data not shown).
FIG. 3.
Expression levels of RIG-I and SeV infection in cells expressing NS1. (A) Expression level of RIG-I in cells expressing NS1. 293T cells were transfected with 0.5 μg of the pHISG54-GFP/CAT reporter plasmid (lanes 1 to 6), 0.25 μg of the pCAGGS-RIG-I (top panel, lanes 3 to 6) or pCAGGS-RIG-IN (bottom panel, lanes 3 to 6) plasmid, and 2 μg of empty plasmid (lanes 1 to 4) or NS1 expression plasmid (lanes 5 and 6). At 24 h posttransfection, cells were mock infected (lanes 1, 3, and 5) or infected with SeV (lanes 2, 4, and 6). Cells were collected and lysed in SDS-PAGE gel loading buffer at 24 h postinfection. Proteins were detected by Western blotting by using monoclonal anti-Flag M2 and monoclonal NS1 antibodies. An unspecific weak band can be seen in the top panel between Flag-RIG-I and NS1 signals. (B through E) SeV infection levels in cells expressing NS1. Cells transfected with empty plasmid (B and C) or NS1 expression plasmid (D and E) were mock infected (B and D) or infected with SeV (C and E). At 24 h postinfection, cells were fixed and viral proteins were detected by indirect immunofluorescence assays with SeV-specific antibodies.
The inhibitory activity of NS1 on the RIG-I-mediated induction of IFN-β could be indirectly mediated by a reduction in RIG-I expression. However, Fig. 1 shows that NS1 expression does not appreciably affect the expression level of RIG-I. For further confirmation, the expression levels of RIG-I and RIG-IN in the presence or absence of NS1 were examined by Western blotting (Fig. 3A). NS1 expression did not affect expression levels of RIG-I or RIG-IN, indicating that inhibition of RIG-I-mediated induction of IFN-β by NS1 was not caused by differences of RIG-I expression levels. Of note, the expression levels of RIG-IN are much lower than those of wild-type RIG-I (Fig. 3A).
To show that NS1 does not affect the replication of SeV, 293T cells transfected with either empty plasmid or the NS1 expression plasmid were infected with SeV and then stained with a pool of monoclonal antibodies against SeV (Fig. 3B to E). Cells transfected with empty (Fig. 3B and C) or NS1 (Fig. 3D and E) plasmids show the same level of SeV viral replication (Fig. 3C and E), indicating that NS1-mediated inhibition of SeV activation of IFN expression is not due to a general inhibition of SeV replication by the presence of the influenza virus protein NS1.
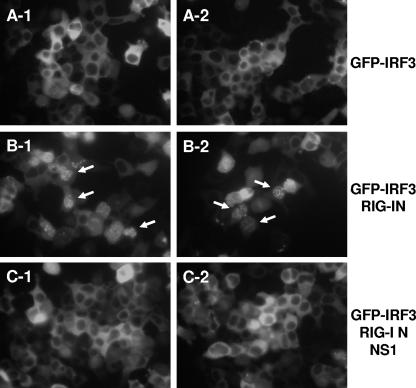
Influenza A virus NS1 inhibits nuclear translocation of IRF-3 induced by RIG-I.
We have previously demonstrated that the NS1 protein of influenza A virus inhibits nuclear translocation of IRF-3 after viral infection (49). To examine the effect of NS1 on the nuclear translocation of GFP-IRF-3 caused by RIG-I, 293T cells were cotransfected with a GFP-IRF-3 expression plasmid and the RIG-I amino-terminal mutant (RIG-IN) expression plasmid with or without the NS1 plasmid. In the absence of RIG-IN, GFP-IRF-3 accumulated solely within the cytoplasm (Fig. 4, panels A-1 and A-2). Expression of RIG-IN induced the nuclear translocation of GFP-IRF-3, as expected (Fig. 4, panels B-1 and B-2). The nuclear translocation of GFP-IRF-3 was impaired in cells cotransfected with expression plasmids for RIG-IN and influenza A virus NS1 (Fig. 4, panels C-1 and C-2).
FIG. 4.
Inhibition of RIG-IN-induced nuclear translocation of GFP-IRF-3 by NS1. 293T cells were transfected with 250 ng of the pEGFP-IRF-3 expression plasmid in the absence (A) or presence of 250 ng of RIG-IN expression plasmid alone (B) or with 2 μg of NS1 expression plasmid (C) as indicated. Nuclear translocation of GFP-IRF-3 was monitored at 48 h posttransfection under a fluorescence microscope. Representative cells with nuclear translocation of GFP-IRF-3 are indicated with arrows. Two representative fields per transfection are shown.
NS1 inhibits activation of the IFN-β promoter by IPS-1.
IPS-1/MAVS/VISA/CARDIF has been identified as a downstream mediator of RIG-I signaling through its CARD (19). We tested the effect of NS1 expression on IFN-β promoter activation by IPS-1. 293T cells were transfected with pIFNβ-GFP/CAT in the presence or absence of an IPS-1 expression plasmid and/or NS1 expression plasmid, and IFN-β promoter activity was monitored by examining the expressions of the GFP reporter gene (Fig. 5). Overexpression of IPS-1 induced the activation of the IFN-β promoter, similar to what was found with RIG-IN. Consistent with the results in Fig. 2, expression of NS1 inhibited the activation of the IFN-β promoter by RIG-I or RIG-IN. The activation by IPS-1 was also inhibited by NS1 expression. Even when RIG-I or RIG-IN was coexpressed with IPS-1, NS1 inhibited the activation of IFN-β promoter (Fig. 5).
FIG. 5.
Inhibition of IPS-1 by NS1. (Upper panel) Fluorescent reporter assays. 293T cells were transfected with 0.5 μg of pIFN-β-GFP/CAT and 0.5 μg of pCAGGS-Luc, 250 ng of pCAGGS-Flag-RIG-I or pCAGGS-Flag-RIG-IN (as indicated at the left), 0.25 μg of pCAAGGS-HA-IPS-1 (as indicated in the top), and 2.5 μg of pCAGGS-NS1 (as indicated in the top) by using CaPO4. The total DNA amount was normalized with empty pCAGGS plasmid. Expression of GFP driven by the IFN-β promoter was observed by fluorescence microscopy. (Lower panel) Enzymatic reporter assays. After the fluorescence was observed, cells were collected and lysed in reporter lysis buffer (Promega). The lysates were cleared by centrifugation and supernatants were subjected to CAT and luciferase assays. Relative CAT activity was normalized to the values obtained for luciferase activity. The y axis indicates activation (n-fold) of the IFN-β promoter by setting the reporter activity in the absence of IPS-1, RIG-I, RIG-IN, and NS1 at 1. The reporter assays were conducted in triplicate. Error bars indicate standard deviations.
NS1, RIG-I, and IPS-1 are copurified in insoluble complexes.
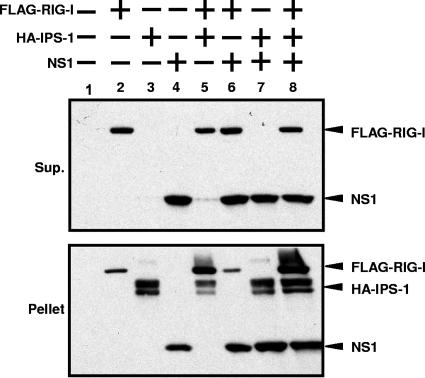
It is known that IPS-1 associates with mitochondrial membranes (44), while RIG-I and NS1 are nonmembrane proteins. Consistent with the Western blot detection of the overexpressed NS1 and RIG-I in the soluble fraction (Fig. 3A), the majority of NS1 and overexpressed RIG-I remained in a soluble fraction after cell lysis with RIPA buffer, while IPS-1 remained in a RIPA buffer-insoluble fraction (Fig. 6). Interestingly, coexpression of IPS-1 with RIG-I and/or NS1 resulted in increased levels of NS1 and RIG-I present in the RIPA-insoluble, IPS-1-associated cellular fraction (Fig. 6). These results strongly suggest that IPS-1, RIG-I, and NS1 become associated in the same cellular complexes.
FIG. 6.
Copurification of IPS-1, RIG-I, and NS1 in detergent insoluble complexes. A total of 2 × 106 293T cells were transfected with 2.5 μg of pCAGGS-FLAG-RIG-I (lanes 2, 5, 6, and 8), 2.5 μg of pCAGGS-HA-IPS-1 (lanes 3, 5, 7, and 8), and/or 2.5 μg of pCAGGS-NS1 (lanes 4, 6, 7, and 8). Amounts of transfected DNA were adjusted with empty pCAGGS plasmid. At 48 h posttransfection, cells were subjected to reversible cross-linking reaction and then lysed in 500 μl of RIPA buffer. After centrifugation at 10,000 × g for 5 min, cell supernatants (Sup.) and pellets were separated. The supernatant was mixed with equal volume of 2 × SDS gel loading buffer. The pellet was resuspended in 50 μl of SDS gel loading buffer. The samples were boiled for 5 min and then subjected to 12% SDS-PAGE. Expression levels of Flag-RIG-I, HA-IPS-1, and NS1 were detected by Western blotting using anti-Flag, anti-HA, and anti-NS1 antibodies, respectively.
RIG-I-dependent induction of IFN-β during influenza virus infection.
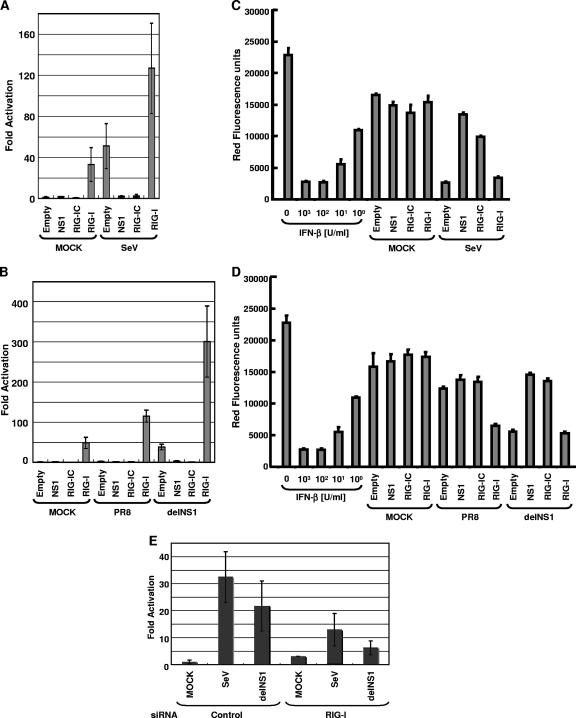
The results in Fig. 2, 4, and 5 indicate that the NS1 protein of influenza virus inhibits the RIG-I/IPS-1-mediated activation of IRF-3. This is likely to result in inhibition of IFN-β production during influenza virus infection. In order to demonstrate that the RIG-I pathway is responsible for induction of IFN-β in 293T cells, we used a RIG-I mutant which lacks CARD in its amino terminus (RIG-IC). This mutant functions as a dominant negative mutant of RIG-I and inhibits RIG-I-mediated production of IFN-β (55). 293T cells were transfected with plasmids expressing NS1, RIG-I, or RIG-IC and a CAT reporter gene under the control of the IFN-β promoter and then infected with SeV or PR8 or delNS1 virus. The IFN-β promoter was potently activated in cells infected with SeV (Fig. 7A). In contrast, the NS1 gene needed to be deleted in order for influenza A virus infection to effectively activate the IFN-β promoter (Fig. 7B), which is consistent with our previous results (15). The activation of the IFN-β promoter was suppressed in cells expressing RIG-IC (Fig. 7A and B), and in cells transfected with siRNA specific for RIG-I (Fig. 7E), indicating that the RIG-I pathway is critical for the induction of IFN-β by Sendai and influenza viruses in this system. A critical role of RIG-I in the induction of IFN by influenza virus was also recently demonstrated by other groups (23, 35). Interestingly, overexpression of wild-type RIG-I in wild-type influenza PR8 virus-infected cells overcomes the inhibition of IFN induction by this virus (Fig. 7B). Thus, overexpression of RIG-I can titrate out the NS1 inhibitory action (effect) on IFN induction during influenza virus infection. IFN bioassays performed using culture medium supernatants indicated that the activity of the IFN-β promoter reporter gene correlated with the levels of endogenous IFN induced in these cells (Fig. 7C and D).
FIG. 7.
Involvement of RIG-I in SeV- and influenza A virus-induced IFN-β production. (A and B) RIG-IC inhibits activation of IFN-β promoter by viral infection. 293T cells were transfected with 0.5 μg of pIFNβ-CAT, 0.5 μg of pCAGGS-Luc, and 0.5 μg of pCAGGS, pCAGGS-NS1, pCAGGS-Flag-RIG-IC, or pCAGGS-Flag-RIG-I, as indicated, using CaPO4. The reporter assays were conducted in triplicate. At 24 h posttransfection, cells were mock infected or infected with SeV, influenza A/PR/8/34 virus, or delNS1 virus at an MOI of 1. Cells were collected at 24 h postinfection and subjected to CAT assays. Relative CAT activity was normalized against relative luciferase activity. The y axis indicates activation (n-fold) of the IFN-β promoter by setting the reporter activities of empty pCAGGS-transfected and mock-infected cells at 1. (C and D) Suppression of virus-induced IFN-β production by RIG-IC and NS1. Before cells were collected, culture media were harvested and subjected to a bioassay for determination of IFN levels/production based on RFP expression in NDV-mRFP-infected Vero cells. The y axis indicates the red fluorescence produced by NDV-mRFP. (E) siRNA for RIG-I inhibits activation of IFN-β promoter by viral infection. To knock down the expression of RIG-I, RNA interference was performed with siRNA (see Materials and Methods). Expression of RIG-I was downregulated in Stealth_1199-transfected cells but not in Stealth_control_1199-transfected cells (data not shown), and that of β-actin was not affected by the transfection of both Stealth_1199 and Stealth_control_1199 (data not shown). After incubation, cells were mock infected or infected with SeV or delNS1. After the infection, cells were incubated for 24 h and then cells were collected and subjected to CAT assay. The y axis indicates activation (n-fold) of IFN-β promoter by setting the reporter activity of Stealth_control_1199 and the reporter plasmid-transfected and mock-infected cells at 1.
DISCUSSION
We previously found that the NS1 protein of influenza A virus inhibits the induction of IFN-β during influenza virus infection (15). Expression of only the amino-terminal dsRNA-binding domain of NS1 appears sufficient to prevent the induction of IFN-β (51). In agreement, mutations of the amino acids R38 and K41 responsible for binding to dsRNA result in mutant NS1 proteins with impaired abilities to inhibit the induction of IFN-β (11, 49). A possible mechanism then for inhibition of IFN induction by the NS1 protein is the sequestration of dsRNA induced during influenza virus infection. However, several lines of evidence suggest that additional mechanisms are involved in the NS1 inhibitory properties on IFN production. First, dsRNA-binding-deficient NS1 proteins are not completely deficient in inhibition of IFN induction (11). Second, the IFN antagonist functions of the NS1 protein from some viral strains are more efficient than others in inhibiting the IFN system, depending on the host species (16). This host specificity cannot be explained by merely sequestration of viral dsRNA and most likely reflects NS1 interactions with host factors. Third, dsRNA is likely not to be the only molecule produced during viral infection to trigger the IFN system; in fact, little dsRNA is detected in cells infected with influenza viruses (53). Therefore, we postulated that, in addition to inhibiting dsRNA binding, the NS1 protein of influenza A virus may inhibit the induction of IFN-β by interacting with host factors involved in IFN-β expression.
The inhibition of IFN-α/β production by the NS1 protein is mediated by an inhibition of the activation of transcription factors involved in induction of the IFN-β promoter, including IRF-3, a key mediator of IFN-β production (49). IRF-3 can be activated in response to different stimuli, including the activation of TLR pathways, but more recently, it was found that induction of IFN-β by RNA virus infection is mediated in most cell systems by a TLR-independent pathway based on activation of cellular RNA helicases, such as RIG-I and Mda-5 (23, 36). In fact, RIG-I was recently demonstrated to be essential for the induction of IFN-β by influenza virus in a mouse knockout system (23). Our results with a dominant negative RIG-I and with RIG-I-specific siRNA are also consistent with this notion (Fig. 7). It remains to be seen whether the NS1 protein can inhibit the activation of IFN in specialized cells that use TLR-7/8 pathways to induce IFN in response to RNA viruses, such as plasmacytoid dendritic cells (22).
Based on the proposed central role for RIG-I as a cellular sensor of viral products resulting in activation of IRF-3 and induction of IFN-β and also based on the fact that the NS1 of influenza virus inhibits the activation of IRF-3, we investigated the possibility that NS1 and RIG-I interact. This was readily seen in coimmunoprecipitation experiments (Fig. 1). Importantly, NS1 expression inhibited the RIG-I-mediated activation of IRF-3 and of the IFN-β promoter even when a constitutively activated mutant form of RIG-I, RIG-IN, was used (Fig. 2 and 4). Wild-type RIG-I is believed to be activated by binding of its C-terminal helicase domain to dsRNA. Binding of dsRNA results in a conformational change within RIG-I that exposes an N-terminal domain containing a CARD that acts as a protein-protein-interacting module, thus allowing the recruitment of downstream cellular factors and the induction of IRF-3 activation (55). Expression of this RIG-I CARD or RIG-IN bypasses the requirement for dsRNA for activation and results in constitutive induction of IRF-3 and IFN-β. Since NS1 expression can inhibit signaling mediated by RIG-IN, it suggests a mechanism of action of NS1 independent of dsRNA binding. Future characterization of the domains of NS1 responsible for binding to and inhibiting RIG-I may shed light on the structural requirements for the IFN-antagonistic functions of the influenza A virus NS1 protein. In this respect, it is worth mentioning that the crystal structure of the N-terminal region of NS1 is known (29) and that the structure of the C-terminal region of NS1 has recently been solved (8). Once these domains are determined, the use of reverse genetics will allow the generation of mutant viruses expressing NS1 proteins impaired in RIG-I binding in order to study the phenotypic impact of this NS1 property.
Although we could demonstrate an interaction between RIG-I and NS1 in coimmunoprecipitation experiments, we cannot exclude the possibility that this interaction is not direct and could be mediated by a second cellular bridging protein. In fact, when we used NS1 and RIG-I purified from bacteria, we were unable to detect an interaction (data not shown). The downstream cellular effector of RIG-I is known to be a mitochondrial resident protein with a cytoplasmic CARD, known as IPS-1, MAVS, VISA, or CARDIF (25, 37, 44, 54). Interestingly, when we overexpressed IPS-1, we observed that both RIG-I and NS1 levels are enriched in a RIPA buffer-insoluble cellular fraction that is highly abundant in IPS-1. Overexpression of RIG-I did not affect the levels of NS1 recovered in the insoluble fraction when coexpressed with IPS-1, indicating that RIG-I does not compete with NS1 for binding to IPS-1. In fact, the results suggest that NS1 localizes in the same complex with RIG-I and IPS-1 during viral infection, resulting in inhibition of further downstream signaling to the IRF-3 kinases. We are currently exploring this possibility.
The RIG-I/IPS-1 pathway appears to be targeted by different viruses to achieve inhibition of the IFN-β system. One of the best characterized examples of such a virus is hepatitis C virus, whose viral protease, NS3-4A, cleaves IPS-1 and renders this cellular protein inactive (28, 30, 37). Several paramyxoviruses encode a viral protein, the V protein, that interacts with mda-5, a cellular protein with high similarity to RIG-I and which has also been implicated in antiviral functions by triggering IFN-β (3). In the case of influenza A virus, the NS1 protein is required for IFN-β antagonism, and this appears to be mediated by two independent mechanisms, one involving sequestration of dsRNA (11) and the second one most likely mediated through an interaction with RIG-I (this study). Moreover, NS1 has also acquired mechanisms to inhibit the IFN response at steps after IFN transcription, including inhibitory effects of cellular RNA processing (26) and inhibition of two important IFN antiviral effectors, PKR (5, 27) and OAS (38). The presence of multiple mechanisms in the NS1 protein of influenza virus to inhibit the IFN-α/β system illustrates the importance of this system in the fight against virus infections in general and influenza virus infections in particular.
Despite the presence of inhibitors of IFN-β production in many viruses, it is also evident that hosts are able to induce IFN-β after viral infection. Influenza viruses are not an exception to this observation. Natural infection with influenza A virus results in IFN-α/β production (20), and in fact, global patterns of host gene expression during influenza virus infection are characterized by an IFN signature with an upregulation of many IFN-α/β-responsive genes (21). However, the absence of NS1 renders influenza virus an even higher IFN-α/β inducer, and this greatly limits its replication in vivo, resulting in attenuation (13, 43, 46). Thus, hosts and viruses have evolved a very intricate and delicate balance of responses and counter responses, and the IFN-α/β system exemplifies many of these processes as a central mediator of innate and adaptive immunity (14). A better understanding of these processes may help us in the design of novel therapeutic and prophylactic strategies.
In summary, we found that the NS1 protein of influenza virus interacts with RIG-I and inhibits the RIG-I pathway, preventing the activation of IFN-β by the cell. Targeting of the RIG-I/IPS-1 pathway leading to the activation of IFN appears to be a strategy shared by several viruses that we are just starting to understand. Our results are likely to stimulate more research on the interactions of different viruses with the RIG-I/IPS-1 pathway and research on how these interactions may modulate virulence and pathogenesis.
Acknowledgments
We thank Roger Tsien and Jonathan Yewdell for kindly providing mRFP cDNA and NS1 antibodies, respectively. We thank Ben Chen for the 96-well microplate reader. We thank Randy Albrecht for valuable discussions. We also thank Richard Cádagan for excellent technical assistance.
These studies were partially supported by NIH grant R01AI46954 to A.G.-S. and NIH grants AI060389 and AI40035 (M.G. and Y.-M.L.). W.B.C. is supported by a fellowship awarded by Northeast Biodefence Center—Lipkin, PI (AI057158).
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Akira, S., and K. Takeda. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499-511. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 3.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. García-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann, M., A. García-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biron, C. A., and G. C. Sen. 2001. Interferons and other cytokines, p. 321-351. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott-Raven, Philadelphia, PA.
- 7.Bluyssen, H. A., R. J. Vlietstra, A. van der Made, and J. Trapman. 1994. The interferon-stimulated gene 54 K promoter contains two adjacent functional interferon-stimulated response elements of different strength, which act synergistically for maximal interferon-alpha inducibility. Eur. J. Biochem. 220:395-402. [DOI] [PubMed] [Google Scholar]
- 8.Bornholdt, Z. A., and B. V. Prasad. 2006. X-ray structure of influenza virus NS1 effector domain. Nat. Struct. Mol. Biol. 13:559-560. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diebold, S. S., T. Kaisho, H. Hemmi, S. Akira, and C. Reis e Sousa. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303:1529-1531. [DOI] [PubMed] [Google Scholar]
- 11.Donelan, N., C. F. Basler, and A. García-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding defective NS1 protein induces high levels of IFN-β and is attenuated in mice. J. Virol. 77:13257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelmann, K. H., S. Richardson-Burns, L. Alexopoulou, K. L. Tyler, R. A. Flavell, and M. B. Oldstone. 2004. Does Toll-like receptor 3 play a biological role in virus infections? Virology 322:231-238. [DOI] [PubMed] [Google Scholar]
- 13.Ferko, B., J. Stasakova, J. Romanova, C. Kittel, S. Sereinig, H. Katinger, and A. Egorov. 2004. Immunogenicity and protection efficacy of replication-deficient influenza A viruses with altered NS1 genes. J. Virol. 78:13037-13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.García-Sastre, A., and C. A. Biron. 2006. Type 1 interferons and the virus-host relationship: a lesson in détente. Science 312:879-882. [DOI] [PubMed] [Google Scholar]
- 15.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 16.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. García-Sastre. 2002. Cellular transcriptional profiling in influenza A virus infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: Induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heil, F., H. Hemmi, H. Hochrein, F. Ampenberger, C. Kirschning, S. Akira, G. Lipford, H. Wagner, and S. Bauer. 2004. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303:1526-1529. [DOI] [PubMed] [Google Scholar]
- 19.Hiscott, J., R. Lin, P. Nakhaei, and S. Paz. 2006. MasterCARD: a priceless link to innate immunity. Trends Mol. Med. 12:53-56. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser, L., R. S. Fritz, S. E. Straus, L. Gubareva, and F. G. Hayden. 2001. Symptom pathogenesis during acute influenza: interleukin-6 and other cytokine responses. J. Med. Virol. 64:262-268. [DOI] [PubMed] [Google Scholar]
- 21.Kash, J. C., C. F. Basler, A. García-Sastre, V. Carter, R. Billharz, D. E. Swayne, R. M. Przygodzki, J. K. Taubenberger, M. G. Katze, and T. M. Tumpey. 2004. Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J. Virol. 78:9499-9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, H., S. Sato, M. Yoneyama, M. Yamamoto, S. Uematsu, K. Matsui, T. Tsujimura, K. Takeda, T. Fujita, O. Takeuchi, and S. Akira. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19-28. [DOI] [PubMed] [Google Scholar]
- 23.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 24.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 25.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 26.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181-189. [DOI] [PubMed] [Google Scholar]
- 27.Li, S., J. Y. Min, R. M. Krug, and G. C. Sen. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13-21. [DOI] [PubMed] [Google Scholar]
- 28.Lin, R., J. Lacoste, P. Nakhaei, Q. Sun, L. Yang, S. Paz, P. Wilkinson, I. Julkunen, D. Vitour, E. Meurs, and J. Hiscott. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 80:6072-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, J., P. A. Lynch, C. Y. Chien, G. T. Montelione, R. M. Krug, and H. M. Berman. 1997. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat. Struct. Biol. 4:896-899. [DOI] [PubMed] [Google Scholar]
- 30.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 103:6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López, C. B., B. Moltedo, L. Alexopoulou, L. Bonifaz, R. A. Flavell, and T. M. Moran. 2004. TLR-independent induction of dendritic cell maturation and adaptive immunity by negative-strand RNA viruses. J. Immunol. 173:6882-6889. [DOI] [PubMed] [Google Scholar]
- 32.Ludwig, S., X. Wang, C. Ehrhardt, H. Zheng, N. Donelan, O. Planz, S. Pleschka, A. García-Sastre, G. Heins, and T. Wolff. 2002. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 76:11166-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lund, J. M., L. Alexopoulou, A. Sato, M. Karow, N. C. Adams, N. W. Gale, A. Iwasaki, and R. A. Flavell. 2004. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA 101:5598-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malmgaard, L., J. Melchjorsen, A. G. Bowie, S. C. Mogensen, and S. R. Paludan. 2004. Viral activation of macrophages through TLR-dependent and -independent pathways. J. Immunol. 173:6890-6898. [DOI] [PubMed] [Google Scholar]
- 35.Matikainen, S., J. Siren, J. Tissari, V. Veckman, J. Pirhonen, M. Severa, Q. Sun, R. Lin, S. Meri, G. Uze, J. Hiscott, and I. Julkunen. 2006. Tumor necrosis factor alpha enhances influenza A virus-induced expression of antiviral cytokines by activating RIG-I gene expression. J. Virol. 80:3515-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melchjorsen, J., S. B. Jensen, L. Malmgaard, S. B. Rasmussen, F. Weber, A. G. Bowie, S. Matikainen, and S. R. Paludan. 2005. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J. Virol. 79:12944-12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 38.Min, J. Y., and R. M. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo(A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA 103:7100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakaya, T., J. Cros, M.-S. Park, Y. Nakaya, H. Zheng, A. Sagrera, E. Villar, A. García-Sastre, and P. Palese. 2001. Recombinant Newcastle disease virus as a vaccine vector. J. Virol. 75:11868-11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 41.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. García-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park, M. S., J. Steel, A. García-Sastre, D. Swayne, and P. Palese. 2006. Engineered viral vaccine constructs with dual specificity: avian influenza and Newcastle disease. Proc. Natl. Acad. Sci. USA 103:8203-8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quinlivan, M., D. Zamarin, A. García-Sastre, A. Cullinane, T. Chambers, and P. Palese. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 79:8431-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 45.Smith, E. J., I. Marié, A. Prakash, A. García-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 46.Solórzano, A., R. J. Webby, K. M. Lager, B. H. Janke, A. García-Sastre, and J. A. Richt. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 48.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. García-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. García-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 97:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, X., C. F. Basler, B. R. G. Williams, R. H. Silverman, P. Palese, and A. García-Sastre. 2002. Functional replacement of the carboxy-terminal two thirds of the influenza A virus NS1 protein with short heterologous dimerization domains. J. Virol. 76:12951-12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. García-Sastre. 2000. Influenza A virus NS1 protein prevents the activation of NF-κB and induction of type I IFN. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber, F., V. Wagner, S. B. Rasmussen, R. Hartmann, and S. R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 55.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]