Abstract
Flock House virus (FHV; Nodaviridae) is a positive-strand RNA virus that encapsidates a bipartite genome consisting of RNA1 and RNA2. We recently showed that specific recognition of these RNAs for packaging into progeny particles requires coat protein translated from replicating viral RNA. In the present study, we investigated whether the entire assembly pathway, i.e., the formation of the initial nucleating complex and the subsequent completion of the capsid, is restricted to the same pool of coat protein subunits. To test this, coat proteins carrying either FLAG or hemagglutinin epitopes were synthesized from replicating or nonreplicating RNA in the same cell, and the resulting particle population and its RNA packaging phenotype were analyzed. Results from immunoprecipitation analysis and ion-exchange chromatography showed that the differentially tagged proteins segregated into two distinct populations of virus particles with distinct RNA packaging phenotypes. Particles assembled from coat protein that was translated from replicating RNA contained the FHV genome, whereas particles assembled from coat protein that was translated from nonreplicating mRNA contained random cellular RNA. These data demonstrate that only coat proteins synthesized from replicating RNA partake in the assembly of virions that package the viral genome and that RNA replication, coat protein translation, and virion assembly are processes that are tightly coupled during the life cycle of FHV.
Flock House virus (FHV) is a nonenveloped, icosahedral insect virus of the family Nodaviridae. It has a bipartite positive-strand RNA genome that is packaged into a single virion (3, 11, 20). The viral replication cycle, which is confined to the cytoplasm of infected cells, gives rise to three RNA species: RNA1 (3.1kb), RNA2 (1.4kb), and RNA3, a subgenomic RNA that corresponds to the 3′-terminal 387 nucleotides of RNA1. Only RNA1 and RNA2 are packaged into virions (7). RNA1 encodes protein A (112 kDa), the RNA-dependent RNA polymerase, which is located on the outer mitochondrial membrane in infected cells (14, 15). RNA2 encodes protein alpha, the capsid precursor protein (43 kDa). Protein alpha and the two genomic RNAs initially assemble into a noninfectious precursor particle called a provirion (7, 8). Provirions mature and acquire infectivity by spontaneous cleavage of protein alpha into proteins beta (38 kDa) and gamma (5 kDa) (8, 19).
Assembly of FHV particles is thought to begin with a nucleating step in which a coat protein subunit, or a multimer thereof, interacts with an encapsidation signal on the viral RNAs to form a specific nucleoprotein complex (17). This complex is then thought to be propagated into a spherically closed particle by accretion of coat protein subunits that are guided into the growing shell by interactions with secondary structural elements on the RNA (21). RNA and coat protein regions that are critical for specific packaging of the FHV genome have been identified previously (13, 18, 25). In addition, we recently demonstrated that specific genome packaging requires coat protein translated from replication-competent RNA2 templates (24). Coat protein translated from nonreplicating mRNA packages cellular RNA, even when replicating RNA1 and RNA2 are present in the same cell. These results suggested a tight linkage between viral RNA replication and translation on one hand and specific genome recognition on the other. However, they did not address the question of whether this type of linkage extended to particle assembly. The objective of this study was to determine whether the completion of the capsid subsequent to the formation of the nucleation complex is also restricted to subunits derived from replicating RNA2 or whether coat proteins that are translated from a nonreplicating mRNA could participate in this process.
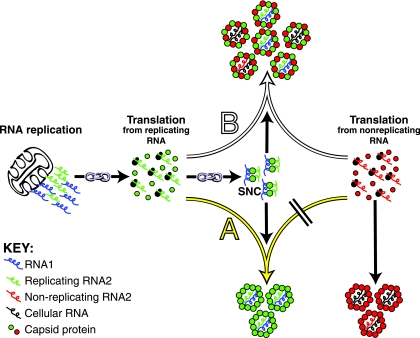
We rationalized that if the link between RNA replication/translation and specific genome packaging extended to particle assembly, the synthesis of coat proteins from both replicating and nonreplicating RNAs in the same cell would result in the formation of two distinct populations of virus particles (schematically illustrated as pathway A in Fig. 1). One of these particle populations would be made up of coat proteins that were translated from replicating RNA and would contain viral RNA. The other population would be made up of coat proteins that were translated from nonreplicating RNA and would contain cellular RNA. Alternatively, if the link between RNA replication/translation and specific genome packaging did not extend to assembly, then the two differentially translated coat proteins would form a single population of “mosaic” virus particles (schematically illustrated as pathway B in Fig. 1).
FIG. 1.
Two possible outcomes of assembly when coat proteins are coexpressed from replicating and nonreplicating RNAs in the same cell. Arrows with superimposed chain links represent the previously described coupling between RNA replication, coat protein translation, and specific genome recognition (24). The exact composition and structure of the specific nucleoprotein complex (SNC) are not yet known. The yellow arrow (A) points to an assembly pathway that would be followed if RNA replication/translation were coupled to specific genome recognition and virion assembly. This pathway leads to the segregation of the coexpressed coat proteins into two independent populations of virus particles with distinct RNA packaging characteristics. The white arrow (B) points to an alternate pathway that would be followed if virion assembly were not coupled to replication/translation. This pathway leads to the formation of a single population of mosaic particles with a mixed RNA packaging phenotype.
To test this, we took advantage of a baculovirus expression system for the expression of FHV proteins from either replicating or nonreplicating viral RNA (24). In this system, the replication of RNA1 is launched upon infection of Sf21 cells with the recombinant baculovirus AcR1δ (10). Translation of the primary RNA1 transcripts yields protein A, which in turn replicates the RNA1 message and subgenomic RNA3 to high levels. To synthesize coat protein from replicating RNA2 in these cells, they are transfected 24 h later with in vitro-synthesized RNA2 transcripts. RNA2 is then replicated by protein A, and the coat proteins translated from this RNA efficiently package viral RNAs 1 and 2 during assembly. To synthesize coat protein from nonreplicating RNA2, AcR1δ-infected Sf21 cells are coinfected with a recombinant baculovirus, which expresses RNA2 lacking authentic 5′ and 3′ untranslated regions (−5′3′UTR). These RNA2 transcripts are not recognized by protein A as templates for replication (1, 2, 12), and coat protein synthesized from these RNAs packages a heterogeneous mix of cellular RNAs, including the RNA2 coat protein message (10, 16, 24).
Here we show that the coexpression of coat proteins from replicating and nonreplicating RNA2 leads to the formation of two distinct populations of virus particles with different RNA packaging phenotypes. These results demonstrate that genome recognition and assembly are both coupled to the synthesis of coat protein produced from replication-competent RNA2 templates and that artificially generated coat protein is excluded from this pool of proteins. This may constitute a safety mechanism for the virus to ensure efficient and accurate formation of progeny virions during the viral replication cycle.
MATERIALS AND METHODS
Cells.
Spodoptera frugiperda cells (line IPLB-Sf21) (23) were propagated in TC100 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml amphotericin B at 27°C as described previously (16).
Construction of transfer vectors for the synthesis of epitope-tagged coat proteins from nonreplicating RNA.
pBP9R2(−5′3′UTR) is a transfer vector for the generation of the recombinant baculovirus AcR2(−5′3′UTR), which was previously used to synthesize FHV coat protein from nonreplicating RNA (24). In this study, pBP9R2(−5′3′UTR) was modified to enable baculovirus expression of CP[FG]-NR and CP[HA]-NR (coat proteins synthesized from nonreplicating RNA that contain FLAG [FG] and hemagglutinin [HA] epitopes, respectively). In the transfer vector for CP[FG]-NR, a 545-bp SpeI-HpaI fragment from plasmid p2BS(+)-Δ31/FLAG (10) was used to replace a corresponding 521-bp fragment in pBP9R2(−5′3′UTR). This 545-bp fragment encoded a partial FHV coat protein sequence with the FG (DYKDDDDK) epitope inserted between residues 205 and 206. Similarly, in the transfer vector for CP[HA]-NR, a 554-bp SpeI-HpaI DNA fragment that was generated by overlap extension PCR (9) was used to replace a corresponding 521-bp fragment in pBP9R2(−5′3′UTR). This 545-bp fragment encoded a partial coat protein sequence with the HA epitope (YPYDVPDYA) and a GSSG linker inserted between residues 206 and 209. DNA sequencing confirmed that the desired mutations were present in the CP[FG]-NR and CP[HA]-NR transfer vectors and that no errors were introduced by PCR.
Construction of plasmid that enables the synthesis of FG-tagged coat proteins from replicating RNA.
Capped FHV RNA2 transcripts that are replication competent in established FHV expression systems can be generated using the plasmid p2BS(+)-wt (10, 18). The coding sequence for wild-type (wt) coat protein carried on this plasmid was modified to that of CP[FG]-NR by use of a cloning strategy identical to the one described above for the CP[FG]-NR transfer vector. The resultant plasmid (pCP[FG]-R) enabled the synthesis of CP[FG]-R, an FG-tagged coat protein synthesized from replicating RNA. DNA sequencing confirmed that the desired mutation was present in pCP[FG]-R.
Generation of recombinant baculoviruses.
Recombinant baculoviruses for the expression of CP[FG]-NR and CP[HA]-NR (designated AcCP[FG]-NR and AcCP[HA]-NR, respectively) were generated following the protocols of the manufacturer (BD Biosciences). In brief, each transfer vector was transfected into Sf21 cells together with Bsu36I-linearized BacPAK6 baculovirus DNA (BD Biosciences), and cell supernatants were harvested 3 days posttransfection. Putative AcCP[FG]-NR and AcCP[HA]-NR were purified by plaquing the cell supernatants once on Sf21 cell monolayers and amplified following confirmation of the expression of the inserted genes.
Infection of Sf21 cells.
Monolayers consisting of 1.5 × 106 Sf21 cells in six-well plates were infected at a multiplicity of infection of 5 PFU per cell by the addition of a 0.5-ml sample containing recombinant baculovirus stocks. This was followed by a 1-h incubation at room temperature (with rocking), the removal of the unattached baculovirus, and the addition of 2 ml of complete growth medium to each well. Incubation was continued at 27°C for 2 to 5 days without agitation.
Infection/transfection of Sf21 cells.
An infection/transfection protocol for the production of FHV particles that efficiently package genomic RNA was described previously (10). In brief, monolayers consisting of 1.5 × 106 Sf21 cells in six-well plates were infected at a multiplicity of infection of 5 PFU per cell with AcR1δ, a recombinant baculovirus, for the expression of FHV RNA1. In some instances the cells were coinfected with AcR1δ and AcCP[HA]-NR. Following infection or coinfection, Cellfectin (Invitrogen) was used for the transfection of 100 ng of capped CP[FG]-R RNA2 at 24 h postinfection. This RNA was synthesized by in vitro transcription from XbaI-linearized pCP[FG]-R by use of a technique identical to the one described for the synthesis of wt RNA2 from XbaI-linearized p2BS(+)-wt (18). The transfected cells were incubated without agitation for an additional 1 to 4 days at 27°C.
Purification of epitope-tagged FHV particles by sucrose gradient centrifugation.
Epitope-tagged FHV particles were purified from Sf21 cells 5 days postinfection by pelleting through a 30% (wt/wt) sucrose cushion followed by sedimentation though a 10 to 40% (wt/wt) sucrose gradient as described previously (10). The particles were harvested from the sucrose gradient by needle puncturing.
Immunoprecipitation of HA-tagged FHV particles.
A Roche immunoprecipitation kit (protein A) was used in conjunction with 20 μl of rabbit anti-HA antibodies (Sigma) to precipitate HA-tagged particles from sucrose gradient preparations containing 2 μg of FHV particles. The immunoprecipitation and washing conditions were identical to those described in the protocol of the manufacturer (Roche). In brief, the particles were immunoprecipitated overnight at 4°C on a rocking platform. Immune complexes were collected by centrifugation and washed four times with buffers containing different salt and detergent concentrations. The first supernatant fraction from each experiment was concentrated approximately fivefold with a Savant SpeedVac concentrator and subjected, together with the immunoprecipitated particles, to immunoblot analysis with either mouse anti-FLAG M2 antibodies or mouse anti-HA antibodies (both from Sigma).
Chromatographic separation of FG- and HA-tagged FHV particles.
To separate FG- and HA-tagged FHV particles, sucrose gradient-purified particles were subjected to ion-exchange chromatography (IEC), using an Uno Q column (Bio-Rad) linked to a Bio-Rad Biologic DuoFlow system. The column was preequilibrated in 50 mM HEPES (pH 7)-40 mM NaCl, and a linear 40 to 1,000 mM gradient was used (at 1 ml/min for 20 ml). The particles were eluted at 4°C, and fractions were diluted 1:1 in 50 mM HEPES (pH 7)-40 mM NaCl subsequent to collection.
Immunoblot analysis.
Protein samples were run on NuPAGE 4 to 12% Bis-Tris gels (Invitrogen) and subjected to immunoblot analysis as described previously (4).
RNA analysis.
RNA was extracted from IEC-purified FHV particles by means of phenol-chloroform extraction as described previously (18) and analyzed by electrophoresis through a nondenaturing 2% Seakem LE agarose (FMC Corp.) gel in TAE (Tris-acetate-EDTA) buffer.
RESULTS
Assembly of epitope-tagged FHV particles from coat proteins that are translated from either replicating or nonreplicating RNA.
To determine if coat proteins translated from replicating and nonreplicating RNAs in the same cell assemble into two independent populations of virus particles, a means to distinguish between the differentially translated proteins was required. To this end, we utilized coat protein mutants displaying different epitope tags depending on what kind of template they were derived from. Specifically, an FG epitope and a HA epitope were inserted into an FHV coat protein loop that is prominently exposed on the surface of assembled virions (Fig. 2A) (6). Coat protein carrying the FG epitope was produced from replicating templates and is referred to as CP[FG]-R, while coat protein containing the HA epitope was produced from nonreplicating templates and is referred to as CP[HA]-NR. A recombinant baculovirus for the synthesis of FG-tagged coat protein from nonreplicating RNA, CP[FG]-NR, was also generated and used in control experiments.
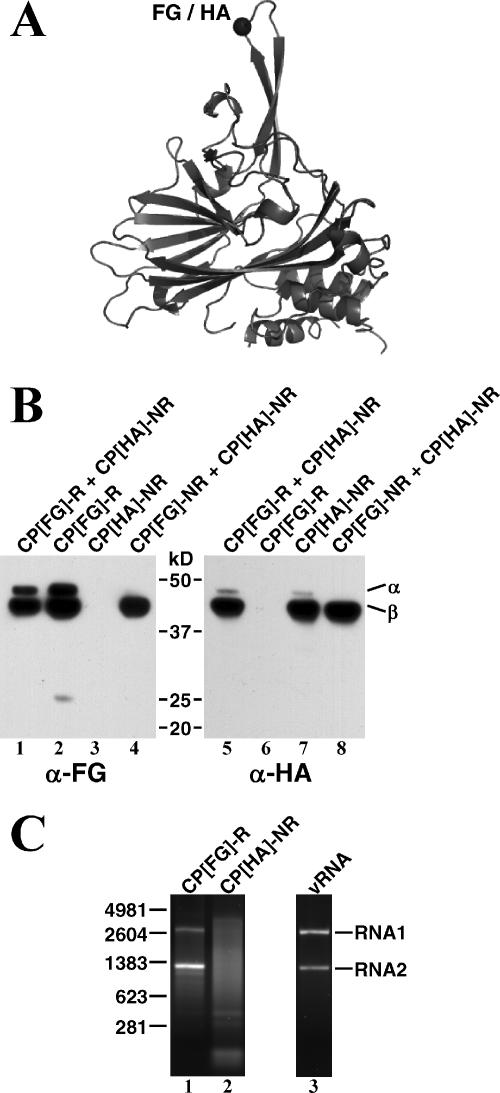
FIG. 2.
Assembly of epitope-tagged FHV particles. (A) Ribbon diagram of the FHV coat precursor protein (protein alpha [α]) showing the point of insertion for FG or HA peptides into an extended loop that is on the surface of the capsid. β, protein beta. (B) Immunoblot analysis on sucrose gradient-purified particles assembled from the indicated epitope-tagged coat proteins. FG- and HA-tagged coat proteins were detected using monoclonal antibodies to FG (α-FG) and HA (α-HA), respectively. (C) Electrophoretic analysis of RNA packaged by CP[FG]-R and CP[HA]-NR when these proteins are expressed separately in the presence of replicating RNA1. RNA was extracted from sucrose gradient-purified particles, and 1-μg aliquots were run on a nondenaturing agarose gel. RNA from authentic FHV particles (vRNA) was run for comparison in lane 3. The molecular sizes of RNA markers (in nucleotides) are indicated on the left.
In preliminary studies, CP[FG]-R and CP[HA]-NR were expressed separately to confirm that the mutant proteins assembled into FG- and HA-tagged virus particles, respectively. Sf21 cells infected with AcR1δ and transfected with CP[FG]-R RNA contained particles that cosedimented with wt FHV particles on sucrose gradients (data not shown). Immunoblot analysis of these particles with monoclonal anti-FG antibodies revealed the presence of both protein alpha and cleavage product beta (Fig. 2B, lane 2). The second coat protein cleavage product, gamma (4 kDa), was not detected under these conditions, as it does not contain the FG epitope. Similar results were obtained for the purification of HA-tagged particles from cells that were coinfected with AcR1δ and AcCP[HA]-NR. In this case, monoclonal anti-HA antibodies were used to demonstrate that particle preparations from these cells contained HA-tagged proteins alpha and beta (Fig. 2B, lane 7). In both experiments, the anti-FG and anti-HA antibodies showed no detectable cross-reactivity with the differentially tagged coat proteins (Fig. 2B, lanes 3 and 6, respectively).
The maturation cleavage of HA-tagged alpha coat protein into beta and gamma was more efficient than that of the FG-tagged protein (Fig. 2B, compare lanes 2 and 7). Upon prolonged incubation, however, the extents of cleavage in both types of particles were identical, suggesting a difference in the rates of maturation (data not shown). The reason for this difference is not known, but variation in maturation efficiency has been observed previously upon insertion of heterologous peptides or protein domains into the FHV coat protein (our unpublished data). It was proposed (26) that the autocatalytic cleavage reaction is driven by local strain between the coat protein subunits and that the relief of this strain during the maturation process lowers the overall rate of the cleavage reaction. It is possible that the insertion of different epitopes affects subunit-subunit strain differentially, thus explaining the observed variation in the levels of alpha and beta protein in the FG- and HA-tagged proteins. Since the cleavage occurs after particle formation, variations in maturation cleavage do not reflect differences in the ability of the proteins to assemble.
We next confirmed that the presence of the epitopes did not alter the RNA packaging phenotype of the mutant coat protein, i.e., that CP[FG]-R and CP[HA]-NR packaged viral RNA and random cellular RNA, respectively (24). To this end, RNA extracted from the two types of particles was analyzed on a nondenaturing 2% agarose gel. As anticipated, both viral RNAs 1 and 2 were packaged by CP[FG]-R, and random cellular RNA was packaged by CP[HA]-NR (Fig. 2C). It is known from previous studies that this random cellular RNA includes the message of the FHV coat protein (10, 24).
Native FHV particles contain equimolar amounts of the two genomic segments, but particles assembled from CP[FG]-R contained more RNA2 than RNA1 (Fig. 2C, lanes 1 and 3). The reason for this anomaly is currently unknown and subject to further investigation. Taken together, our results demonstrated that CP[FG]-R and CP[HA]-NR assemble into epitope-tagged FHV particles in a manner that is analogous to what has been described for wt coat proteins (24).
Immunoprecipitation of HA-tagged particles from a mixture of FG- and HA-tagged particles.
The FG and HA epitopes had been inserted into the FHV coat protein at a position that is displayed on the surface of the virion following assembly. The next objective was to demonstrate that antibodies against either HA or FG could be used to immunoprecipitate whole virus particles and that they would specifically precipitate the particle with the appropriate antigen. This was tested by first mixing purified FG- and HA-tagged particles in a 1:1 molar ratio and then subjecting the mixture to immunoprecipitation with polyclonal rabbit anti-HA antibodies. The resultant immunoprecipitate as well as protein remaining in the supernatant were loaded into separate lanes on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel for subsequent immunoblot analysis with mouse monoclonal anti-FG and anti-HA antibodies. As shown in Fig. 3A (lanes 1 to 3), HA-tagged coat proteins were detected exclusively in the immunoprecipitate, whereas FG-tagged protein was detected only in the supernatant (Fig. 3B, lanes 1 to 3). In Fig. 3B, the levels of FG-tagged protein observed for the supernatant fraction (lane 3) are slightly increased from those for the total fraction (lane 1). This irregularity was not consistently observed, and it was most probably caused by overconcentration of the supernatant prior to immunoblot analysis.
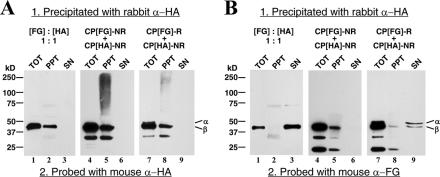
FIG. 3.
Immunoprecipitation analysis of particles purified from cells expressing CP[FG]-NR + CP[HA]-NR (lanes 4 to 6) or CP[FG]-R + CP[HA]-NR (lanes 7 to 9). Immunoprecipitation was carried out using polyclonal anti-HA antibodies (rabbit α-HA) as described in Materials and Methods. Total input protein (TOT) and precipitated protein (PPT), as well as protein remaining in the supernatant (SN), were then subjected to immunoblot analysis with monoclonal antibodies to HA (mouse α-HA; panel A) and FG (mouse α-FG; panel B). In a control experiment, individually expressed FG- and HA-tagged particles were mixed in a 1:1 molar ratio prior to immunoprecipitation analysis (lanes 1 to 3). Note that the immunoreactive protein species with molecular masses lower than that of protein beta (β) visible for CP[FG]-NR + CP[HA]-NR and CP[FG]-R + CP[HA]-NR (lanes 4 to 9) were also detected for the 1:1 mixture of individually expressed FG- and HA-tagged particles (lanes 1 to 3) following prolonged exposure of the blots to film (data not shown). α, protein alpha.
Taken together, these results confirmed that the HA epitope, and by extension the FG epitope, was effectively displayed on the surface of the particles. In addition, because of the specificity of the HA antibodies, this result suggested that the immunoprecipitation approach could be used to determine if distinct populations of FG- and HA-tagged particles were assembled from CP[FG]-R and CP[HA]-NR.
Assembly of mosaic particles from FG- and HA-tagged coat proteins.
In order to use the epitope-tagged coat proteins to investigate what types of particles result by coexpression of coat proteins from replicating and nonreplicating templates (Fig. 1), it was important to establish that, in principle, mosaic virus particles could be assembled from FG- and HA-tagged coat proteins. There was a slight possibility that the presence of the epitopes themselves would cause the proteins to segregate into different populations independently of their translation from replicating or nonreplicating RNA. To exclude this possibility, Sf21 cells were coinfected with recombinant baculoviruses for the expression of CP[FG]-NR and CP[HA]-NR. Particles were purified from these cells by sucrose gradient centrifugation, and the presence of CP[FG]-NR and CP[HA]-NR in these particles was confirmed by immunoblotting (Fig. 2B, lanes 4 and 8). In addition, the particles were immunoprecipitated with anti-HA antibodies, and both the precipitate and the supernatant were subjected to immunoblot analysis to determine if CP[FG]-NR coprecipitated with CP[HA]-NR. This would be the case if the proteins had formed FG/HA mosaics. As expected, CP[HA]-NR was detected in the immunoprecipitate but not in supernatant (Fig. 3A, lanes 4 to 6). Moreover, CP[FG]-NR was also detected exclusively in the immunoprecipitate (Fig. 3B, lanes 4 to 6). This result suggested that mosaic virus particles could indeed be formed from FG- and HA-tagged coat proteins when they were cosynthesized from nonreplicating RNA templates. In addition, this result indicated that the population contained few, if any, particles that contained only one type of coat protein.
In addition to FG- and HA-tagged coat proteins, protein species that were either smaller or larger than FHV coat protein were detected on immunoblots (Fig. 3A and B). The smaller proteins probably represented coat protein breakdown products, which are also commonly detected for wt FHV, while nonspecific interactions between the mouse monoclonal antibodies used for immunodetection and rabbit anti-HA antibodies used for immunoprecipitation resulted in the detection of the larger protein species.
Independent confirmation for the assembly of mosaic particles was obtained by IEC. Preliminary IEC analyses showed that the difference between the charge properties of the FG and HA epitopes resulted in a clear difference in the retention times for FG- and HA-tagged particles upon elution with a linear salt gradient. FG-tagged particles eluted at 380 mM NaCl, while HA-tagged particles eluted at 470 mM NaCl (Fig. 4A). In contrast, mosaic particles eluted at 425 mM NaCl, in a peak that was located exactly halfway between the peaks for FG- and HA-tagged particles (Fig. 4A). Taken together, these results demonstrated that mosaic particles could be easily distinguished from FG- or HA-tagged particles. In addition, the mosaic particles probably contained nearly equivalent amounts of the differentially labeled proteins, given that they eluted halfway between HA- and FG-tagged particles.
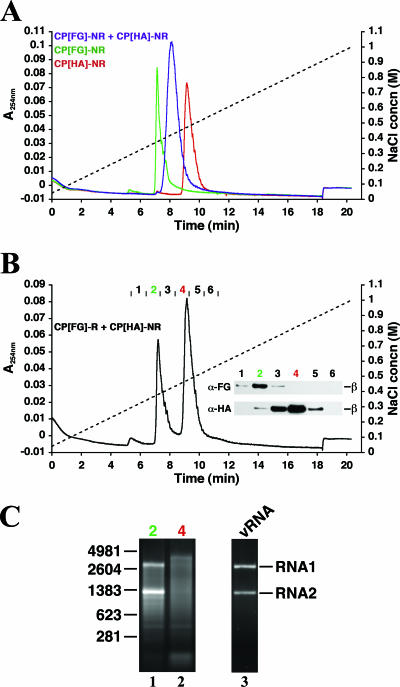
FIG. 4.
Chromatographic separation of different epitope-tagged particles. (A) IEC of particles purified from cells expressing CP[FG]-NR (green profile), CP[HA]-NR (red profile), or CP[FG]-NR + CP[HA]-NR (purple profile). The A280 elution profiles (colored lines) and the relative NaCl concentration (dashed line) are shown. (B) IEC of particles purified from cells expressing CP[FG]-R + CP[HA]-NR. (Inset) Immunoblot analysis of fractions comprising the peaks resolved by IEC with monoclonal antibodies to HA (α-HA) and FG (α-FG). (C) Electrophoretic analysis of RNA extracted from the epitope-tagged particles in fractions 2 and 4 (panel B). RNA from authentic FHV particles (vRNA) was run for comparison in lane 3. The molecular sizes of RNA markers (in nucleotides) are indicated on the left.
Assembly of two independent populations of epitope-tagged FHV particles with distinct RNA packaging characteristics.
The results obtained up to this point in our study indicated that differentially tagged proteins could be used to distinguish between the possibilities that coat proteins coexpressed from replicating and nonreplicating RNAs would form mosaic particles or segregate into two distinct populations of viral capsids. To address this issue, Sf21 cells were first coinfected with AcR1δ and AcCP[HA]-NR and then transfected 24 h later with CP[FG]-R RNA transcripts. Four days posttransfection, particles were gradient purified, and their protein composition and RNA packaging phenotype analyzed.
Immunoblot analysis confirmed that both CP[FG]-R and CP[HA]-NR were present in the population of purified particles (Fig. 2B, lanes 1 and 5). Immunoprecipitation of particles with anti-HA antibodies revealed that HA-tagged coat protein was present exclusively in the precipitate together with trace amounts of FG-tagged protein (Fig. 3A and B, lanes 7 to 9). The majority of the detectable FG-tagged protein, however, was present in the supernatant after immunoprecipitation (Fig. 3B, lanes 7 to 9). These results implied that CP[HA]-NR-containing particles were selectively precipitated from a mixture that also contained CP[FG]-R particles.
Corroborating evidence for the existence of two distinct populations was obtained by IEC, which resolved the particles into two peaks. One peak was detected at the same position as FG-tagged particles, whereas the other was detected at the position of HA-tagged particles (compare elution profiles in Fig. 4B to those in Fig. 4A). Immunoblot analysis of protein in fractions comprising the two peaks showed that the material eluting earlier represented CP[FG]-R, while the later material represented CP[HA]-NR (Fig. 4B, insert).
To determine the RNA packaging phenotypes of the IEC-purified FG- and HA-tagged particles, RNA was extracted from fractions 2 (FG-tagged particles; Fig. 4B) and 4 (HA-tagged particles; Fig. 4B) and analyzed electrophoretically (Fig. 4C). While FG-tagged particles had packaged viral RNAs 1 and 2 (Fig. 4C, lane 1), HA-tagged particles contained a heterogeneous mix of cellular RNAs (Fig. 4C, lane 2). These results demonstrated that the packaging of distinct RNA species accompanied the segregation and assembly of CP[FG]-R and CP[HA]-NR into independent virus particle populations.
DISCUSSION
In this study, coat proteins displaying either FG or HA peptides were used to distinguish between proteins derived from replicating or nonreplicating RNA, respectively. Remarkably, two independent populations of virus particles that predominately packaged viral RNA and random cellular RNA, respectively, were detected upon coexpressing the FG- and HA-tagged proteins. This was manifested in the selective immunoprecipitation of HA-tagged particles from FG-tagged particles with anti-HA antibodies (Fig. 3) and by exploiting the different charge properties of FG and HA peptides to separate two populations of FHV particles by IEC (Fig. 4B).
Two lines of evidence demonstrate that the segregation of the FG- and HA-tagged proteins in vivo was not caused by an inability of these proteins to form mosaic particles. First, results from this study show that the coexpression of FG- and HA-tagged coat proteins from nonreplicating RNA leads to the assembly of a single population of FG/HA mosaic particles (Fig. 3 and 4A). Second, results from a previous study show that it is also possible to produce mosaic particles when coat proteins with different epitopes are coexpressed from replicating RNA (10). Moreover, the segregation of the FG- and HA-tagged proteins cannot be attributed to the synthesis of these proteins at different periods in time from replicating and nonreplicating RNAs, respectively. FHV coat protein is maximally produced from nonreplicating RNA between 24 and 48 h postinfection (16), which matches the period during which a high level of FG-tagged coat protein was synthesized from replicating RNA in this study (data not shown). Therefore, when all these factors are considered, it can be concluded that our results can be attributed only to the source of RNA from which the coat proteins were translated.
FHV particles produced from baculovirus vectors yielding replicating RNA1 and RNA2 always include a minor fraction of particles that contain random cellular RNA (10). In light of our new data, these results can now be explained by the fact that primary transcription from the baculovirus genome continues throughout the experiment, in parallel with the replication of FHV RNAs by protein A. Presumably, not all primary baculovirus-derived transcripts become subject to replication, particularly early in the experiment, when few FHV replication complexes had been established. These nonreplicating transcripts would give rise to coat protein subunits that package cellular RNA. At later times in the experiment, the majority of the RNA1 and RNA2 transcripts would be generated by FHV replication, and the fraction of coat protein subunits that specifically package the viral genome would predominate.
The complete segregation of differentially translated coat proteins in an individual cell provides further support for our hypothesis that coat protein translation from replicating RNA, as opposed to nonreplicating RNA, takes place in distinct cellular compartments (24). According to this hypothesis, coat protein translation from replicating RNA is confined to a cellular compartment adjacent to the mitochondrial sites of FHV RNA synthesis. The resultant spatial coordination between coat proteins and cellular pools of viral RNA would explain why only the proteins that are synthesized from replicating RNA partake in specific genome recognition and virion assembly. However, we cannot rule out a more complex scenario in which translation from replicating RNA2 somehow enables an FHV-specific trafficking mechanism linking newly synthesized coat proteins to the cellular compartments for virion assembly. The mechanism by which viral proteins are translated from the nonpolyadenylated genomic RNAs of FHV is still unclear. It is known, however, that the poly(A)-independent translation of reo- and alfamovirus genomic RNAs requires a set of translation initiation factors that is different from that used by the host (5, 22). It is therefore possible that a putative translation factor(s) specific for the translation of FHV coat proteins from replicating RNA2 could be required for the posttranslational trafficking of this protein to cellular compartments for virion assembly. Coat proteins translated from nonreplicating RNAs, which are polyadenylated subsequent to nuclear transcription, would therefore be unable to traffic to these compartments and would assemble around random cellular RNAs.
What is the advantage of coupling synthesis of FHV RNA and coat protein tightly to genome packaging and the formation of progeny virions? One advantage may be the increased efficiency of particle assembly when all the components are confined to a restricted compartment in the infected cell. Another likely facet of spatially coordinating the assembly of virions with the site of RNA and protein synthesis is the need to ensure that coat protein selects viral RNA for packaging. FHV coat protein is exceptionally basic, and our results show that it assembles around random cellular RNA with efficiency similar to that with which it assembles around viral RNA. Such aberrant packaging must be prevented during the viral infection cycle, and this could be achieved, in principle, by keeping the coat protein confined to a location of the cell where newly synthesized viral RNAs accumulate.
Current studies in our laboratory are directed at identifying the sites of RNA and coat protein synthesis from replicating and nonreplicating RNAs in FHV-infected cells and examining whether coat protein is indeed synthesized at the site of particle assembly.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM53491).
Footnotes
Published ahead of print on 1 November 2006.
Manuscript 18190-MB from the Scripps Research Institute.
REFERENCES
- 1.Albarino, C. G., L. D. Eckerle, and L. A. Ball. 2003. The cis-acting replication signal at the 3′ end of Flock House virus RNA2 is RNA3-dependent. Virology 311:181-191. [DOI] [PubMed] [Google Scholar]
- 2.Ball, L. A. 1995. Requirements for the self-directed replication of flock house virus RNA 1. J. Virol. 69:720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dearing, S. C., P. D. Scotti, P. J. Wigley, and S. D. Dhana. 1980. A small RNA virus isolate from the grass grub Costelytra zealandica (Coleoptera: Scarabaeidae). N. Z. J. Zool. 7:267-269. [Google Scholar]
- 4.Dong, X. F., P. Natarajan, M. Tihova, J. E. Johnson, and A. Schneemann. 1998. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J. Virol. 72:6024-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreher, T. W., and W. A. Miller. 2006. Translational control in positive strand RNA plant viruses. Virology 344:185-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher, A. J., and J. E. Johnson. 1993. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 361:176-179. [DOI] [PubMed] [Google Scholar]
- 7.Friesen, P. D., and R. R. Rueckert. 1981. Synthesis of black beetle virus proteins in cultured Drosophila cells: differential expression of RNAs 1 and 2. J. Virol. 37:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallagher, T. M., and R. R. Rueckert. 1988. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 62:3399-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 10.Krishna, N. K., D. Marshall, and A. Schneemann. 2003. Analysis of RNA packaging in wild-type and mosaic protein capsids of flock house virus using recombinant baculovirus vectors. Virology 305:10-24. [DOI] [PubMed] [Google Scholar]
- 11.Krishna, N. K., and A. Schneemann. 1999. Formation of an RNA heterodimer upon heating of nodavirus particles. J. Virol. 73:1699-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindenbach, B. D., J. Y. Sgro, and P. Ahlquist. 2002. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 76:3905-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall, D., and A. Schneemann. 2001. Specific packaging of nodaviral RNA2 requires the N-terminus of the capsid protein. Virology 285:165-175. [DOI] [PubMed] [Google Scholar]
- 14.Miller, D. J., and P. Ahlquist. 2002. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 76:9856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneemann, A., R. Dasgupta, J. E. Johnson, and R. R. Rueckert. 1993. Use of recombinant baculoviruses in synthesis of morphologically distinct viruslike particles of flock house virus, a nodavirus. J. Virol. 67:2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneemann, A., T. M. Gallagher, and R. R. Rueckert. 1994. Reconstitution of Flock House provirions: a model system for studying structure and assembly. J. Virol. 68:4547-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneemann, A., and D. Marshall. 1998. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J. Virol. 72:8738-8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneemann, A., W. Zhong, T. M. Gallagher, and R. R. Rueckert. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 66:6728-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scotti, P. D., S. Dearing, and D. W. Mossop. 1983. Flock House virus: a nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae). Arch. Virol. 75:181-189. [DOI] [PubMed] [Google Scholar]
- 21.Tihova, M., K. A. Dryden, T. V. Le, S. C. Harvey, J. E. Johnson, M. Yeager, and A. Schneemann. 2004. Nodavirus coat protein imposes dodecahedral RNA structure independent of nucleotide sequence and length. J. Virol. 78:2897-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varani, G., and F. H. Allain. 2002. How a rotavirus hijacks the human protein synthesis machinery. Nat. Struct. Biol. 9:158-160. [DOI] [PubMed] [Google Scholar]
- 23.Vaughn, J. L., R. H. Goodwin, G. J. Tompkins, and P. McCawley. 1977. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera: Noctuidae). In Vitro 13:213-217. [DOI] [PubMed] [Google Scholar]
- 24.Venter, P. A., N. K. Krishna, and A. Schneemann. 2005. Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. J. Virol. 79:6239-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong, W., R. Dasgupta, and R. Rueckert. 1992. Evidence that the packaging signal for nodaviral RNA2 is a bulged stem-loop. Proc. Natl. Acad. Sci. USA 89:11146-11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zlotnick, A., V. S. Reddy, R. Dasgupta, A. Schneemann, W. J. Ray, Jr., R. R. Rueckert, and J. E. Johnson. 1994. Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single aspartic acid residue. J. Biol. Chem. 269:13680-13684. [PubMed] [Google Scholar]