Abstract
In a companion paper (D. Ostertag, T. M. Hoblitzell-Ostertag, and J. Perrault, J. Virol. 81:492-502, 2007), we provided indirect evidence that cell-type-specific growth restriction of the vesicular stomatitis virus (VSV) polR mutants may be due to enhanced production of double-stranded RNA (dsRNA). We show here that polR growth in mouse L-929 cells was rescued by vaccinia virus coinfection and that sole expression of the vaccinia virus dsRNA-binding E3L protein, via coinfection with an engineered VSV minigenome, also restored polR growth. Expression of dsRNA-binding protein NS1A or NS1B from influenza virus, but not C protein from Sendai virus, which does not bind dsRNA, likewise effected polR rescue. The N-terminal dsRNA-binding domain of NS1A, only 73 amino acids in length, but not a full-size mutant NS1A lacking dsRNA-binding activity, restored polR growth. Both key aspects of polR growth restriction, namely inhibition of genome replication and release of low-infectivity virus particles, were countered by expression of the dsRNA-binding proteins. We tested the effects of overproducing dsRNA in wild-type VSV infections by coinfecting cells with a VSV recombinant expressing the sense strand of the enhanced green fluorescent protein gene (VSV-GFP) and one expressing the antisense strand (VSV-PFG). These coinfections mimicked all aspects of polR restriction, including host range, lack of effect on transcription, reduced virus particle infectivity, and insensitivity to inhibition of host gene transcription or dsRNA-activated protein kinase activity. We conclude that, for some cell types, overproduction of dsRNA during VSV infection triggers an immediate and constitutive host cell antiviral effector response independent of interferon induction or signaling.
Vesicular stomatitis virus (VSV), a prototypic member of the Rhabdoviridae RNA virus family, is exceptionally sensitive to the effects of interferon (IFN) and has long been used in assays to detect the presence of this antiviral activity (44). But in the absence of an IFN response, VSV grows readily to high titers in most vertebrate cell types. Virally produced double-stranded RNA (dsRNA) plays a crucial role in inducing type I interferon (IFN-α/β) production in these cells (for reviews, see references 29 and 34). Once IFN signaling establishes an antiviral state, viral dsRNA again plays a critical role by turning on two key cellular antiviral effector proteins, namely, dsRNA-activated protein kinase (PKR) and dsRNA-activated 2′,5′-oligoadenylate synthetase (OAS), both of which are upregulated by type I IFN (for a review, see reference 48). PKR activation leads to α subunit of eukaryotic initiation factor 2 (eIF-2α) phosphorylation and shutoff of protein translation while 2′,5′-oligo(A) activates RNase L and viral mRNA degradation. In a companion paper (44a), we established that the cell-type-specific growth restriction of the VSV polR mutant is due to a block in viral genome replication and decreased infectivity of released virus particles. Since eIF-2α phosphorylation is enhanced in polR-infected L-929 cells, we surmised that restriction might be due to a host cell antiviral response triggered by aberrant polR virus RNA synthesis. Our findings, however, ruled out a role for PKR or OAS activation as well as host gene induction in polR restriction. In this paper, we show that enhanced viral dsRNA production is nevertheless responsible for this phenomenon and that wild-type (wt) VSV is subject to a similar restriction when excess dsRNA from a nonviral source is generated during infection.
VSV serves as a model for understanding basic aspects of viral RNA synthesis in all nonsegmented negative-strand RNA viruses (Mononegavirales) (reviewed in reference 47). The polymerase of these viruses consists of two virus-encoded subunits: the L protein (catalytic subunit) and the P protein, the latter serving as a template-binding subunit and a cofactor for polymerization. All viral RNA synthesis takes place in the cytoplasm. The P/L polymerase complex functions solely with viral RNA templates fully encapsidated with the nucleocapsid protein (N or NP). Transcription involves sequential copying of virus genes in the order found on the genome. For VSV, this entails five genes: N, P, M, G, and L. Replication of viral genomes requires synthesis of antigenomes that serve as templates for progeny genome synthesis. All replicative synthesis is tightly coupled to the assembly of nascent strands by the N protein.
VSV polR mutants display unique alterations in viral RNA synthesis, and these pertain to the distinction between genome transcription and genome replication (13, 45). These changes are caused by a single amino acid substitution in the N protein of the polR VSV mutant (Arg179 to His). The companion paper offers a more detailed description of the two modes of VSV RNA synthesis and the effects of the polR mutation (44a). With respect to the present paper, the salient polR virus alteration is a partial uncoupling of replication from encapsidation, resulting in synthesis of unencapsidated leader-N gene read-through transcripts of varying sizes. These illegitimate replication products are produced in abundance, along with normal transcription products, by the polR virion-associated template-polymerase complex in vitro (45) but are much less abundant in polR VSV-infected cells (44a).
The polR N protein mutation is also responsible for the unique virus host range growth defect of these mutants (12, 44a). polR grows well in many established cell lines, such as BHK and HeLa, but displays growth restriction in a number of others, notably, mouse L-929 and rat 3Y1 cells, where infectious virus yields are ∼100-fold- and ∼40-fold-lower than wt virus, respectively. In these restricted host cells, polR genome replication is inhibited 6- to 10-fold, and the infectivity of released virus particles is reduced 5- to ≥10-fold. polR mRNA synthesis, however, is not compromised, and viral protein translation is only moderately reduced (two- to threefold) in both permissive and restricted cells. Even though inhibition of viral protein accumulation is clearly not the basis for restriction, polR nonetheless causes a threefold increase in eIF-2α phosphorylation compared to the wt in L-929 cells at a time when most viral protein synthesis has already taken place. These results suggest that polR infections activate PKR more effectively, possibly due to overproduction of dsRNA. But neither 2-aminopurine (2-AP), a PKR inhibitor, nor actinomycin D (act D), a cellular transcription inhibitor, affect polR restriction (44a). The cellular response to polR thus appears to be independent of IFN induction and signaling.
Two different types of cellular receptors have recently been implicated in sensing the presence of virally produced dsRNA and initiating the signaling cascades that result in induction of host cell antiviral genes: the transmembrane Toll-like receptor 3 (TLR3), predominantly expressed in cells that play a central role in innate and adaptive immunity, and two DexD/H box RNA helicases, retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated antigen 5 (MDA5), which are found in the cytoplasm of most, if not all, cell types (for reviews, see references 34, 51, and 52). Although the earliest steps in the TLR3 and RIG-I/MDA5 signaling pathways clearly involve distinct components, both pathways nonetheless converge downstream to activate crucial transcription factors, including IFN regulatory factor 3 (IRF-3) and IRF-7, ATF2/cJun, and NF-κB, required for induction of IFN-β and other cellular antiviral defense genes (reviewed in reference 43).
Many viruses encode antiviral antagonists that block IFN induction and/or IFN signaling (for reviews, see references 23, 29, 30, and 33). The particular step(s) targeted by these antagonists varies from one virus to another, but dsRNA-binding proteins constitute an important class that can potentially block the earliest step of IFN induction as well as dsRNA activation of antiviral effector proteins. Of particular interest for the present study are the vaccinia virus E3L protein and the influenza virus NS1 proteins. Both can prevent IFN induction and PKR activation by sequestering dsRNA and/or by direct binding to PKR (5, 40, 46, 58, 60). E3L is at least partially responsible for vaccinia virus resistance to IFN and the broad host range of this virus in mammalian cells (38). Likewise, influenza A virus lacking the NS1A gene grows very poorly in IFN-competent host cells (24). In contrast, VSV does not encode a dsRNA-binding protein but generally counters IFN production by rapidly shutting down host cell functions, including transcription, nuclear-cytoplasmic transport, and translation (1, 7, 14, 15, 20, 21).
It has long been assumed that IFN induction requires only very small amounts of virally produced dsRNA. Marcus and Sekellick showed decades ago that a single VSV defective virus particle containing a so-called snap-back dsRNA molecule is sufficient to induce maximum IFN production in hyperresponsive aged chicken embryo fibroblasts (41). But aside from this study, almost nothing is known regarding amounts of dsRNA needed for IFN induction or direct activation of antiviral effector proteins in various cell types. As noted above, negative-strand RNA viruses like VSV do not generate dsRNA intermediates during replication, so what might be responsible for generating the small amounts needed for triggering antiviral responses is unclear. Nonetheless, PKR activation and, by inference, virally produced dsRNA, clearly plays a critical role in VSV sensitivity to type I IFN. For example, VSV growth is enhanced in PKR-null cultured cells (3), and sensitivity to VSV infection is dramatically increased in at least some strains of PKR-null mice (3, 19, 56). Moreover, it was shown long ago that type I IFN blocks VSV growth in L-929 cells via PKR activation and translational shutdown and that vaccinia virus coinfection rescues VSV from this inhibition (61). The rescuing activity provided by vaccinia virus was subsequently shown to be attributable to the dsRNA-sequestering activity of its E3L protein (53).
We show here that vaccinia virus coinfection likewise rescues polR growth in L-929 cells and that expression of E3L protein alone or expression of influenza NS1A or NS1B proteins counters polR virus restriction as well. Furthermore, a truncated version of NS1A containing only the dsRNA-binding domain appears sufficient for rescue. Moreover, we demonstrate that coinfection of cells with wt VSV-derived recombinants, engineered to produce dsRNA from a green fluorescent protein (GFP) reporter gene, leads to the same cell-type-specific growth restriction. On the basis of these findings, we propose that enhanced dsRNA accumulation during virus infection of some cell types triggers a constitutive antiviral activity independent of host gene transcription and the classical IFN response.
(The work presented here formed part of a Ph.D. thesis submitted to the University of California—San Diego and San Diego State University by D. Ostertag and part of an M.S. thesis submitted to San Diego State University by T. M. Hoblitzell-Ostertag.)
MATERIALS AND METHODS
Cell culture and virus sources.
Growth of BHK-21, L-929, and 3Y1 cells and generation of the recombinant wt VSV and recombinant polR VSV used throughout this study are described in a companion paper (44a). Generation of recombinant VSV-GFP and VSV-PFG is described below. Titers of all VSV stocks were determined by plaque assay in BHK cells. Stocks of the recombinant vaccinia-T7 virus (vTF7.3) (see reference 22), simply referred to as vaccinia in this study, were grown in HeLa cells, and titers were determined by plaque assay in the same cell line.
Generation of recombinant minigenomes expressing heterologous genes.
The VSV minigenome plasmid (pBS-GMMG) containing the VSV M and G genes (55) was kindly provided by Michael Whitt (University of Tennessee Health Science Center). The unique NcoI site in the G gene was first removed by introducing a silent mutation in the Ser199 codon (UCC to UCU) using standard oligonucleotide-directed mutagenesis. An oligonucleotide cassette containing cloning sites (SmaI, NcoI, and PmlI), flanked by VSV transcription start and stop sequences, was then inserted into the ClaI site between the M and G genes of pBS-GMMG grown in the dam− GM2163 Escherichia coli host. The cassette was constructed by annealing the following two oligonucleotides: 5′-CTCATCGATCTCTGTTAGTTTTTTTCATACACGTGTTCCATGGTCCCGGGATCGATCTC-3′ (minus-sense transcription start and stop sequences are in boldface type, and ClaI and cloning sites are underlined) and 5′-CTCATCGATCTCTGTTAG-3′. The ends of the annealed product were first blunted using Klenow DNA polymerase before being digested with ClaI. The resulting plasmid, designated pBS-GMMG(MCS), served as starting material for insertion of the various antiviral antagonist genes.
Antiviral antagonist genes were inserted into pBS-GMMG(MCS) as follows. The vaccinia E3L gene (WR strain) was first excised from plasmid pMT2/Va− (kindly provided by Bertram Jacobs, Arizona State University) with EcoRI/PstI digestion and cloned into the corresponding sites of the pSP73 vector (Promega). The PvuII/EcoRV fragment containing the E3L gene was then recovered and inserted into the SmaI site of pBS-GMMG(MCS) to yield pBS-MG-E3L. The wild-type influenza virus NS1A gene (PR8 strain) and derivative mutants were recovered from plasmids pCAGGS-NS1(SAM), pCAGGS-NS1(1-73)HA, and pCAGGS-NS1R38AK41A(SAM), all kindly provided by Aldolfo Garcia-Sastre, Mount Sinai School of Medicine (57). The full-length NS1A gene was cut from pCAGGS-NS1(SAM) using EcoRI/XhoI digestion, blunted with Klenow DNA polymerase, and inserted into the SmaI site of pBS-GMMG(MCS). The truncated NS1(1-73) gene was likewise excised from pCAGGS-NS1(1-73)HA and inserted into the corresponding sites of the pTM1 plasmid (kindly provided by Bernard Moss, NIAID) to generate pTM1-NS1A(1-73). The NcoI/StuI fragment of pTM1-NS1A was then inserted into the NcoI/PmlI sites of pBS-GMMG(MCS). An identical strategy was used to insert the NS1 R38AK41A gene into pBS-GMMG(MCS). The NS1B gene was recovered from the pGEM4-NS1B plasmid (kindly provided by Robert Krug, University of Texas at Austin) by PCR amplification using primers 5′-TTAATAGGAACCATGGCGGACAACATGACC-3′ and 5′-AATGTCGCCATGGTAATTGTCTCCCTCTTC-3′ (NcoI sites are underlined). Following NcoI digestion, the PCR product was cloned into the corresponding site of pBS-GMMG(MCS). Sendai C genes in plasmids pTM1-CM (C gene from M strain) and pTM1-CS (C gene from H strain with stop codon) were kindly provided by Daniel Kolakofsky, University of Geneva School of Medicine. These were digested with NcoI and StuI, and the C gene fragments were inserted into the NcoI and PmlI sites of pBS-GMMG(MCS). All of the pBS-GMMG(MCS)-derived plasmids containing the various antiviral antagonist genes were verified by sequencing to confirm the presence of the correct inserts.
Recovery of minigenome particles from the various minigenome plasmid constructs was carried out essentially as described previously (55), except as noted below. In brief, BHK cells were first infected with vaccinia-T7 at a multiplicity of infection (MOI) of 10 PFU/cell and then cotransfected with optimal amounts of minigenome templates and pGEM-L, pGEM-P, and pGEM-N support plasmids (31, 54). Cytosine arabinoside at 40 μg/ml was included in the recovery protocol in lieu of filtration to minimize vaccinia virus-induced cytopathic effect and replication. Following incubation at 37°C for 36 to 48 h, released minigenome particles from the cell supernatants were amplified through two successive rounds in fresh BHK cells under the same conditions, except for the omission of minigenome plasmids and addition of the supernatants at 3 to 6 h posttransfection. The resulting supernatants were clarified by low-speed centrifugation to remove cell debris and stored frozen at −80°C. Minigenome particles in these working stocks were quantified relative to known amounts of infectious wt VSV genomes by Northern blot hybridization using a plus-sense M gene T7 transcript probe (see below).
Western blot analysis.
Cytoplasmic extracts of infected L-929 cells (see Fig. 2A and B) were prepared at ∼12 h postinfection (p.i.) in cell extraction buffer (Biosource) with the addition of protease cocktail inhibitor (Sigma) and 1 mM phenylmethylsulfonyl fluoride (Sigma). Samples were analyzed on a 10 to 20% Tricine gel (Invitrogen) in XT running buffer (Bio-Rad) and transferred in Nupage buffer (Invitrogen) plus 20% methanol onto a polyvinylidene difluoride membrane (Millipore). Membranes were blocked in 5% nonfat dried milk (NFDM)-Tris-buffered saline (TBS) for 1 h before the addition of primary rabbit polyclonal anti-E3L or anti-NS1A (kindly provided by Bertram Jacobs, Arizona State University, and Adolfo Garcia-Sastre, Mount Sinai School of Medicine, respectively) at 1:1,000 dilution in 1% OVA-TBS followed by incubation overnight at 4°C. The blots were then washed three times in TBS plus 0.5% Tween before the addition of Alexa Flour 680 goat anti-rabbit immunoglobulin G secondary antibody (Invitrogen) in 1% OVA-TBS, being washed three times in TBS, and being scanned with the Odyssey infrared imaging system using the manufacturer's suggested protocol (Li-Cor Biosciences). Probing of blots shown in Fig. 2C with a 1:2,500 dilution of rabbit polyclonal anti-C (kindly provided by Dominique Garcin, University of Geneva School of Medicine) was carried out as detailed in the companion paper (44a).
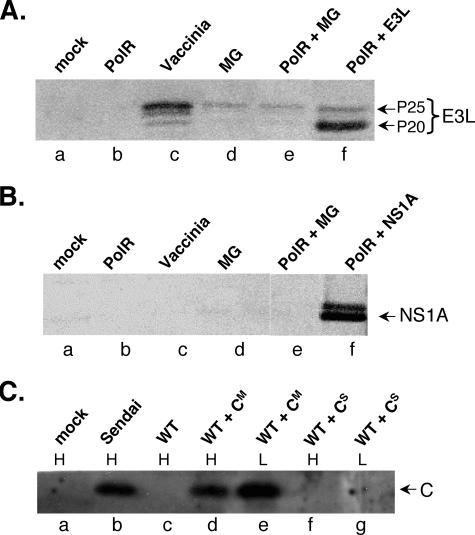
FIG. 2.
Western blot analysis of E3L (A), NS1A (B), and Sendai C (C) protein expressed in the VSV minigenome system. All samples originated from L-929 cells, except for lanes labeled H in panel C, which were derived from HeLa cells. Infections were carried out at an effective MOI of 10 PFU/cell for wt and polR VSV and a fourfold-higher multiplicity of minigenome particles (lanes d to f in panels A and B and lanes d to g in panel C). MG refers to control minigenome particles encoding VSV M and G proteins only. The control CS minigenome particles contained a C gene with a premature stop codon (see text). All extracts (∼12 h p.i. in the case of infected samples) were obtained from approximately equivalent numbers of cells. Vaccinia-T7-infected L-929 samples (MOI of 25 PFU/cell) and Sendai virus-infected HeLa cells (a gift from Dominique Garcin, University of Geneva School of Medicine) were included as controls.
Generation of recombinant VSV-GFP and VSV-PFG.
The EcoRV/SpeI fragment containing the enhanced green fluorescent protein gene was excised from the pBI-EGFP plasmid (Clonetech), blunted with Klenow DNA polymerase, and inserted at the SmaI site of pBS-GMMG(MCS). Both sense (pGMMG-GFP) and antisense orientations (pGMMG-PFG) of the resulting plasmids were recovered. To incorporate the GFP and PFG sequences into the full-length pVSVFL+ plasmid (39), the EcoRV fragment from the latter plasmid (containing the P gene) was first inserted into the corresponding site of pGMMG-GFP and pGMMG-PFG. The XbaI/NheI fragments from the resulting plasmids were then used to replace the corresponding ones in pVSVFL+. Infectious recombinant VSV-GFP and VSV-PFG viruses were then recovered as described previously (44a).
Coinfections with vaccinia virus, VSV minigenome particles, and VSV-GFP/VSV-PFG.
All VSV wt and VSV polR infections were carried out at an effective MOI of 10 (see reference 44a). For vaccinia coinfections, monolayers of BHK or L-929 cells (95% confluent) were first infected with vaccinia at an MOI of 10 PFU/cell for 2 h at 37°C prior to VSV infection. All such coinfections included 2 μg/ml act D from the time of addition of vaccinia virus. Independent experiments showed similar results when act D was added at the time of VSV infection. Coinfections of VSV minigenome particles were carried out simultaneously with wt or polR VSV (effective MOI of 10 PFU/cell) in amounts ranging from equivalent to infectious virus to a fourfold excess based on Northern blots of particle RNA. Coinfections with VSV-GFP and VSV-PFG were carried out at the indicated MOIs. Infected cell culture supernatants were collected 24 to 30 h p.i. and clarified by centrifugation (600 × g, 5 min) before determination of infectious virus yields and released particle content. The same clarified supernatants were also used for quantitation of released particles by Northern blot analysis of genomes as in the companion paper (44a), except for using a plus-sense M gene riboprobe copied from a pGEM plasmid encoding the entire VSV M gene. The same probe was used for Northern analysis of replication products in infected cells.
Analysis of intracellular viral RNAs and released virus particle RNAs.
Preparation of cytoplasmic extracts, RNA extractions, and analysis of transcription and replication products was carried out as described previously (44a), except the following Northern blot probes were used. A T7 transcript containing the entire plus-sense M gene sequence was used to detect minus-sense replication products, and an SP6 transcript containing the entire minus-sense N gene sequence was used to detect N transcripts. GFP sense transcripts were likewise probed using a minus-sense T7 transcript containing the entire gene sequence.
RESULTS
Coinfection with vaccinia virus relieves polR VSV growth restriction in L-929 cells.
wt VSV growth is blocked in L-929 cells pretreated with type I IFN, but coinfection with vaccinia virus relieves this block by preventing cellular PKR activation (61). Even in the absence of IFN pretreatment, vaccinia coinfection was reported to increase wt VSV yields 10- to 20-fold in L-929 cells (62). Some of the preliminary findings obtained in our lab several years ago also indicated that vaccinia virus coinfection stimulates polR VSV growth in L-929 cells (11), suggesting a parallel between polR restriction and the type I IFN-mediated antiviral response. We therefore set out to characterize the effects of vaccinia coinfection on recombinant polR growth in more detail. As in the accompanying paper (44a), BHK and L-929 cells were infected with polR VSV at an effective MOI of 10 PFU/cell but, in this case, with or without coinfection with vaccinia virus also at an MOI of 10 PFU/cell. All infections also included 2 μg/ml act D to inhibit vaccinia virus late functions, as in the earlier IFN rescue studies. act D has no significant effect on polR restriction (44a). In the nonrestricted BHK cells, vaccinia coinfection reduced wt VSV yields from ∼8,000 PFU/cell to ∼3,400 PFU/cell and polR yields from ∼3,700 PFU/cell to ∼1,000 PFU/cell (Fig. 1A, left panel). In L-929 cells, coinfection increased wt virus yields, as previously reported, but the effect here was more modest (∼150 PFU/cell to ∼520 PFU/cell). More importantly, coinfection in L-929 cells caused a dramatic increase in polR yields, i.e., from ∼0.5 PFU/cell to ∼220 PFU/cell (Fig. 1B, left panel). Restriction of polR infectious virus yields in L-929 cells is thus efficiently countered by vaccinia virus coinfection.
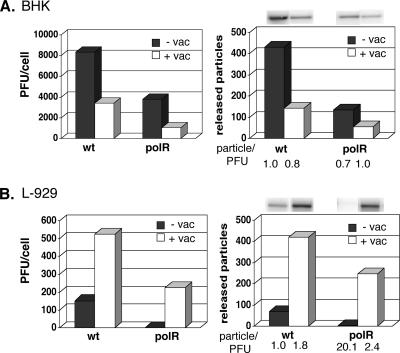
FIG. 1.
Effects of vaccinia virus coinfection on VSV wt and VSV polR growth in BHK (A) and L-929 (B) cells. All infections were done at an effective MOI of 10 PFU/cell for both VSV and vaccinia (vac) in the presence of 2 μg/ml act D. Infectious VSV yields (left panels) and released virus particle amounts (right panels) were determined at ∼24 h (BHK) or ∼30 h (L-929) p.i. by plaque assay in BHK cells and Northern blot analysis of virus genomes. +, with vaccinia virus coinfection; −, without vaccinia virus coinfection.
As documented in the preceding paper, polR restriction in L-929 cells is due in part to release of virus particles that are less infectious than those released by wt virus (≤10-fold). To test whether vaccinia virus coinfection also restored infectivity of released polR virus particles, the infected cell supernatants for which titers were determined above for PFU yields were also probed for the presence of virus genomes by Northern blotting as before. For comparative purposes, wt virus particle/PFU ratios in the absence of coinfection were arbitrarily given a value of 1.0. In BHK cells, vaccinia coinfection decreased wt and polR virus particle release by about the same amount as PFU and thus had little or no effect on particle/PFU ratios (Fig. 1A, right panel). In L-929 cells, coinfection increased wt particle release slightly more than PFU (∼8-fold versus ∼4-fold) but boosted polR particle release 62-fold (Fig. 1B, right panel), which restored particle/PFU ratios to near wt virus levels (from 20.1 to 2.4). Coinfection with vaccinia thus not only rescues polR virus particle release in L-929 cells but also restores their infectivity.
polR virus genome replication but not transcription is reduced compared to wt virus in infected L-929 cells (44a). Additional experiments showed that coinfection with vaccinia virus also restored the polR deficit in genome replication (data not shown, see also results below). We therefore conclude that vaccinia coinfection reverses all previously established blocks associated with polR virus growth restriction in L-929 cells. Moreover, based on previous findings showing rescue of VSV from an IFN-induced block under identical conditions, these findings suggest that polR virus restriction is in fact due to a cellular antiviral response.
Expression of dsRNA-binding proteins encoded by vaccinia virus and influenza viruses rescues polR virus restriction.
Previous work identified the vaccinia-encoded dsRNA-binding protein E3L as the critical component responsible for wt VSV growth rescue in IFN-treated L-929 cells (53). E3L is thought to prevent cellular PKR activation at least in part by sequestering dsRNA. To test whether polR rescue by vaccinia coinfection is due to E3L, we expressed this protein in a defective VSV minigenome that could be used in place of vaccinia for coinfection. This strategy ensured that E3L expression would occur in concert with polR infection in all cells, as in the vaccinia coinfection experiments. The defective minigenome approach also avoided possible biosafety concerns resulting from incorporation of the E3L gene in recombinant infectious VSV. We thus employed a well-characterized VSV minigenome, encoding only the virus M and G proteins (55), and engineered an E3L-encoding transcription unit between the two, using appropriate VSV transcriptional start and stop sequences (50). Particles containing the modified minigenome were recovered and amplified as previously described using the vaccinia-T7 expression system to provide trans-acting VSV functions (55).
As expected, coinfection of L-929 cells with polR virus and E3L minigenomes resulted in E3L protein expression (Fig. 2A). The amounts produced were comparable to a vaccinia-T7 infection, except that the P20 rather than the P25 form of the protein preferentially accumulated in the minigenome system (Fig. 2A, lane f versus lane c). However, both species of E3L are known to display similar dsRNA-binding and antiviral activities (38). Trace amounts of E3L were also evident in cells coinfected with control minigenomes (lanes d and e), which is due to small amounts of vaccinia-T7 virus remaining in the amplified minigenome preparations (see below).
L-929 cells were infected with wt and polR virus as described above and coinfected either with control minigenome particles (MG) or E3L-expressing minigenome particles (E3L). The latter were used at varying multiplicities, ranging from amounts equivalent to infectious virus particles (based on Northern blots of particle RNA) to four times as many. Figure 3 illustrates the effects on infectious yields at the highest input of minigenomes. For wt virus infections (Fig. 3A), MG particle coinfections had negligible effects on PFU yields, while E3L-expressing particles caused a relatively small decrease (∼ 60%). MG particles and E3L-expressing particle effects on wt yields in L-929 cells varied from experiment to experiment, ranging from slight inhibition to slight stimulation, typically within the range of accuracy of our plaque assays, i.e., twofold or less. For polR infections (Fig. 3B), MG particles increased virus yields ∼4-fold but E3L particles boosted PFU yields dramatically (46-fold). Coinfection with lower multiplicities of minigenome particles showed a dose-dependent response, but at higher multiplicities, infectious virus yields generally decreased (not shown), an effect presumably due to competition of minigenomes with standard virus for replication (see below). Additional studies established that the moderate increase in polR yields with MG coinfection, which also varied from experiment to experiment, was due to small amounts of vaccinia-T7 virus remaining in the amplified minigenome particle preparations (Fig. 2). We conclude from these results that rescue of polR infectious virus growth by vaccinia virus coinfection is mediated to a large extent by the E3L protein.
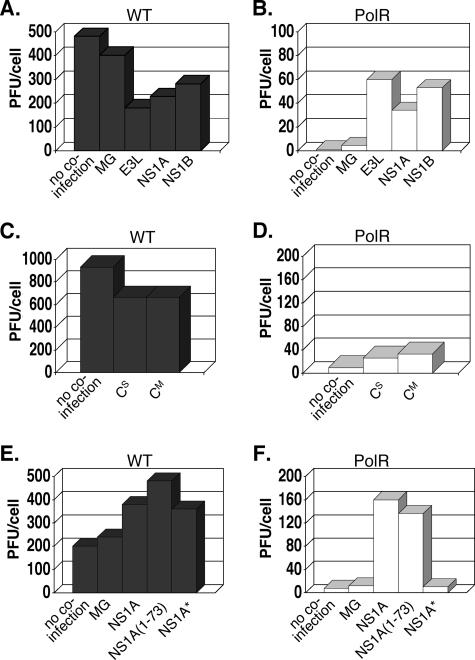
FIG. 3.
Effects of coinfections with minigenomes expressing various antiviral antagonist proteins on wt (A, C, and E) and polR (B, D, and F) infectious virus yields in L-929 cells. MG refers to control minigenome particles expressing only VSV M and G proteins. E3L, NS1A, NS1B, and CM designate minigenome particles expressing full-length wt antagonist proteins from vaccinia, influenza A, influenza B, and Sendai virus, respectively. CS is identical to CM except that it contains a stop codon preventing C protein expression. NS1A(1-73) expresses only the N-terminal RNA-binding domain of NS1A, while NS1A* expresses a mutant protein (R38A/K41A) lacking dsRNA-binding activity.
Although the dsRNA-binding activity of E3L is clearly implicated in wt VSV rescue from type I IFN-treated L-929 cells, it seemed possible that some other property of E3L was involved in polR rescue. To address this, we used the minigenome expression system to test the effects of other viral dsRNA-binding proteins known to function as antagonists of cellular antiviral defenses. NS1A protein from influenza A and NS1B protein from influenza B are distantly related to each other and unrelated to E3L but also bind to dsRNA (see the introduction). NS1A protein expression from minigenomes is shown in Fig. 2B (no antibody was available to us to confirm NS1B expression). The effects of NS1A and NS1B expression were tested in parallel to E3L in the experiment for which results are shown in Fig. 3. Coinfection with NS1A- or NS1B-expressing minigenome particles had little or no effect on wt virus yields (Fig. 3A) but boosted polR yields 35-fold and 55-fold, respectively (Fig. 3B). These results clearly show that the ability to rescue polR growth is not unique to E3L but is also displayed by other viral antagonist proteins that bind dsRNA.
To further test whether the above rescues correlated with the dsRNA-binding property of the proteins rather than some other antiviral antagonist property, we tested the effects of expressing Sendai virus C protein in the minigenome system. Sendai C lacks dsRNA-binding activity and functions as an antiviral antagonist by inhibiting STAT1 protein signaling required for IFN-induced gene activation (27, 28, 32). The control minigenome used in this case (CS) contained a C gene rendered nonfunctional by virtue of a translational stop codon (26). Western blot analysis confirmed that minigenome particles encoding CM but not CS expressed the C protein (Fig. 2C). Coinfection with either control CS particles or CM particles expressing a functional C protein showed only a slight inhibitory effect (∼30% decrease) on wt virus yields (Fig. 3C). With polR infections (Fig. 3D), both types of particles showed only slight increases in virus yields (two- to threefold). Western blot analysis of infected extracts revealed that CM was in fact produced in this minigenome system in amounts comparable to Sendai virus-infected cells (Fig. 2C). These results strongly suggest that rescue of polR growth restriction requires a protein with dsRNA-binding properties and that STAT1-mediated signaling is not required for restriction.
NS1A protein rescue activity maps to its dsRNA-binding domain.
While E3L, NS1A, and NS1B all share the ability to bind dsRNA, these proteins possess additional effector domains thought to play a role in their antiviral antagonist activity (8, 18, 35-37). To establish beyond a doubt that the dsRNA-binding activity of these proteins is solely responsible for polR rescue, we tested two mutant NS1A proteins: a truncated protein containing only the N-terminal dsRNA-binding domain (10), designated here as NS1A(1-73), and a full-size protein containing two points mutations (R38A/K41A) that destroy its dsRNA-binding property (17), designated here as NS1A*. The results comparing the effects of these mutant proteins versus full-size NS1A on virus yields are shown in Fig. 3E and F. For wt virus, all three NS1A constructs caused a small increase (2- to 2.5-fold) in wt virus yields. For polR infections, only the full-size NS1A protein and the truncated version NS1A(1-73) rescued polR virus yields significantly (20- and 17-fold, respectively). These results clearly show that the dsRNA-binding domain of NS1A, only 73 amino acids long, is sufficient for rescuing polR growth in L-929 cells.
Antiviral antagonist proteins rescue all aspects of polR virus growth restriction.
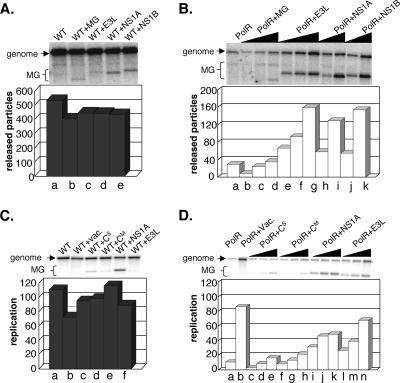
As for vaccinia virus coinfection, it was important to demonstrate that viral dsRNA-binding proteins did not simply boost virus yields but also restored the specific defects associated with polR restriction. Effects of minigenome coinfection in L-929 cells on wt and polR virus particle release, measured as described before, are shown in Fig. 4. All Northern blot samples of released particles were analyzed in parallel on the same gel but are shown separately for wt (Fig. 4A) and polR (Fig. 4B) to better illustrate the effects of minigenome coinfections. Note that both minigenomes and full-size genomes were observed in released particles, as expected, and the plots shown reflect the sum of the two. With wt virus, few minigenome particles of any kind were released (Fig. 4A, lanes b to e), even though these were present in fourfold excess in the inoculum. Cells infected with polR virus without coinfection released ∼17-fold-fewer full-size genomes than wt, as expected (Fig. 4B, lane a). This release was minimally affected by coinfection with increasing amounts of MG control minigenomes (Fig. 4B, lanes b to d). In contrast, coinfection with increasing amounts of E3L-, NS1A-, or NS1B-expressing particles led to a significant boost in particle release (lanes e to k). Moreover, in this case, particles containing full-size genomes as well as particles containing minigenomes were released. Note that the boost in particle release at the highest input of E3L was ∼5.5-fold relative to polR only (Fig. 4B, lane g versus a) which is about 11-fold less than that observed above with vaccinia virus coinfection (Fig. 1D). However, minigenome coinfection experiments involve a complex competition between replication of full-size genomes and minigenomes, with the latter providing a complementing function for polR replication. This is evident from the major increase in minigenome particle release in polR versus wt infections (Fig. 4B versus A), which is consistent with the requirement for expression of the rescuing proteins in the polR case. Nonetheless, these results clearly show that all three viral dsRNA-binding proteins substantially stimulate virus particle release. In contrast, coinfection with a minigenome expressing the Sendai C protein showed no significant increase in polR virus particle release (data not shown).
FIG. 4.
Effects of coinfections with minigenomes expressing various antiviral antagonist proteins in L-929 cells on wt virus particle release (A), polR virus particle release (B), wt intracellular replication products (C), and polR intracellular replication products (D), which were all probed by Northern blotting (see text). All released particle samples (panels A and B) and all intracellular replication samples taken at 8 h p.i. (panels C and D) were analyzed in parallel on the same gel and are shown separately to better illustrate effects of minigenome coinfections. Coinfection with vaccinia virus (vac) was included for comparison in panels C and D, lanes b.
We next tested whether the antiviral antagonist proteins also rescued the infectivity of released virus particles. Infectious virus yields and released particle data from the experiment for which results are shown in Fig. 4 were used to calculate a relative particle/PFU ratio, in this case using only full-size genome values (Table 1). As expected, released virus particles from wt infections showed little or no change in particle/PFU ratio upon coinfection with any of the minigenome particles. With polR infections, the control minigenome significantly lowered the particle/PFU ratio of released particles (∼3-fold), again likely due to low levels of vaccinia virus in the minigenome preparation but, more importantly, the viral antagonist-expressing minigenomes lowered the particle/PFU ratio to levels essentially identical to those of the wt (0.9 to 1.3). We conclude that expression of the antiviral antagonist proteins is sufficient to restore the infectivity of released polR particles.
TABLE 1.
Effect of minigenome coinfections on infectivity of released VSV particles from L-929 cellsa
| Virus and protein | Particle/PFU ratio |
|---|---|
| wt | 1.0 |
| wt + MG | 0.9 |
| wt + E3L | 2.2 |
| wt + NS1A | 1.7 |
| wt + NS1B | 1.3 |
| polR | 15.6 |
| polR + MG | 4.5 |
| polR + E3L | 1.3 |
| polR + NS1A | 1.2 |
| polR + NS1B | 0.9 |
The particle/PFU ratio of wt virus was arbitrarily set to 1.0.
Since inhibition of polR genome replication is also a hallmark of polR growth restriction, we examined the effects of expressing E3L, NS1A, or Sendai C protein on intracellular accumulation of wt and polR replication products in L-929 cells at 8 h p.i. (Fig. 4C and 4D). Vaccinia virus coinfection was included in this experiment for comparative purposes. Replication values in the plots shown refer to the sum of full-size genomes and minigenomes. Neither vaccinia coinfection nor coinfection with any of the minigenomes caused significant changes in overall genome replication in wt virus-infected cells (Fig. 4C). Note that E3L and Sendai C minigenomes replicated poorly in wt virus-infected cells (Fig. 4C, lanes c, d, and f), in agreement with particle release data, but NS1A minigenomes replicated more efficiently (lane e). This phenomenon was confirmed repeatedly in additional experiments, but the basis for the enhanced replicative ability of the NS1A-expressing minigenome remains unclear. More importantly, the low level of polR genome replication was stimulated 4.6-fold at the highest input of NS1A minigenomes and 6.4-fold at the highest input of E3L minigenomes (Fig. 4D, lanes a versus k and n) compared to an 8.1-fold stimulation obtained with vaccinia coinfection (lane b). In contrast, the highest input of CM-expressing and Cs-expressing minigenomes (lanes e and h) stimulated replication no more than twofold, as expected from the released particle data of Fig. 3. We conclude from these results that the block in intracellular genome replication characteristic of polR restriction in L-929 cells is also relieved by expression of dsRNA-binding antiviral antagonist proteins to levels approaching vaccinia coinfection. All aspects of polR restriction are thus rescued by expression of these proteins.
Overproduction of dsRNA in wt virus infection causes a host range growth restriction similar to polR virus.
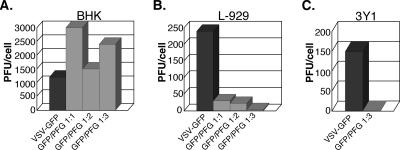
The above results clearly implicated enhanced production of dsRNA, or some highly structured RNA, as the root cause for polR restriction. If so, we wondered whether production of any kind of dsRNA during wt virus infections might lead to the same host range growth defect. To test this, we devised a simple strategy to enhance dsRNA production in wt virus infection while making no changes in viral proteins. Accordingly, we engineered two recombinant viruses, one with an additional transcription unit expressing the GFP reporter gene between the viral M and G genes, and one expressing the identical GFP sequence in the opposite orientation. Coinfection of cells with the two recombinant viruses, designated here VSV-GFP and VSV-PFG, would then be expected to generate large amounts of dsRNA due to annealing of complementary reporter gene sequences. Initial experiments showed that the growth properties of VSV-GFP and VSV-PFG were identical to those of the wt and produced similar yields of infectious virus and that each virus generated the expected sense and antisense GFP transcripts (data not shown). Coinfection experiments were then carried out in BHK and L-929 cells, as well as 3Y1 cells, which restrict polR growth in the same manner as L-929 cells (see reference 44a) to determine the effects on infectious virus yields. Addition of increasing amounts of VSV-PFG relative to VSV-GFP in the inoculum had little or no effect on PFU yields in BHK cells but dramatically inhibited wt virus yields in L-929 cells and 3Y1 cells (Fig. 5). Growth inhibition increased from ∼10-fold at 1:1 ratio of infecting viruses to ∼80-fold at a 3:1 ratio in L-929 cells and >100-fold at a 3:1 ratio in 3Y1 cells. We presume the increase in restriction at a 3:1 virus ratio compared to a 1:1 ratio reflects an increase in the number of coinfected cells. As expected, yields from infections with VSV-GFP virus only were not affected by increasing the MOI from 5 to 20 PFU/cell (data not shown). HeLa cells, which do not restrict polR virus growth (12), behaved similarly to BHK cells in this assay (data not shown). These striking results not only provide very strong support for the hypothesis that enhanced dsRNA production is responsible for polR growth restriction but also show that no specific viral sequence or other polR virus property is involved in restriction. In addition, these data suggest that growth restriction may be quantitatively similar when a relatively small excess of dsRNA is produced (polR infections) and when a large excess is produced (VSV-GFP/PFG). However, we have not directly measured amounts of dsRNA accumulating in polR infections versus VSV-GFP/PFG coinfections.
FIG. 5.
Effect of VSV-GFP/VSV-PFG coinfection on infectious virus yields in BHK (A), L-929 (B), and 3Y1 (C) cells. Infection with VSV-GFP was carried out at an effective MOI of 10 PFU/cell (BHK) and 5 PFU/cell (L-929 and 3Y1). For coinfections, VSV-PFG was added at the same MOI as VSV-GFP (1:1) or at twofold higher (1:2) or threefold higher (1:3).
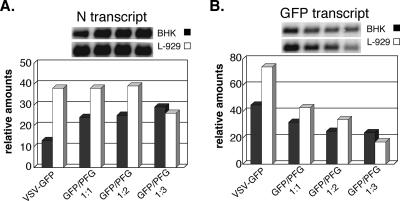
To further substantiate whether the same restriction phenomenon is involved in the wt GFP/PFG virus coinfection as for polR, we carried out Northern blot analysis of VSV N transcripts (Fig. 6A) and GFP coding-sense transcripts (Fig. 6B) in both permissive BHK cells and restrictive L929 cells at 8 h p.i. N transcript amounts increased slightly (∼2.5-fold) with increasing ratios of VSV-PFG/VSV-GFP in BHK cells. In L-929 cells, N transcript amounts were ∼3-fold higher than in BHK cells when infected with VSV-GFP only and remained essentially unchanged with coinfection with VSV-PFG, despite the precipitous drop in infectious virus yields. This lack of effect on genome transcription is a hallmark of polR restriction. GFP sense transcripts also accumulated in these coinfections, as expected, but in contrast to viral N transcripts, their amounts decreased with increasing VSV PFG/GFP ratios (Fig. 6B). These results are consistent with formation of dsRNA due to annealing of GFP and PFG transcripts and known effects of cellular DICER activity on dsRNA (6). Note that degradation of GFP transcripts was more pronounced in L-929 than in BHK cells (∼5-fold versus ∼2-fold decrease), which may indicate a difference in the amounts and/or activity of DICER in these two cell types.
FIG. 6.
Effect of VSV-GFP/VSV-PFG coinfection on viral N transcript and GFP transcript accumulation in BHK and L-929 cells. Infections were carried out as described for Fig. 5. Samples for Northern blotting were taken at 6 h p.i. and 8 h p.i. for BHK and L-929 cells, respectively.
We next tested whether VSV-GFP/VSV-PFG coinfection also involved release of virus particles with reduced infectivity. Ratios of PFU/genome in released particles showed no significant change for VSV-GFP versus coinfections with VSV-GFP/VSV-PFG (≤10%) in BHK cells. In contrast, coinfections with a threefold excess of VSV-PFG over VSV-GFP in L-929 cells reduced released particle infectivity ∼20-fold (data not shown), again mimicking polR results.
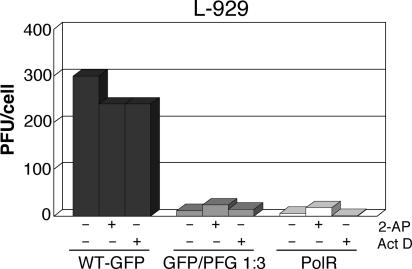
Lastly, we wanted to test if wt VSV restriction mediated by dsRNA is also independent of PKR activation and host gene transcriptional activation as for polR (44a). The effects of 2-AP and act D on yields from VSV GFP/PFG coinfections and polR in L-929 cells are shown in Fig. 7. In the absence of either inhibitor, virus growth restriction in VSV-GFP/PFG coinfections was similar to polR infections, i.e., ∼25- and 50-fold, respectively. In the presence of 2-AP, VSV-GFP growth was essentially unaffected, while polR yields were slightly increased (∼3-fold), in agreement with previous results (44a), and GFP/PFG coinfection titers increased ∼2-fold. Blocking host gene transcription with act D likewise had little or no effect on VSV-GFP/PFG coinfections or polR infection (Fig. 7). These results leave little doubt that wt virus growth restriction caused by enhanced dsRNA production occurs by the same mechanism as polR virus restriction.
FIG. 7.
Effect of 10 mM 2-AP and 5 μg/ml act D on infectious virus yields from VSV-GFP infection or VSV-GFP/VSV-PFG coinfection of L-929 cells at the same MOI as used for Fig. 5. polR infection (effective MOI of 10 PFU/cell) was included for comparison. Inhibitors were present throughout infection. +, inhibitor present; −, inhibitor absent.
DISCUSSION
We showed in an accompanying paper (44a) that the VSV polR mutant displays a unique growth defect in several established cell lines. Under single-cycle infection conditions, mouse L-929 cells yield ∼100-fold less infectious polR virus than the wt, while rat 3Y1 cells yield ∼40-fold less. In contrast, BHK cells yield only slightly less mutant virus than the wt. The nature of this host range phenotype is remarkable in that only two specific steps in virus multiplication are targeted: genome replication and infectivity of released virus particles. We hypothesized that polR restriction might be due to a cellular antiviral response based mainly on two observations: (i) polR infection results in more extensive eIF-2α phosphorylation than wt in L-929 cells and (ii) polR virus RNA synthesis is anomalous and generates unencapsidated replication products that could possibly lead to enhanced levels of dsRNA. We tested this hypothesis here by first taking a lead from earlier studies showing that vaccinia virus coinfection rescues wt VSV growth from the inhibitory effects of type I IFN in L-929 cells by sequestering dsRNA and averting cellular PKR activation (53). Our results showed that vaccinia coinfection restored polR virus genome replication and infectivity of released particles to wt VSV levels in L-929 cells under conditions identical to those used earlier for wt VSV rescue in IFN-pretreated cells (61). We then proceeded to show that expression of the vaccinia virus E3L protein alone, via coinfection with an engineered defective VSV minigenome, also rescued polR VSV growth in L-929 cells.
Although the C-terminal dsRNA-binding domain of E3L has been implicated in VSV rescue from a type I IFN-mediated block (53), recent studies suggest that the N-terminal Z-DNA binding domain of this protein also contributes to its antiviral antagonist properties (8, 36, 37). We therefore tested other dsRNA-binding proteins for their effects on polR restriction. Both NS1A and NS1B proteins from influenza A and influenza B viruses, respectively, rescued polR growth in L-929 cells. These two antiviral antagonist proteins share a structurally similar N-terminal RNA-binding domain but their C-terminal domains, which also participate in blocking the host cell antiviral response, are unrelated to each other or to E3L (35). The NS1A C-terminal domain inhibits host cell mRNA processing (9), while that from NS1B blocks the functioning of the cellular ubiquitin-like ISG15 protein (66). In contrast to E3L and influenza NS1 proteins, expression of Sendai virus C protein, an antiviral antagonist protein that lacks dsRNA-binding activity, had no effect on polR restriction. Notably, Sendai C can also rescue VSV from a type I IFN-mediated block but does so by virtue of inhibiting STAT1 protein activity, which is required for IFN signaling (25, 27). The inability of Sendai C to rescue polR is thus consistent with our evidence that restriction is not due to IFN-mediated signaling (44a).
The effects of E3L, NS1A, and NS1B proteins on polR restriction implicate sequestering of dsRNA or some highly structured RNA as the likely mechanism for rescue. To further substantiate this conclusion, we showed here that the N-terminal RNA-binding domain of NS1A, only 73 amino acids in length (10), rescued polR, while a full-size NS1A protein harboring two amino acid substitutions that abolish RNA binding (17) failed to do so. These results leave little doubt that dsRNA sequestering is the basis for polR rescue.
If overproduction of viral dsRNA is solely responsible for polR restriction, then wt virus might be expected to show the same phenomenon if more dsRNA is produced during infection. We showed that this is in fact the case by coinfecting cells with two infectious recombinant viruses, one expressing the sense strand of the enhanced GFP gene (VSV-GFP) and one expressing the antisense strand (VSV-PFG). Remarkably, coinfection with these viruses mimicked all aspects of the polR restriction phenotype. Infectious virus yields were unaffected by coinfection in BHK or HeLa cells but were reduced 100-fold or more in L-929 and 3Y1 cells. This restriction had no effect on viral mRNA accumulation, and released virus particles displayed ∼20-fold-lower infectivity than those produced by infection with VSV-GFP alone. Moreover, as for polR, neither act D nor 2-AP rescued the block in virus growth. Notably, GFP transcript accumulation was reduced in coinfected cells, as might be expected from DICER-mediated degradation of dsRNA (6). We conclude from these results that dsRNA need not originate from a VSV sequence to cause growth restriction and that enhanced production of unrelated dsRNA, in the absence of the polR N mutation, is sufficient for triggering this constitutive host cell antiviral activity. For brevity, we refer to this phenomenon as the CAVE (constitutive antiviral effector) response.
Interestingly, what we already know about the changes in viral RNA synthesis caused by the polR mutation suggests an obvious candidate source for enhanced levels of dsRNA. The mutant virus synthesizes substantial amounts of unencapsidated plus-sense replication products in vitro, although this illegitimate synthesis lacks processivity and generates heterogeneous size transcripts ∼200 to 400 nucleotides long (45). Small amounts of these dead-end products are detectable in polR-infected cells when assayed by RNase protection of a 3′-end-labeled genome probe (unpublished). This phenomenon is not unique to VSV polR, as analogous abortive replication products terminating heterogeneously within the N gene have also been shown to accumulate in cells infected with Sendai virus Z strain (59). What is important here, however, is that unencapsidated replication products of both plus and minus sense very likely accumulate in these virus infections. This can be inferred from our earlier studies showing that polR virus readily reads through the minus-sense leader termination site of copy-back defective interfering particle templates in vitro in the absence of encapsidation (45). Accumulation of unencapsidated minus-sense replication products in infected cells, even if only a few hundred nucleotides long, should produce dsRNA by annealing to plus-sense L gene mRNA transcripts whose 3′ ends terminate only 59 nucleotides from the 5′ end of the genome. We propose that this annealing reaction not only accounts for enhanced dsRNA production in polR-infected cells but also serves as the minimal source of dsRNA necessary for triggering the classical cellular antiviral transcriptional response in wt virus-infected cells. This same annealing phenomenon likely applies to other nonsegmented negative-strand RNA viruses.
Marcus and Sekellick (41) reported long ago that a single molecule of preformed dsRNA within a defective interfering VSV particle is sufficient for full induction of type I IFN in aged chicken embryo fibroblasts. Although this important finding may be somewhat unique to the cell and virus system used, it nonetheless underscores the notion that extremely small amounts of dsRNA suffice to trigger IFN induction. But our findings here show that dsRNA accumulation in excess of what is normally seen in VSV-infected cells triggers an IFN-independent antiviral response in at least some cell types. The threshold level of dsRNA required for this CAVE response is as yet ill defined and could well differ between cell types and under varying growth conditions. However, we have shown previously that polR revertant viruses display varying levels of restriction that correlate with the extent of abortive replication product synthesis (12). These findings suggest that the amount of dsRNA generated by the revertant viruses affects the magnitude of the CAVE response, at least beyond the threshold found in wt-infected cells. Nonetheless, the amounts produced in polR-infected L-929 cells appear to be nearly saturating for this antiviral response, since the extent of restriction is roughly similar to that of VSV-GFP/VSV-PFG coinfections, where much larger amounts of dsRNA are undoubtedly generated (likely equivalent to G transcript levels in wt infections).
The nature of the constitutive antiviral effector(s) triggered by overproduction of dsRNA remains unclear. PKR is known to play a central role in the classical antiviral response to VSV in cell culture as well as in the innate immune response to VSV in mice (3, 19, 56). PKR is upregulated by type I IFN and, upon binding to dsRNA, phosphorylates eIF-2α, which effectively shuts down protein translation (4, 48, 63). But the CAVE response is insensitive to 2-AP, a well-known inhibitor of PKR activity, and viral protein synthesis inhibition is not responsible for polR restriction (44a). Likewise, involvement of the dsRNA-activated OAS pathway can also be ruled out, since no degradation of viral mRNAs takes place. A role for cellular adenosine deaminase, also directly activated by dsRNA (48), is unlikely, since antiviral activity in this case would require that viral dsRNA function as an intermediate in replication. Several other cellular proteins display dsRNA-binding properties, but none of these have so far been shown to possess antiviral effector activity (29, 49). Given that restriction involves both an inhibition of genome replication as well as reduced virus particle infectivity, more than one antiviral effector activity is likely triggered by overproduction of dsRNA, perhaps in response to a single upstream signaling component with low affinity for dsRNA.
Recent studies have identified two cytoplasmic RNA helicases, RIG-I and MDA5, that act as immediate sensors of virally produced dsRNA and elicit the signaling cascade required for the classical type I IFN induction response (2, 65). In fact, the role of RIG-I in type I IFN induction was first uncovered in VSV-infected L-929 cells (64), but in our studies here and in the previous study (44a), neither act D nor 2-AP, both of which inhibit type I IFN induction in L-929 cells (16, 42), showed a significant effect on polR or VSV-GFP/PFG coinfection restriction in L-929 cells. These results argue against a role for RIG-I or MDA5 in triggering the CAVE response. Nonetheless, in the absence of transcription inhibitors, this classical signaling pathway may well lead to host gene transcriptional activation in the presence of enhanced levels of dsRNA, even if this activation plays no role in restricting virus growth under the single-cycle infections examined here. The same considerations also apply to TLR-3, a transmembrane receptor found predominantly in cells involved in the immune response (52), which also triggers the host cell antiviral transcriptional response when it binds dsRNA originating from extracellular sources. We are currently addressing whether RIG-I, MDA5, or TLR3 activation or downstream activation of key transcription factors such as IRF-3 might have roles in addition to host gene induction and somehow lead to activation of constitutive antiviral effectors.
Lastly, could the constitutive antiviral response described here play some role in natural virus infections? As mentioned above, natural strains of Sendai virus differ in their ability to generate unencapsidated replication products in infected cells, so it seems very likely that the extent of viral dsRNA accumulation in infected cells varies depending on the particular virus source and perhaps also infected cell type, at least as far as nonsegmented negative-strand viruses are concerned. Available evidence so far indicates that such viruses do not encode dsRNA-binding proteins. These agents thus have the potential to produce elevated amounts of dsRNA and possibly trigger a CAVE response. This constitutive cellular antiviral activity might serve to limit production of infectious virus in initially infected cells, reduce spread to adjacent cells or tissues, and even facilitate more efficient induction of the classical IFN response. Enhanced dsRNA production by other types of viruses could conceivably also trigger a CAVE response, or some other IFN-independent antiviral response, at least under some conditions. Our studies suggest novel ways of influencing dsRNA production during viral infections and determining possible effects on antiviral responses in a variety of contexts.
Acknowledgments
This investigation was supported in part by a grant from NIAID (RO1-AI21572) to J.P. and by a scholarship from the ARCS Foundation, Inc., to D.O.
We thank Michael Whitt, Bertram Jacobs, Adolfo Garcia-Sastre, Robert Krug, Bernard Moss, and Daniel Kolakofsky for providing reagents. We are also grateful to Joseph Chuang for early contributions to this work.
Footnotes
Published ahead of print on 25 October 2006.
REFERENCES
- 1.Ahmed, M., M. O. McKenzie, S. Puckett, M. Hojnacki, L. Poliquin, and D. S. Lyles. 2003. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J. Virol. 77:4646-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balachandran, S., P. C. Roberts, L. E. Brown, H. Truong, A. K. Pattnaik, D. R. Archer, and G. N. Barber. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129-141. [DOI] [PubMed] [Google Scholar]
- 4.Barber, G. N. 2005. The dsRNA-dependent protein kinase, PKR and cell death. Cell Death Differ. 12:563-570. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann, M., A. Garcia-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 7.Black, B. L., and D. S. Lyles. 1992. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J. Virol. 66:4058-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandt, T., M. C. Heck, S. Vijaysri, G. M. Jentarra, J. M. Cameron, and B. L. Jacobs. 2005. The N-terminal domain of the vaccinia virus E3L-protein is required for neurovirulence, but not induction of a protective immune response. Virology 333:263-270. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien, C. Y., Y. Xu, R. Xiao, J. M. Aramini, P. V. Sahasrabudhe, R. M. Krug, and G. T. Montelione. 2004. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry 43:1950-1962. [DOI] [PubMed] [Google Scholar]
- 11.Chuang, J. L. 1997. Vesicular stomatitis virus transcription and replication; insights derived from the poIR mutants. Ph.D. thesis. University of California—San Diego and San Diego State University.
- 12.Chuang, J. L., R. L. Jackson, and J. Perrault. 1997. Isolation and characterization of vesicular stomatitis virus PoIR revertants: polymerase readthrough of the leader-N gene junction is linked to an ATP-dependent function. Virology 229:57-67. [DOI] [PubMed] [Google Scholar]
- 13.Chuang, J. L., and J. Perrault. 1997. Initiation of vesicular stomatitis virus mutant polR1 transcription internally at the N gene in vitro. J. Virol. 71:1466-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connor, J. H., and D. S. Lyles. 2005. Inhibition of host and viral translation during vesicular stomatitis virus infection. eIF2 is responsible for the inhibition of viral but not host translation. J. Biol. Chem. 280:13512-13519. [DOI] [PubMed] [Google Scholar]
- 15.Connor, J. H., and D. S. Lyles. 2002. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 76:10177-10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daigneault, L., A. Haggarty, Q. H. Meng, and D. Skup. 1992. Two distinct pathways of interferon induction as revealed by 2-aminopurine. Nucleic Acids Res. 20:2749-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donelan, N. R., C. F. Basler, and A. Garcia-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77:13257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donelan, N. R., B. Dauber, X. Wang, C. F. Basler, T. Wolff, and A. Garcia-Sastre. 2004. The N- and C-terminal domains of the NS1 protein of influenza B virus can independently inhibit IRF-3 and beta interferon promoter activation. J. Virol. 78:11574-11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durbin, R. K., S. E. Mertz, A. E. Koromilas, and J. E. Durbin. 2002. PKR protection against intranasal vesicular stomatitis virus infection is mouse strain dependent. Viral Immunol. 15:41-51. [DOI] [PubMed] [Google Scholar]
- 20.Enninga, J., D. E. Levy, G. Blobel, and B. M. Fontoura. 2002. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science 295:1523-1525. [DOI] [PubMed] [Google Scholar]
- 21.Faria, P. A., P. Chakraborty, A. Levay, G. N. Barber, H. J. Ezelle, J. Enninga, C. Arana, J. van Deursen, and B. M. Fontoura. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 17:93-102. [DOI] [PubMed] [Google Scholar]
- 22.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 25.Garcin, D., J. Curran, M. Itoh, and D. Kolakofsky. 2001. Longer and shorter forms of Sendai virus C proteins play different roles in modulating the cellular antiviral response. J. Virol. 75:6800-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcin, D., J. B. Marq, F. Iseni, S. Martin, and D. Kolakofsky. 2004. A short peptide at the amino terminus of the Sendai virus C protein acts as an independent element that induces STAT1 instability. J. Virol. 78:8799-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hengel, H., U. H. Koszinowski, and K. K. Conzelmann. 2005. Viruses know it all: new insights into IFN networks. Trends Immunol. 26:396-401. [DOI] [PubMed] [Google Scholar]
- 31.Jackson, R. L., D. Spadafora, and J. Perrault. 1995. Hierarchal constitutive phosphorylation of the vesicular stomatitis virus P protein and lack of effect on P1 to P2 conversion. Virology 214:189-197. [DOI] [PubMed] [Google Scholar]
- 32.Kato, A., Y. Ohnishi, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2001. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol. 75:3802-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 34.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 35.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181-189. [DOI] [PubMed] [Google Scholar]
- 36.Kwon, J. A., and A. Rich. 2005. Biological function of the vaccinia virus Z-DNA-binding protein E3L: gene transactivation and antiapoptotic activity in HeLa cells. Proc. Natl. Acad. Sci. USA 102:12759-12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langland, J. O., and B. L. Jacobs. 2004. Inhibition of PKR by vaccinia virus: role of the N- and C-terminal domains of E3L. Virology 324:419-429. [DOI] [PubMed] [Google Scholar]
- 38.Langland, J. O., and B. L. Jacobs. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133-141. [DOI] [PubMed] [Google Scholar]
- 39.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 92:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, S., J. Y. Min, R. M. Krug, and G. C. Sen. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13-21. [DOI] [PubMed] [Google Scholar]
- 41.Marcus, P. I., and M. J. Sekellick. 1977. Defective interfering particles with covalently linked [+/−]RNA induce interferon. Nature 266:815-819. [DOI] [PubMed] [Google Scholar]
- 42.Marcus, P. I., and M. J. Sekellick. 1988. Interferon induction by viruses. XVI. 2-Aminopurine blocks selectively and reversibly an early stage in interferon induction. J. Gen. Virol. 69:1637-1645. [DOI] [PubMed] [Google Scholar]
- 43.McWhirter, S. M., B. R. Tenoever, and T. Maniatis. 2005. Connecting mitochondria and innate immunity. Cell 122:645-647. [DOI] [PubMed] [Google Scholar]
- 44.Meager, A. 2002. Biological assays for interferons. J. Immunol. Methods 261:21-36. [DOI] [PubMed] [Google Scholar]
- 44a.Ostertag, D., T. M. Hoblitzell-Ostertag, and J. Perrault. 2007. Cell-type-specific growth restriction of vesicular stomatitis virus polR mutants is linked to defective viral polymerase function. J. Virol. 81:492-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrault, J., G. M. Clinton, and M. A. McClure. 1983. RNP template of vesicular stomatitis virus regulates transcription and replication functions. Cell 35:175-185. [DOI] [PubMed] [Google Scholar]
- 46.Romano, P. R., F. Zhang, S. L. Tan, M. T. Garcia-Barrio, M. G. Katze, T. E. Dever, and A. G. Hinnebusch. 1998. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol. Cell. Biol. 18:7304-7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rose, J. K., and M. A. Whitt. 2001. Rhabdoviridae: the viruses and their replication, p. 1221-1244. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 48.Samuel, C. E. 1991. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology 183:1-11. [DOI] [PubMed] [Google Scholar]
- 49.Saunders, L. R., and G. N. Barber. 2003. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 17:961-983. [DOI] [PubMed] [Google Scholar]
- 50.Schnell, M. J., L. Buonocore, M. A. Whitt, and J. K. Rose. 1996. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J. Virol. 70:2318-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schroder, M., and A. G. Bowie. 2005. TLR3 in antiviral immunity: key player or bystander? Trends Immunol. 26:462-468. [DOI] [PubMed] [Google Scholar]
- 52.Sen, G. C., and S. N. Sarkar. 2005. Transcriptional signaling by double-stranded RNA: role of TLR3. Cytokine Growth Factor Rev. 16:1-14. [DOI] [PubMed] [Google Scholar]
- 53.Shors, S. T., E. Beattie, E. Paoletti, J. Tartaglia, and B. L. Jacobs. 1998. Role of the vaccinia virus E3L and K3L gene products in rescue of VSV and EMCV from the effects of IFN-alpha. J. Interferon Cytokine Res. 18:721-729. [DOI] [PubMed] [Google Scholar]
- 54.Spadafora, D., D. M. Canter, R. L. Jackson, and J. Perrault. 1996. Constitutive phosphorylation of the vesicular stomatitis virus P protein modulates polymerase complex formation but is not essential for transcription or replication. J. Virol. 70:4538-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stillman, E. A., J. K. Rose, and M. A. Whitt. 1995. Replication and amplification of novel vesicular stomatitis virus minigenomes encoding viral structural proteins. J. Virol. 69:2946-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stojdl, D. F., N. Abraham, S. Knowles, R. Marius, A. Brasey, B. D. Lichty, E. G. Brown, N. Sonenberg, and J. C. Bell. 2000. The murine double-stranded RNA-dependent protein kinase PKR is required for resistance to vesicular stomatitis virus. J. Virol. 74:9580-9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan, S. L., and M. G. Katze. 1998. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J. Interferon Cytokine Res. 18:757-766. [DOI] [PubMed] [Google Scholar]
- 59.Vidal, S., and D. Kolakofsky. 1989. Modified model for the switch from Sendai virus transcription to replication. J. Virol. 63:1951-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, W., and R. M. Krug. 1996. The RNA-binding and effector domains of the viral NS1 protein are conserved to different extents among influenza A and B viruses. Virology 223:41-50. [DOI] [PubMed] [Google Scholar]
- 61.Whitaker-Dowling, P., and J. S. Youngner. 1983. Vaccinia rescue of VSV from interferon-induced resistance: reversal of translation block and inhibition of protein kinase activity. Virology 131:128-136. [DOI] [PubMed] [Google Scholar]
- 62.Whitaker-Dowling, P., and J. S. Youngner. 1988. Vaccinia virus stimulates the growth of vesicular stomatitis virus at the level of protein synthesis in mouse L cells. Virus Res. 10:215-224. [DOI] [PubMed] [Google Scholar]
- 63.Williams, B. R. 2001. Signal integration via PKR. Sci. STKE 2001:RE2. [DOI] [PubMed]
- 64.Yoneyama, M., and T. Fujita. 2004. RIG-I: critical regulator for virus-induced innate immunity. Tanpakushitsu Kakusan Koso 49:2571-2578. (In Japanese.) [PubMed] [Google Scholar]
- 65.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 66.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]