Abstract
Viral pathogenesis depends on a suitable milieu in target host cells permitting viral gene expression, propagation, and spread. In many instances, viral genomes can be manipulated to select for propagation in certain tissues or cell types. This has been achieved for the neurotropic poliovirus (PV) by exchange of the internal ribosomal entry site (IRES), which is responsible for translation of the uncapped plus-strand RNA genome. The IRES of human rhinovirus type 2 (HRV2) confers neuron-specific replication deficits to PV but has no effect on viral propagation in malignant glioma cells. We report here that placing the critical γ134.5 virulence genes of herpes simplex virus type 1 (HSV) under translation control of the HRV2 IRES results in neuroattenuation in mice. In contrast, IRES insertion permits HSV propagation in malignant glioma cell lines that do not support replication of HSV recombinants carrying γ134.5 deletions. Our observations indicate that the conditions for alternative translation initiation at the HRV2 IRES in malignant glioma cells differ from those in normal central nervous system (CNS) cells. Picornavirus regulatory sequences mediating cell type-specific gene expression in the CNS can be utilized to target cancerous cells at the level of translation regulation outside their natural context.
Herpes simplex virus type 1 (HSV) and poliovirus (PV) are neuropathogens that target cells in the central nervous system (CNS). Both use cellular receptors belonging to the nectin family: nectin-1 or the PV receptor-related molecule 1 for HSV (15) and CD155 or the PV receptor for PV (35). Both viruses have been genetically engineered to attenuate neuropathogenicity. Nonneurovirulent variants of both HSV and PV selectively propagate in malignant glioma cells and are being investigated as oncolytic agents against glioblastoma multiforme (GBM).
The HSV double-stranded DNA (dsDNA) genome of ∼152,250 bp encodes at least 84 gene products. Attenuation was achieved by deletion of the γ134.5 gene, abolishing neurovirulence in mice (2, 5) and primates (21). Moreover, Δγ134.5 HSVs, either with or without a mutation in another nonessential gene, have been safely administered to GBM patients (32, 48). A key function of the γ134.5 gene product, infected-cell protein 34.5 (ICP34.5), is to oppose host antiviral responses that culminate in protein synthesis shutoff. HSV infection results in dsRNA-dependent protein kinase (PKR) activation, which unchecked, arrests translation via phosphorylation of the α subunit of eukaryotic initiation factor (eIF-2α) (4, 6). ICP34.5 redirects cellular protein phosphatase 1α to dephosphorylate eIF-2α, maintaining a pool of active eIF-2α (19). Δγ134.5 mutants are unable to counter PKR, and viral growth is therefore inhibited in the CNS (4, 6). However, these mutants propagate efficiently in tumor cells with defective or suppressed PKR responses (1), spurring clinical application as oncolytic agents (32, 48).
Δγ134.5 growth is impeded in some cancer cell lines (39, 50), indicating that ICP34.5 is required for robust propagation in these cell types. HSVs that conditionally express ICP34.5 in GBM would resolve this impasse, selectively supplying the protein in tumor cells resistant to Δγ134.5 viruses. This strategy has been pursued by selection for second-site suppressor mutants capable of PKR inhibition yet attenuated for neurovirulence (52) or by placing γ134.5 under transcriptional control of glioma-specific promoters (24, 25, 43).
We report here engineering of HSVs specifically targeting GBM at the level of translation control. This was achieved by replacing noncoding regions driving translation of the γ134.5 transcript with picornavirus 5′ and 3′ untranslated regions (UTRs) conferring active gene expression in GBM, while thwarting ICP34.5 synthesis in the CNS.
Neurovirulence of PV, a plus-strand RNA virus of the Picornaviridae family associated with paralytic poliomyelitis, depends on its internal ribosomal entry site (IRES). The ∼7,400-nucleotide (nt) PV genome consists of a single open reading frame (ORF) (27), which is translated in a 5′ end-, cap-independent fashion by an IRES in the ∼750-nt 5′UTR (22, 45). The PV IRES codetermines pathogenicity, because substitution with the corresponding segment of human rhinovirus type 2 (HRV2) eliminates neurovirulence in human PV receptor transgenic mice (16) and in primates (17). The HRV2 IRES produces neuron-specific growth defects (3, 16) but does not affect propagation in established (18) and primary (36) GBM cell lines. PV/HRV2 chimeras efficiently reduce the tumor burden in GBM xenograft animal models (18, 44). Specificity for GBM is attributed to neuronal repressors of HRV2 IRES function, the dsRNA binding protein 76 (DRBP76) and nuclear factor 45 (NF45) (37, 38). DRBP76 forms a heterodimer with NF45 in neuronal cells (but not in GBM cells) (38), which binds to the HRV2 IRES and produces a nonperforming ribonucleoprotein (RNP) complex (38). The heterodimer associates with the translation apparatus exclusively in neuronal cells and inhibits translation initiation at the HRV2 IRES (38).
Placing the HSV γ134.5 transcript under translation control of the HRV2 IRES leads to efficient ICP34.5 expression in HRV2 IRES-permissive cell lines. Insertion of picornavirus UTR sequences does not compromise the integrity of the γ134.5 transcript in HSV-infected cells. HSV recombinants expressing ICP34.5 under HRV2 IRES control display vastly improved growth in glioma cells resistant to Δγ134.5 virus. Neurovirulence assessment in mice indicates that ICP34.5 is not expressed or is expressed at very low levels in neuronal cells, as the recombinant virus was safely administered to mice over a range of doses that were lethal with wild-type (wt) HSV.
MATERIALS AND METHODS
Cells and growth curves.
Vero and U373 cells (ATCC, Manassas, VA) were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with penicillin, streptomycin, and amphotericin B and 10% fetal bovine serum (FBS). SMA-560 cells (a gift from D. Bigner, Duke University) were propagated in 1:1 DMEM-Ham's F-12 medium (Invitrogen, Carlsbad, CA) containing 10% FBS and penicillin, streptomycin, and amphotericin B. Cells were maintained at 37°C in 5% CO2. For multicycle growth curves, 106 cells were plated in 35-mm dishes. After 24 h, medium was replaced with DMEM containing 1% FBS (DMEM-1% FBS), and 104 PFU of virus was applied. After a 1-h incubation period at 37°C, cells were refed with DMEM-1% FBS. Following incubation at 37°C for 24, 72, or 120 h, cells were lysed by two freeze-thaw cycles, and titers were determined by plaque assay on Vero cells following established protocols (40).
Construction of virus recombinants.
The HSV (Patton) strain is used as the backbone for all recombinants described in this study; viral nucleotide numbers refer to the HSV strain 17 sequence (GenBank accession number X14112). To generate recombinant HSVs, foreign sequences were cloned into a plasmid containing the HSV (Patton) BamHI SP fragment. First, an XbaI restriction site was engineered, including the stop codon of the γ134.5 ORF, by ligating fragments from SexAI/BstXI-digested plasmid, SexAI/XbaI-digested PCR product generated with primers (i) (5′-CCACCTGGTGGTCTGGGCCTCGGC-3′) and (ii) (5′-CGTCTAGACCGAGTTCGCCGGGCC-3′), and XbaI/BstXI-digested PCR product obtained with primers (iii) (5′-GTCTAGACGTTACACCCGAGGCGGCCT-3′) and (iv) (5′-CCCCAGGGGAGTGGTTACGCGC-3′). An XbaI/ClaI fragment containing the PV 3′UTR (8) was ligated to annealed complementary oligonucleotides (v) (5′(phospho)-CGATAATAAAAGATCTTTATTTTCATTACATCTGTGTGTTGGTTTTTTGTGTG-3′) and (vi) (5(phospho)′-TCGACACACAAAAAACCAACACACACAGATGTAATGAAAATAAAGATCTTTTATTAT-3′) comprising a synthetic polyadenylation signal sequence (30) to generate an XbaI/SalI fragment. This fragment was 3′ blunted and cloned into BstXI/XbaI-digested BamHI SP plasmid along with a BstXI/StuI fragment from the BamHI SP plasmid to yield plasmid BamX3. A SacI/XbaI-digested fragment from BamX3 spanning the γ134.5 5′UTR/ORF was subcloned into SacI/XbaI-digested pUC19 (New England Biolabs). The resulting plasmid was digested with DraIII and XbaI to remove a portion of the 5′UTR and the entire γ134.5 ORF. The digested vector was ligated with a DraIII/NcoI-digested fragment containing the PV cloverleaf (CL) and HRV2 IRES derived from PCR with primers (vii) (5′-GCTTTAAAGCGGTGGCGGCGGGCAGCCTTAAAACAGCTCTG GGGT-3′) and (viii) (5′-CATGTGCGCCCACTTTCTGTG-3′) from a PV1(RIPO) template (16) and an NcoI/XbaI-digested BamX3 fragment containing the γ134.5 ORF. The SacI/XbaI fragment from the resulting plasmid, which includes the PV CL, HRV2 IRES, and γ134.5 ORF, was ligated back into BamX3 to yield plasmid BamIR. ATGmut-34.5 was obtained by ligation of an EcoRI/BsaAI-digested PCR product generated with primers (ix) (5′-CGCCTGTTTTATACTCCCTTCCC-3′) and (x) (5′-GTTACGTGCTCTAGCTGCAGGGTTAAGGTTAGCCACGTTCAG-3′) from PV1(RIPO) template cDNA with pUC19 vector containing the HRV2 IRES and γ134.5 ORF EcoRI/XbaI fragment from BamIR. The HpaI/XbaI fragment from this plasmid was then ligated back into BamIR to yield BamIR/ATGmut.
To construct PV-34.5, a SacI site was introduced at the start of the γ134.5 ORF via PCR from plasmid BamIR template DNA with primers (xi) (5′-ATGGGAGCTCGCCGCCGCCGCCATCGCG-3′) and (ii). The SacI/XbaI-digested PCR product, encompassing the γ134.5 ORF, was cloned into SacI/XbaI-digested vector pPV1(M)-Luc-PVA50 (3) to insert the ORF downstream of the PV IRES. To repair the SacI site insertion, this plasmid was digested with SacI, blunt ended with Klenow fragment, cleaved near the 3′ end of the PV IRES with MunI, and ligated with annealed complementary oligonucleotides (xii) (5′(phospho)-AATTGTATCATAATGGCC-3′) and (xiii) (5′(phospho)-GGCCATTATGATAC-3′). The KpnI/XbaI restriction fragment from the resulting plasmid, which contained a portion of the PV CL, the entire PV IRES, and the γ134.5 ORF, was cloned into KpnI/XbaI-digested BamIR to yield plasmid BamPV.
Full-length recombinant viruses were generated as described previously (41) by homologous recombination of the BamIR, BamIR/ATGmut, or BamPV plasmid with SPBg5e viral DNA, an HSV mutant in which the ORFs in both γ134.5 copies are replaced with β-glucuronidase-coding sequences (40).
Southern blot.
Following two rounds of plaque purification on Vero cells, the genetic structures of virus recombinants were assessed by Southern blot analysis. Viral DNA was isolated and prepared for Southern blotting as described previously (40). Briefly, viral DNAs were digested with BamHI and EcoRI (HRV-34.5/ATGmut-34.5) or BamHI alone (PV-34.5), fractionated by electrophoresis in 1% agarose gels, transferred to nylon membranes, and incubated with labeled probe. Digoxigenin-labeled probes were prepared by random priming with the DIG High Prime kit (Roche, Indianapolis, IN) from (i) the BamHI-BstXI segment of BamHI SP, which hybridizes to HSV genomic sequences downstream of the γ134.5 ORF; (ii) PV CL/HRV2 IRES DNA amplified by PCR using primers (vii) and (viii) from PV1(RIPO) cDNA; and (iii) PV CL/PV IRES DNA amplified by PCR with primers (vii) and (viii) from BamPV plasmid DNA. Probe (i) identifies ∼6-kb and ∼2.8-kb fragments in HSV (Patton) viral DNA corresponding to the genomic BamHI SP and S fragments, respectively. Insertion of the IRES to engineer HRV-34.5 and ATGmut-34.5 introduces an EcoRI site so that probe (i) recognizes an identical ∼2.8-kb segment from both the BamHI SP and S fragments. Probe (ii) is derived from HRV2 IRES cDNA and hybridizes only to a ∼2.8-kb segment in the HRV-34.5 and ATGmut-34.5 genomes. Due to the presence of a BamHI site within the PV IRES, probes (i) and (iii) both hybridize to a ∼2.8-kb fragment following BamHI digestion of PV-34.5 viral DNA.
Northern blot.
U373 cells were infected at a multiplicity of infection (MOI) of 10, and total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) 18 h postinfection (p.i.). Glyoxal-denatured RNA (8 μg per lane) was separated by electrophoresis on 1% agarose gels and transferred to nylon membranes. DNA probes were generated by random-primed labeling (Roche, Indianapolis, IN) with [α-32P]dCTP (GE Healthcare, Piscataway, NJ) from template DNA derived from PCR with primers (i) and (iv) on plasmid BamHI SP or BamIR. Prehybridization and hybridization were each carried out for 1 h at 70°C in ExpressHyb buffer (Clontech, Mountain View, CA). The membranes were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.05% sodium dodecyl sulfate (SDS) for 40 min at 25°C and in 0.1 × SSC-0.1% SDS for 40 min at 52°C. Bands were visualized by exposure to X-ray film.
Analysis of viral protein synthesis and immunoblotting.
Metabolic labeling of infected cells to monitor late viral protein synthesis was carried out as described previously (40). For immunoblot analysis, Vero cells were infected at an MOI of 3 and lysed at various times postinfection. Cell lysis, SDS-polyacrylamide gel electrophoresis (PAGE), and Western blotting were carried out as described before (8) with rabbit polyclonal anti-ICP34.5 or anti-ICP27 sera (42).
Neurovirulence assay and histopathology.
Four-week-old female BALB/c mice were procured (Charles River Laboratories), anesthetized, and injected intracranially (i.c.) with 20 μl of virus diluted in DMEM-1% FBS via a 28.5-gauge syringe (insuject; Becton-Dickinson, Franklin Lakes, NJ). All mice were closely observed following the inoculation event to exclude neurological deficits as a result of the injection procedure. Survival of the animals was monitored for 28 days; mice exhibiting symptoms of neurological illness were sacrificed, and their brains were dissected and processed for paraffin embedding by standard procedures (10) for further tests. NCSS software (NCSS, Kaysville, UT) was used for statistical analysis of survival data with the nonparametric log rank test. All animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee. Paraformaldehyde-fixed, paraffin-embedded mouse brains were processed into 8-μm sections. Deparaffinized and cleared slides were treated to block endogenous peroxidases with 0.6% H2O2 for 8 min. Slides were immunostained with primary rabbit polyclonal anti-HSV-1 antibody (B0114; Dako, Carpinteria, CA), secondary anti-species biotinylated antibody and horseradish peroxidase-streptavidin conjugate using the EnVision + Dual Link kit (Dako, Carpinteria, CA) following the manufacturer's instructions. The slides were counterstained with hematoxylin and mounted for microscopic analysis.
RESULTS
Construction of recombinant HSVs.
Picornavirus UTRs were inserted into the HSV genome to regulate expression of the γ134.5 gene. We generated constructs containing the 5′UTRs of wt PV type 1 (Mahoney) and PV-RIPO (16), respectively (Fig. 1). Both feature the 5′ terminal cloverleaf of PV preceding either the HRV2 (HRV-34.5) or PV (PV-34.5) IRES (Fig. 1C and E). In addition, the PV 3′UTR was included in this design, since it is implicated in the neuron-specific defect of PV-RIPO (8). The group of IRES-containing HSVs described in this report will be referred to collectively as IRES-34.5. The native γ134.5 5′UTR was replaced by the heterologous picornavirus 5′UTR, which was inserted immediately downstream of the γ134.5 transcriptional start site (Fig. 1C to E). The PV 3′UTR and a synthetic polyadenylation signal sequence were positioned downstream of the γ134.5 stop codon in place of the cognate 3′UTR (Fig. 1C to E). All genetic manipulations were carried out with a plasmid containing the BamHI SP fragment of the HSV genome (see Materials and Methods). We confirmed the genetic structure of the plaque-purified recombinant viruses PV/HRV-34.5 by Southern blotting using IRES-specific probes as well as the BamHI-BstXI segment from the BamHI SP fragment (see Materials and Methods; Fig. 2A and B).
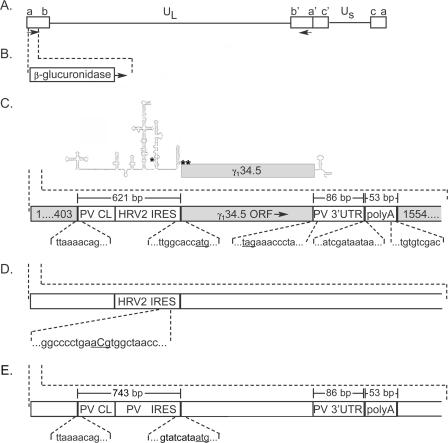
FIG. 1.
Genetic structure of recombinant viruses. (A) The wild-type HSV genome features unique long (UL) and unique short (US) regions each flanked by repetitive regions (rectangles), containing both γ134.5 copies (arrows). (B) The Δγ134.5 variant SPBg5e contains β-glucuronidase ORFs in place of the γ134.5 coding regions (shown only for one locus). (C) HRV-34.5 was engineered by inserting the PV CL and HRV2 IRES downstream of the γ134.5 transcriptional start site and immediately upstream from the initiation codon. The sequence context of the initiation codon (underlined) is shown. The PV 3′UTR and a synthetic polyadenylation signal sequence were positioned directly downstream of the γ134.5 stop codon (underlined). Numbers in boxes correspond to HSV nucleotides. The size of inserts is shown in brackets, and junction nucleotide sequence is indicated below. The predicted structure of the HRV-34.5 transcript is shown at the top of the panel (the locations of the initiation codon [**] and the location of the point mutation in ATGmut-34.5 [*] are indicated). (D) ATGmut-34.5 is identical to HRV-34.5 except for a point mutation within the IRES (T450⇒C). Nucleotide numbers are from the numbering of the wt HRV2 genome sequence (GenBank accession number X02316). (E) PV-34.5 is identical to HRV-34.5 except for the presence of the PV IRES instead of its HRV2 counterpart.
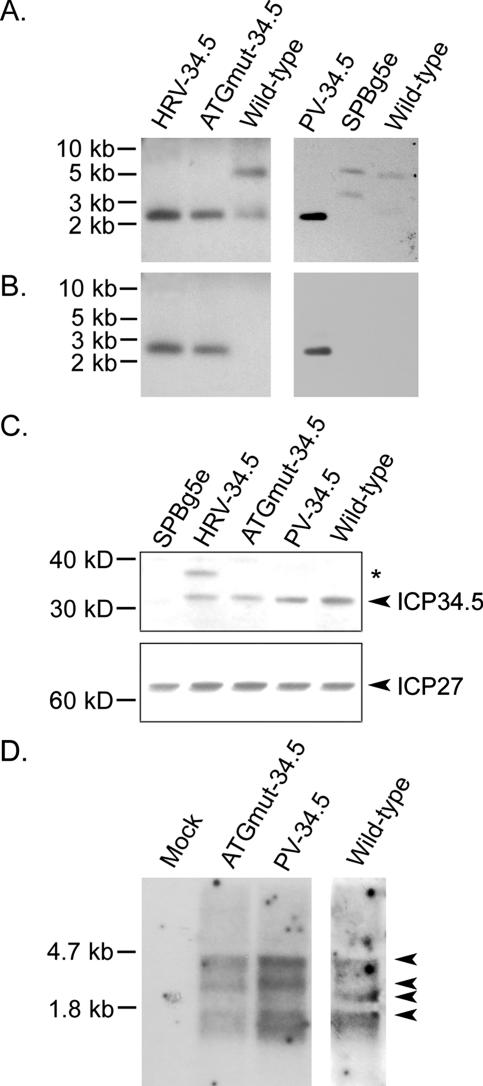
FIG. 2.
Mapping the IRES insert and ICP34.5 expression of recombinant viruses. (A and B) Southern blots of recombinant HSV genomes; (C) Western blot of ICP34.5 from infected cells; (D) Northern blot of γ134.5 mRNA from infected cells. (A) Probe corresponding to the BamHI-BstXI segment hybridizes downstream of the γ134.5 gene and yields the expected segments in all viral genomes (see Materials and Methods). (B) Probe derived from HRV2 (left) or PV IRES (right) cDNA uniquely detects the expected ∼2.8-kb fragments in the HRV-34.5, ATGmut-34.5, and PV-34.5 lanes. (C) Vero cells were infected at an MOI of 3, and the cells were lysed in SDS buffer 18 h p.i. (Top) ICP34.5 (arrowhead) was detected by immunoblotting. A higher-molecular-weight protein (asterisk) produced during HRV-34.5 infection is not detected after infection with ATGmut-34.5 or PV-34.5. (Bottom) Expression of ICP27 detected in Vero cells infected with the indicated recombinant. (D) Total RNA was isolated from U373 cells 18 h p.i. and subjected to Northern blot analysis with DNA probes specific for the PV 3′UTR-containing γ134.5 transcript (mock, ATGmut-34.5, and PV-34.5 lanes) or for the cognate 3′UTR-containing γ134.5 transcript (wt HSV lane).
IRES-driven ICP34.5 expression in recombinant HSV-infected cells.
To determine whether the IRESs drive γ134.5 expression during infection with recombinant HSVs, we performed immunoblot assays for detection of ICP34.5 (Fig. 2C). Production of ICP34.5 is evident following infection of Vero cells with all recombinants, indicating that the inserted foreign regulatory elements do indeed mediate translation of the IRES-34.5 mRNAs in permissive cells (Fig. 2C). Insertion of heterologous UTRs into the HSV genome does not affect general HSV gene expression, as ICP27 was expressed at wt levels by all recombinant viruses (Fig. 2C).
Unexpectedly, in addition to ICP34.5, our antibody detected a higher-molecular-weight protein in cells infected with HRV-34.5 (Fig. 2C). The presence of this protein suggested translation initiation at an upstream cryptic AUG in frame with the true initiation codon (at position 622 of the 5′UTR [Fig. 1C]). This may be due to scanning of the capped HRV-34.5 message or irregular cap-independent initiation at a normally silent AUG. An in-frame AUG in poor Kozak context within IRES stem-loop domain V (nt 449 to 451; Fig. 1C) without a downstream in-frame stop codon, if used, could give rise to the enlarged ICP34.5 polypeptide. To eliminate synthesis of this enlarged variant, we changed the sequence in HRV-34.5 [T450⇒C (Fig. 1D)] to generate ATGmut-34.5 (Fig. 1D). This eliminated biosynthesis of the higher-molecular-weight protein (Fig. 2C). We employed both the HRV-34.5 and ATGmut-34.5 viruses in subsequent studies.
Interestingly, the cryptic AUG giving rise to the aberrant translation product with HRV-34.5 is conserved in the PV IRES and in frame with the true PV initiation codon. Yet, we did not detect enlarged ICP34.5 proteins generated at the PV IRES (Fig. 2C). This discrepancy could be due to different conditions for initiation at upstream AUGs mediated by diverse higher-order structures of the PV and HRV2 IRESs. Translation products generated by aberrant initiation at the equivalent upstream AUGs in the PV and HRV2 IRESs would differ by 41 amino acids in size (due to the larger size of the former); this may influence recognition by antibodies used in the ICP34.5 immunoblot. Last, control of aberrant initiation at picornavirus IRESs in vitro has been assigned to IRES trans-acting factors (34). Different RNP complexes forming at the PV and HRV2 IRESs in cells infected with HSV recombinants may affect initiation codon choice. Since PV-34.5 generates a single ICP34.5 protein of the correct predicted size, we used this construct for further studies.
Efficient synthesis of ICP34.5 suggests that the IRES-driven γ134.5 message is stable and translation competent in infected cells. Nevertheless, to exclude effects of heterologous UTRs on template stability, we evaluated the integrity of IRES-34.5 mRNAs in infected cells. Previous studies reported Northern blot detection of specific γ134.5 transcripts of ∼1.3, 4, and 5.2 kb in infected Vero or neuroblastoma cells (7, 29). We isolated total RNA from U373 glioblastoma cells infected with ATGmut-34.5 or PV-34.5 18 h p.i. Using a probe that hybridizes to the 3′ 185 nt of the γ134.5 ORF and the 3′UTR, we observed γ134.5-specific transcripts of approximately 1.3 and 4 kb in length (Fig. 2D). In addition, we detected a third band of approximately 2.5 to 3 kb. Further studies would be required to determine the exact nature and significance of these various transcripts. Importantly, γ134.5 transcript integrity and stability following infection with ATGmut-34.5 or PV-34.5 do not appear to differ from wt HSV-infected cells, and overt transcript degradation was not apparent under the conditions tested with any of the viruses (Fig. 2D).
Replication of recombinant HSV in glioma cells.
ICP34.5 is produced during infection of Vero cells with HSV IRES-34.5 and ATGmut-34.5 recombinants at reduced levels than those of wt virus infection (Fig. 2C). To evaluate whether γ134.5 gene expression in our recombinant viruses occurs at levels sufficient to counter the PKR-induced shutoff of protein synthesis, we measured viral protein synthesis at late times of infection. We evaluated universally permissive Vero cells, the human glioma cell line U373, which is resistant to Δγ134.5 HSVs (52), and murine SMA-560 glioma cells.
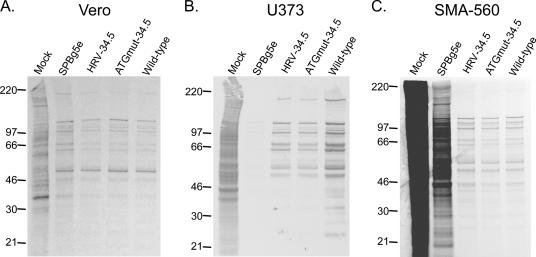
Cells were infected with virus at an MOI of 3 and labeled for 1 hour with 35S-labeled amino acids 18 h p.i. Cell lysates were fractionated by SDS-PAGE, and protein expression was detected autoradiographically. Late viral protein synthesis is roughly equivalent for all viruses tested in Vero cells, which are permissive for growth of the Δγ134.5 virus SPBg5e (Fig. 3A). However, as previously shown (52) SPBg5e protein expression is thwarted in U373 cells (Fig. 3B). In contrast, robust viral protein synthesis was evident after infection with HRV-34.5 or ATGmut-34.5 in U373 cells. These findings suggest that ICP34.5 production was sufficient to antagonize the PKR response at late times in HRV-34.5- or ATGmut-34.5-infected U373 cells (Fig. 3B). Similar results were obtained in SMA-560 cells (Fig. 3C). Unexpectedly, there is incomplete shutoff of host protein synthesis during SPBg5e infection of SMA-560 cells, and viral gene products could not be clearly discerned (Fig. 3C). In contrast, viral gene expression levels in HRV-34.5- or ATGmut-34.5-infected SMA-560 cells were at wt levels (Fig. 3C). We did not evaluate PV-34.5 in this assay, since it exhibited a propagation rate similar to those of HSV (Patton) and the other IRES-34.5 recombinants in U373 and SMA-560 cells (Table 1).
FIG. 3.
Analysis of late viral protein synthesis. Vero (A), U373 human glioma (B), or SMA-560 mouse astrocytoma (C) cells were mock infected or infected at an MOI of 3. Eighteen hours p.i., proteins were pulse-labeled for 1 h with 35S-labeled amino acids. Whole-cell lysates were fractionated by SDS-PAGE, and the fixed and dried gels were exposed to X-ray film. The positions of molecular mass standards (in kilodaltons) are indicated to the left of the gels.
TABLE 1.
Replication of the recombinant viruses in glioma cellsa
| Cell and virus | Titer (PFU/ml)
|
||
|---|---|---|---|
| Day 1 p.i. | Day 3 p.i. | Day 5 p.i. | |
| U373 cells | |||
| SPBg5e | 3 × 103 | 1.8 × 104 | 3.6 × 104 |
| HRV-34.5 | 6 × 104 | 5 × 107 | 4 × 107 |
| ATGmut-34.5 | 2.8 × 105 | 5.4 × 107 | 3 × 107 |
| PV-34.5 | 4 × 105 | 8 × 107 | 8 × 107 |
| Wild-type virus | 4 × 105 | 1.6 × 108 | 1.4 × 108 |
| SMA-560 cells | |||
| SPBg5e | 400 | 1 × 103 | 34 |
| HRV-34.5 | 5 × 104 | 3 × 105 | 1.2 × 105 |
| ATGmut-34.5 | 1.8 × 105 | 6 × 105 | 1 × 105 |
| PV-34.5 | 6 × 105 | 2 × 106 | 3 × 105 |
| Wild-type virus | 1.2 × 106 | 5 × 106 | 4 × 106 |
U373 or SMA-560 cells (1 × 106) were infected with 104 PFU of virus. Cells were lysed at 1, 3, and 5 days p.i., and viral titers were determined by plaque assay on Vero cells.
We performed multicycle growth curve analyses to determine the efficiency of IRES-34.5 recombinant replication in human and murine glioma cells. U373 and SMA-560 cells were infected at an MOI of 0.01, and viral titers were quantified at various times p.i. SPBg5e replicates exceedingly poorly, reaching titers of only 3.6 × 104 PFU/ml and 34 PFU/ml after 5 days in U373 and SMA-560 cells, respectively (Table 1). In contrast, the IRES-containing recombinant viruses grew very efficiently, with titers >1,000- to 30,000-fold above those of SPBg5e at 3 and 5 days p.i. (Table 1). The titers of IRES-34.5 viruses were only ∼4- to 40-fold below those achieved by wt HSV (Table 1).
Thus, translation control elements in IRES-34.5 recombinants permit productive infection of both mouse and human glioma cells resistant to Δγ134.5 HSV. Replication of HRV-34.5, ATGmut-34.5, and PV-34.5 did not significantly differ in either cell line. We conclude that diverse picornavirus IRESs driving HSV ICP34.5 expression mediate efficient viral gene expression and propagation in U373 glioma cells and that there are no significant species-specific effects on the performance of HRV- or PV-derived IRESs in murine glioma cells.
Neuropathogenicity of HSV IRES recombinants in mice.
HRV-34.5 and ATGmut-34.5 HSVs propagate efficiently in glioma cells, but do they recapitulate in vivo the neuron-specific translation deficit of the HRV2 IRES observed with PV-RIPO? This question was addressed by neurovirulence assessment of the recombinant viruses, as well as SPBg5e and HSV (Patton) by intracranial inoculation of BALB/c mice. Groups of four female 4-week-old mice received intracerebral injections of the virus strain to be tested into the right caudate nucleus. The mice were observed, and their neurological status was monitored over a 28-day period. Injection with HSV (Patton) virus resulted in death of all four mice at the highest dose (10,000 PFU) within 24 h p.i. and was fatal at even the lowest dose (10 PFU) by 7 days p.i. (Table 2). Confirming prior observations (5), SPBg5e is severely attenuated, and accordingly, no deaths occurred in the animals injected with SPBg5e at any dose in the range tested (Table 2). Similarly, HRV-34.5 and ATGmut-34.5 were significantly attenuated compared to HSV (Patton) at all doses (P < 0.05). A single mouse inoculated with HRV-34.5 at the highest dose (10,000 PFU) succumbed on day 3, but all remaining mice injected with HRV-34.5 and ATGmut-34.5 survived (Table 2). Attenuation of HRV-34.5 and ATGmut-34.5 is not due to host range defects of HRV2 IRES function in murine cells, because in contrast to SPBg5e, these viruses exhibited efficient gene expression (Fig. 3C) and growth in mouse SMA-560 glioma cells (Table 1). Our data indicate that placing the γ134.5 gene under translation control of PV-RIPO regulatory genetic elements results in attenuation of neurovirulence.
TABLE 2.
Neurovirulence attenuation of recombinant virusesa
| Virus | No. of surviving mice/no. of mice injected with the following dose (PFU):
|
|||
|---|---|---|---|---|
| 10,000 | 1,000 | 100 | 10 | |
| SPBg5e | 4/4 | 4/4 | 4/4 | NDc |
| HRV-34.5 | 3/4 (3)b | 4/4 | 4/4 | ND |
| ATGmut-34.5 | 4/4 | 4/4 | 4/4 | ND |
| PV-34.5 | 0/4 (2, 3, 4, 5) | 0/4 (6, 6, 6, 7) | 4/4 | ND |
| Wild-type | 0/4 (1) | 0/4 (1, 2, 3, 3) | 0/4 (4) | 0/4 (6, 6, 7, 7) |
BALB/c mice were mock infected or injected intracranially with various doses of virus, and survival was monitored over 28 days. Four mock-infected animals survived.
The time to death in days is shown in parentheses.
ND, not determined.
Evaluation of PV-34.5 in vivo revealed a significant difference in neurovirulence potential compared to ATGmut-34.5 (at 10,000 and 1,000 PFU; P < 0.05) and HRV-34.5 (at 1,000 PFU; P < 0.05). This suggests that, in contrast to the HRV2 IRES, the PV IRES is capable of mediating viral gene expression and growth in the CNS, resulting in considerable neurovirulence: all animals inoculated with 1,000 PFU and beyond succumbed to the infection (Table 2). The fact that HSV (Patton) was lethal at doses lower than those of PV-34.5 suggests that the PV IRES insert slightly lowers HSV propagation and/or invasion, at least in the CNS.
Immunohistopathology of HSV in mouse brain.
In addition to attenuating neurovirulence, placing γ134.5 under translation control of picornavirus UTRs could in principle redirect HSV cell type specificity in the infected mouse brain. To evaluate this possibility, we conducted immunohistopathological analyses of the brains of mice infected i.c. with ATGmut-34.5 and HSV (Patton). Mice infected with 1,000 PFU of either virus were sacrificed 3 days p.i., and their brains were dissected and histologically processed. The entire brain of sacrificed animals was cut into sections 8 μm thick, and a series of 20 sections containing overt lesions corresponding to the inoculation site and adjacent areas were chosen for further analysis (Fig. 4). We detected unambiguous HSV gene expression only in the brains of mice infected with HSV (Patton) (Fig. 4). We identified four main sites of HSV gene expression: the ependymal cell layer (Fig. 4A to D), sporadic neurons in the hippocampal formation (Fig. 4E to H), the meninges (Fig. 4I and J), and in the vicinity of the inoculation site (Fig. 4K and L). This pattern of viral gene expression is reminiscent of earlier observations regarding HSV encephalitis after intracerebral inoculation into BALB/c mice (26, 33).
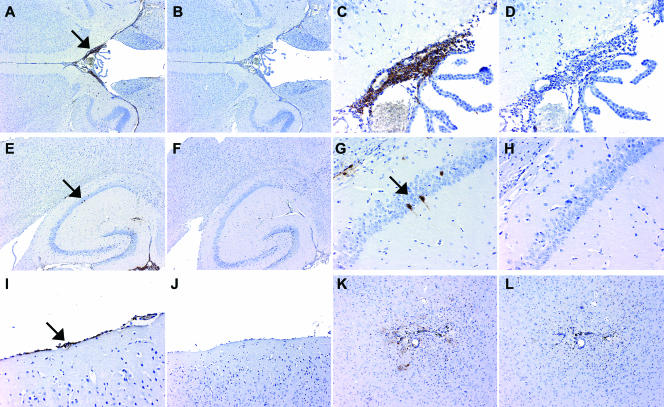
FIG. 4.
Immunohistopathology of HSV (Patton) in mouse brain. Adjacent sections were stained with anti-HSV polyclonal antiserum (A, C, E, G, I, and K) or isotype-controlled nonspecific rabbit immunoglobulin G (B, D, F, H, J, and L). (A and B) Ependymal lining of the third ventricle. HSV-positive cells line the entire surface of the ventricle and are particularly abundant adjacent to the choroid plexus (indicated by the arrow in panel A). There was no staining with control antibody (B). (C and D) Detail of panels A and B. Higher-power magnification demonstrates the distribution of HSV-infected cells with relation to the choroid plexus. (E and F) Overview of the hippocampal formation. The arrow points at two individual hippocampal neurons staining positive for HSV antigen. Note prominent stain of the ependymal cell layer (lower right). (G and H) Detail of panels E and F. Solitary neurons within the hippocampal formation and their axonal/dendritic extensions stain positive for HSV. (I and J) HSV-positive cells in the pia mater. (K and L) The immediate vicinity of the inoculation site.
By far the most obvious and widespread site of HSV gene expression was the ependymal cell layer. We observed widely distributed infection of cells in the ependymal lining of all ventricles (Fig. 4A to D). Occasionally, HSV-positive staining was detected in subependymal clusters of cells adjacent to affected ependymal areas (data not shown). Productive infection of ependymal cells has been observed before with HSV (1716), a Δγ134.5 variant, as well as with wt HSV (26). However, in this study, ependymal cells were affected only after intraventricular, but not parenchymal, injection of virus (26). Intracranial inoculations in our study were targeted to the basal ganglia (Fig. 4K) in a location not communicating with the ventricular system. The fact that we did observe ependymal cell infection may reflect either enhanced neuropathogenicity/invasiveness of wt HSV (Patton) compared to HSV (17) (which was studied in reference 26) or unintended ventricular spread (which commonly occurs after i.c. inoculation in mice, due to the small size of their brain). We did observe a low level of staining in parts of the ependymal layer in the brains of mice infected with ATGmut-34.5. However, the signal was marginally above background observed with isotype-matched control immunoglobulin G (data not shown).
We observed sporadic HSV antigen expression in neurons located in the hippocampal formation. The total number of neurons affected in individual brains was low; the example shown (two neurons in the hippocampal formation; Fig. 4E to H) were the only neurons identified in 20 consecutive sections in the animal tested. The location of the infected neurons bears no apparent relation to the inoculation site (in the caudate nucleus) or confirmed sites of HSV infection (e.g., the ependymal cell layer). On the basis of these findings, we assume that infection of solitary neurons may be a chance phenomenon. HSV (Patton)-infected animals were sacrificed in a preterminal state. Further expansion of HSV infection to neighboring neurons may be curtailed by the rapid death of affected animals. We did not observe HSV antigen in any neurons or glia in ATGmut-34.5-infected mice. Detection of HSV in ependymal cells and the hippocampus could indicate a preference for HSV propagation in dividing CNS cells (26); both structures are known to harbor mitotically active populations in the adult brain (14, 49).
Mice infected with HSV (Patton) showed signs of meningeal viral gene expression (Fig. 4I and J). Positive stain was visible in cells of the pia mater (Fig. 4I) and occasionally in small clusters of cells in the underlying cortex (data not shown). Finally, there was evidence of viral gene expression in the vicinity of the inoculation site. Generally, the signal was diffuse and difficult to assign to defined populations of cells (Fig. 4K and L). We did not detect viral gene expression at the site of inoculation with ATGmut-34.5 (data not shown). Our findings suggest that expression of ICP34.5 under HRV2 IRES control does not redirect HSV cell type specificity in the mouse brain and that HSV gene expression in the main structures affected by HSV (Patton) is substantially reduced or absent with ATGmut-34.5.
DISCUSSION
We demonstrated that HSV neuropathogenicity in mice can be attenuated by modifying translation regulation of key virulence genes in the brain. Moreover, the principle of targeting translation control in GBM, first discovered with PV replicating under control of a heterologous IRES, can be applied to direct HSV gene expression and cytotoxicity specifically to malignant glioma cells.
HRV2 IRES function in the CNS exhibits similar cell type-specific restrictions in the context of PV, an RNA virus that depends on cap-independent translation, and HSV, a DNA virus that does not. Translation regulation of the HSV γ134.5 gene by the HRV2 IRES results in profound neuroattenuation, while PV IRES-controlled ICP34.5 expression retains a high level of neurovirulence inherent in wt HSV.
Parallel cell type-specific function of picornavirus IRESs within their native and an artificial HSV context is surprising, since both PV and HSV cause profound and fundamentally different perturbations of translation control in their hosts. The artificial IRES-γ134.5 transcript may thus be at an inherent disadvantage in HSV-infected cells lacking PV-induced alterations of the host cell translation machinery.
Enterovirus (e.g., PV) and HRV IRES efficiency may be affected by alterations of translation initiation factors in infected host cells: enteroviruses and rhinoviruses express proteases that cleave the eukaryotic initiation factor 4G (eIF4G) (11) and poly(A) binding protein (PABP) (23). We recently reported that enterovirus IRES function is remarkably robust in the absence of eIF4G or PABP cleavage and that viral IRES- and cap-driven translation efficiencies are comparable in uninfected cells (9). The fact that the HRV2 IRES mediates efficient ICP34.5 expression in glioma cells in the absence of enterovirus proteases and their effects on the translation apparatus corroborates these findings.
Posttranscriptional control of host and viral gene expression during lytic HSV infection is in part determined by the virion host shutoff (Vhs) protein, which associates with eIF4F and endoribonucleolytically degrades mRNAs (12, 13, 51). It has been proposed that VP16 binding to Vhs prevents degradation of viral mRNAs to protect viral gene expression during lytic infection (28). Studies with encephalomyocarditis virus IRES-containing mRNAs in rabbit reticulocyte lysate suggested that Vhs may target IRESs for destruction in vitro (31). It is thus conceivable that the IRES-γ134.5 transcript, containing complex structured foreign UTRs, is prone to degradation by Vhs in HSV-infected cells. Our data exclude this scenario: IRES-γ134.5 transcripts were intact in HSV-infected cells and the IRES-34.5 constructs grow with near wt efficiency in U373 and SMA-560 glioma cells, while SPBg5e hardly grows at all. If Vhs preferentially degraded IRES-containing messages, both constructs would exhibit similar growth properties.
Our prior investigations have revealed different conditions for translation initiation in GBM versus neuronal cells. We reported that the DRBP76:NF45 heterodimer binds to the HRV2 IRES and prevents polysome assembly at the PV-RIPO RNA specifically in neuronal cells (37, 38). Our observation that the HRV2 IRES exhibits identical cell type-specific function in a diverse context reinforces the notion of a different environment for translation initiation at the HRV2 IRES in GBM and the normal CNS. It has been suggested that Ras/Akt oncogenic signaling, which transforms normal glial precursors and produces a GBM-like phenotype (20), affects gene expression patterns through differential polysome recruitment of existing mRNAs (46, 47). It was speculated that RNA binding proteins with cell type-specific function or distribution may selectively recruit certain mRNA subsets into polysomes (47). Genetically engineered HSVs with translationally controlled virulence genes provide a precedent for this hypothesis and implicate the DRBP76:NF45 heterodimer in differential translation initiation in normal and transformed cells in the CNS.
Acknowledgments
This work is supported by Public Health Service awards NS20023 (M.G.) and GM056927 (I.M.). S. A. Campbell is the recipient of a Susan G. Komen Dissertation Research Award. We gratefully acknowledge a Seth Harris Feldman Award from the Brain Tumor Society (M.G.) and the support of the Irma T. Hirschl Charitable Trust (I.M.).
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Andreansky, S., L. Soroceanu, E. R. Flotte, J. Chou, J. M. Markert, G. Y. Gillespie, B. Roizman, and R. J. Whitley. 1997. Evaluation of genetically engineered herpes simplex viruses as oncolytic agents for human malignant brain tumors. Cancer Res. 57:1502-1509. [PubMed] [Google Scholar]
- 2.Bolovan, C. A., N. M. Sawtell, and R. L. Thompson. 1994. ICP34.5 mutants of herpes simplex virus type 1 strain 17syn+ are attenuated for neurovirulence in mice and for replication in confluent primary mouse embryo cell cultures. J. Virol. 68:48-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, S., J. Lin, E. Dobrikova, and M. Gromeier. 2005. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J. Virol. 79:6281-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou, J., J. J. Chen, M. Gross, and B. Roizman. 1995. Association of a M(r) 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with gamma 134.5- mutants of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 92:10516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou, J., E. R. Kern, R. J. Whitley, and B. Roizman. 1990. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science 250:1262-1266. [DOI] [PubMed] [Google Scholar]
- 6.Chou, J., and B. Roizman. 1992. The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA 89:3266-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou, J., and B. Roizman. 1986. The terminal a sequence of the herpes simplex virus genome contains the promoter of a gene located in the repeat sequences of the L component. J. Virol. 57:629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobrikova, E., P. Florez, S. Bradrick, and M. Gromeier. 2003. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc. Natl. Acad. Sci. USA 100:15125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobrikova, E., R. Grisham, J. Lin, and M. Gromeier. 2006. Competitive translation efficiency at the picornavirus type 1 internal ribosomal entry site facilitated by viral cis and trans factors. J. Virol. 80:3310-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dufresne, A. T., and M. Gromeier. 2004. A nonpolio enterovirus with respiratory tropism causes poliomyelitis in intercellular adhesion molecule 1 transgenic mice. Proc. Natl. Acad. Sci. USA 101:13636-13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etchison, D., S. C. Milburn, I. Edery, N. Sonenberg, and J. W. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806-14810. [PubMed] [Google Scholar]
- 12.Feng, P., D. N. Everly, Jr., and G. S. Read. 2005. mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J. Virol. 79:9651-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng, P., D. N. Everly, Jr., and G. S. Read. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 75:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gage, F. H., J. Ray, and L. J. Fisher. 1995. Isolation, characterization, and use of stem cells from the CNS. Annu. Rev. Neurosci. 18:159-192. [DOI] [PubMed] [Google Scholar]
- 15.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 16.Gromeier, M., L. Alexander, and E. Wimmer. 1996. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. USA 93:2370-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gromeier, M., B. Bossert, M. Arita, A. Nomoto, and E. Wimmer. 1999. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J. Virol. 73:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gromeier, M., S. Lachmann, M. R. Rosenfeld, P. H. Gutin, and E. Wimmer. 2000. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc. Natl. Acad. Sci. USA 97:6803-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, B., M. Gross, and B. Roizman. 1997. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland, E. C., J. Celestino, C. Dai, L. Schaefer, R. E. Sawaya, and G. N. Fuller. 2000. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat. Genet. 25:55-57. [DOI] [PubMed] [Google Scholar]
- 21.Hunter, W. D., R. L. Martuza, F. Feigenbaum, T. Todo, T. Mineta, T. Yazaki, M. Toda, J. T. Newsome, R. C. Platenberg, H. J. Manz, and S. D. Rabkin. 1999. Attenuated, replication-competent herpes simplex virus type 1 mutant G207: safety evaluation of intracerebral injection in nonhuman primates. J. Virol. 73:6319-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang, S. K., H. G. Krausslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joachims, M., P. C. Van Breugel, and R. E. Lloyd. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kambara, H., H. Okano, E. A. Chiocca, and Y. Saeki. 2005. An oncolytic HSV-1 mutant expressing ICP34.5 under control of a nestin promoter increases survival of animals even when symptomatic from a brain tumor. Cancer Res. 65:2832-2839. [DOI] [PubMed] [Google Scholar]
- 25.Kanai, R., H. Tomita, A. Shinoda, M. Takahashi, S. Goldman, H. Okano, T. Kawase, and T. Yazaki. 2006. Enhanced therapeutic efficacy of G207 for the treatment of glioma through Musashi1 promoter retargeting of gamma34.5-mediated virulence. Gene Ther. 13:106-116. [DOI] [PubMed] [Google Scholar]
- 26.Kesari, S., T. M. Lasner, K. R. Balsara, B. P. Randazzo, V. M. Lee, J. Q. Trojanowski, and N. W. Fraser. 1998. A neuroattenuated ICP34.5-deficient herpes simplex virus type 1 replicates in ependymal cells of the murine central nervous system. J. Gen. Virol. 79:525-536. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura, N., B. L. Semler, P. G. Rothberg, G. R. Larsen, C. J. Adler, A. J. Dorner, E. A. Emini, R. Hanecak, J. J. Lee, S. van der Werf, C. W. Anderson, and E. Wimmer. 1981. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature 291:547-553. [DOI] [PubMed] [Google Scholar]
- 28.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, L. Y., and P. A. Schaffer. 1998. A virus with a mutation in the ICP4-binding site in the L/ST promoter of herpes simplex virus type 1, but not a virus with a mutation in open reading frame P, exhibits cell-type-specific expression of γ134.5 transcripts and latency-associated transcripts. J. Virol. 72:4250-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levitt, N., D. Briggs, A. Gil, and N. J. Proudfoot. 1989. Definition of an efficient synthetic poly(A) site. Genes Dev. 3:1019-1025. [DOI] [PubMed] [Google Scholar]
- 31.Lu, P., H. A. Saffran, and J. R. Smiley. 2001. The vhs1 mutant form of herpes simplex virus virion host shutoff protein retains significant internal ribosome entry site-directed RNA cleavage activity. J. Virol. 75:1072-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markert, J. M., M. D. Medlock, S. D. Rabkin, G. Y. Gillespie, T. Todo, W. D. Hunter, C. A. Palmer, F. Feigenbaum, C. Tornatore, F. Tufaro, and R. L. Martuza. 2000. Conditionally replicating herpes simplex virus mutant, G207 for the treatment of malignant glioma: results of a phase I trial. Gene Ther. 7:867-874. [DOI] [PubMed] [Google Scholar]
- 33.Markovitz, N. S., D. Baunoch, and B. Roizman. 1997. The range and distribution of murine central nervous system cells infected with the γ134.5− mutant of herpes simplex virus 1. J. Virol. 71:5560-5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendelsohn, C. L., E. Wimmer, and V. R. Racaniello. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855-865. [DOI] [PubMed] [Google Scholar]
- 36.Merrill, M. K., G. Bernhardt, J. H. Sampson, C. J. Wikstrand, D. D. Bigner, and M. Gromeier. 2004. Poliovirus receptor CD155-targeted oncolysis of glioma. Neuro-oncology 6:208-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merrill, M. K., E. Dobrikova, and M. Gromeier. 2006. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA binding protein 76. J. Virol. 80:3147-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merrill, M. K., and M. Gromeier. 2006. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosomal entry site. J. Virol. 80:6936-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohr, I. 2005. To replicate or not to replicate: achieving selective oncolytic virus replication in cancer cells through translational control. Oncogene 24:7697-7709. [DOI] [PubMed] [Google Scholar]
- 40.Mohr, I., and Y. Gluzman. 1996. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 15:4759-4766. [PMC free article] [PubMed] [Google Scholar]
- 41.Mohr, I., D. Sternberg, S. Ward, D. Leib, M. Mulvey, and Y. Gluzman. 2001. A herpes simplex virus type 1 γ34.5 second-site suppressor mutant that exhibits enhanced growth in cultured glioblastoma cells is severely attenuated in animals. J. Virol. 75:5189-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulvey, M., J. Poppers, D. Sternberg, and I. Mohr. 2003. Regulation of eIF2α phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J. Virol. 77:10917-10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura, H., H. Kasuya, J. T. Mullen, S. S. Yoon, T. M. Pawlik, S. Chandrasekhar, J. M. Donahue, E. A. Chiocca, R. Y. Chung, and K. K. Tanabe. 2002. Regulation of herpes simplex virus gamma(1)34.5 expression and oncolysis of diffuse liver metastases by Myb34.5. J. Clin. Investig. 109:871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochiai, H., S. Campbell, G. E. Archer, T. A. Chewning, E. Dragunsky, A. Ivanov, M. Gromeier, and J. H. Sampson. 2006. Targeted therapy of human glioblastoma multiforme neoplastic meningitis with intrathecal delivery of an oncolytic recombinant poliovirus. Clin. Cancer Res. 12:1349-1354. [DOI] [PubMed] [Google Scholar]
- 45.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 46.Rajasekhar, V. K., and E. C. Holland. 2004. Postgenomic global analysis of translational control induced by oncogenic signaling. Oncogene 23:3248-3264. [DOI] [PubMed] [Google Scholar]
- 47.Rajasekhar, V. K., A. Viale, N. D. Socci, M. Wiedmann, X. Hu, and E. C. Holland. 2003. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell 12:889-901. [DOI] [PubMed] [Google Scholar]
- 48.Rampling, R., G. Cruickshank, V. Papanastassiou, J. Nicoll, D. Hadley, D. Brennan, R. Petty, A. MacLean, J. Harland, E. McKie, R. Mabbs, and M. Brown. 2000. Toxicity evaluation of replication-competent herpes simplex virus (ICP 34.5 null mutant 1716) in patients with recurrent malignant glioma. Gene Ther. 7:859-866. [DOI] [PubMed] [Google Scholar]
- 49.Reynolds, B. A., and S. Weiss. 1992. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255:1707-1710. [DOI] [PubMed] [Google Scholar]
- 50.Smith, K. D., J. J. Mezhir, K. Bickenbach, J. Veerapong, J. Charron, M. C. Posner, B. Roizman, and R. R. Weichselbaum. 2006. Activated MEK suppresses activation of PKR and enables efficient replication and in vivo oncolysis by Δγ134.5 mutants of herpes simplex virus 1. J. Virol. 80:1110-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taddeo, B., W. Zhang, and B. Roizman. 2006. The UL41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc. Natl. Acad. Sci. USA 103:2827-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taneja, S., J. MacGregor, S. Markus, S. Ha, and I. Mohr. 2001. Enhanced antitumor efficacy of a herpes simplex virus mutant isolated by genetic selection in cancer cells. Proc. Natl. Acad. Sci. USA 98:8804-8808. [DOI] [PMC free article] [PubMed] [Google Scholar]