Abstract
The vaccinia virus-encoded D5 protein is an essential ATPase involved in viral DNA replication. We have expanded the genotypic and phenotypic analysis of six temperature-sensitive (ts) D5 mutants (Cts17, Cts24, Ets69, Dts6389 [also referred to as Dts38], Dts12, and Dts56) and shown that at nonpermissive temperature all of the tsD5 viruses exhibit a dramatic reduction in DNA synthesis and virus production. For Cts17 and Cts24, this restriction reflects the thermolability of the D5 proteins. The Dts6389, Dts12, and Dts56 D5 proteins become insoluble at 39.7°C, while the Ets69 D5 protein remains stable and soluble and retains the ability to oligomerize and hydrolyze ATP when synthesized at 39.7°C. To investigate which structural features of D5 are important for its biological and biochemical activities, we generated targeted mutations in invariant residues positioned within conserved domains found within D5. Using a transient complementation assay that assessed the ability of D5 variants to sustain ongoing DNA synthesis during nonpermissive Cts24 infections, only a wtD5 allele supported DNA synthesis. Alleles of D5 containing targeted mutations within the Walker A or B domains, the superfamily III helicase motif C, or the AAA+ motif lacked biological competency. Furthermore, purified preparations of these variant proteins revealed that they all were defective in ATP hydrolysis. Multimerization of D5 appeared to be a prerequisite for enzymatic activity and required the Walker B domain, the AAA+ motif, and a region located upstream of the catalytic core. Finally, although multimerization and enzymatic activity are necessary for the biological competence of D5, they are not sufficient.
Vaccinia virus is the prototypic member of the poxvirus family, whose most infamous member is the causative agent of smallpox, variola virus. Vaccinia virus encodes ∼200 gene products and replicates solely within the cytoplasm of the infected cell. Vaccinia virus is thought to encode most, if not all, of the proteins involved in the faithful and robust duplication of its 192-kb DNA genome. Genetic, genomic, and biochemical studies have elucidated five proteins as being essential for vaccinia virus DNA replication. These include the viral DNA polymerase (E9 [2, 3, 25-27, 38, 40, 41, 45]), the heterodimeric processivity factor (A20/D4 [16, 32, 39]), a serine/threonine protein kinase (B1 [34, 35, 43]), and a DNA-independent nucleoside triphosphatase (NTPase; D5 [9, 10]). D5 is a 90-kDa protein that is made at early times postinfection, with the peak synthesis coinciding with the onset of viral DNA synthesis (10). We have previously described a protocol for in vivo overexpression and subsequent purification of D5 and demonstrated that it possesses nucleic acid-independent NTPase activity (9). From the three original collections of vaccinia virus temperature-sensitive (ts) mutants, the Condit collection (4), the Ensinger collection (8), and the Dales collection (5), six mutants with lesions that map to the D5 open reading frame (ORF) have been described: Cts17, Cts24, Ets69, Dts6389 (also referred to as Dts38), Dts12, and Dts56 (the letter preceding each name denotes which collection they are from; for example, Cts17 is from the Condit collection). Preliminary phenotypic analyses revealed that these mutants were defective in accumulating viral DNA sequences when infections were maintained at high temperature, as assessed by failure to incorporate [3H]thymidine or accumulate Hoechst-staining cytoplasmic viral DNA factories (8, 11, 28). Temperature shift experiments indicated that the tsD5 mutants have a “fast-stop” DNA minus phenotype, with DNA synthesis ceasing within 5 min after cultures were moved to the nonpermissive temperature (28). This finding is suggestive of the D5 protein being involved directly at the replication fork (11, 28). Additional, albeit indirect, evidence suggestive of D5's positioning at the viral replication fork comes from the two-hybrid screen which uncovered an interaction between D5 and the A20 protein, a component of the viral polymerase processivity factor (24). However, we do not believe that D5 is essential for the assembly of a processive DNA polymerase complex per se, since cytoplasmic extracts prepared from nonpermissive tsD5 infections are fully competent for the processive replication of a singly primed, single-stranded DNA template (32). We would therefore postulate that D5 is involved at the replication fork and that its engagement at the fork is required for the replication of a double-stranded viral genome.
While D5 is highly conserved among chordo- and entomopoxviruses, extensive database searching and complex sequence comparisons have failed to reveal a cellular homolog for D5. Comparative analysis of protein sequences encoded by four diverse families of nucleocytoplasmic large eukaryotic DNA viruses (NCLDV, which encompasses Poxviridae, Asfarviridae, Iridoviridae, and Phycodnaviridae) has demonstrated that D5 is one of only five proteins that are absolutely conserved among this unique group of viruses (17). Comparative sequence analysis revealed the presence of a “D5 family-specific motif” spanning amino acid residues 405 to 492, which is located upstream of the predicted ATPase domain. This D5 family-specific motif was found to be encoded by the NCLDV D5 orthologs as well as certain bacteriophages, but not by any other prokaryotic or eukaryotic proteins (17). Furthermore, iterative database searches have positioned D5 as a peripheral member of the AAA+ (ATPases associated with various cellular activities) family of ATPases (17, 30). This diverse family of prokaryotic, eukaryotic, and viral proteins is characterized by the presence of a 200- to 250-amino-acid core that consists of the Walker A and Walker B domains as well as an AAA+ minimal consensus domain (ATNRPDxxDxALxR, where x represents any amino acid [30]). While the functions of the cellular AAA+ proteins are highly diverse, some notable members of this family that function during DNA replication include three members of the origin recognition complex, members of the minichromosome maintenance protein complex, and members of the replication factor C complex (30). Within the AAA+ class of proteins, D5 is further subcharacterized as a member of the superfamily III (SFIII) helicase family (17), which consists of (predicted and true) helicases from small RNA and DNA viruses (13, 21). PSI-BLAST searches seeded with the putative catalytic domain of D5 revealed a statistically significant sequence similarity between D5 and a bona fide SFIII helicase, the papillomavirus E1 protein, in the fifth iteration (17). SFIII helicases are defined by the presence of three conserved domains: Walker A and Walker B boxes and a motif C that consists of an invariant asparagine residue preceded by a run of hydrophobic residues (13, 21).
In this study, we have extended the genotypic and phenotypic analysis of the tsD5 mutants and characterized the biochemical activities of the tsD5 proteins. We also established the first transient complementation assay designed to enable structure/function analysis of an early vaccinia virus protein. These studies have revealed that invariant residues positioned within each of the highly conserved motifs found in the carboxy-terminal portion of D5 are essential for the NTPase activity of the protein. Multimerization of D5, which requires a region located upstream of the catalytic domain, is also required for catalytic activity. Finally, we show that the NTPase activity of D5 is necessary, but not sufficient, for fulfillment of its essential contribution to viral DNA replication.
MATERIALS AND METHODS
Reagents.
Restriction endonucleases, Escherichia coli polymerase I, Klenow fragment of E. coli DNA polymerase, T4 DNA ligase, calf intestinal phosphatase (CIP), pancreatic RNase, deoxynucleoside triphosphates, PCR-grade, high-fidelity Taq polymerase, Taq polymerase, and DNA molecular weight standards were purchased from Roche Diagnostics (Indianapolis, Ind.) and were used as specified by the manufacturer. 32P-labeled nucleoside triphosphates were purchased from Perkin-Elmer Life Sciences (Boston, Mass.). 3XFLAG peptide and EZview Red anti-FLAG M2 affinity gel beads were obtained from Sigma (St. Louis, Mo.). Lipofectamine 2000 and 14C-labeled protein molecular weight markers were acquired from Invitrogen (Carlsbad, Calif.). Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, Iowa).
Cells and virus.
Monolayer cultures of African green monkey BSC40 cells or mouse L929 cells were maintained in Dulbecco modified Eagle medium (Invitrogen) containing 5% fetal calf serum. Wild-type (wt) vaccinia virus (WR or IHD-W strains) and the tsD5 mutants (Cts17, Cts24, Ets69, Dts6389, Dts12, and Dts56) were grown in BSC40 or L929 cells. The IHD-W wt strain and the Cts17, Cts24, Dts12, and Dts56 viruses were kindly provided by Richard Condit (University of Gainesville, Gainesville, Fla.). Ets69 was provided by Marcia Ensinger, and Dts6389 was originally provided by Grant McFadden (University of Gainesville). The viral recombinant expressing T7 RNA polymerase, vTF7.5 (7), was originally provided by Bernard Moss (NIH). The viral recombinant enabling T7-mediated overexpression of D5 has been previously described (9). Viral stocks were prepared from cytoplasmic lysates of infected cells by ultracentrifugation through 36% sucrose; titers were determined on BSC40 cells. For virological studies, 31.5°C and 39.7°C were used as the permissive and nonpermissive temperatures, respectively.
Determination of 24-hour viral yield.
Confluent 35-mm dishes of BSC40 cells were infected at a multiplicity of infection (MOI) of 2 or 15 and maintained at 31.5°C or 39.7°C. Cells were harvested at 24 h postinfection (hpi), collected by centrifugation, and resuspended in 1 mM Tris (pH 9.0). The cells were disrupted by three cycles of freeze-thawing and two 15-second bursts of sonication. Viral yields were assessed in plaque assays performed on BSC40 cells at 31.5°C. Several independent experiments were performed; each was titrated in duplicate, and the standard deviation is shown.
Southern dot blot hybridization to assess viral DNA accumulation.
Confluent 35-mm dishes of BSC40 cells were infected at an MOI of 5 and incubated at 31.5°C or 39.7°C. Cells were harvested at the indicated time points (3, 6, 9, 12, and 24 hpi) and subjected to Southern dot blot analysis as previously described (9, 44). Hybridization was performed using a radiolabeled probe representing the HindIII E and G fragments of the vaccinia virus genome; the dot blots were visualized by autoradiography and quantitated on a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The experiment was performed in triplicate, each sample was spotted in quadruplicate, and the data were averaged and plotted with standard deviations.
Immunoblot analysis of D5 expression, stability, and solubility.
BSC40 or L929 cells were infected at an MOI of 10 and incubated at 31.5°C or 39.7°C for 8 h. To detect D5 in whole-cell lysates, cells were washed with phosphate-buffered saline and disrupted by the addition of protein sample buffer (final concentrations, 1% sodium dodecyl sulfate [SDS], 1% β-mercaptoethanol, 50 mM Tris [pH 6.8], 10% glycerol). For detection of soluble D5 in a postnuclear supernatant (PNS), cells were washed in ice-cold isotonic buffer (10 mM Tris-HCl [pH 8], 150 mM NaCl, 5 mM EDTA), incubated on ice for 10 min in ice-cold hypotonic buffer (10 mM Tris-HCl [pH 7.8], 10 mM KCl, 5 mM EDTA), and disrupted by Dounce homogenization. The nuclei and insoluble aggregates were removed by centrifugation at 1,000 × g for 10 min at 4°C. Samples were then resolved on SDS-10% acrylamide gels and transferred electrophoretically to nitrocellulose filters (Schleicher & Schuell, Keene, N.H.). The blots were analyzed by incubation with a polyclonal serum directed against D5 (10), followed by a horseradish peroxidase-conjugated secondary antiserum (Bio-Rad). After development with chemiluminescent Super Signal WestPico reagents (Pierce, Rockford, Ill.), immunoreactive proteins were visualized on Kodak MR film or captured by exposure on an AlphaImager documentation system and quantitated using FluorChem 8900 software.
Preparation of genomic viral DNA and determination of the sequence of the D5 alleles.
One confluent 15-cm dish of BSC40 cells was infected with the virus of interest at an MOI of 0.5 and maintained at 31.5°C for 72 h. Virions were purified from the PNS by centrifugation and treated with 0.1% proteinase K, 1% SDS, 14.3 mM β-mercaptoethanol, 200 mM NaCl at 37°C for 90 min; genomic DNA was retrieved by organic extraction and ethanol precipitation. Two independent PCRs were performed for each D5 allele using the D5 5′ BamHI and D5 3′ primers (Table 1); the sequence of each amplified allele was determined in duplicate.
TABLE 1.
Primers used in these studies
| Primer | Sequencea |
|---|---|
| OL-UP | 5′-GAGGATCCGATATCCAACCATTAAC-3′ |
| OL-DP | 5′-GAATTCGAGCTCGGTACC-3′ |
| K509A-5′ | 5′-CAACTGGAGCGTCGACAACCAAACGTTT-3′ |
| K509A-3′ | 5′-GTTGTCGACGCTCCAGTTGCAGTTTCTC-3′ |
| E557Q-5′ | 5′-TCTGTAGCCAACTACCTGATTTTGCC-3′ |
| E557Q-3′ | 5′-AGGTAGTTGGCTACAGAATACAGATC-3′ |
| N605D-5′ | 5′-TCGATACTGATTACAAACCTGTTTTT-3′ |
| N605D-3′ | 5′-TTTGTAATCAGTATCGATAATGATTG-3′ |
| R619A,R620A-5′ | 5′-CATTAATGGCAGCAATTGCCGTCGTGCGAT-3′ |
| R619A,R620A-3′ | 5′-CGGCAATTGCTGCCATTAATGCGTTATCTA-3′ |
| 413-5′ | 5′-GAGGATCCCATATGCTTCCGTTTAAAAATGGTG-3′ |
| 301-5′ | 5′-GAGGATCCCATATGGGTGCTATTAGAATTTAC-3′ |
| D5 5′ BamHI | 5′-GAGGATCCATGGATGCGGCTATTAG-3′ |
| D5 5′ NdeI | 5′-GAGGATCCATATGGATGCGGCTATTAG-3′ |
| D5 3′ | 5′-GAGGATCCTTACGGAGATGAAATATC-3′ |
Relevant restriction enzyme sites are indicated in bold type. Initiation and termination codons are underlined. Site-directed mutations are indicated in italics.
Marker rescue.
Confluent monolayers of BSC40 cells in 35-mm dishes were infected with Dts12 or Dts56 at an MOI of 0.03 at 31.5°C. At 4 hpi, 5 μg of linearized pTM1 (empty vector), pBR322-HindIIID, pTM1-D5, or pTM1-D4 DNA was applied to cells as CaPO4 precipitates. At 7 hpi, cultures were shifted to 39.7°C and maintained under these conditions until 72 hpi. Cells were then harvested, and the yield of temperature-insensitive virus was determined by titration at 39.7°C. Experiments were performed and titrated in triplicate, and the results were averaged.
Construction of the pInt-D5 constructs for transient complementation.
The pInt vector contains 150 bp of the vaccinia virus G8R promoter and upstream region cloned into the XbaI and BamHI sites of the pUC19 vector (46). Initially, wtD5 was amplified from viral genomic DNA using the primers D5 5′ BamHI and D5 3′ (Table 1). This PCR product was digested with BamHI prior to ligation into pInt DNA that had been similarly digested and treated with CIP. Alleles of D5 containing site-directed mutations within the conserved motifs positioned in the 3′-half of the ORF were generated by overlap PCR using the pInt-wtD5 DNA as the template. Table 1 details the primers used to generate the various D5 mutants. Primer OL-UP contains an EcoRV site at its 5′ terminus, which corresponds to the internal EcoRV site starting at nucleotide 1401 of the D5 sequence. The sequence of primer OL-DP corresponds to nucleotides 396 to 413 of the pUC19 plasmid (GenBank accession number L09137); the primer contains an EcoRI site that is found within the pUC19 multicloning site. Two separate rounds of PCR were performed to generate products that overlapped by 17 bp. In the first round, two separate reactions using primers OL-UP plus 3′ or OL-DP plus 5′ were performed. A mixture of the two reaction mixtures served as the template for a third PCR, which was performed using the OL-UP and OL-DP primers. The final PCR product was digested with EcoRV and EcoRI and ligated into the pInt-wtD5 vector (see above) that had been similarly digested and treated with CIP. Plasmids were prepared from E. coli transformants using the alkaline lysis procedure (15).
Transient complementation assay.
Transient complementation assays were performed as previously described with minor modifications (29, 33). Replicate 35-mm dishes of BSC40 cells were infected with Cts24 at an MOI of 5 and incubated at 31.5°C. At 3 hpi, 1 μg of supercoiled DNA (pInt or pInt:D5 constructs) was introduced using the Lipofectamine 2000 reagent. At 5 hpi, cells were shifted to 39.7°C. At 5, 8, and 24 hpi, cells were harvested and analyzed for the accumulation of viral DNA sequences by Southern dot blot hybridization (44). At 24 hpi, a replicate set of samples was harvested and whole-cell lysates were examined for D5 accumulation by immunoblot analysis.
Generation of epitope-tagged D5 alleles and purification of the 3XFLAG-D5 proteins. (i) Generation of pTM1-3XFLAG tsD5 constructs.
Genomic viral DNA served as a template for amplification of the 2,358-bp wt and tsD5 ORFs using high-fidelity Taq polymerase (Invitrogen). The upstream primer, D5 5′ NdeI, and the downstream primer, D5 3′, introduced NdeI and BamHI sites prior to and after the translational initiation and termination codons, respectively (Table 1). After restriction enzyme cleavage, the D5 alleles were cloned into pTM1-3XFLAG vector DNA (33) that had been similarly digested and treated with CIP. The sequence of the various constructs was confirmed by automated DNA sequencing.
(ii) Generation of pTM1-3XFLAG site-directed D5 constructs.
The pInt-D5 constructs (see above) were used as the template for amplification of the site-directed D5 alleles. Again, high-fidelity Taq polymerase was used in conjunction with the D5 5′ NdeI and D5 3′ PCR primers (Table 1) to amplify the site-directed mutants for transfer into the pTM1-3XFLAG vector. Clones were prepared as described above; all constructs were verified by automated DNA sequencing.
(iii) Expression and purification of pTM1-3XFLAG D5 proteins.
Confluent 10-cm dishes of BSC40 cells were infected with vTF7.5 (7) at an MOI of 5 and incubated at 37°C. At 3 hpi, 12 μg of supercoiled DNA (pTM1-3XFLAG:D5 constructs) was introduced using the Lipofectamine 2000 reagent, and cultures were shifted to 31.5°C. At 24 hpi, cells were harvested and incubated in FLAG lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mM phenylmethylsulfonyl fluoride; 1 ml/107 cells) at 4°C for 20 min with end-over-end mixing. Clarified lysates were then incubated with EZview Red anti-FLAG M2 affinity gel beads for 16 h at 4°C; the beads were washed repeatedly with Tris-buffered saline (50 mM Tris [pH 7.4], 150 mM NaCl), and bound proteins were eluted in Tris-buffered saline by the addition of buffer containing 150 ng/μl 3XFLAG peptide (Sigma). Eluates were subjected to analysis by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining to assess purity, and the concentrations of D5 were quantitated using FluorChem 8900 software.
ATP binding and hydrolysis assays.
ATP binding and ATPase assays were preformed as previously described (9). Briefly, in order to assay the ATP binding activity of the protein samples, approximately 150 ng of the 3XFLAG-D5 proteins was incubated with 3.3 μM [α-32P]dATP (6 Ci/μmol) in the presence of MgCl2 on ice under a UV lamp. These samples were then resolved electrophoretically and visualized by autoradiography. To analyze ATPase activity, approximately 150 to 300 ng of the 3XFLAG-D5 proteins was incubated in a reaction mixture containing 1 mM [α-32P]dATP (6 Ci/μmol) for 1 h at 37°C. One microliter of the reaction mixture was spotted onto a polyethylenimine-cellulose F plate, and the substrate and products were resolved by ascending chromatography in 0.8 M CH3COOH-0.8 M LiCl. Reaction products were visualized by autoradiography and quantitated on a Storm PhosphorImager using ImageQuant software.
Generation of the 301-785 and 413-785 constructs.
The truncated D5 alleles were amplified using genomic viral DNA as a template and either the 301-5′ or 413-5′ primer in conjunction with the D5 3′ primer (Table 1). The PCR products were either digested with BamHI prior to ligation into pInt DNA that had been similarly digested and treated with CIP or digested with NdeI and BamHI prior to ligation into the pTM1-3XFLAG DNA that had been similarly digested and treated with CIP. The sequences of the constructs were confirmed by automated DNA sequencing.
Assessment of D5 multimerization by copurification of 3XFLAG-D5 and untagged D5.
Confluent 10-cm dishes of BSC40 cells were infected with vTF7.5 (7) with or without vTMD5 (9) (MOI of 2 for each virus) at 37°C. At 3 hpi, 12 μg of supercoiled DNA (12 μg of pTM1-3XFLAG:D5 or 3 μg of pTM1-3XFLAG:D5 plus 9 μg of pTM1:D5) was introduced using the Lipofectamine 2000 reagent, and cultures were shifted to 31.5°C. At 24 hpi, cells were harvested and clarified lysates were prepared. The 3XFLAG-D5 protein, and also any associated proteins, was subjected to affinity purification using the EZview Red anti-FLAG M2 affinity resin as described above. Eluates were resolved on SDS-8% acrylamide gels; the 3XFLAG-D5 and D5 proteins were visualized by silver staining and quantitated using FluorChem 8900 software. The amount of each protein present in the “input” sample was analyzed by immunoblot analysis using the anti-D5 antibody.
Preparation of digital figures.
Original data were scanned on an Epson Expression 1680 scanner (U.S. Epson Inc., Long Beach, Calif.) and were adjusted with Adobe Photoshop software (Adobe Systems Inc., San Jose, Calif.). Data from the dot blot hybridization and ATPase assay were acquired on a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and quantitated using ImageQuant software (Molecular Dynamics). Graphs were plotted using SigmaPlot (SPSS Science, Chicago, Ill). Images from the immunoblot analysis were acquired using the AlphaImager documentation system (Alpha Innotech, San Leandro, Calif.). Final figures were assembled and labeled with Canvas software (Deneba Systems, Miami, Fla.).
RESULTS
Genetic mapping of the tsD5 mutants.
As an initial approach to dissecting the biochemical and biological functions of D5, we undertook a phenotypic analysis of ts mutants possessing lesions within the D5 ORF. Originally, four tsD5 viruses, Cts17, Cts24, Ets69, and Dts6389 (now designated as Dts38), were defined as having a defect in viral DNA synthesis at the nonpermissive temperature (8, 11, 28). Two additional viruses, Dts12 and Dts56, were found to belong to this complementation group during the recent reevaluation of the Dales mutant collection (22). The genetic lesions within the Cts17, Cts24, and Ets69 D5 ORFs have been previously mapped (11); the Cts17 and Cts24 lesions are located in the 5′ portion of the ORF, while the Ets69 lesion resides in the 3′ portion (Fig. 1). To map the lesion within the remaining three ts mutants, viral genomic DNA was prepared from cells infected with Dts6389, Dts12, Dts56, and the parental IHD-W wt virus, and the D5 ORF was amplified and subjected to DNA sequencing. The D5 ORF within the Dts12 virus possessed a G348→A transition that would lead to a met116→ile substitution; in Dts56, a T635→C transition was observed that would result in a val212→ala substitution. Marker rescue analyses (not shown) confirmed that the temperature sensitivity of these viruses is indeed linked to these mutations, since recombination between the viral genomes and a wtD5 ORF generated temperature-insensitive virus. The D5 ORF of Dts6389 contained two lesions: a C482→T transition, resulting in a ser161→phe substitution, and a G847→A transition, resulting in an ala283→thr substitution. The first of these two lesions is identical to that found in the Cts17 genome; whether the second lesion contributes to the ts phenotype has not been addressed. These studies also revealed that the ORFs encoded by the wt IHD-W and WR strains differed in nine nucleotide positions, resulting in a single amino acid substitution (thr437→met) in an area of the vaccinia virus D5 protein that does not display a high degree of conservation when compared to orthologs in other poxviruses.
FIG. 1.
Predicted sequence of the D5 protein and identification of conserved domains and sites affected in temperature-sensitive and site-directed mutants. The predicted amino acid sequence of the vaccinia virus D5 protein (Western Reserve strain; protein database entry P04305) is shown. The residues altered in the various tsD5 strains are circled; those shown in gray circles have been previously mapped (T143I in Cts24, S161F in Cts17, and P682S in Ets69) (11). The residues shown in white type within black circles indicate lesions that were mapped in this study: M116I in Dts12, V212A in Dts56, and S161F and A283T in Dts6389. The four conserved motifs identifying D5 as a member of the SFIII helicase subgroup of the AAA+ family of proteins (17) are boxed. These motifs include a Walker A box/P-loop (white box, black type), a Walker B box (light gray box, black type), the motif C of SFIII proteins (dark gray box, white italic type), and the AAA+ family motif (black box, white type). The invariant residues targeted for site-directed mutagenesis within these motifs are underlined (K509A, E557Q, N605D, and R619A,R620A). The inverted triangles mark the beginning of the 301-D5 (filled) or 413-D5 (empty) fragment. While the majority of the ts lesions map to the N-terminal portion of the protein, the C-terminal portion of D5 contains the putative enzymatic/functional domains.
The tsD5 mutants exhibit a tight temperature-sensitive phenotype.
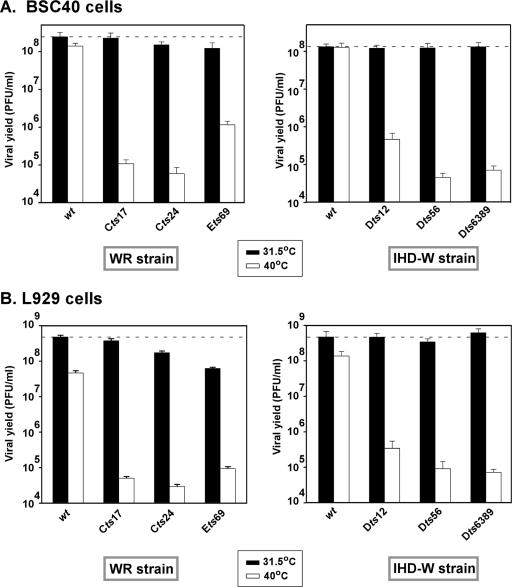
The 24-h viral yield from a single infectious cycle was determined by infecting either primate BSC40 (Fig. 2A) or murine L929 (Fig. 2B) cells with the various wt or tsD5 strains. Confluent monolayers of each cell type were infected at an MOI of 2 and incubated for 24 h at either permissive or nonpermissive temperature (Fig. 2). Upon harvesting, the total yield of cell-associated virus was determined by titration at 31.5°C. In BSC40 cells, wild-type viruses of both the WR and IHD strains produced equivalent yields of virus at both temperatures, as shown in Fig. 2A. Comparable viral yields were also produced when cells were infected with the tsD5 viruses at the permissive temperature. However, the tsD5 viruses produced 300- to 2,500-fold less virus when infections were performed at the nonpermissive temperature. Cts24, Cts17, Dts6389 and Dts56 displayed the tightest ts phenotype, whereas Ets69 and Dts12 were significantly leakier.
FIG. 2.
The tsD5 viruses exhibit a temperature-sensitive reduction in the production of infectious virus during a single round of infection. Confluent 35-mm dishes of BSC40 (A) or L929 (B) cells were infected with the WR (wt, Cts17, Cts24, and Ets69) (left graphs) and IHD-W (wt, Dts12, Dts56, and Dts6389) (right graphs) strains at an MOI of 2 and incubated for 24 h at either 31.5°C or 39.7°C. The yield of cell-associated virus was quantitated by titration on BSC40 cells at 31.5°C. The dashed line denotes the amount of infectious virus made during a wt infection at 31.5°C. Each infection was performed in duplicate, and each assay was titrated in duplicate. Error bars denote standard deviations.
In L929 cells, in contrast, even the wt viruses produced 3- to 10-fold less virus at the nonpermissive temperature than at the permissive temperature; the L929 cells themselves appear to be less heat tolerant than the BSC40 cells. Two of the tsD5 viruses were somewhat impaired even at the permissive temperature, producing three- to eightfold less virus than did the wt WR parent (Fig. 2B). At the nonpermissive temperature, the tsD5 viruses produced 700 to 8,500-fold less virus than they did at low temperature. Similar results were obtained when infections were performed at a high MOI (MOI of 15) (data not shown).
Viral DNA replication is impaired during nonpermissive tsD5 infections.
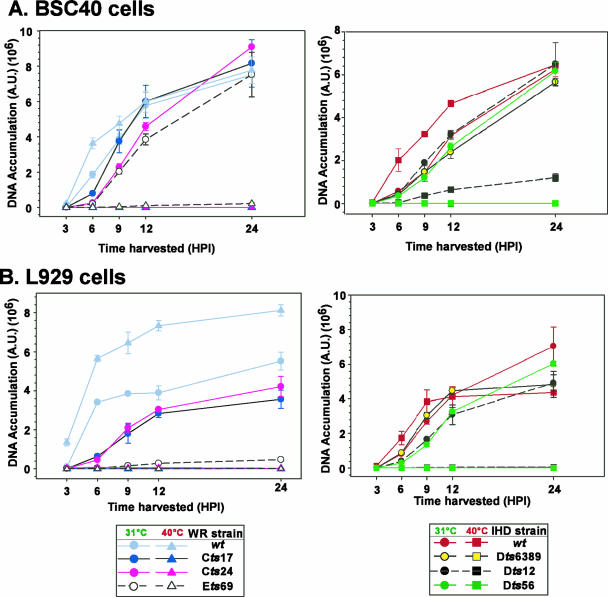
In earlier studies, the DNA replication defect of the tsD5 viruses was studied either by measuring the incorporation of [3H]thymidine into nascent viral DNA (11) or by visualizing the accumulation of Hoechst-staining cytoplasmic factories via immunofluorescence (8). For this study, we sought to quantitate the DNA synthesis defect of all of the tsD5 mutants using the Southern dot blot hybridization technique (44). BSC40 or L929 cells were infected with the wt or tsD5 viruses and maintained at 31.5°C or 39.7°C for 3, 6, 9, 12, and 24 h. Lysates were prepared at each time point, and the levels of viral DNA that had accumulated were assessed; the data shown in Fig. 3 are from a representative experiment. Experiments performed in BSC40 cells are shown in Fig. 3A, with viruses from the WR strain shown in the left graph and those from the IHD strain shown in the right graph. Figure 3B show the results of similar experiments performed in L929 cells.
FIG. 3.
tsD5 viruses show a defect in viral DNA synthesis at high temperature. Confluent 35-mm dishes of BSC40 (A) or L929 (B) cells were infected with the WR (left plots) or IHD (right plots) strain at an MOI of 5 and maintained at 31.5°C or 39.7°C. Individual cultures were harvested at 3, 6, 9, 12, or 24 hpi, and the levels of accumulated viral DNA were determined by Southern dot blot hybridization. The average of the data obtained from a representative experiment, which was analyzed in quadruplicate, is shown with the standard deviation. The legends are shown at the bottom of the figure; note that the scales for the y axes differ among the panels.
During permissive infections of BSC40 cells performed with the tsD5 viruses of the WR strain, the accumulation of viral DNA was somewhat delayed compared to wt infections. For Cts17, DNA accumulation returned to wt levels by 9 hpi; for Cts24 and Ets69, viral DNA accumulation did not reach wt levels until 24 hpi. Nevertheless, by 24 hpi, approximately equal amounts of viral DNA were seen in all samples. For the IHD viruses, all of the permissive infections accumulated DNA with similar kinetics (Fig. 3A, right panel).
In contrast, even after 24 h of infection at the nonpermissive temperature (39.7°C), only the two wt strains (WR and IHD) directed the accumulation of significant amounts of viral DNA. For Cts17, Cts24, Dts6389, and Dts56, <1% of wt levels of DNA were seen; Ets69 (Fig. 3A, left panel) directed the accumulation of <3% of wt levels of DNA, whereas Dts12 (Fig. 3A, right panel) directed the accumulation of ∼18% of wt levels of DNA. The modest reduction in DNA accumulation seen with Dts12 is puzzling, since it correlates poorly with the nearly 2.5-log reduction seen in viral yield.
During permissive infections of L929 cells, we again noted a delay in DNA accumulation with the tsD5 WR strains compared to wt infections (Fig. 3B, left panel). Moreover, reduced levels of DNA were seen at 24 hpi, with Cts17, Cts24, and Ets69 accumulating 65%, 76%, and <8% of the levels of DNA seen during wt infections, respectively. The dramatic reduction in viral DNA accumulation observed during permissive Ets69 infection of L cells is also puzzling, since it does not correlate well with the more modest 10-fold reduction observed for viral yield (Fig. 2B). The IHD tsD5 strains, Dts12 and Dts56, exhibited an ∼3-h delay in viral DNA accumulation (Fig. 3B, right panel), but by 24 hpi wt levels of DNA were seen. Most importantly, a strict DNA-minus phenotype was observed for all of the tsD5 viruses during nonpermissive infections of L929 cells: no detectable accumulation of DNA was seen.
Status of tsD5 proteins at high temperature.
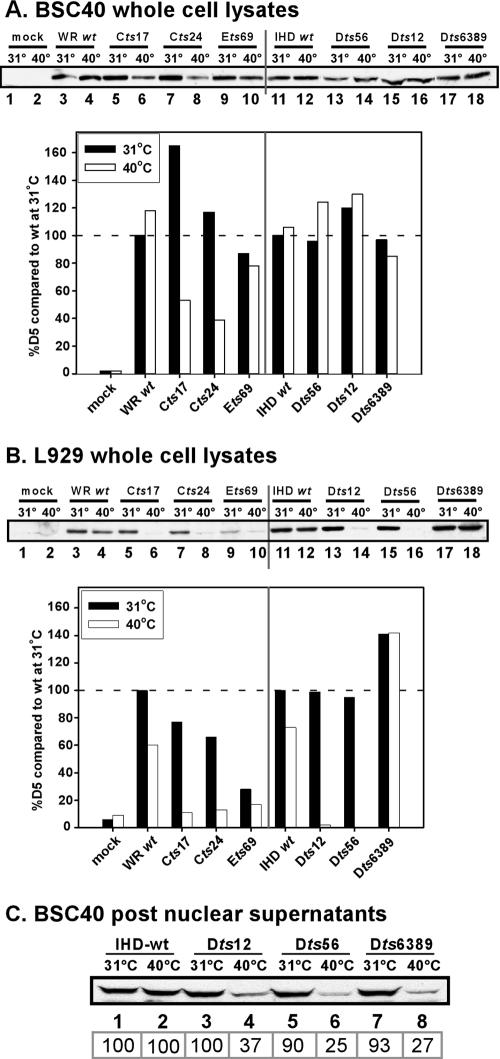
The phenotype associated with ts mutations can reflect changes to the affected protein that alter its solubility, stability, or function at the high temperature. Initially, we compared the first two parameters for the wt and tsD5 proteins. BSC40 or L929 cells were left uninfected or infected at an MOI of 10 for 8 h at either 31.5°C or 39.7°C. The total levels of D5 were assessed by immunoblot analysis; all data were normalized to the levels of D5 seen during infections with the corresponding wt strain at 31.5°C. Data from a representative experiment are presented in Fig. 4.
FIG. 4.
Analysis of accumulation and solubility of tsD5 proteins during infections performed at permissive and nonpermissive temperatures. BSC40 (A) or L929 (B) cells were either mock infected or infected with the various WR and IHD strains at an MOI of 10 and maintained at either 31.5°C or 39.7°C for 8 h prior to harvesting. Whole-cell lysates were subjected to immunoblot analysis with an anti-D5 antiserum; the level of D5 in each sample was normalized to the amount of D5 that accumulated during infection with the appropriate wt strain at 31.5°C. Infections were performed in duplicate, and each infected cell lysate was analyzed in duplicate. A representative blot is shown. (C) BSC40 cells were infected with the various IHD viruses at an MOI of 10 and maintained at either the permissive (odd lanes) or nonpermissive (even lanes) temperature for 8 h. Postnuclear supernatants were prepared and analyzed by immunoblotting with the anti-D5 antiserum; the boxed numbers below the lanes represent the relative levels of the tsD5 proteins compared to the amount of wtD5 that accumulated at the respective temperature.
During BSC40 infections, both the WR and IHD wtD5 proteins accumulated to nearly equal levels at both low and high temperatures (Fig. 4A, lanes 3 and 4 and lanes 11 and 12). However, accumulation of the Cts17 and Cts24 D5 proteins was temperature sensitive; indeed, although higher-than-wt levels of protein accumulated at 31.5°C, much less protein was seen at 39.7°C. For Cts17, only 32% as much D5 accumulated at 39.7°C than at 31.5°C; for Cts24, this value was 33%. None of the other tsD5 mutants encoded proteins that were thermolabile in BSC40 cells. Since both Cts17 and Dts6389 share the same S161F substitution, this observation suggests that the second substitution found in Dts6389, A283T, may suppress the thermolability of the mutant protein.
In L929 cells, we observed significantly more variability in the levels of D5 protein that accumulated (Fig. 4B). The wt proteins showed some thermolability, in keeping with our finding that these cells show some temperature sensitivity for virus production (Fig. 2); for the WR and IHD strains, the levels of protein that accumulated at 39.7°C were ∼60% and 73% of those that accumulated at 31.5°C, respectively. Low levels of the Ets69 D5 protein accumulated even at the permissive temperature (28% of wt levels), consistent with our observation that this virus only produced low levels of DNA in L929 cells (Fig. 3). The Cts17 and Cts24 D5 proteins, which are thermolabile in BSC40 cells, retained their thermolability in L929 cells; in addition, the D5 proteins encoded by Dts56 and Dts12 were highly thermolabile in L929 cells, with <5% of the wt levels of protein remaining at 8 hpi. Of the ts mutants, only Dts6389 encodes a D5 protein that is thermostable in L929 cells.
The thermolability of the D5 proteins encoded by Cts17 and Cts24 provides a plausible explanation for the DNA-minus phenotype observed for these mutants in BSC40 cells. However, the other tsD5 proteins were shown to be stable at 39.7°C in this cell line, although the mutants did not replicate their DNA at this temperature. To test whether these proteins might be insoluble at the nonpermissive temperature, we prepared postnuclear supernatants from BSC40 cells infected for 8 h at either 31°C or 39.7°C and quantitated the levels of D5 by immunoblot analysis (Fig. 4C). During wt (lanes 1 and 2) and Ets69 (not shown) infections, equivalent levels of soluble D5 were observed in the two lysates. However, for Dts12, Dts56, and Dts6389, only 25 to 37% as much soluble D5 was present at 39.7°C than at 31.5°C. Thus, the inability of these viruses to replicate may be due to the presence of the D5 protein in a misfolded and hence insoluble form.
Generation, expression, and purification of 3XFLAG-tsD5 proteins: tsD5 proteins retain the ability to bind and hydrolyze ATP.
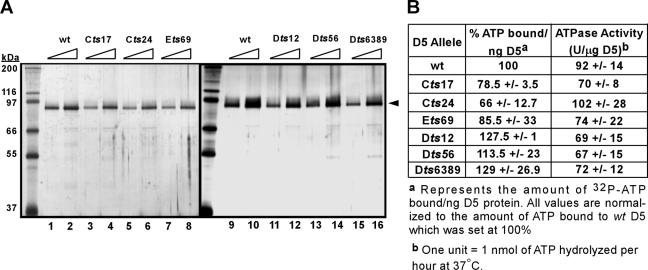
D5 has been shown to bind and hydrolyze ATP (9); therefore, we were interested in determining if any of the tsD5 proteins were defective in these activities, especially the Ets69 D5 protein, since we had found that it is stable and soluble during nonpermissive BSC40 infections. To avoid complications of lability or insolubility, we expressed these proteins at 31.5°C; we and others have observed that the proteins encoded by ts alleles commonly show a biochemical defect in vitro even when synthesized at the permissive temperature (35). At 24 hpi, cells were harvested and the 3XFLAG-D5 proteins were affinity purified; the purified preparations are shown in Fig. 5A.
FIG. 5.
Comparative analysis of the ATP binding and hydrolysis activities of wt and mutant 3XFLAG-D5 proteins. (A) 3XFLAG-D5 proteins were purified to near homogeneity. Cells were infected so as to overexpress the various 3XFLAG-D5 proteins at permissive temperature. The D5 proteins were affinity purified from lysates prepared at 24 hpi; a silver-stained gel of the purified proteins is shown in panel A (for each preparation, 1/10 and 1/20 of the yield from 1 × 107 cells are shown in adjacent lanes). The black arrowhead indicates the 3XFLAG-D5 proteins; protein standards are indicated on the left, with their masses shown in kilodaltons. (B) tsD5 proteins retain near-wt activity for ATP binding and ATP hydrolysis. ATP binding and ATP hydrolysis assays were carried out as described in Materials and Methods. The average of the data obtained from several replicates is shown with the standard deviation.
We initially assessed the ability of the various tsD5 proteins to bind ATP in vitro. An ∼150-ng aliquot of the various D5 proteins, or ∼1.5 μg of the Klenow fragment of Escherichia coli DNA polymerase as a control, was incubated with [α-32P]dATP in the presence of MgCl2 on ice under a UV germicidal lamp. Reaction mixtures were resolved electrophoretically and analyzed by autoradiography and phosphorimager analysis; the data are tabulated in Fig. 5B. Although some variability was observed, all of the tsD5 proteins retained the ability to bind ATP; visualization of this binding was dependent on UV cross-linking, as shown previously (9).
We next addressed whether the tsD5 proteins retained NTPase activity. An ∼150- to 300-ng aliquot of the 3XFLAG-D5 proteins was incubated with [α-32P]dATP, and the enzymatic conversion of ATP to ADP was revealed by thin-layer chromatography, autoradiography, and phosphorimager analysis. The specific activities of the wt and tsD5 proteins are tabulated in Fig. 5B. The first thing to note is that the specific activity of the 3XFLAG wtD5 is comparable to what was previously described for conventionally purified, untagged wtD5 (92 U/μg 3XFLAG-D5 versus 79 U/μg conventionally purified D5 [9]), confirming that the epitope tag does not adversely affect D5's activity. The other conclusion that emerged from this analysis was that all of the tsD5 proteins retained near-wt levels of ATPase activity (Fig. 5B). Lastly, we directed expression of 3XFLAG-Ets69-D5 at 31.5°C or 39.7°C. We observed that the two protein preparations had nearly the same specific activity, suggesting that enzymatic activity is also not affected by the Ets69 mutation (data not shown).
Establishment of a transient complementation assay: rescue of the DNA-minus phenotype of Cts24 by expression of D5 from an intermediate promoter.
Transient complementation assays have become a useful tool for structure/function analyses of essential vaccinia virus proteins. Cells are infected with a conditionally lethal virus under nonpermissive infections (e.g., high temperature or the absence of an inducer), plasmids expressing the wt or variant alleles of the gene of interest are introduced by infection, and their ability to restore a wt phenotype is assayed. To date, this analysis has been limited to proteins encoded by late viral genes (29, 33, 47). A major limitation to the application of this approach to early proteins is the fact that early mRNAs are transcribed within the viral core and, therefore, the early transcription machinery, which is sequestered within this compartment, is not accessible to transfected DNA. To circumvent this problem, we took advantage of the observation that tsD5 mutants rapidly cease further DNA synthesis when they are shifted to the nonpermissive temperature in the midst of DNA replication (5 hpi). The D5 ORF was therefore placed under the transcriptional control of a promoter derived from an intermediate gene, the G8R gene (46), and we assessed whether expression of D5 from this plasmid would complement the fast-stop phenotype of Cts24.
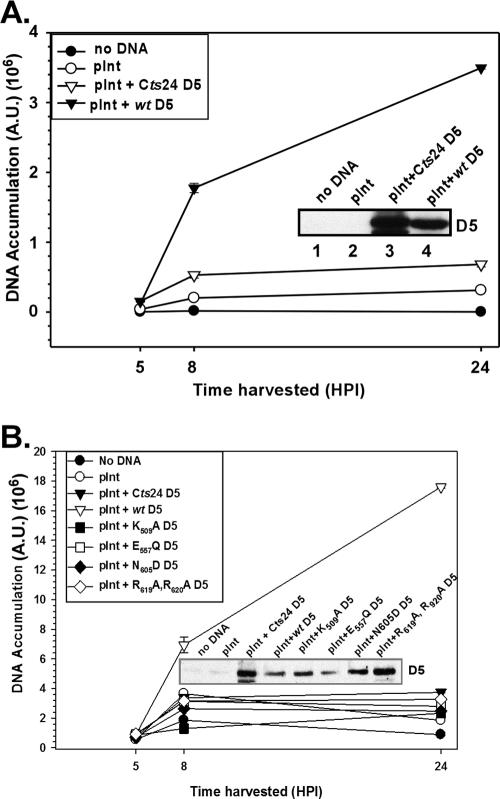
BSC40 cells were infected with Cts24 at 31.5°C, and at 3 hpi cells were left untransfected, transfected with empty vector (pInt), or transfected with plasmids containing the wt or Cts24 D5 alleles. At 5 hpi, cells were either harvested or shifted to 39.7°C and maintained at that temperature until 8 or 24 hpi, as shown. Samples were analyzed for the accumulation of D5 protein and/or viral DNA. As can be seen in Fig. 6A, expression of wtD5 from the G8 promoter did sustain ongoing viral DNA synthesis when Cts24-infected cultures were shifted to 39.7°C. In fact, at 24 hpi the levels of DNA that accumulated in cultures receiving pInt-D5 were 11-fold higher than those seen in cultures receiving empty vector (pInt). Viral DNA synthesis was stimulated only twofold by the introduction of vector encoding Cts24 D5. Immunoblot analysis of whole-cell lysates confirmed that the wt and Cts24 D5 proteins were expressed to comparable levels (Fig. 6A inset, lanes 3 and 4, respectively). Some stimulation of DNA synthesis was seen even when empty vector DNA was introduced (compare pInt to no DNA), although the mechanism underlying this phenomenon is not understood. (We believe that nicked and/or linear DNA stimulates DNA synthesis in a nonspecific manner; minimizing this effect was accomplished by using high-quality DNA preparations that were prepared with the alkaline lysis method and were predominantly in the intact, supercoiled form.) Establishment of this transient complementation system, which allowed us to distinguish between biologically competent and incompetent forms of D5, enabled us to initiate a structure/function analysis of D5.
FIG. 6.
Transient expression of wtD5 enables sustained viral DNA synthesis when Cts24 infections are shifted to 39.7°C in the midst of DNA replication. (A) Transient expression of wtD5, but not Cts24 D5, rescues viral DNA synthesis. BSC40 cells were infected with Cts24 at an MOI of 5 at 31.5°C; at 3 hpi, cells were mock transfected (no DNA) or transfected with supercoiled empty vector (pInt) or pInt:D5 constructs. At 5 hpi, cells were shifted to 39.7°C for the remainder of the experiment. At 5, 8, and 24 hpi, cells were harvested, and the levels of accumulated viral DNA were quantitated by Southern dot blot hybridization; the analysis was performed in quadruplicate, and the average value is shown with error bars. The inset shows an immunoblot that confirms the accumulation of the D5 proteins. (B) D5 alleles containing targeting mutations within conserved motifs are unable to rescue DNA replication during Cts24 infections. The transient complementation assay described for panel A was applied to the analysis of pInt:D5 constructs encoding D5 proteins containing the indicated mutations.
The invariant residues within the conserved domains of D5 are crucial for the ability of D5 to complement the Cts24 viral DNA synthesis defect.
Bioinformatic comparisons led to the characterization of D5 as a member of the SFIII DNA helicase subgroup of AAA+ proteins (17). Despite this characterization and the empirical demonstration that D5 is an NTPase, the biochemical contribution of D5 to DNA replication is not known. We therefore wanted to determine which structural feature(s) of the D5 protein was important for its biological function. Our mutagenesis strategy reflected studies of other SFIII helicases (19, 49) and AAA+ proteins (14) as well as the computer-assisted alignments presented by Iyer et al. (17). Overlap PCR was used to alter invariant residues found within conserved motifs: the Walker A domain (K509A), the Walker B domain (E557Q), the SFIII motif C region (N605D), and the defining motif of the AAA+ proteins (R619A,R620A) (Fig. 1). Each altered allele was cloned into the pInt vector and used in the transient complementation assay described above.
As seen in Fig. 6B, while introduction of either empty pInt or pInt-Cts24D5 resulted in only a very weak stimulation of viral DNA synthesis, expression of wtD5 resulted in a significant augmentation of viral DNA synthesis at 39.7°C (at 24 hpi, 9.5 times more viral DNA was detected in cells receiving this plasmid compared to pInt). Interestingly, when any of the site-directed D5 mutants was transfected into the Cts24-infected cells, little or no stimulation of DNA synthesis was seen (1.2 to 1.7 times the amount of viral DNA at 24 hpi compared to empty vector). Immunoblot analysis confirmed that each of the D5 proteins was expressed at comparable levels (Fig. 6B, inset). These data indicate that each of the conserved motifs associated with the characterization of D5 as a member of the SFIII helicase subfamily of the AAA+ proteins is essential for the ability of D5 to support viral DNA replication.
Biochemical characterization of D5 site-directed alleles.
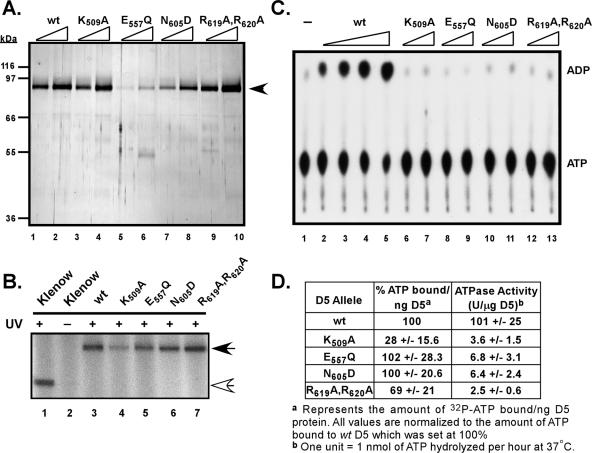
We wished to determine whether the loss of biological competence seen for the site-directed D5 mutants correlated, as expected, with a loss of enzymatic activity. The various D5 alleles were subcloned into the pTM1-3XFLAG vector, and epitope-tagged proteins were expressed and affinity purified to near homogeneity (Fig. 7A). All the proteins were expressed to equal levels with the exception of the E557Q D5 protein, for which the yield was reduced ∼2-fold. This poor yield may reflect a folding defect, which has been described for other SFIII proteins bearing a mutation at this site (49).
FIG. 7.
Biochemical analysis of site-directed D5 mutants. (A) Expression and purification of 3XFLAG-D5 proteins containing site-directed mutations in conserved motifs. The 3XFLAG-D5 proteins with site-directed mutations were expressed, purified, and visualized as described for Fig. 5A. The black arrowhead indicates the 3XFLAG-D5 proteins; protein standards are indicated on the left. (B) ATP binding activity of the site-directed mutated D5 proteins. An ∼150-ng aliquot of the 3XFLAG-D5 proteins, or 1.5 μg of the Klenow fragment of E. coli DNA polymerase I, was incubated with [α-32P]dATP on ice and UV irradiated (or left on ice without UV exposure [lane 2]). The samples were resolved by electrophoresis on an SDS-10% acrylamide gel, and bound [α-32P]dATP was visualized by autoradiography. (C) Mutagenesis of the invariant residues within the conserved D5 motifs ablates ATPase activity. Reaction mixtures containing increasing concentrations of 3XFLAG-D5 proteins (wt at 30, 75, 150, and 300 ng/reaction mixture [lanes 2 to 5]; all others were at 150 and 300 ng/reaction mixture [lanes 6 to 13]) or no protein (lane 1) and 1 mM [α-32P]dATP were analyzed by ascending chromatography. ADP and ATP were visualized by autoradiography. (D) Quantitative summary of the ATP binding and hydrolysis activity of the wt and mutant D5 proteins. The average activity obtained for each protein preparation was calculated after phosphorimager analysis of replicate experiments and is shown with the standard deviation.
Next, ATP binding studies were conducted on the various site-directed mutants. Mutations within the Walker B (E557Q) domain or the SFIII motif C (N605D) had no effect on the specific activity of ATP binding (Fig. 7B, lanes 5 and 6, and D). However, the ATP binding activity of the Walker A mutant (K509A) was reduced to approximately 28% of that observed for wtD5. This result was not surprising, since the Walker A domain, or P-loop, is thought to bind directly to the α-phosphate moiety of the ATP molecule (48). The mutant protein altered in the AAA+ signature (R619A,R620A) exhibited a reduction in ATP binding activity to 69% of wt levels.
ATPase assays were also performed with all of the mutant forms of D5: alteration of any of the invariant residues within the conserved motifs resulted in the nearly complete loss of ATPase activity (Fig. 7C). When the specific activities of the various D5 proteins were compared, it was apparent that the site-directed mutants had only 2 to 6% of the activity observed for wtD5. These results are consistent with the findings obtained in mutational analyses of other AAA+ proteins (see references found within reference 14).
In conjunction, the biochemical characterization of the mutant forms of D5 and the data obtained from the transient-complementation assays establish that the ATPase activity of D5 is essential for its biological competency.
Analysis of the multimerization of the wt and mutant D5 proteins.
A common feature of the diverse members of the family of AAA+ proteins is their assembly into biologically active oligomers, generally hexameric ring structures (reviewed in reference 14). Biochemical and structural studies have revealed that adapting this configuration allows the conserved arginine residue within the “arginine finger” motif of an individual subunit to insert itself into the ATP binding pocket of the Walker A domain on an adjacent subunit. This juxtaposition allows for an interaction between the “arginine finger” and the γ-phosphate of ATP bound in the adjacent subunit, thus providing a basis for the stabilization and ATP-driven oligomerization of multimeric structures (31). Mutational studies of several members of this family of proteins have confirmed that altering the conserved arginine residue within the arginine finger results in the loss of ATP hydrolysis as well as oligomerization (31). While we do know that D5 is a member of the AAA+ protein family that has the capacity to form higher-ordered structures (9), we do not know if the invariant residues positioned at the end of the AAA+ motif are necessary for oligomerization. Therefore, in the absence of crystallographic data, we can only hypothesize that the two invariant arginine residues positioned at the end of the AAA+ motif (R619 and R620) could potentially serve as the “arginine finger.”
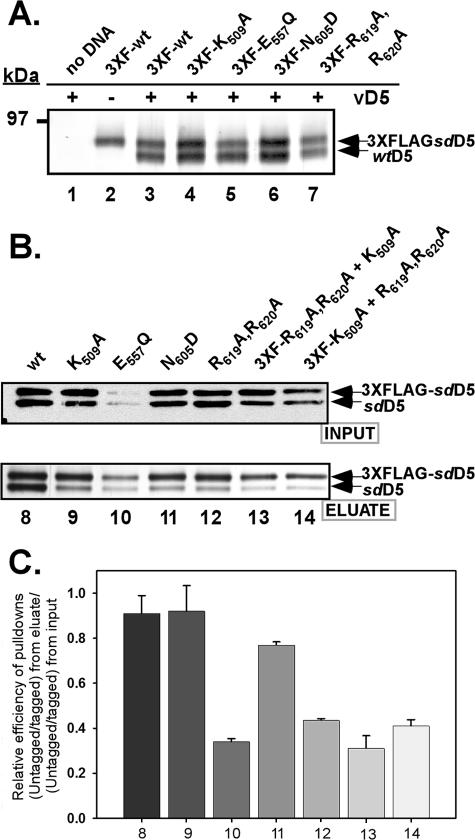
To test if these arginine residues or any of the other invariant residues were involved in ATP-driven oligomer formation, we performed affinity pull-down assays. Briefly, BSC40 cells were infected with recombinant vaccinia viruses so as to overexpress wtD5 (vTF7.5 and vTMD5) and then transfected with pTM1-3XFLAG plasmids encoding the site-directed D5 mutants. At 24 hpi, lysates were prepared and the 3XFLAG-tagged D5 proteins and any stably interacting proteins were retrieved on an affinity matrix. Eluates were resolved by 8% acrylamide SDS-PAGE and silver staining; under these conditions, the 3XFLAG-D5 (predicted molecular mass of ∼89.6 kDa) can be resolved from the untagged D5 protein (predicted molecular mass of ∼87.1 kDa). As can be seen in Fig. 8A, when untagged wtD5 and 3XFLAG:wtD5 were cooverexpressed, both were retrieved by the anti-FLAG resin (lane 3), consistent with the association of D5 molecules into a higher-order structure (greater than or equal to dimer). As expected, untagged D5 was not retrieved on the anti-FLAG resin when 3XFLAG-D5 was not present (lane 1). We observed that each of the mutant forms of 3XFLAG-D5 was able to interact with untagged wtD5 in this assay (lanes 4 to 7).
FIG. 8.
Participation of conserved family domains in D5 oligomerization. (A) The site-directed D5 mutants retained the ability to interact with wt D5. BSC40 cells were coinfected with the vTF7.5 and vTMD5 (vD5) viruses (lane 2 received only vTF7.5 infection) each at an MOI of 2. At 3 hpi, cells were left untransfected (lane 1, no DNA) or transfected with supercoiled pTM1-3XFLAG:D5 constructs. The 3XFLAG proteins and any tightly associated untagged D5 were affinity purified on anti-FLAG resin. The eluates were analyzed by fractionation on an SDS-8% acrylamide gel; only the relevant portion of the silver-stained gel is shown. (B) Some of the site-directed mutants showed a decreased ability to self-associate. BSC40 cells were infected with vTF7.5 and, at 3 hpi, cotransfected with supercoiled pTM-1 containing the untagged and 3XFLAG-tagged versions of the same (lanes 8 to 12) or different (lanes 13 and 14) D5 alleles. A portion of the clarified cytoplasmic lysates was removed prior to affinity purification (input, top panel); the remainder was used for affinity purification of the 3XFLAG-D5 and any associated untagged D5 (eluate, bottom panel). Input and eluate samples were resolved electrophoretically; the input samples were subjected to immunoblot analysis with an anti-D5 antiserum, and the eluates were visualized by silver staining. (C) Graphic presentation of the relative efficiency of copurification of untagged D5 with 3XFLAG-D5 protein. The data shown in panel B were quantitated and used to obtain a ratio of the relative efficiency of pull down. For each sample, the ratio within the eluates of the untagged D5 to the 3XFLAG-tagged D5 was compared to the ratio of untagged D5 to tagged D5 present in the input sample. The average of replicate experiments is shown with the standard deviation.
Knowing that the altered D5 proteins retained the ability to interact with wtD5, we next wanted to determine if the mutant D5 proteins could self-associate and/or associate with each other. BSC40 cells were infected and transfected so as to express the untagged and 3XFLAG-tagged versions of the same (Fig. 8B, lanes 8 to 12) or different (lanes 13 and 14) D5 mutant proteins. Prior to incubation with the anti-FLAG resin, a portion of the “input” sample was retained for immunoblot analysis using the anti-D5 antibody (Fig. 8B, top panel) in order to assess the relative abundance of each of the proteins. This assay was designed to achieve equivalent expression of 3XFLAG-tagged and untagged D5 proteins within each cotransfection; however, in some cases, the tagged protein accumulated to higher levels than the untagged protein (Fig. 8B, top panel, lanes 9, 11, 13, and 14). Furthermore, we again observed that the E557Q D5 protein was diminished in its accumulation (Fig. 8B, top panel, lane 10). The remainder of the cell lysate was used for affinity purification of the tagged (and associated untagged) proteins, and the eluates were analyzed by electrophoresis and visualized by silver staining (Fig. 8B, bottom panel). We calculated the relative pull-down efficiency for each protein, taking into account the levels of input and retrieved proteins (Fig. 8C). This analysis revealed that mutations within the invariant residues of the Walker B domain (E557Q) and the AAA+ motif (the putative “arginine finger,” R619A,R620A) affected the self-association of these proteins (Fig. 8C, bars 10 and 12, which correspond to B, bottom panel, lanes 10 and 12). Furthermore, we saw a reciprocal reduction in association between the R619A,R620A D5 (arginine finger) mutant and the K509A D5 (Walker A) mutant (Fig. 8B, bottom panel, lanes 13 and 14, and C, bars 13 and 14). These data imply that the Walker B domain and the putative “arginine finger” motif of D5 play some role in facilitating or stabilizing the multimerization of D5.
Coupling of oligomerization and ATPase activity.
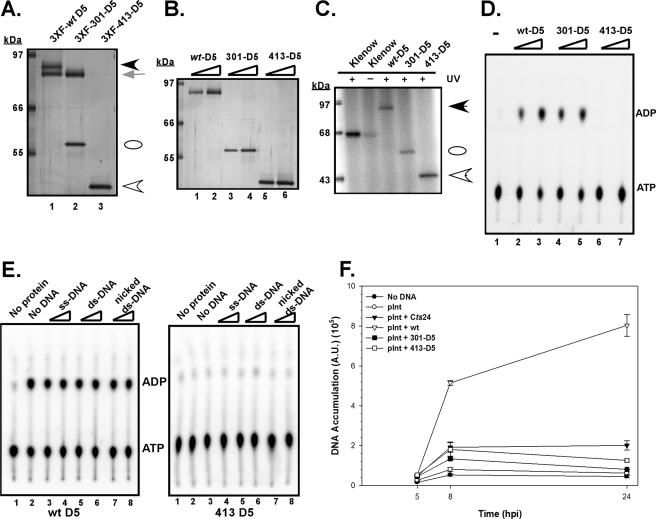
While several invariant residues positioned in the conserved domains of D5's catalytic core participate in the stabilization of multimeric structures, evidence from other model systems suggests that a domain outside of the catalytic core is likely to mediate multimerization. Indeed, for simian virus 40 (SV40) T antigen and human papillomavirus (HPV) E1, both notable SFIII/AAA+ proteins, a helical domain positioned immediately upstream of the enzymatic domain is required for oligomerization (1, 12, 23, 42). Furthermore, formation of higher-ordered structures is crucial for the NTPase activity of these proteins (12, 37). Therefore, we were interested in determining if regions outside of D5's catalytic core mediate multimerization and if multimerization is necessary for enzymatic activity. We therefore performed a serial deletion analysis of the N terminus of the D5 protein. The various truncated D5 mutants were transferred into the pTM1-3XFLAG vector, and FLAG affinity pull-down assays were performed on cell lysates that were programmed to overexpress full-length untagged wtD5 in the context of the deletion mutants (data not shown). We observed that sequences within amino acids (aa) 301 to 413 were required for oligomerization of D5 (Fig. 9A). While a fragment lacking the first 300 aa (301-D5; lane 2) retained the ability to pull down untagged D5, a deletion mutant missing the first 412 aa (413-D5; lane 3) could not. The latter observation is interesting because residues 413 to 785 of the vaccinia virus D5 protein have been characterized as a “D5 conserved domain” (http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid = pfam03288), and it is this portion of D5 that encompasses the catalytic motifs. To examine the biochemical properties of these deletion mutants, we expressed and purified 3XFLAG-tagged wt, 301, and 413 D5 proteins (Fig. 9B, lanes 1 to 6). While both the 3XFLAG-301 D5 and 3XFLAG-413 D5 proteins retained the ability to be cross-linked to ATP (Fig. 9C), only the 3XFLAG-301 D5 protein retained ATPase activity (Fig. 9D). The 3XFLAG-413 D5 protein, despite containing all of the catalytic motifs, was enzymatically inactive.
FIG. 9.
Preliminary mapping of the oligomerization/enzymatic domain. (A) A domain located between amino acids 301 and 413 is required for stoichiometric association with full-length wtD5. Cells were programmed to overexpress a full-length untagged version of D5 in the context of full-length 3XFLAG-tagged D5 (lane 1) or N-terminal truncation D5 mutants 3XFLAG-301-D5 (lane 2) or 3XFLAG-413-D5 (lane 3). Affinity purification of the tagged protein and any associated, untagged D5 was performed as previously described; the eluates were resolved electrophoretically and visualized by silver staining. While both the full-length 3XFLAG-wtD5 (black arrowhead) and the 3XFLAG-301-D5 (white oval) were able to retrieve full-length untagged D5 (gray arrow), the 3XFLAG-413-D5 (white arrowhead) was deficient in this activity. (B) Expression and purification of wt and N-terminal-truncated 3XFLAG-tagged D5 proteins. The 3XFLAG-wt, 3XFLAG-301-D5, and 3XFLAG-413-D5 proteins were expressed and purified, and increasing amounts of the purified preparations were resolved electrophoretically and visualized by silver staining. These affinity-purified proteins were used in biochemical analyses. (C) Both N-terminal truncation mutants retain ATP binding activity. ATP binding studies were conducted as described previously. The black arrowhead marks the migration of full-length 3XFLAG-D5, the white oval marks the migration of the 3XFLAG-301-D5 protein, and the white arrowhead marks the migration of the 3XFLAG-413-D5 protein. (D) Only the 301-D5 protein, which is capable of multimerization, retains enzymatic activity. Reaction mixtures containing either no protein (lane 1) or 150 to 300 ng of the 3XFLAG-wt (lanes 2 and 3), 3XFLAG-301-D5 (lanes 4 and 5), or 3XFLAG-413-D5 (lanes 6 and 7) proteins were incubated with [α-32P]dATP, and the conversion of ATP to ADP was monitored by ascending chromatography. (E) The 413-D5 protein lacks intrinsic ATPase activity which cannot be restored by the addition of exogenous nucleic acid. Reaction mixtures containing either no protein (lanes 1) or ∼150 ng of the 3XFLAG-wt or 3XFLAG-413-D5 proteins were incubated with [α-32P]dATP in the absence of DNA (lanes 2) or in the presence of 250 (lanes 3, 5, and 7) or 500 (lanes 4, 6, and 8) ng of DNA. ssM13mp10 was the source of single-stranded DNA (ss-DNA; lanes 3 and 4), salmon sperm DNA was the source of double-stranded DNA (ds-DNA; lanes 5 and 6), and activated salmon sperm was the source of nicked double-stranded DNA (lanes 7 and 8). (F) Neither the 301- nor the 413-D5 protein can rescue viral DNA synthesis during nonpermissive Cts24 infection. The transient complementation assay previously described was applied to pInt constructs expressing either 301-D5 or 413-D5; neither protein retained the ability to restore DNA synthesis.
Unlike most NTPases involved in DNA replication, the ATPase activity of full-length D5 is DNA independent. We considered the possibility that the N′ terminus of D5, which is missing in 413-D5, might confer this nucleic acid independence, and we therefore tested whether the addition of a variety of nucleic acids would activate the ATPase activity of the truncated protein (Fig. 9E). However, 413-D5 remained enzymatically inert even in the presence of these cofactors. It was not surprising, therefore, to find that the 413-D5 ORF was unable to compensate for the loss of D5 in our transient complementation assay (Fig. 9F), since we had previously shown that NTPase activity is required for biological complementation (Fig. 7).
The 301-D5 ORF was also tested in our transient complementation assay. Because the protein encoded by this ORF can multimerize and is enzymatically active, it was perhaps unexpected that we found it unable to rescue the Cts24 DNA replication defect. Although the N′ terminus of D5 appears to be dispensable for the assembly of an enzymatically active form of D5, it must perform some other role that is essential for biological competency.
DISCUSSION
Characterization of tsD5 viruses and proteins.
The vaccinia virus-encoded D5 NTPase plays an integral and indispensable role in the complex process of viral DNA replication. Although tsD5 viruses have been known to have a DNA-minus phenotype at the nonpermissive temperature for some time (8, 11, 28), the precise role(s) played by the D5 protein during replication has remained elusive. Although bioinformatic analyses have suggested that the D5 protein possesses all of the hallmarks of a DNA helicase, we have been unable to observe helicase activity using traditional oligonucleotide unwinding assays. The studies described herein were performed to gain further insight into the structure and function of the D5 protein and to assess whether the motifs suggestive of DNA's identity as a helicase were in fact required for its biological function. Our data are summarized in Table 2.
TABLE 2.
Summary of genetic and biochemical properties of the D5 mutant proteins
| D5 | Affected residue(s) | Protein status at 39.7°Ca | ATP bindingb | ATPase activityc | Multimerizationd | Viral DNA replicatione |
|---|---|---|---|---|---|---|
| wt | Stable | + | + | + | + | |
| Cts17 | S161F | Unstable | + | + | + | − |
| Cts24 | T143I | Unstable | + | + | + | − |
| Ets69 | P682S | Stable, soluble | + | + | + | − |
| Dts6389 | S161F,A283T | Insoluble | + | + | + | − |
| Dts12 | M116I | Insoluble | + | + | + | − |
| Dts56 | V212A | Insoluble | + | + | + | − |
| Walker A | K509A | ND | Reduced | − | + | − |
| Walker B | E557Q | ND | + | − | +/− | − |
| SFIII motif C | N605D | ND | + | − | + | − |
| AAA+ motif | R619A,R620A | ND | + | − | +/− | − |
| 413-D5 | Δ1-412 | ND | + | − | − | − |
| 301-D5 | Δ1-300 | ND | + | + | + | − |
Whole-cell lysates or PNS were prepared from infected BSC40 cells at 8 hpi and analyzed using an anti-D5 antiserum. The D5 protein is referred to as “unstable” if less D5 was detected in whole-cell lysate prepared from nonpermissive infections than wt infection at permissive temperature; the “insoluble” moniker was given to those D5 proteins that were not detected in the PNS fraction from nonpermissive infections. D5 stability is host cell specific (Fig. 4), since all but one (Dts6389) of the tsD5 proteins are thermolabile during nonpermissive infections in L929 cells. ND, not determined.
+, ATP binding was observed at ≥65% of wt activity. The K509A mutation in the Walker A domain diminished ATP binding to ∼30% of wt activity.
+, ATPase activity was ≥65% of wt activity; −, ATPase activity was <10% of wt activity.
BSC40 cells were programmed to overexpress untagged full-length wtD5 along with 3XFLAG-tagged versions of the various D5 proteins. Affinity purification of the tagged protein and any associated untagged D5 was performed and examined by silver staining. +, the 3XFLAG protein associated with the untagged protein at near-stoichiometric levels; +/−, reduction in relative pull-down efficiency; −, complete inability of the tagged protein to retrieve untagged D5.
BSC40 or L929 cells infected with the ts viruses failed to accumulate viral DNA at high temperature as determined by Southern dot blot hybridization on cytoplasmic viral DNA harvested at various times postinfection. When introduced by transfection, plasmids encoding the site-directed or truncated D5 mutants were unable to support ongoing viral DNA synthesis (determined at 8 and 24 hpi) when permissive Cts24 infections were shifted to nonpermissive temperature in the midst of DNA replication (5 hpi).
Initially, we extended the analysis of the six known tsD5 viruses. All of these viruses show a defect in DNA synthesis at the nonpermissive temperature, and all but one carry lesions within the N-terminal region of the D5 protein, which lacks any defining motifs or homology to other cellular or nonpoxvirus proteins.
We attempted to identify the basis of the ts phenotype of these viruses. During nonpermissive infection of BSC40 cells, the Cts17 and Cts24 D5 proteins are thermolabile, and the Dts6389, Dts12, and Dts56 D5 proteins are insoluble. These altered physical characteristics are probably sufficient to explain the defect in DNA synthesis. The Ets69 D5 protein, however, is both stable and soluble at the nonpermissive temperature. Moreover, the Ets69 D5 protein synthesized at high temperature retains oligomerization capability and NTPase activity (not shown). Cumulatively, these data suggest that the Ets69 lesion affects the D5 protein in some other manner, perhaps by diminishing its competence in establishing protein-protein or protein-DNA interactions. It is also worth noting that the phenotype of the tsD5 viruses varies according to the host cell. The ts phenotype of the viruses is stricter in L929 than in BSC40 cells, with 10- to 50-fold less virus being produced at the nonpermissive temperature. None of the tsD5 viruses synthesized appreciable levels of viral DNA at high temperature in L929 cells. Furthermore, all but one of the tsD5 proteins were thermolabile in L929 cells. Lastly, even during permissive infection of L929 cells, the Ets69 D5 protein was unstable, resulting in only 8% of wt levels of DNA synthesized by 24 hpi. The observation that the tsD5 mutants behave somewhat differently in different cell types is in keeping with what has been observed for other complementation groups of vaccinia virus mutants (34).
Functional consequence of mutating the ATP binding and hydrolysis domains, the SFIII motif, and the AAA+ region.
As described above, bioinformatic analysis of the D5 protein sequence has identified several functional motifs within the C-terminal portion of the protein (17). D5 contains both a Walker A and a Walker B box, which are functionally required to bind and hydrolyze ATP, respectively (48), consistent with the ATP binding and hydrolysis activity previous reported for D5 (9). The Walker B domain of D5 is classified as “nontraditional” (17) because one, rather than two, glutamic acid residues are found; however, other SFIII helicases are known to display considerable variability in this motif (20). D5 contains two additional family defining motifs: the signature SFIII motif C as well as the AAA+ motif/arginine finger (17). To probe whether the conserved motifs were required for the biochemical and biological functions of D5, we generated a family of D5 alleles encoding proteins in which the invariant residues within these motifs were altered: K509A, E557Q, N605D, and R619A,R620A. In other catalytically active members of the AAA+ class of proteins, these residues have been shown to interact with the ATP molecule (14). While the UV cross-linking of purified D5 to [α-32P]ATP was not affected by mutations within the Walker B or SFIII motif C (E557Q D5 and N605D D5, respectively) (Fig. 7), alterations of the Walker A box or the AAA+ motif (K509A and R619A,R620A, respectively) (Fig. 7) reduced the ATP binding activity of D5. Additionally, each of the mutations rendered the D5 proteins essentially unable to hydrolyze ATP (Fig. 7). These data are consistent with the results obtained from studies of other SFIII/AAA+ proteins (14, 19, 49). Specifically, the adeno-associated virus (AAV) type 2 Rep68 protein (49), the minute virus of mice major nonstructural protein (NS-1) (19), and the HPV E1 protein (36, 50) have each been subjected to detailed analyses. In general, a K→A alteration in the Walker A domain inhibits ATP binding and hydrolysis and, consequently, the associated biochemical activities. For each of these proteins, an E→Q substitution in the Walker B domain impaired ATP hydrolysis activity. Mutations within the SFIII motif C of either NS-1 (N486D or N486H) or E1 (N586A) resulted in a reduction in ATPase activity and, for the NS-1 protein, a concomitant loss of helicase activity. Lastly, an R→A substitution in the arginine finger impaired ATP hydrolysis in most AAA+ proteins (reviewed in reference 14).
Establishment of a transient complementation assay for an early viral protein.
The data described above validate the prediction that the conserved domains within the C terminus of D5 play important and predictable roles in the ATP binding and ATP hydrolysis activities of the protein. To assess the biological relevance of these activities, we established a transient complementation assay to monitor the ability of D5 protein variants to restore DNA replication during nonpermissive tsD5 infections. Transfection of a wt copy of D5 under the transcriptional control of a viral intermediate promoter enabled DNA synthesis to continue when Cts24 infections were shifted to nonpermissive temperature in the midst of DNA replication. This effect was not seen when empty vector or a plasmid encoding the Cts24 D5 protein was introduced, indicating that we were observing specific complementation. Most importantly, none of the enzymatically deficient mutants of D5, despite being stably expressed, retained complementation activity. These data strongly support the conclusion that the ATPase activity of D5 is essential for its biological competency.
Relationship between oligomerization and catalytic activity for AAA+ proteins.
Biochemical and structural studies of numerous AAA+ proteins have indicated that oligomerization is a prerequisite for enzymatic function and biological activity (14). Structural analysis of two SFIII helicases (the SV40 T antigen and a variant of the AAV2 Rep68/78 protein) has confirmed that they form homo-hexameric ring structures (18, 23). The domains required for oligomerization of the SV40 T antigen (12, 23) and HPV11 E1 (1, 42) have been mapped. For the SV40 T antigen, a helical zinc finger domain is absolutely required for T antigen oligomerization (23); although monomeric T antigen possesses an intact ATP binding and hydrolysis domain, it completely lacks ATPase and helicase activities (12). For HPV11 E1, a helical domain positioned just upstream of the ATP binding module mediates E1-E1 homotypic interactions (1, 42), and ATPase and helicase activities are dependent upon E1 oligomerization into hexameric structures (37).
For the D5 protein, we have shown that the C-terminal region of D5 (amino acids 413 to 785), which contains all of the catalytic motifs (Walker A and B, SFIII motif C, and AAA+) and is both stable and soluble, cannot oligomerize and lacks ATPase activity (Fig. 9). These data suggest that D5 possesses an upstream oligomerization domain. Indeed, a variant of D5 containing amino acids 301 to 785 regained the ability to oligomerize and concomitantly regained ATPase activity. These data imply that the NTPase activity of D5, like other SFIII/AAA+ proteins, requires oligomer formation. Determination of whether D5 assembles into a hexameric ring is certainly worth pursuing.
While oligomerization of AAA+ proteins is required for catalytic activity, the interplay between protomer subunits has also been shown to be crucial for the propagation of hydrolysis-dependent conformational changes. In this respect, it is the “arginine finger,” an invariant arginine residue strategically positioned at the end of the helical bundle 4 (α4), that extends into the ATP binding pocket of the adjacent protomer and interacts with the γ-phosphate of bound ATP (14, 31). Mutation of the arginine finger in a variety of AAA+ proteins (reviewed in references 14 and 31) often impairs ATP hydrolysis without eliminating oligomerization. That is what we observed for D5. While the hydrolytic activity of D5 is dependent upon the presence of an intact putative arginine finger (Fig. 7), we observed that formation of D5 oligomers is only mildly diminished by its absence (Fig. 8).
Although the D5 variant containing amino acid residues 301 to 785 could oligomerize and possessed wild-type levels of ATPase activity, it was not competent to sustain ongoing DNA replication during a nonpermissive Cts24 infection. Thus, the N terminus of D5 must confer additional properties and/or activities on D5, such as the ability to interact with DNA or other components of the replication machinery. Understanding how the N terminus of D5 contributes to its activity will be a focus of future study. Given the significant homology of D5 to other replicative helicases, our demonstration that ATPase activity is required for D5's biological function, and the known interaction of D5 with a component of the viral polymerase processivity factor, it is enticing to speculate that D5 acts as the replicative helicase in the context of the assembled replicative machinery. It is also provocative that in all three kingdoms (prokaryote, eukaryote, and archaea) the replication “initiator” or origin binding proteins are AAA+ proteins whose nucleotide hydrolysis activity is essential for origin remodeling. Moreover, three distinct viral origin binding proteins, SV40 Tag, HPV E1, and AAV Rep, are SFIII helicases/AAA+ proteins. Although vaccinia virus replication has been predicted to initiate within the telomeric regions (6), no origin or origin binding protein has been defined. The studies described herein lay the groundwork for future analysis of how the essential D5 protein contributes to the complex process of vaccinia virus DNA replication.
Acknowledgments
We thank the members of the Traktman lab for their insightful discussions regarding this project as well as critical review of the manuscript. We also thank R. Wedge for excellent technical assistance.
This work was supported by a grant to P.T. from the National Institute of Allergy and Infectious Diseases (NIH 2 R01 AI21758).
Footnotes
Published ahead of print on 8 November 2006.
REFERENCES
- 1.Abbate, E. A., J. M. Berger, and M. R. Botchan. 2004. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 18:1981-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Challberg, M. D., and P. T. Englund. 1979. Purification and properties of the deoxyribonucleic acid polymerase induced by vaccinia virus. J. Biol. Chem. 254:7812-7819. [PubMed] [Google Scholar]
- 3.Challberg, M. D., and P. T. Englund. 1979. The effect of template secondary structure on vaccinia DNA polymerase. J. Biol. Chem. 254:7820-7826. [PubMed] [Google Scholar]
- 4.Condit, R. C., A. Motyczka, and G. Spizz. 1983. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology 128:429-443. [DOI] [PubMed] [Google Scholar]
- 5.Dales, S., V. Milovanovitch, B. G. Pogo, S. B. Weintraub, T. Huima, S. Wilton, and G. McFadden. 1978. Biogenesis of vaccinia: isolation of conditional lethal mutants and electron microscopic characterization of their phenotypically expressed defects. Virology 84:403-428. [DOI] [PubMed] [Google Scholar]
- 6.Du, S., and P. Traktman. 1996. Vaccinia virus DNA replication: two hundred base pairs of telomeric sequence confer optimal replication efficiency on minichromosome templates. Proc. Natl. Acad. Sci. USA 93:9693-9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elroy-Stein, O., T. R. Fuerst, and B. Moss. 1989. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc. Natl. Acad. Sci. USA 86:6126-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ensinger, M. J. 1982. Isolation and genetic characterization of temperature-sensitive mutants of vaccinia virus WR. J. Virol. 43:778-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, E., N. Klemperer, R. Ghosh, and P. Traktman. 1995. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 69:5353-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, E., and P. Traktman. 1987. Molecular genetic analysis of a vaccinia virus gene with an essential role in DNA replication. J. Virol. 61:3152-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans, E., and P. Traktman. 1992. Characterization of vaccinia virus DNA replication mutants with lesions in the D5 gene. Chromosoma 102:S72-S82. [DOI] [PubMed] [Google Scholar]
- 12.Gai, D., D. Li, C. V. Finkielstein, R. D. Ott, P. Taneja, E. Fanning, and X. S. Chen. 2004. Insights into the oligomeric states, conformational changes, and helicase activities of SV40 large tumor antigen. J. Biol. Chem. 279:38952-38959. [DOI] [PubMed] [Google Scholar]
- 13.Gorbalenya, A. E., E. V. Koonin, and Y. I. Wolf. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262:145-148. [DOI] [PubMed] [Google Scholar]
- 14.Hanson, P. I., and S. W. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519-529. [DOI] [PubMed] [Google Scholar]
- 15.Ish-Horowicz, D., and J. F. Burke. 1981. Rapid and efficient cosmid cloning. Nucleic Acids Res. 9:2989-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishii, K., and B. Moss. 2001. Role of vaccinia virus A20R protein in DNA replication: construction and characterization of temperature-sensitive mutants. J. Virol. 75:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer, L. M., L. Aravind, and E. V. Koonin. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720-11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James, J. A., C. R. Escalante, M. Yoon-Robarts, T. A. Edwards, R. M. Linden, and A. K. Aggarwal. 2003. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure 11:1025-1035. [DOI] [PubMed] [Google Scholar]
- 19.Jindal, H. K., C. B. Yong, G. M. Wilson, P. Tam, and C. R. Astell. 1994. Mutations in the NTP-binding motif of minute virus of mice (MVM) NS-1 protein uncouple ATPase and DNA helicase functions. J. Biol. Chem. 269:3283-3289. [PubMed] [Google Scholar]
- 20.Koonin, E. V. 1993. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 21:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin, E. V. 1993. A highly conserved sequence motif defining the family of MutT-related proteins from eubacteria, eukaryotes and viruses. Nucleic Acids Res. 21:4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lackner, C. A., S. M. D'Costa, C. Buck, and R. C. Condit. 2003. Complementation analysis of the Dales collection of vaccinia virus temperature-sensitive mutants. Virology 305:240-259. [DOI] [PubMed] [Google Scholar]
- 23.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512-518. [DOI] [PubMed] [Google Scholar]
- 24.McCraith, S., T. Holtzman, B. Moss, and S. Fields. 2000. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. USA 97:4879-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald, W. F., N. Klemperer, and P. Traktman. 1997. Characterization of a processive form of the vaccinia virus DNA polymerase. Virology 234:168-175. [DOI] [PubMed] [Google Scholar]
- 26.McDonald, W. F., and P. Traktman. 1994. Overexpression and purification of the vaccinia virus DNA polymerase. Protein Expr. Purif. 5:409-421. [DOI] [PubMed] [Google Scholar]
- 27.McDonald, W. F., and P. Traktman. 1994. Vaccinia virus DNA polymerase. In vitro analysis of parameters affecting processivity. J. Biol. Chem. 269:31190-31197. [PubMed] [Google Scholar]
- 28.McFadden, G., and S. Dales. 1980. Biogenesis of poxviruses: preliminary characterization of conditional lethal mutants of vaccinia virus defective in DNA synthesis. Virology 103:68-79. [DOI] [PubMed] [Google Scholar]
- 29.Mercer, J., and P. Traktman. 2003. Investigation of structural and functional motifs within the vaccinia virus A14 phosphoprotein, an essential component of the virion membrane. J. Virol. 77:8857-8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neuwald, A. F., L. Aravind, J. L. Spouge, and E. V. Koonin. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27-43. [PubMed] [Google Scholar]
- 31.Ogura, T., S. W. Whiteheart, and A. J. Wilkinson. 2004. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 146:106-112. [DOI] [PubMed] [Google Scholar]
- 32.Punjabi, A., K. Boyle, J. DeMasi, O. Grubisha, B. Unger, M. Khanna, and P. Traktman. 2001. Clustered charge-to-alanine mutagenesis of the vaccinia virus A20 gene: temperature-sensitive mutants have a DNA-minus phenotype and are defective in the production of processive DNA polymerase activity. J. Virol. 75:12308-12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Punjabi, A., and P. Traktman. 2005. Cell biological and functional characterization of the vaccinia virus F10 kinase: implications for the mechanism of virion morphogenesis. J. Virol. 79:2171-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rempel, R. E., M. K. Anderson, E. Evans, and P. Traktman. 1990. Temperature-sensitive vaccinia virus mutants identify a gene with an essential role in viral replication. J. Virol. 64:574-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rempel, R. E., and P. Traktman. 1992. Vaccinia virus B1 kinase: phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J. Virol. 66:4413-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocque, W. J., D. J. Porter, J. A. Barnes, E. P. Dixon, D. C. Lobe, J. L. Su, D. H. Willard, R. Gaillard, J. P. Condreay, W. C. Clay, C. R. Hoffman, L. K. Overton, G. Pahel, T. A. Kost, and W. C. Phelps. 2000. Replication-associated activities of purified human papillomavirus type 11 E1 helicase. Protein Expr. Purif. 18:148-159. [DOI] [PubMed] [Google Scholar]
- 37.Sedman, J., and A. Stenlund. 1998. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 72:6893-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sridhar, P., and R. C. Condit. 1983. Selection for temperature-sensitive mutations in specific vaccinia virus genes: isolation and characterization of a virus mutant which encodes a phosphonoacetic acid-resistant, temperature-sensitive DNA polymerase. Virology 128:444-457. [DOI] [PubMed] [Google Scholar]
- 39.Stanitsa, E. S., L. Arps, and P. Traktman. 2006. Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J. Biol. Chem. 281:3439-3451. [DOI] [PubMed] [Google Scholar]
- 40.Taddie, J. A., and P. Traktman. 1991. Genetic characterization of the vaccinia virus DNA polymerase: identification of point mutations conferring altered drug sensitivities and reduced fidelity. J. Virol. 65:869-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taddie, J. A., and P. Traktman. 1993. Genetic characterization of the vaccinia virus DNA polymerase: cytosine arabinoside resistance requires a variable lesion conferring phosphonoacetate resistance in conjunction with an invariant mutation localized to the 3′-5′ exonuclease domain. J. Virol. 67:4323-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Titolo, S., A. Pelletier, A. M. Pulichino, K. Brault, E. Wardrop, P. W. White, M. G. Cordingley, and J. Archambault. 2000. Identification of domains of the human papillomavirus type 11 E1 helicase involved in oligomerization and binding to the viral origin. J. Virol. 74:7349-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traktman, P., M. K. Anderson, and R. E. Rempel. 1989. Vaccinia virus encodes an essential gene with strong homology to protein kinases. J. Biol. Chem. 264:21458-21461. [PubMed] [Google Scholar]
- 44.Traktman, P., and K. Boyle. 2004. Methods for analysis of poxvirus DNA replication. Methods Mol. Biol. 269:169-186. [DOI] [PubMed] [Google Scholar]
- 45.Traktman, P., M. Kelvin, and S. Pacheco. 1989. Molecular genetic analysis of vaccinia virus DNA polymerase mutants. J. Virol. 63:841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Traktman, P., K. Liu, J. DeMasi, R. Rollins, S. Jesty, and B. Unger. 2000. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J. Virol. 74:3682-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unger, B., and P. Traktman. 2004. Vaccinia virus morphogenesis: A13 phosphoprotein is required for assembly of mature virions. J. Virol. 78:8885-8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker, S. L., R. S. Wonderling, and R. A. Owens. 1997. Mutational analysis of the adeno-associated virus type 2 Rep68 protein helicase motifs. J. Virol. 71:6996-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White, P. W., A. Pelletier, K. Brault, S. Titolo, E. Welchner, L. Thauvette, M. Fazekas, M. G. Cordingley, and J. Archambault. 2001. Characterization of recombinant HPV6 and 11 E1 helicases: effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J. Biol. Chem. 276:22426-22438. [DOI] [PubMed] [Google Scholar]