Abstract
To model human immunodeficiency virus (HIV) perinatal transmission, we studied infection of simian-human immunodeficiency virus (SHIV) SF162P3 in 10 pregnant Macaca nemestrina females and their offspring. Four of nine infants born to and suckled by these dams had evidence of infection, a transmission rate of 44.4% (95% confidence interval, 13.7% to 78.8%). We quantified transplacentally acquired and de novo Env-specific immunoglobulin G (IgG), IgM, and neutralizing antibodies in newborns. Transmission of escape variants was confirmed. In utero infection (n = 1) resulted in high viremia, depletion of peripheral CD4+ T cells, and rapid evolution of env in blood and tissues. Peripartum or postpartum SHIV infection (n = 3) resulted in postacute viral control that was undetectable by very sensitive multiplex PCR, despite increasing antibodies. Seropositive infants with highly controlled viremia had homogeneous peripheral blood env sequences, and their tissues had <3 copies per million cells. A high incidence of seropositive virus-low or -negative SHIV infection in infant macaques has implications for HIV type 1 perinatal transmission and detection.
Human immunodeficiency virus type 1 (HIV-1) infection results in variable times to pathogenesis and disease. Typically, acute viremia is accompanied by the development of antiviral humoral and cell-mediated responses, followed by partial resolution to a “steady-state” virus load. AIDS results after long-term chronic HIV infection and accompanying immunologic failure, with loss of CD4+ T cells to below 200 cells/μl of plasma, making the patient susceptible to opportunistic infections. The majority of HIV-1-infected patients—typical progressors—follow the classic clinical pattern of 5 to 10 years to disease. Some progress to AIDS faster (rapid progressors, <5 years), while others progress to disease slowly (slow progressors, or long-term nonprogressors, >10 years). Exposed seronegatives (ES) are individuals who are continuously exposed yet remain negative for serum antibodies (Abs) and detectable virus. In many cases, ES have been shown to be positive for mucosal immunoglobulin A (IgA) (7, 26, 32, 33), proliferative responses, and cytotoxic-T-lymphocyte responses to HIV antigens (22, 41, 44). These individuals can also harbor HIV-1 DNA in resting CD4+ T cells at extremely low levels, suggesting a highly controlled infection (63). More controversial is the pattern of transiently viremic individuals who are intermittently positive for virus load or immune responses. Early evidence for transient viremia in pediatric cases was ruled out due to sample mislabeling (12), but there is additional evidence for low-level or transient viremia prior to acute HIV viremia (10) or in the context of antiretroviral treatment (18). Infection of macaques with many simian immunodeficiency virus (SIV) or simian-human immunodeficiency virus (SHIV) isolates also results in variable infection patterns, and transient or low-level infections have occurred following low-dose challenges (31, 35, 52, 55). These macaques mount detectable lymphoproliferative and cytotoxic-T-lymphocyte responses to SIV antigens and weak antibody responses (34). Similar to observations in HIV-1 ES, the presence of very low-level SIV infection in macaques has been established (64).
HIV-infected newborns and children can experience rapid disease progression, making antiviral-drug intervention critical. Perinatal transmission rates for HIV-1 are 35 to 44%, with approximately 16% greater risk of acquisition if the child is breastfed (36). Rates have plummeted with the utilization of antiretroviral drugs to limit viral replication at the time of birth. Macaque models hold potential for investigating aspects of mother-to-child transmission that are difficult to assess in human studies (19). We describe a maternal-transmission model in which pregnant macaques (dams) were infected intravenously with pathogenic SHIV in their second trimester. We sought to quantify transplacental transfer of maternal binding antibodies and neutralizing antibodies (NAbs) and to follow virological and immunological outcomes in the newborns, born vaginally in most cases and allowed to suckle (20). These pregnant dams progressed to term, delivered vaginally unless complications ensued, and suckled their infants postpartum. This model differs from other perinatal models in that (i) pregnancy was established prior to infection and (ii) the virus was SHIV-SF162P3, a mucosally transmitted, CCR5 coreceptor-utilizing isolate that does not cause drastic depletion in peripheral CD4+ T cells in juvenile macaques (16, 20, 27). We observed transmission of neutralization-resistant viruses and a high incidence of persistently seropositive low-level infection in infants in this model.
MATERIALS AND METHODS
Animals.
Macaques (Macaca nemestrina) were housed at the Washington National Primate Research Center (WaNPRC) under protocols approved by the University of Washington IACUC (20). Dams and infants were necropsied 6 months after birth; the organs and tissues were examined by histology.
Viral stock.
The stock of SHIV-SF162P3 (NIH), derived from SHIV-SF162P3 (16), was provided by Ranajit Pal. This stock was titered in vivo intravenously (103 50% macaque infectious doses [MID50]/ml) in juvenile macaques (20) and orally in newborns (0.5 to 1 MID50/ml).
Virus load and immunophenotyping.
Viral loads in plasma and peripheral blood mononuclear cells (PBMC) were determined by real-time PCR (20). The detection limits were 100 copies of RNA/ml of plasma and 3 copies/μg of DNA. Lymphocyte subsets were determined by flow cytometry.
Genomic-DNA and viral-RNA isolation.
PBMC and plasma at particular times postinfection in mothers and postbirth in babies were separated from blood by gradient centrifugation using lymphocyte separation medium (Fisher Scientific), followed by two washes with RPMI 1640 medium. PBMC suspensions were stored in liquid nitrogen in 10% dimethyl sulfoxide-90% fetal bovine serum until the genomic DNA was isolated. Plasma was stored at −20°C until the viral RNA was isolated. Genomic-DNA extraction from uncultured PBMC and viral-RNA extraction from plasma samples were performed for one sample per day in a separate DNA isolation containment hood using unidirectional work flow and other cleaning precautions to avoid sample cross-contamination. PBMC genomic DNA was extracted using the QIAGEN QiaAmp DNA blood minikit according to the manufacturer's instructions. Cell-free viral RNA from the SHIV-SF162P3 challenge stock was isolated using a QIAGEN viral-RNA minikit according to the manufacturer's instructions. Two-step reverse transcription-PCR of plasma viral RNA was performed using an Invitrogen two-step reverse transcription-PCR kit and Superscript III to generate cDNA, 2 μl of which was then processed similarly to the PCR of tissue genomic DNA, as mentioned below.
SHIV gp120 cloning and sequencing.
Since HIV-1 envelope is the only viral protein on the virion surface that is subject to immune pressure from neutralizing antibodies, we wanted to determine the evolution of the gene in mother-infant pairs. To this end, the C1-to-C5 region of gp120 was amplified from PBMC of mother-infant pairs by nested PCR using Roche Expand Hi-Fidelity Taq DNA polymerase (Roche Diagnostics, Indianapolis, IN) from 200 ng of genomic DNA. To avoid amplification bias, five replicates per sample were amplified and pooled prior to cloning. Specific primers matched to the database SHIV-SF162P3 sequence (accession number AF536757) were used. The first-round primers were EO (TAGAGCCCTGGAAGCATCCAGGAAGTCAGCCTA) and ED12 (AGTGCTTCCTGCTGCTCCCAAGAACCCAAG) (10). The second-round gp120 primers were 627L NheI, to insert an NheI site at the 5′ end of gp120 to fuse with the tissue-type plasminogen activator (t-PA) signal (GATGTTGATGATCTGTGCTAGCGTAGAAAAATTGTGGGTCA), and ED8 ClaI, to introduce a ClaI site in the C5 region of gp120 (ATATTTATATAAATCGATTCTCCAATTGTCCCTCATATCTCC). First-round and second-round PCRs were performed with 10 pmol of the sense and antisense primers, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate, and 3.5 U Expand High-Fidelity enzyme, and the cycling conditions were as follows: denaturation at 95°C for 5 min; 10 cycles of 94°C for 40 s and 50°C for 30 s; 68°C for 3 min 15 s; 10 cycles of 94°C for 40 s and 55°C for 30 s; 68°C for 3 min 15 s; 15 cycles of 94°C for 40 s and 60°C for 30 s; 68°C for 3 min 15 s; and a final elongation at 68°C for 10 min. Positive controls included genomic DNA containing 1, 10, and 100 copies of HIV-1 DNA. Negative controls included nuclease-free water and SHIV-negative DNA controls. PCR products from five replicate reactions were pooled and purified using a QIAGEN Qiaquick PCR purification kit according to the manufacturer's instructions. The purified PCR products were then ligated into TOPO-TA 2.1 vector (Invitrogen, Carlsbad, CA) using 4 μl of PCR product and 1 μl of vector for 30 min at room temperature. Two microliters of the ligation mixture was then transformed into Invitrogen's TOP10F′ chemically competent cells. The transformed cells were selected with ampicillin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. Ten to 12 colonies from each sample were picked, and plasmid DNA was isolated using a QIAGEN miniprep kit according to the manufacturer's instructions. Inserts (1.35-kb gp120) were confirmed by double digestion with NheI (New England Biolabs) and ClaI (New England Biolabs). Gp120 clones were sequenced in both directions with primers ED5P3mod (ATGGGATCAAAGTCTAGAGCCATGTG), KK1 (GCACAGTACAATGTACACATGGAA), 218 (ATCATTACACTTTAGAATCGC), and ED3 (TTACAGTAGAAAAATTCCCC) (10) using Prism Dye terminator kits (ABI, Foster City, CA) on an Applied Biosystems 3730XL Genetic Analyzer.
Autologous pseudovirus construction.
To determine the neutralization susceptibilities of viral variants from the infected babies B1 and B2 to their paired maternal and concurrent plasmas, single-cycle competent pseudoviruses were generated. First, full-length envelope gp160 was amplified from B1 and B2 PBMC very early postbirth by nested PCR using Roche Expand Hi-Fidelity Taq DNA polymerase (Roche Diagnostics, Indianapolis, IN) from 200 ng of genomic DNA. To avoid amplification bias, five replicates per sample were amplified and pooled prior to cloning. Specific primers matched to the database SHIV-SF162P3 sequence were used. The gp160 first-round primers were EO (TAGAGCCCTGGAAGCATCCAGGAAGTCAGCCTA) and EO1 (TCCAGTCCCCCCTTTTCTTTTAAAAA). The first-round cycling conditions for gp160 PCR were as follows: denaturation at 95°C for 5 min; 35 cycles of 94°C for 40 s and 61°C for 30 s; 68°C for 3 min 15 s; and elongation at 68°C for 10 min. The second-round primers for gp160 amplification were NheI gp160 P3 F, to insert an NheI site at the 5′ end of gp120 to fuse with the t-PA signal (GCGGCGGCGGCTAGCGTAGAAAAATTGTGGGTCAC), and P3 gp160 R ClaI, to insert a ClaI site at the 3′ end of gp41(GCCGCCGCCATCGATTTATAGCAAAGCCCTTTC). The second-round cycling conditions for gp160 were as follows: 95°C for 5 min; 10 cycles of 94°C for 40 s and 50°C for 30 s; 68°C for 3 min 15 s min; 25 cycles of 94°C for 40 s and 60°C for 30 s; 68°C for 3 min 15 s; and a final elongation at 68°C for 10 min. Positive controls included genomic DNA containing 1, 10, and 100 copies of HIV-1 DNA. Negative controls included nuclease-free water and SHIV-negative DNA controls. PCR products from five replicate reactions were pooled and cloned into TOPO-TA 2.1 vector (Invitrogen, Carlsbad, CA) as mentioned above. Inserts (2.5-kb gp160) were confirmed by double digestion with NheI and ClaI. gp160 clones were sequenced in both directions with the gp120 primers mentioned above and two additional primers for the gp41 region of gp160, env6R (CTTGCCCACTTATCCAATTC) and env8R (CACAATCCTCGCTGCAATCAAG), using Prism Dye terminator kits (ABI, Foster City, CA) on an Applied Biosystems 3730XL Genetic Analyzer. Unique gp160 clones with no missense mutations were subcloned into an expression vector, pEMC*, following restriction digestion with NheI and ClaI and ligation with a Roche Rapid DNA ligation kit (Roche Diagnostics, Indianapolis, IN). Five to 10 ng of the ligation reaction mixture was used to transform MAX Efficiency Stbl2 competent cells (Invitrogen, Carlsbad, CA) in accordance with the manufacturer's instructions, and the cells were grown at 30°C for 24 h. Five or six transformants were selected, plasmid purified with a QIAGEN miniprep kit, and sequenced again for verification of the insert. 293T cells were transfected with Fugene 6 (Roche, Indianapolis, IN) in accordance with the manufacturer's instructions. The cells were plated to 50% confluence in a T75 tissue culture flask; 2 μg total DNA with a 20:1 backbone/Env ratio was prepared in a 6-μl Fugene-94-μl Dulbecco's modified Eagle's medium, 10% fetal calf serum, 1% l-glutamine, 1% penicillin/streptomycin mixture. The virus was harvested 48 h later, spun at 2,000 rpm for 10 min, and stored at −70°C until it was used. The pseudovirus was titered on Tzm-bl cells, and 200 50% tissue culture infective doses (TCID50) were added to each neutralization assay as described below.
Nested multiplex PCR.
The first round was a multiplex PCR; the second round was a nested single-gene amplification of either nef or p15. The first-round primers for nef were F2 Nef (ATTGGAGTCAGGAACTAAAGAATAGT) and R1 Nef (CAGTGATATTTATACATCAAGAAA), and those for amplifying gag-p15 were F3 p15 (CCAGACGGCGTGAGGAGCGGGAGAGGAA) and R1 p17 (TACCCAGGCATTTAATGTTCTC). The second-round primers for nef were F3 Nef (ATGAGGCGGTCCAGGCCGTCTGGAG) and R3 Nef (TCAGCCATGTTAAGAAGGCCTCTTGCGGTTAG), and for p15 they were F2 p17 (ATGGGCGTGAGAAACTCCGT) and R2 p17 (GCTTAATGGCAGGTGGACATAGTTA). The cycling conditions (first round) were denaturation at 95°C for 5 min and 40 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 2 min, with elongation at 72°C for 10 min. The second-round cycling conditions differed only in having 45 cycles instead of 40. Positive controls included genomic DNA containing 1, 10, and 100 copies of HIV-1 DNA. Negative controls included nuclease-free water and SHIV-negative DNA controls. SHIV copy numbers were calculated based on multiple independent limiting-dilution PCRs on each sample (62-64) and using the QUALITY program (http://ubik.microbiol.washington.edu/computing/) (43).
Antibody assays.
IgM and IgG antibodies in plasma that were directed to recombinant envelope glycoproteins (HIVSF162 gp120 and HIV89.6 gp160) were measured by enzyme-linked immunosorbent assay and normalized to standard reference sera (15, 20). Briefly, Immunosorb plates (Nalge Nunc, Rochester, NY) were coated with 2 μg/ml of recombinant HIVSF162 gp120 (a gift from Chiron Corp.) or HIV89.6 gp160 (a gift of Shiu-Lok Hu at the WaNPRC) in carbonate/bicarbonate buffer (10 mM Na2CO3, 40 mM NaHCO3, pH 9.6) at 4°C overnight. The plates were blocked with 1× phosphate-buffered saline (PBS)-2% bovine serum albumin-5% fetal bovine serum for 1 h and washed with 0.2% Tween 20 in PBS. The plasma samples were serially diluted in disruption buffer (1× PBS-5% fetal bovine serum-2% bovine serum albumin-1% Triton X-100) and transferred to the coated plate. After a 1-h incubation at room temperature, the plates were washed five times, followed by a 1-h incubation with 1:200 goat anti-human IgM or 1:5,000 goat anti-monkey IgG conjugated to horseradish peroxidase (ICN-Cappel), and washed five times, followed by a 5-min final wash. The plates were developed with 100 μl TMB (Sigma) for 7 min and stopped with 100 μl 1 N H2SO4. Absorbance at 450 nm was read on a Vmax microplate reader (Molecular Devices Co., Sunnyvale, CA). The endpoint titer was defined as the reciprocal of the highest dilution that gave an absorbance value more than three times the deviation from the mean of preimmune serum at the corresponding dilution. NAb assays against HIVSF162 were performed using the cMAGI assay (20). Neutralization of the stock SHIV-SF162P3 and autologous viral variants was performed with the Tzm-bl assay (6, 58) using cells provided by N. Landau via the NIH AIDS Reagent Repository. Briefly, 200 50% tissue culture infective doses of virus were added to twofold serial dilutions of sera in the presence of 7.5 μg/ml DEAE-dextran in a total volume of 150 μl Dulbecco's modified Eagle's medium, 10% fetal calf serum, 1% l-glutamine, 1% penicillin/streptomycin for 1 h at 37°C. Each well received 100 μl of Tzm-bl cells resuspended in medium at 1 × 105 cells/ml. Forty-eight hours later, the cells were lysed for 2 min directly in the neutralization plate, using 100 μl of Bright-Glo luciferase assay substrate (Promega, Madison, WI), and immediately analyzed for luciferase activity on a luminometer. The reciprocal dilution of serum necessary to achieve 50% neutralization was reported. Neutralization values greater than three times the prebleed background level were considered significant. The lowest dilution of prebleed sera tested (1:50) never reached 50% neutralization; therefore, values above 150 were considered significant. All values were calculated with respect to virus only: [(value for virus only − value for cells only) − (value for serum only − value for cells only)]/(value for virus only − value for cells only).
Percent divergence and diversity calculations.
Proviral nucleotide sequences were edited and contigs were assembled with Sequencher 4.5 (Gene Codes Corporation, Ann Arbor, MI) and aligned using CLUSTAL X (8). Sequences with missense mutations were excluded from the analyses. The full-length env gene (gp160) from the SHIV-SF162P3 challenge stock was amplified according to the methods described above. A consensus (P3 CON) was created from the five unique sequenced clones obtained using the Los Alamos National Laboratory database consensus maker (http://www.hiv.lanl.gov/content/hiv-db/CONSENSUS/CONSENSUS_TOOL/SimpCon.html). Percent diversity was defined as the sequence heterogeneity at a particular time point in a tissue sample from one animal. Percent diversity was calculated with MEGA3.0 (24), using the Kimura two-parameter model with pairwise deletions (transition-to-transversion ratio [κ] = 2). For analysis of divergence, each clone sequence from a tissue was compared to the inoculum consensus (P3 CON), and divergence was measured with MEGA3.0, using the Kimura two-parameter model with pairwise deletions (κ = 2). The average divergence at each time point for each animal was reported.
Maximum likelihood analyses.
Codon-aligned nucleotide sequences trimmed to the same starting and ending nucleotides were analyzed by maximum likelihood using PAUP*4.0 (D. L. Swofford, Sinauer Associates, Inc., Sunderland, MA) (6, 51). The trees were rooted on the SHIV-SF162P3 consensus sequence (P3 CON) created from five unique inoculum clones. Maximum likelihood trees (κ = 2) were created using the HKY85 model with a subtree-pruning-regrafting-branch-swapping algorithm. The starting tree was obtained by neighbor joining, and the starting branch lengths were obtained using the Rogers-Swofford approximation method.
Analysis of glycosylation.
AminoTrack 3TM was used to identify potential N-linked glycosylation (PNG) sites within each sequence. Protein sequences were deduced (Sequencher 4.5) and aligned (CLUSTAL X) using HIV-HXB2 (accession no. K03455) for reference. This freely available web-based software (http://apps.sbri.org/AminoTrack/; 28) provides outputs that denote the amino acid positions at which a PNG site is present. The presence of a PNG is indicated by “1”; “0” means it is absent. Sequence motifs of NXS/TX, where X is any amino acid except proline, are recognized by the program as a PNG. PNG site positions were noted, and the number of PNG sites per autologous virus variant were tallied.
Genotyping via microsatellite DNA analyses.
The identities of PBMC and tissue samples were verified by multiplex-PCR analysis using coded samples (University of California macaque microsatellite typing service [40]). Macaque microsatellite DNAs were scored for 100% genetic identity at 23 loci for samples from the same animal, a 50% match in the alleles for a paired dam-infant, and no match between unrelated animals.
Statistical analyses.
To compare percent divergence between concurrent time points in mother-infant pairs 1 and 2, two-tailed Mann-Whitney U tests were used (Graphpad Prism Software version 4.03). To assess the accuracy of reported proportions, exact binomial 95% confidence intervals (CI) were calculated.
Nucleotide sequence accession numbers.
The nucleotide sequences are available under GenBank accession numbers DQ862866 to DQ863102.
RESULTS
SHIV viral loads and disease outcomes in titration animals and in mother-infant pairs.
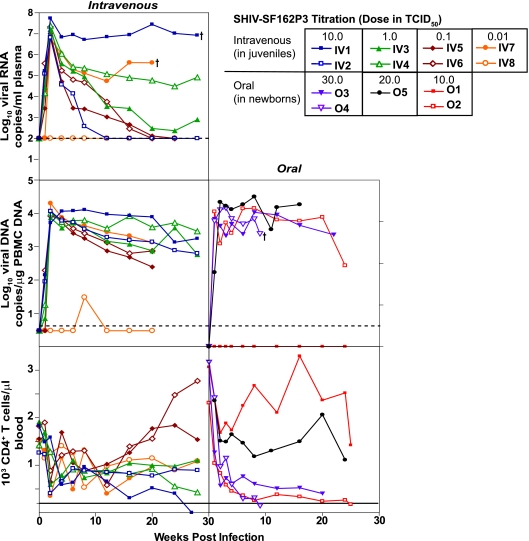
The overall goal of this study was to develop a macaque model of HIV-1 mother-to-child transmission and to evaluate virological and immunological parameters of transmission in the context of the same mucosally infecting strain, SHIV-SF162P3. This virus is a CCR5-utilizing strain that is transmitted mucosally by the vaginal route (16). The virus stock, SHIV-SF162P3 (NIH), was obtained during acute viremia after one additional in vivo passage in Macaca mulatta. To determine its infectiousness in Macaca nemestrina, it was titered intravenously in eight juveniles and orally in five newborns to obtain the MID50 (Fig. 1). We measured the viral loads in the plasmas and PBMC of juvenile titration animals and pregnant dams (Fig. 2A) and only in PBMC in the babies to conserve the plasma for serology. Seven of the eight juveniles had high acute plasma viremia (107 to108 RNA copies/ml plasma), varying levels of postacute RNA virus, and loss of CD4+ T cells in the acute phase that recovered. IV1 in the highest-inoculation-dose group had the highest virus load and lost CD4 cells by week 25, while IV8 in the lowest-inoculation-dose group had delayed, controlled viremia (Fig. 1). Orally inoculated SHIV-SF162P3 established high, persistent viremia (104 copies/μg) in four of the five newborns and rapid CD4 depletion in three of the five; it failed to infect one of the five at the lowest inoculation dose (Fig. 1). The 50% MID50 titers were observed to be 0.01 TCID50 by the intravenous route and 10.0 TCID50 by the oral route, a difference of 1,000-fold by the mucosal route that has been previously documented with other SIV and SHIV isolates.
FIG. 1.
Longitudinal follow-up of virus loads in plasma and PBMC and CD4+ T-cell counts in SHIV-SF162P3 titration animals. (Left panels) Intravenous titration of SHIV-SF162P3 in juvenile M. nemestrina macaques. Groups of two juvenile macaques were exposed to 10-fold-decreasing concentrations of challenge stock, with the highest dose of 10.0 TCID50 being the undiluted challenge stock. (Right panels) Oral titration of SHIV-SF162P3 in newborn M. nemestrina macaques. Groups of two newborn macaques were exposed to increasing concentrations of challenge stock, with the lowest dose of 10.0 TCID50 being the undiluted challenge stock. Newborns exposed to 20.0 and 30.0 TCID50 were exposed twice and three times, respectively, at 1-hour intervals to 1 ml of undiluted virus stock. Plasma virus loads in juveniles were monitored by real-time quantitative RNA PCR. PBMC DNA virus loads in juveniles and newborns were estimated by real-time DNA PCR. The limits of detection of real-time RNA and DNA PCRs were 100 copies/ml plasma and 3 copies/μg DNA, respectively. The solid lines in the CD4+ T-cell graphs indicate a CD4+ T-cell count of 250, the level below which AIDS is reported to occur. A dagger indicates death of the macaque due to AIDS.
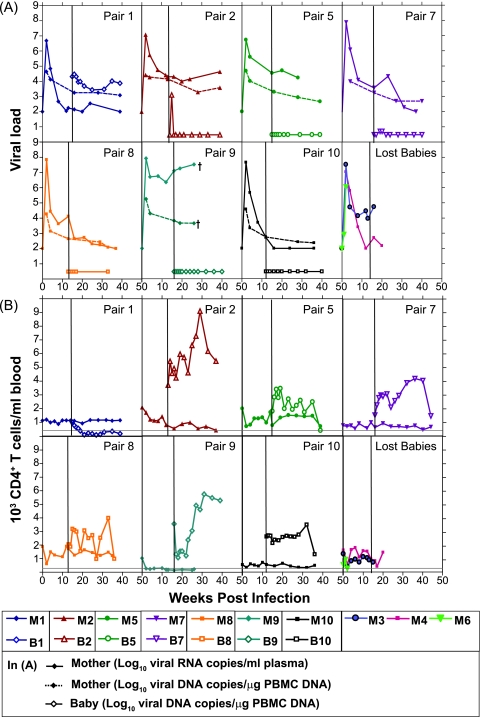
FIG. 2.
Paired viral loads (A) and CD4+ T-cell counts (B) in mother-infant pairs. Viral loads and CD4+ T-cell counts for the babies (B1 to B10; open symbols) are plotted with respect to their paired mothers (M1 to M10; solid symbols) during the course of SHIV infection. The solid vertical line in each panel indicates the birth of the infant. The maternal viral loads were determined by real-time RNA PCR in plasma (solid lines) and real-time DNA PCR in PBMC (dotted lines). Real-time DNA PCR in PBMC determined the virus levels in the babies. The bottom-right graphs in panels A and B show the viral loads and CD4+ T-cell counts in the mothers who lost their babies. The solid gray horizontal line in each graph in panel B indicates a CD4+ T-cell count of 250, the level below which AIDS is reported to occur. A dagger indicates the death of the macaque due to AIDS. The limits of detection of real-time RNA and DNA PCRs are 100 copies/ml plasma and 3 copies/μg DNA.
Ten adult M. nemestrina females (M1 to M10) were inoculated intravenously 10 to 12 weeks into pregnancy with 100 MID50, and term infants (B1 to B10) were born vaginally unless complications arose (20). Pregnant dams were allowed to suckle their infants for 6 months postbirth. SHIV-SF162P3 replicated to 107 to108 RNA copies/ml plasma during acute viremia in pregnant dams (Fig. 2A) and then resolved to varying levels, as was seen in juveniles in the virus titration. The levels of provirus in infected cells in PBMC in the dams resolved by week 4 to between 103 and 104 copies/μg and were more tightly clustered than plasma RNA copies. Postacute maternal viral-DNA and -RNA set points varied and were stable by parturition. M9 had the highest viral load at delivery and was euthanized at week 26 with AIDS-like symptoms. Pregnancy in M6 resulted in abortion, with peak plasma viremia not different from that of other dams. Eight of the 10 infected dams showed evidence of stable CD4+ T cells in the periphery in 9 months of observation (Fig. 2B), consistent with infection with the virus in M. mulatta (16, 17). Two mothers (M6 and M9) showed drastic peripheral depletion of CD4+ T cells to below 250, a definition for AIDS in most macaque lentivirus models (15). M9 was a classic rapid progressor, with increasing viral load (Fig. 2A) and decreasing CD4+ T cells (Fig. 2B).
Babies B3 and B4, born to M3 and M4, respectively, died on the day of birth of causes unrelated to SHIV infection. Only one infant, B1, was positive at birth (by C section), indicating in utero transmission (11%; 95% CI, 0.3% to 48.2%) (20). The viral load in B1 was consistently high, with corresponding rapid and permanent depletion of peripheral CD4+ T cells (Fig. 2). B2 was positive only at week 2 postbirth (1,277 copies/μg PBMC DNA), indicating an intrapartum or early postpartum mode of transmission. B5, B8, B9, and B10 were delivered vaginally and were negative for viral DNA at all time points tested. B7, delivered by C section, was weakly positive by nested-set and quantitative PCRs at weeks 3 and 4 postbirth. In contrast to oral titration of SHIV-SF162P3 in newborns, levels of CD4+ T cells varied in the babies exposed to virus from their mothers. B1, with a high persistent virus load from birth, experienced a rapid decline in peripheral CD4+ T cells by week 8 (week 20 in M1) (Fig. 2B). At necropsy (week 24 postbirth), B1 had enlarged lymph nodes, severe dehydration, and diarrhea, symptoms of AIDS in macaque lentivirus models. B2 had high and increasing CD4+ T-cell counts. Peripheral CD4+ T cells also increased in B9 from week 8 postbirth, concomitant with an increase in CD8+ T cells (data not shown). CD4+ T-cell levels varied normally in all the remaining babies (B5, B7, B8, and B10). Necropsy and histology data revealed that mothers M2, M5, and M9 and the in utero-infected B1, with viral loads of greater than 104 copies, had moderate to severe follicular hyperplasia in the axillary and inguinal lymph nodes typical of experimental SIV/SHIV infection in macaques, while the remaining animals had mild follicular hyperplasia (data not shown).
SHIV envelope-specific IgM and IgG antibody responses.
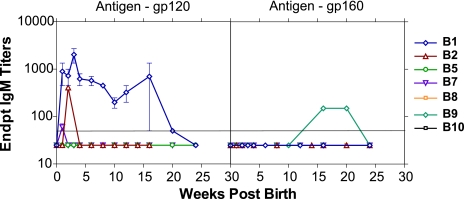
To differentiate between de novo antibody responses to SHIV infection and passively transferred maternal IgG, we monitored plasma for the development of viral Env-specific antibodies of the IgM isotype, comparing responses using recombinant envelope gp120 and gp160. None of the mothers had detectable Env-specific IgM during the last trimester, and none of the babies had IgM on the day of birth (data not shown). Plasma from babies positive for a DNA viral load for at least one of the time points tested (B1, B2, and B7) developed IgM against the homologous Env gp120SF162 (Fig. 3). B1 had the highest levels of IgM, followed by B2 and, finally, B7. Surprisingly, plasma from B9, which was negative for a DNA viral load at all time points tested and negative for IgM against gp120, had IgM against heterologous Env gp16089.6, denoting responses to gp41 (Fig. 3). Nine of 10 SHIV-infected dams seroconverted by 4 weeks postinfection, and IgG titers to Env gp120SF162 plateaued by the time of delivery (Fig. 4A). M9 failed to seroconvert prior to delivery, typical of a rapid progressor. Mothers with babies lost to follow-up (M3, M4, and M6) had Env-specific IgG titers that were not significantly different (P = 0.27; two-tailed Mann-Whitney test; 95% CI) from other mothers. Plasmas from six of the remaining seven infants had high levels of maternal IgG at birth, indicating passive transfer of maternal IgG. Infants negative for virus and Env-specific IgM (B5, B8, and B10) had decaying maternal IgG. In contrast, we measured de novo responses (increase after the decay of maternal IgG) in B1, B2, and B9. B1, infected in utero, had the highest de novo IgG response. B2, positive for IgM and virus load only at week 2, had a small increase in IgG, followed by a prolonged decay. B9, virus negative at all the time points tested, had Env-specific IgM and developed increasing de novo IgG at weeks 8 to 10 (Fig. 4A), consistent with viral replication in this infant and concomitant with increases in CD4+ (Fig. 2B) and CD8+ T cells. M9 was seronegative during the time points that IgG was first detected in B9, further substantiating the development of IgG in B9 as a de novo response. Maternal IgG decayed in B7, weakly IgM and virus positive.
FIG. 3.
Envelope-specific IgM antibody responses in infants born to SHIV-infected dams. IgM endpoint titers were determined against (A) rgp120 from HIVSF162 and (B) rgp160 from HIV89.6. The solid horizontal line indicates plasma dilutions of 1:50, the assay detection limit. Ab titers of <50 are graphed as 25. Endpt, endpoint. The error bars represent standard deviations of IgM titers from three independent experiments.
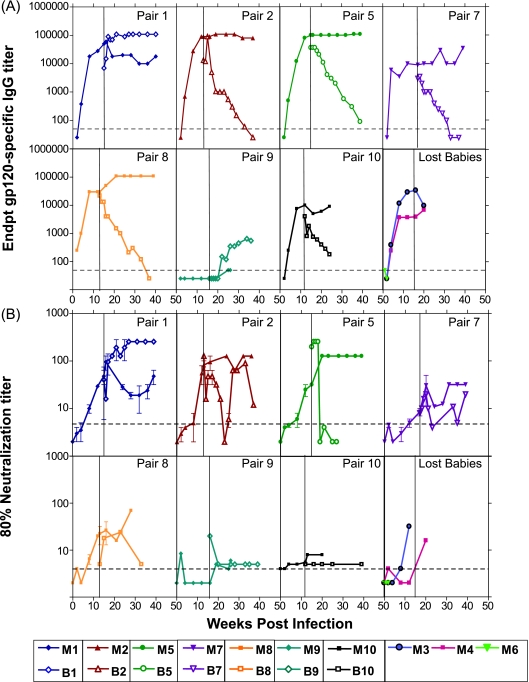
FIG. 4.
Humoral immune responses. (A) Envelope-specific binding Ab titers versus rgp120 from HIV-SF162. (B) Neutralizing-Ab titers versus HIV-SF162 in mother-infant pairs. Babies (open symbols) are plotted with respect to their paired mothers (solid symbols) during the course of SHIV infection. The solid vertical line in each graph indicates the birth of the infant. The dotted lines indicate plasma dilutions of 1:50 (A) and 1:4 (B), the assay detection limits. IgG titers of <50 are graphed as 25. NAb titers of <4 are graphed as 2. Median IgG and NAb titers from three independent experiments are shown. Endpt, endpoint.
Neutralizing-antibody responses in mother-infant pairs.
NAb titers were followed longitudinally in mother-infant pairs using HIVSF162 in order to determine the de novo neutralization profiles in these animals. The env gene in HIVSF162 is 99% identical to that in the infecting SHIV. Seven of 10 mothers had 80% NAb titers ranging between 10 and 100 against HIVSF162 at the time of delivery, and varying levels of NAb were detected in infant plasmas on the day of birth (Fig. 4B). The in utero-infected B1 showed rapid de novo NAb response to infection. In B2, maternal NAbs decayed to baseline and rapidly increased at week 16 (maternal week 25). Of all the infants followed to date, B5 had the highest maternal NAbs at birth, 10-fold higher than in M5, which decayed rapidly in the absence of detectable infection. Minimal NAb titers against HIVSF162 were observed in infants B7, B8, B9, and B10. Plasma from B1 and B2 at birth had high levels of maternal NAbs against the infecting strain, SHIV-SF162P3 (Table 1 and Table 2, respectively). These levels were higher than in M1 and M2 plasmas at delivery but within the variation of the assay.
TABLE 1.
Autologous neutralization of transmitted variants in B1
| Baby plasma | SHIV-SF162P3 inoculuma | B1w1 pseudovirusb
|
||||
|---|---|---|---|---|---|---|
| P3 | P6 | P7 | P12 | P15 | ||
| B1w2 | >675 | 162 | <50 | 270 | 58 | <50 |
| B1w6 | 1,048 | <50 | 780 | 215 | 162 | |
| B1w24 | >675 | 127 | <50 | 83 | 58 | 96 |
| M1 plasma at delivery | 480 | <50 | <50 | <50 | <50 | <50 |
| Total PNG no. in gp160c | 23-24 | 22 | 24 | 22 | 24 | 24 |
50% neutralization titers (50% inhibitory concentrations).
Unique, missense mutation-free, full-length envelope sequences (gp160) from babies and paired mothers were cloned, sequenced, and used for pseudovirus production; 50% neutralization titers of these pseudoviruses from mother-infant pairs are reported. B1w1, B1 at 1 week postbirth.
The average number of PNG sites for full-length gp160 was computed using AminoTrack (Seattle Biomedical Research Institute).
TABLE 2.
Autologous neutralization of transmitted variants in B2
| Baby plasma | SHIV-SF162P3 inoculuma | B2w2 pseudovirusb
|
||
|---|---|---|---|---|
| P2 | P8 | P9 | ||
| B2w0 | <50 | 1600 | 158 | |
| B2w2 | >675 | <50 | 800 | <50 |
| B2w4 | >675 | <50 | 620 | <50 |
| B2w6 | <50 | 568 | <50 | |
| B2w24 | >675 | <50 | <50 | <50 |
| M2 plasma at delivery | 200 | <50 | 200 | <50 |
| Total PNG no. in gp160c | 23-24 | 24 | 23 | 24 |
50% neutralization titers (50% inhibitory concentrations).
Unique, missense mutation-free, full-length envelope sequences (gp160) from babies and paired mothers were cloned, sequenced, and used for pseudovirus production; 50% neutralization titers of these pseudoviruses from mother-infant pairs are reported. B2w2, B2 at 2 weeks postbirth.
The average number of PNG sites for full-length gp160 was computed using AminoTrack (Seattle Biomedical Research Institute).
Envelope evolution in mother-infant pairs.
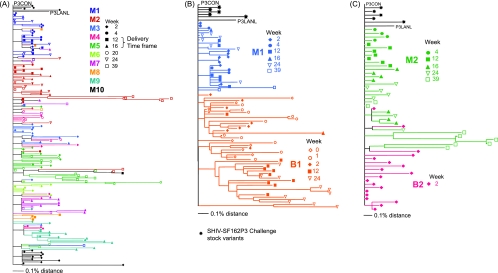
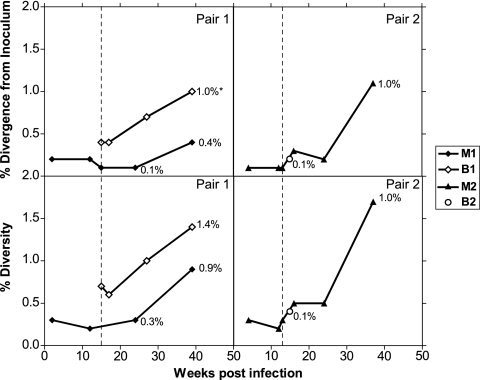
To study the transmission of specific viral variants and env evolution, we determined longitudinal env gp120 sequences in mother-infant pairs. Medians of 9 (range, 4 to 12) unique gp120 sequences free of missense mutations were obtained from the proviral DNA of each animal. Maximum likelihood phylogenetic trees were constructed from the gp120 clones obtained from all the mothers in the study (Fig. 5A), with pair 1 comprising M1 and B1 (Fig. 5B) and pair 2 comprising M2 and B2 (Fig. 5C). The trees were rooted on the consensus SHIV-SF162P3 sequence (P3 CON) from five unique clones from the challenge stock. The published SHIV-SF162P3 gp120 sequence was also included for comparison. Percent divergence, genetic distance from the ancestral sequence, P3 CON and percent diversity, and sequence heterogeneity at a given time point within an animal were quantified. Minimal heterogeneity was observed in the SHIV-SF162P3 inoculum sequences (0.4%). This was expected, as the challenge stock was obtained during acute infection of M. mulatta. env in the pregnant dams was minimally divergent (<0.5%) from the SHIV-SF162P3 inoculum at delivery, weeks 12 to 16 postinfection (Fig. 5A), suggesting that all the neonates in the study were exposed to nearly identical variants. env evolution was observed in some mothers at necropsy (week 39 postinfection), demonstrating that the virus continued to evolve in adult macaques. B1 sequences clustered separately from those of M1 on the maximum likelihood tree (Fig. 5B), suggesting distinct evolution in this infant. M1 sequences were 0.4% divergent from the inoculum and 0.9% diverse at the time of necropsy (Fig. 6). B1 env divergence at birth was greater than that in M1 for the concurrent time point, which supports transmission in this infant prior to birth. Evolution of B1 sequences in utero could not be examined. B1 rapidly accumulated changes in gp120, so that by week 24, B1 env was significantly more divergent than that of M1 for the same time frame (P = 0.005; two-tailed Mann-Whitney test; 95% CI) and had greater sequence heterogeneity (Fig. 5B and 6). In contrast, B2, infected intrapartum and virus positive only at week 2 postbirth, had minimal diversity and divergence (P = 0.4; two-tailed Mann-Whitney test; 95% CI) from the inoculum envelope sequences, showing evidence of unresolved polytomy from M2 sequences on the phylogenetic tree (Fig. 5C and 6). B2 env sequences were more closely related to those of M2 than to those from any other mother (data not shown). We also assessed the samples for genetic identity through microsatellite analyses to rule out sample contamination or switching. Macaque microsatellite DNAs were scored for 100% genetic identity at 23 loci for samples from the same animal, 50% match in the alleles for a paired dam-infant, and no match between unrelated animals. All babies with evidence of infection (B1, B2, B7, and B9) showed 50% genetic compatibility with their mothers, 100% genetic compatibility between samples from the same baby, and 0% compatibility with other mothers and babies at all 23 of the microsatellite loci tested (data not shown).
FIG. 5.
Phylogenetic analyses of individual gp120 sequences, C1 to V5, in (A) SHIV-SF162P3-infected mothers in the study, (B) pair 1 (M1 and B1), and (C) pair 2 (M2 and B2). The maximum likelihood tree is rooted on the SHIV-SF162P3 consensus (P3 CON). P3 CON was created from individual variants cloned and sequenced from the challenge SHIV-SF162P3 stock. Also noted on the phylogenetic tree is the SHIV-SF162P3 sequence from the Los Alamos National Laboratory database (P3 LANL).
FIG. 6.
Envelope sequences from PBMC DNAs of babies (B1 and B2) and their paired mothers (M1 and M2) were obtained at predelivery time points for the mother and postdelivery for mother and baby. For each sample, 8 to 14 clones were sequenced, and missense sequences were excluded from analyses. Percent divergence from the inoculum consensus (P3 CON) and percent diversity within sequences at a time point/tissue were estimated using MEGA3.1 after a CLUSTAL alignment of the sequences. The dotted vertical line in each panel indicates the birth of the infant.
Maternal neutralization escape variants are transmitted to infants.
Due to extremely low levels of virus in infants B7 and B9, we were unable to amplify envelope (gp120 or gp160) to test for autologous NAbs. To analyze for selection of B1 and B2 variants by maternal NAbs, single-cycle competent pseudoviruses from concurrent and early time points were tested against paired longitudinal autologous maternal and baby plasmas. B1 week 1 variants had a range of neutralization susceptibilities by autologous plasma from B1 (Table 1). Of the five variants tested, P6 and P15 were resistant to neutralization by autologous B1 plasma and variant P12 was minimally neutralization sensitive, while variants P3 and P7 were more neutralization sensitive. All of the B1 variants were resistant to plasma from the mother at the time of delivery. Given that B1 at birth demonstrated greater env evolution than M1 at the concurrent time point (Fig. 6), it is highly likely that B1 variants were more evolved than the maternal plasma could recognize. We next tested whether B1 mounted de novo NAbs to transmitted early variants. Autologous NAbs increased against four of five variants by week 6 and waned somewhat by week 24, demonstrating the development of NAbs directed to variants in vivo and showing that these variants were not inherently neutralization sensitive.
B2 env sequences were tested at week 2 for neutralization against autologous plasma and maternal plasma at the time of delivery, the presumed time of transmission. The B2 week 2 variants P2 and P9 were resistant to autologous week 2 plasma from B2 and maternal plasma at delivery, indicating transmission of these neutralization-resistant maternal variants. This neutralization resistance of variants P2 and P9 was maintained longitudinally at weeks 4, 6, and 24 postbirth, while the P9 variant was weakly neutralization sensitive at week 0. In contrast, clone P8 was neutralization sensitive against the autologous B2 week 0, week 2, and maternal plasmas at delivery, with higher NAbs in B2 plasma than in M2 plasma (Table 2). High NAb titers against this variant were maintained at week 6 postbirth and waned by week 24.
We previously analyzed the number and positions of PNG sites in the predicted Env sequence from SHIV-89.6P-infected M. nemestrina macaques, and we have reported that their occurrence in specific locations is constrained, with changes in regions V1, C2, V4, and V5 (6). We therefore analyzed PNGs in the SHIV-SF162P3-infected babies to examine the question of neutralization sensitivities of B1 and B2 variants and glycosylation. The total number of PNG sites in the SHIV-SF162P3 inoculum ranges between 23 and 24, lower than the median number of 31 or 32 PNGs found in the HIV-1 clade B gp160 (61). Most maternal and baby variants, B1-P6, B1-P12, and B1-P15, and B2-P2 and B2-P9, that maintained PNGs were neutralization resistant, and some maintained their neutralization resistance longitudinally (B1-P6, B2-P2, and B2-P9). The two neutralization-sensitive B1 variants, B1-P3 and B1-P7, lacked PNG sites in V1 (positions 139 and 188) and V4 (position 406), while the neutralization-sensitive variant B2-P8 lacked a PNG site at position 339, pointing to potential contributions by the carbohydrates at these key sites to the neutralization resistance of SHIV-SF162P3.
Evidence for extremely low-level infection by highly sensitive multiplex PCR.
De novo antibody responses (IgG and NAb) in babies B2, B7, and B9 and transient viremia in two of these three infants suggested occult infection in the neonates. We tested cells and tissues from B2, B7, and B9 at necropsy and at key time points using standard nested-set PCR techniques but were unable to amplify env from these infants. We therefore employed a highly sensitive multiplex-PCR-based assay (63, 64), amplifying conserved regions from the SIV genome in this SHIV, including nef and gag-p15. The assay was developed and standardized with positive control samples from several dams with known copy numbers to a level that was sensitive to consistently detect two to five copies/μg genomic DNA (Table 3). We screened lymphoid tissues in B2 (tonsil, ileum, thymus, and spleen at week 24) for SHIV nef and p15 copies using multiple replicates to screen up to 20 μg DNA, and we failed to amplify either of these genes. Using this sensitive PCR assay, the number of infected PBMC in B2, B7, and B9 was below the limit of detection, less than three copies/million (less than two to five copies per μg of DNA) in PBMC and tissues, strongly indicative of highly controlled or occult infection at the time points tested (Table 4).
TABLE 3.
Standardization of limiting-dilution nested nef-p15 multiplex PCRa
| Baby | Tissue | Time point (wk) | DNA per reaction (ng)b | No. of replicates | No. positive | Total ng of DNA tested | Estimated copy no./μg DNAc | Estimated copy no./million cellsc,d | Correlation Coefficiente |
|---|---|---|---|---|---|---|---|---|---|
| B1 | Inguinal lymph node | 24 | 20.6000 | 6 | 6 | 123.60000 | 150,403 | 992,662 | 0.97 |
| 4.12000 | 6 | 6 | 24.72000 | ||||||
| 0.82400 | 6 | 6 | 4.90000 | ||||||
| 0.16400 | 6 | 6 | 0.99000 | ||||||
| 0.03300 | 6 | 6 | 0.20000 | ||||||
| 0.00700 | 6 | 6 | 0.04000 | ||||||
| 0.00130 | 6 | 3 | 0.00800 | ||||||
| 0.00030 | 6 | 2 | 0.00200 | ||||||
| 0.00005 | 6 | 0 | 0.00030 | ||||||
| 0.00001 | 6 | 0 | 0.00006 | ||||||
| B1 | Thymus | 24 | 500.000b | 6 | 6 | 3,000.000 | 3,119 | 20,586 | 0.75 |
| 100.000 | 6 | 6 | 600.000 | ||||||
| 20.000 | 6 | 6 | 120.000 | ||||||
| 4.000 | 6 | 6 | 24.000 | ||||||
| 0.800 | 6 | 5 | 4.800 | ||||||
| 0.160 | 6 | 4 | 0.960 | ||||||
| 0.032 | 6 | 0 | 0.192 |
Limit of detection of nested nef-p15 PCR assay: 2 to 5 copies/μg DNA. The limit of detection for the nested multiplex PCR was estimated by performing multiple independent limiting-dilution nested nef-p15 PCRs on a standard panel of templates that contained 1, 10, 100, and 1,000 copies/PCR.
The amount of DNA used in the PCR standardization assays was based on the virus load estimated using real-time DNA PCR. The starting amount was 2,000 copies/μg DNA, with fivefold dilutions to less than 1 theoretical copy/μg DNA.
The copy number per microgram of DNA was estimated using Quality Software (24).
The estimated copy number per microgram DNA was extrapolated to the copy number per million cells based on the amount of DNA per cell (6.6 pg DNA per cell).
Correlation coefficient between viral copies per microgram DNA estimated from Quality Software and the real-time DNA virus load.
TABLE 4.
Cell-associated viral copies in low-level infection subjects: multiple replicate nested nef-p15 multiplex PCRs
| Baby | Tissue | Time point (wk) | DNA per reaction (ng) | No. of replicates | No. positive | Total DNA tested (μg) | Estimated copy no./μg DNAb | Estimated copy no./million cellsb,c |
|---|---|---|---|---|---|---|---|---|
| B2 | PBMC | 4 | 200 | 5 | 0 | 1 | <2.0 | <13.2 |
| PBMC | 24 | 200 | 5 | 0 | 1 | <2.0 | <13.2 | |
| Tonsil | 24 | 200 | 20 | 0 | 4 | <0.5 | <3.0 | |
| Ileum | 24 | 200 | 20 | 0 | 4 | <0.5 | <3.0 | |
| Spleen | 24 | 200 | 20 | 0 | 4 | <0.5 | <3.0 | |
| Thymus | 24 | 200 | 20 | 0 | 4 | <0.5 | <3.0 | |
| B7 | PBMC | 4 | 200 | 10 | 0 | 2 | <1.0 | <6.0 |
| B9 | PBMC | 24 | 200 | 10 | 0 | 2 | <1.0 | <6.0 |
Limit of detection of nested nef-p15 PCR assay: 2 to 5 copies/μg DNA. The limit of detection for the nested multiplex PCR was estimated by performing multiple independent limiting-dilution nested nef-p15 PCRs on a standard panel of templates that contained 1, 10, 100, and 1,000 copies/PCR.
The copy number per microgram of DNA was estimated using Quality Software (24).
The estimated copy number per microgram DNA was extrapolated to the copy number per million cells based on the amount of DNA per cell (6.6 pg DNA per cell).
DISCUSSION
Determining the infection status of newborns exposed to HIV is critically important for the clinical management of both mother and infant, particularly in locales where breastfeeding is crucial to infant survival. Confirmatory tests for HIV infection in neonates are based on viral quantification, because serological tests are confounded by the presence of maternal antibodies.
In this report, we showed virological, serological, and phylogenetic evidence for highly controlled infection in three neonates born to pregnant dams infected with SHIV-SF162P3. SHIV-SF162P3 infection in juvenile and adult macaques results in high acute viremia, followed by resolution to postacute set points (16, 17, 27). In contrast to CXCR4-utilizing SHIVs, such as SHIV-89.6P and SHIV-SF33A, SHIV-SF162P3 infection does not result in drastic irreversible depletion of peripheral CD4+ T cells (17) but rather leads to depletion of gut-associated CD4+ T cells, similar to HIV-1 infection in humans. This aspect of SHIV-SF162P3 makes it more appealing, since the infection and disease profiles are similar to those of HIV-1-infected humans. While there are excellent models emulating HIV-1 breast milk transmission in acute infection of lactating macaques (1, 2, 47) and oral infection of newborn macaques (3, 45, 46, 54, 55, 57), there has been no macaque model that accurately reflects human in utero and intrapartum transmission. To address this, our major goal was to establish a natural transmission model wherein neonatal macaques born to SHIV-SF162P3-infected pregnant dams could be followed for correlates for transmission. Three of four infected infants in our macaque cohort showed de novo IgM, IgG, and NAb responses in the absence of detectable virus in the peripheral blood or tissues examined. The single in utero-infected infant had high viremia and CD4+ T-cell loss, comparable to rapid disease and mortality observed in HIV-1 in utero-infected infants (9), in newborn macaques experimentally infected with SIV (53, 54), and in three of five newborns orally exposed to high doses of this SHIV stock. Surprisingly, three infants controlled their viral loads to an extent seen in only 1/18 juvenile or adult macaques exposed intravenously.
Several lines of evidence support the presence of highly controlled infection in the neonates in this study. In the absence of PCR-detected virus, serology can indicate infection, and indeed, serology is the most widely used HIV test for patients. Detection of SHIV-specific IgM in infants in the perinatal-macaque cohort served as a tool to measure seroconversion in the presence of passively transferred maternal IgG. De novo IgG and NAb responses were observed only in infants positive for virus or IgM. Furthermore, in the low-level-infected animal from which we obtained viral sequences, the phylogenetic evidence supported transmission from its dam. The lack of viral evolution is consistent with replication occurring at extremely low levels. Failure to detect the virus in seropositive infants after the first few weeks using an ultrasensitive multiplex-PCR assay strongly suggests that the SHIV was replicating in sites that we did not assay. The serological evidence, in conjunction with failure to detect virus using the ultrasensitive PCR assay, strongly suggests that very highly controlled SHIV-1 infection can occur in macaque infants.
These observations are unusual but may be consistent with clinical data on “transient” seroconversion or low-level viremia in infants born to HIV-1-infected women and SIV-exposed neonatal macaques (56). Transient infection or occult systemic low-level infection has been found in macaques challenged with low doses of SIV (34, 35, 37, 52). The finding that AIDS can occur in SIV-infected macaques that have controlled viremia for years (5) underscores the importance of studying low-level infection and viral sequestration. The infants in this report were observed for 6 months, and therefore, we do not know when they might have progressed to disease.
Several hypotheses accounting for low-level SHIV infection in these neonates can be proposed, and each needs systematic evaluation in future studies. It is quite possible that occult infection in this macaque model might be strain specific and might be a function of this particular macaque model. Exposure to low viral inoculum in the maternal genital or breast tissues may have reached a minimum threshold to establish latency in lymphoid tissues, but at levels insufficient to result in an overt systemic infection. The infants may have been exposed to attenuated or replication-defective maternal variants, as seen in monkeys inoculated with attenuated SIVmac strains (30). Other host factors, such as genetic defects and distinct HLA haplotypes, may also have shaped this outcome. It has been suggested by a number of authors that the diversity of the quasispecies at initial infection may be a predictor of subsequent progression (14, 21, 23, 25, 38, 48-50). Mothers demonstrated minimal evolution in their envelope sequences up to the time of delivery. It is therefore possible that babies exposed to minimally diverse maternal viral variants at birth and postbirth may have had better prognoses. There is new evidence that maternal NAbs can limit mother-to-child transmission (4). We hypothesize that exposure to maternal SHIV variants with narrow diversity coupled with the presence of maternal NAbs may have contributed to virus control and rapid development of de novo responses in these infants. Escape from autologous NAbs and concurrent evolution of the virus was observed in B1, the infant infected in utero, consistent with recent data supporting the evolution of HIV envelope in response to NAb pressure (6, 42, 60). Neutralization-sensitive variants had PNGs missing at one or two key positions shown previously to affect neutralization of this SHIV (6, 29, 39, 59). In contrast, the apparent establishment of low-level infection in B2 with the sensitive variant P8 raises the possibility that the presence of NAbs effective against this variant prevented widespread and high-level infection. We have previously observed acceleration of de novo NAbs in the presence of passively infused antibodies in juvenile macaques (15). It is also possible that maternal Abs with antibody-dependent cellular cytotoxicity activity may have contributed to controlling the replication and dissemination of neutralization-resistant variants (11, 13). While it is possible that more than one mechanism contributed to virus control, it is more important to resolve whether infants with occult infection are protected from superinfection, subsequently show signs of disease, or transmit infection to others in their sexually active years. As experimental protocols are designed to examine the role of passive monoclonal Ab cocktail treatments for newborns, it will be important to sort out potentially confounding factors of maternal IgG in contributing selection of variants and/or control of viremia in their infants. Determining whether such low-level infections occur in HIV-1-infected infants has important implications for evaluating the true efficacies of therapeutic and vaccine strategies to prevent mother-to-child transmission of HIV-1.
Acknowledgments
We thank N. Doria-Rose and J. Overbaugh and for critical reviews of the manuscript and M. Mahalanabis, C. Ng, W. Sutton, T. Kasprzyk, C. Young, and R. Hotchkiss for technical assistance. We gratefully acknowledge the technical expertise of T. Andrus and H. Zhu for assistance with multiplex PCR; H. Mack and M. Mbowe for hematology and immunophenotyping; and P. Delio for animal care. We thank C. Cheng-Mayer at Aaron Diamond Research Center for the SHIV-SF162P3 virus for in vitro assays, J. Overbaugh at the Fred Hutchinson Cancer Research Center for the viral clone Q23ΔEnv, N. Miller at the Division of AIDS for the SHIV-SF162P3 (NIH) stock, M. Marthas and K. Van Rompay at the California National Primate Research Center for advice on oral virus inoculation, C. Penedo at the California National Primate Research Center for microsatellite analyses, and J. Learn and J. Mullins at the University of Washington for assistance and advice with phylogenetic analyses. Virus stocks and cell lines used in neutralization assays were obtained from the NIH AIDS Research and Reference Reagent Program.
This study was supported by PHS grants R01HD38653 (N.L.H.); R01AI55336, R01AI45402, and R01AI49109 (T.Z.); and RR00166 (D.A., H.B.-O., and S.-L.H.); a grant from the Murdock Charitable Trust (N.L.H.); a fellowship from the Elizabeth Glaser Pediatric AIDS Foundation (D.M.); and donations from the James B. Pendleton Charitable Trust.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Amedee, A. M., N. Lacour, and M. Ratterree. 2003. Mother-to-infant transmission of SIV via breast-feeding in rhesus macaques. J. Med. Primatol. 32:187-193. [DOI] [PubMed] [Google Scholar]
- 2.Amedee, A. M., J. Rychert, N. Lacour, L. Fresh, and M. Ratterree. 2004. Viral and immunological factors associated with breast milk transmission of SIV in rhesus macaques. Retrovirology 1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, T. W., J. Koch, E. S. Mittler, M. Greene, M. Wyand, D. Penninck, and R. M. Ruprecht. 1994. Mucosal infection of neonatal rhesus monkeys with cell-free SIV. AIDS Res. Hum. Retrovir. 10:351-357. [DOI] [PubMed] [Google Scholar]
- 4.Barin, F., G. Jourdain, S. Brunet, N. Ngo-Giang-Huong, S. Weerawatgoompa, W. Karnchanamayul, S. Ariyadej, R. Hansudewechakul, J. Achalapong, P. Yuthavisuthi, C. Ngampiyaskul, S. Bhakeecheep, C. Hemwutthiphan, and M. Lallemant. 2006. Revisiting the role of neutralizing antibodies in mother-to-child transmission of HIV-1. J. Infect. Dis. 193:1504-1511. [DOI] [PubMed] [Google Scholar]
- 5.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 6.Blay, W. M., S. Gnanakaran, B. Foley, N. A. Doria-Rose, B. T. Korber, and N. L. Haigwood. 2006. Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J. Virol. 80:999-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broliden, K., J. Hinkula, C. Devito, P. Kiama, J. Kimani, D. Trabbatoni, J. J. Bwayo, M. Clerici, F. Plummer, and R. Kaul. 2001. Functional HIV-1 specific IgA antibodies in HIV-1 exposed, persistently IgG seronegative female sex workers. Immunol. Lett. 79:29-36. [DOI] [PubMed] [Google Scholar]
- 8.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickover, R. E., M. Dillon, S. G. Gillette, A. Deveikis, M. Keller, S. Plaeger-Marshall, I. Chen, A. Diagne, E. R. Stiehm, and Y. Bryson. 1994. Rapid increases in load of human immunodeficiency virus correlate with early disease progression and loss of CD4 cells in vertically infected infants. J. Infect. Dis. 170:1279-1284. [DOI] [PubMed] [Google Scholar]
- 10.Fiebig, E. W., C. M. Heldebrant, R. I. Smith, A. J. Conrad, E. L. Delwart, and M. P. Busch. 2005. Intermittent low-level viremia in very early primary HIV-1 infection. J. Acquir. Immune Defic. Syndr. 39:133-137. [PubMed] [Google Scholar]
- 11.Forthal, D. N., G. Landucci, and E. S. Daar. 2001. Antibody from patients with acute human immunodeficiency virus (HIV) infection inhibits primary strains of HIV type 1 in the presence of natural-killer effector cells. J. Virol. 75:6953-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frenkel, L. M., J. I. Mullins, G. H. Learn, L. Manns-Arcuino, B. L. Herring, M. L. Kalish, R. W. Steketee, D. M. Thea, J. E. Nichols, S. L. Liu, A. Harmache, X. He, D. Muthui, A. Madan, L. Hood, A. T. Haase, M. Zupancic, K. Staskus, S. Wolinsky, P. Krogstad, J. Zhao, I. Chen, R. Koup, D. Ho, B. Korber, R. J. Apple, R. W. Coombs, S. Pahwa, and N. J. Roberts, Jr. 1998. Genetic evaluation of suspected cases of transient HIV-1 infection of infants. Science 280:1073-1077. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Roman, V. R., L. J. Patterson, D. Venzon, D. Liewehr, K. Aldrich, R. Florese, and M. Robert-Guroff. 2005. Vaccine-elicited antibodies mediate antibody-dependent cellular cytotoxicity correlated with significantly reduced acute viremia in rhesus macaques challenged with SIVmac251. J. Immunol. 174:2185-2189. [DOI] [PubMed] [Google Scholar]
- 14.Greenier, J. L., C. J. Miller, D. Lu, P. J. Dailey, F. X. Lu, K. J. Kunstman, S. M. Wolinsky, and M. L. Marthas. 2001. Route of simian immunodeficiency virus inoculation determines the complexity but not the identity of viral variant populations that infect rhesus macaques. J. Virol. 75:3753-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haigwood, N. L., D. C. Montefiori, W. F. Sutton, J. McClure, A. J. Watson, G. Voss, V. M. Hirsch, B. A. Richardson, N. L. Letvin, S. L. Hu, and P. R. Johnson. 2004. Passive immunotherapy in simian immunodeficiency virus-infected macaques accelerates the development of neutralizing antibodies. J. Virol. 78:5983-5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harouse, J. M., A. Gettie, T. Eshetu, R. C. Tan, R. Bohm, J. Blanchard, G. Baskin, and C. Cheng-Mayer. 2001. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIV(SF162P3). J. Virol. 75:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harouse, J. M., A. Gettie, R. C. Tan, J. Blanchard, and C. Cheng-Mayer. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284:816-819. [DOI] [PubMed] [Google Scholar]
- 18.Hermankova, M., S. C. Ray, C. Ruff, M. Powell-Davis, R. Ingersoll, R. T. D'Aquila, T. C. Quinn, J. D. Siliciano, R. F. Siliciano, and D. Persaud. 2001. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA 286:196-207. [DOI] [PubMed] [Google Scholar]
- 19.Jayaraman, P., and N. L. Haigwood. 2006. Animal models for perinatal transmission of HIV-1. Front. Biosci. 11:2828-2844. [DOI] [PubMed] [Google Scholar]
- 20.Jayaraman, P., D. Mohan, P. Polacino, L. Kuller, N. Sheikh, H. Bielefeldt-Ohmann, B. Richardson, D. Anderson, S. L. Hu, and N. L. Haigwood. 2004. Perinatal transmission of SHIV-SF162P3 in Macaca nemestrina. J. Med. Primatol. 33:243-250. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson, A. C., S. Lindback, H. Gaines, and A. Sonnerborg. 1998. Characterization of the viral population during primary HIV-1 infection. AIDS 12:839-847. [DOI] [PubMed] [Google Scholar]
- 22.Kaul, R., F. A. Plummer, J. Kimani, T. Dong, P. Kiama, T. Rostron, E. Njagi, K. S. MacDonald, J. J. Bwayo, A. J. McMichael, and S. L. Rowland-Jones. 2000. HIV-1-specific mucosal CD8+ lymphocyte responses in the cervix of HIV-1-resistant prostitutes in Nairobi. J. Immunol. 164:1602-1611. [DOI] [PubMed] [Google Scholar]
- 23.Kimata, J. T., L. Kuller, D. B. Anderson, P. Dailey, and J. Overbaugh. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5:535-541. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, J., P. Balfe, C. Arnold, S. Kaye, R. S. Tedder, and J. A. McKeating. 1998. Development of a neutralizing antibody response during acute primary human immunodeficiency virus type 1 infection and the emergence of antigenic variants. J. Virol. 72:8943-8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lizeng, Q., C. Nilsson, S. Sourial, S. Andersson, O. Larsen, P. Aaby, M. Ehnlund, and E. Bjorling. 2004. Potent neutralizing serum immunoglobulin A (IgA) in human immunodeficiency virus type 2-exposed IgG-seronegative individuals. J. Virol. 78:7016-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luciw, P. A., E. Pratt-Lowe, K. E. Shaw, J. A. Levy, and C. Cheng-Mayer. 1995. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl. Acad. Sci. USA 92:7490-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahalanabis, M., J. Blue, and N. L. Haigwood. 2006. AminoTrackTM: automating the entry and analysis of mutations in multiple protein sequences using a spreadsheet format, p. 549-555. In H. R. Arabnia (ed.), Proceedings of the 2006 International Conference on Bioinformatics and Computational Biology (BIOCOMP'06). CSREA Press, Las Vegas, NV.
- 29.Mahalanabis, M., V. M. Hirsch, and N. L. Haigwood. 2005. Infection with a molecularly cloned SIVsm virus elicits high titer homologous neutralizing antibodies with heterologous neutralizing activity. J. Med. Primatol. 34:253-261. [DOI] [PubMed] [Google Scholar]
- 30.Marthas, M. L., R. A. Ramos, B. L. Lohman, K. K. Van Rompay, R. E. Unger, C. J. Miller, B. Banapour, N. C. Pedersen, and P. A. Luciw. 1993. Viral determinants of simian immunodeficiency virus (SIV) virulence in rhesus macaques assessed by using attenuated and pathogenic molecular clones of SIVmac. J. Virol. 67:6047-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marthas, M. L., K. K. van Rompay, M. Otsyula, C. J. Miller, D. R. Canfield, N. C. Pedersen, and M. B. McChesney. 1995. Viral factors determine progression to AIDS in simian immunodeficiency virus-infected newborn rhesus macaques. J. Virol. 69:4198-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazzoli, S., L. Lopalco, A. Salvi, D. Trabattoni, S. Lo Caputo, F. Semplici, M. Biasin, C. Bl, A. Cosma, C. Pastori, F. Meacci, F. Mazzotta, M. L. Villa, A. G. Siccardi, and M. Clerici. 1999. Human immunodeficiency virus (HIV)-specific IgA and HIV neutralizing activity in the serum of exposed seronegative partners of HIV-seropositive persons. J. Infect. Dis. 180:871-875. [DOI] [PubMed] [Google Scholar]
- 33.Mazzoli, S., D. Trabattoni, S. Lo Caputo, S. Piconi, C. Ble, F. Meacci, S. Ruzzante, A. Salvi, F. Semplici, R. Longhi, M. L. Fusi, N. Tofani, M. Biasin, M. L. Villa, F. Mazzotta, and M. Clerici. 1997. HIV-specific mucosal and cellular immunity in HIV-seronegative partners of HIV-seropositive individuals. Nat. Med. 3:1250-1257. [DOI] [PubMed] [Google Scholar]
- 34.McChesney, M. B., J. R. Collins, D. Lu, X. Lu, J. Torten, R. L. Ashley, M. W. Cloyd, and C. J. Miller. 1998. Occult systemic infection and persistent simian immunodeficiency virus (SIV)-specific CD4+-T-cell proliferative responses in rhesus macaques that were transiently viremic after intravaginal inoculation of SIV. J. Virol. 72:10029-10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, C. J., M. Marthas, J. Torten, N. J. Alexander, J. P. Moore, G. F. Doncel, and A. G. Hendrickx. 1994. Intravaginal inoculation of rhesus macaques with cell-free simian immunodeficiency virus results in persistent or transient viremia. J. Virol. 68:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nduati, R., G. John, D. Mbori-Ngacha, B. Richardson, J. Overbaugh, A. Mwatha, J. Ndinya-Achola, J. Bwayo, F. E. Onyango, J. Hughes, and J. Kreiss. 2000. Effect of breastfeeding and formula feeding on transmission of HIV-1: a randomized clinical trial. JAMA 283:1167-1174. [DOI] [PubMed] [Google Scholar]
- 37.Neildez, O., R. Le Grand, P. Caufour, B. Vaslin, A. Cheret, F. Matheux, F. Theodoro, P. Roques, and D. Dormont. 1998. Selective quasispecies transmission after systemic or mucosal exposure of macaques to simian immunodeficiency virus. Virology 243:12-20. [DOI] [PubMed] [Google Scholar]
- 38.Overbaugh, J., and C. R. Bangham. 2001. Selection forces and constraints on retroviral sequence variation. Science 292:1106-1109. [DOI] [PubMed] [Google Scholar]
- 39.Overbaugh, J., and L. M. Rudensey. 1992. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J. Virol. 66:5937-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Penedo, M. C., R. E. Bontrop, C. M. Heijmans, N. Otting, R. Noort, A. J. Rouweler, N. de Groot, N. G. de Groot, T. Ward, and G. G. Doxiadis. 2005. Microsatellite typing of the rhesus macaque MHC region. Immunogenetics 57:198-209. [DOI] [PubMed] [Google Scholar]
- 41.Pinto, L. A., J. Sullivan, J. A. Berzofsky, M. Clerici, H. A. Kessler, A. L. Landay, and G. M. Shearer. 1995. ENV-specific cytotoxic T lymphocyte responses in HIV seronegative health care workers occupationally exposed to HIV-contaminated body fluids. J. Clin. Investig. 96:867-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodrigo, A. G., P. C. Goracke, K. Rowhanian, and J. I. Mullins. 1997. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res. Hum. Retrovir. 13:737-742. [DOI] [PubMed] [Google Scholar]
- 44.Rowland-Jones, S., J. Sutton, K. Ariyoshi, T. Dong, F. Gotch, S. McAdam, D. Whitby, S. Sabally, A. Gallimore, T. Corrah, et al. 1995. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1:59-64. [DOI] [PubMed] [Google Scholar]
- 45.Ruprecht, R. M., T. W. Baba, V. Liska, N. B. Ray, L. N. Martin, M. Murphey-Corb, T. A. Rizvi, B. J. Bernacky, M. E. Keeling, H. M. McClure, and J. Andersen. 1999. Oral transmission of primate lentiviruses. J. Infect. Dis. 179(Suppl. 3):S408-S412. [DOI] [PubMed] [Google Scholar]
- 46.Ruprecht, R. M., F. Ferrantelli, M. Kitabwalla, W. Xu, and H. M. McClure. 2003. Antibody protection: passive immunization of neonates against oral AIDS virus challenge. Vaccine 21:3370-3373. [DOI] [PubMed] [Google Scholar]
- 47.Rychert, J., and A. M. Amedee. 2005. The antibody response to SIV in lactating rhesus macaques. J. Acquir. Immune Defic. Syndr. 38:135-141. [DOI] [PubMed] [Google Scholar]
- 48.Shankarappa, R., P. Gupta, G. H. Learn, Jr., A. G. Rodrigo, C. R. Rinaldo, Jr., M. C. Gorry, J. I. Mullins, P. L. Nara, and G. D. Ehrlich. 1998. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology 241:251-259. [DOI] [PubMed] [Google Scholar]
- 49.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shpaer, E. G., and J. I. Mullins. 1993. Rates of amino acid change in the envelope protein correlate with pathogenicity of primate lentiviruses. J. Mol. Evol. 37:57-65. [DOI] [PubMed] [Google Scholar]
- 51.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 52.Trivedi, P., D. Horejsh, S. B. Hinds, P. W. Hinds II, M. S. Wu, M. S. Salvato, and C. D. Pauza. 1996. Intrarectal transmission of simian immunodeficiency virus in rhesus macaques: selective amplification and host responses to transient or persistent viremia. J. Virol. 70:6876-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Rompay, K. K., C. J. Berardi, S. Dillard-Telm, R. P. Tarara, D. R. Canfield, C. R. Valverde, D. C. Montefiori, K. S. Cole, R. C. Montelaro, C. J. Miller, and M. L. Marthas. 1998. Passive immunization of newborn rhesus macaques prevents oral simian immunodeficiency virus infection. J. Infect. Dis. 177:1247-1259. [DOI] [PubMed] [Google Scholar]
- 54.Van Rompay, K. K., J. L. Greenier, K. S. Cole, P. Earl, B. Moss, J. D. Steckbeck, B. Pahar, T. Rourke, R. C. Montelaro, D. R. Canfield, R. P. Tarara, C. Miller, M. B. McChesney, and M. L. Marthas. 2003. Immunization of newborn rhesus macaques with simian immunodeficiency virus (SIV) vaccines prolongs survival after oral challenge with virulent SIVmac251. J. Virol. 77:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Rompay, K. K., M. L. Marthas, J. D. Lifson, C. J. Berardi, G. M. Vasquez, E. Agatep, Z. A. Dehqanzada, K. C. Cundy, N. Bischofberger, and N. C. Pedersen. 1998. Administration of 9-[2-(phosphonomethoxy)propyl]adenine (PMPA) for prevention of perinatal simian immunodeficiency virus infection in rhesus macaques. AIDS Res. Hum. Retrovir. 14:761-773. [DOI] [PubMed] [Google Scholar]
- 56.Van Rompay, K. K., M. B. McChesney, N. L. Aguirre, K. A. Schmidt, N. Bischofberger, and M. L. Marthas. 2001. Two low doses of tenofovir protect newborn macaques against oral simian immunodeficiency virus infection. J. Infect. Dis. 184:429-438. [DOI] [PubMed] [Google Scholar]
- 57.Van Rompay, K. K., R. P. Singh, L. L. Brignolo, J. R. Lawson, K. A. Schmidt, B. Pahar, D. R. Canfield, R. P. Tarara, D. L. Sodora, N. Bischofberger, and M. L. Marthas. 2004. The clinical benefits of tenofovir for simian immunodeficiency virus-infected macaques are larger than predicted by its effects on standard viral and immunologic parameters. J. Acquir. Immune Defic. Syndr. 36:900-914. [DOI] [PubMed] [Google Scholar]
- 58.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 60.Wu, X., A. B. Parast, B. A. Richardson, R. Nduati, G. John-Stewart, D. Mbori-Ngacha, S. M. Rainwater, and J. Overbaugh. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 80:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, M., B. Gaschen, W. Blay, B. Foley, N. Haigwood, C. Kuiken, and B. Korber. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229-1246. [DOI] [PubMed] [Google Scholar]
- 62.Zhu, T. 2002. HIV-1 in peripheral blood monocytes: an underrated viral source. J. Antimicrob. Chemother. 50:309-311. [DOI] [PubMed] [Google Scholar]
- 63.Zhu, T., L. Corey, Y. Hwangbo, J. M. Lee, G. H. Learn, J. I. Mullins, and M. J. McElrath. 2003. Persistence of extraordinarily low levels of genetically homogeneous human immunodeficiency virus type 1 in exposed seronegative individuals. J. Virol. 77:6108-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu, T., S. L. Hu, F. Feng, P. Polacino, H. Liu, Y. Hwangbo, G. H. Learn, J. I. Mullins, and L. Corey. 2004. Persistence of low levels of simian immunodeficiency virus in macaques that were transiently viremic by conventional testing. Virology 323:208-219. [DOI] [PubMed] [Google Scholar]