Abstract
Viruses in the family Polydnaviridae are symbiotically associated with parasitoid wasps. Wasps inject polydnaviruses (PDVs) when laying an egg into their insect host, and expression of viral gene products causes several physiological alterations, including immunosuppression, that allow the wasp's progeny to develop. As with other PDVs, most Microplitis demolitor bracovirus (MdBV) genes are related variants that form gene families. The largest MdBV gene family includes 13 members that encode predicted proteins related to protein tyrosine phosphatases (PTPs). Sequence analysis during the present study indicated that five PTP family members (PTP-H2, -H3, -N1, and -N2) have fully conserved catalytic domains, whereas other family members exhibited replacements, deletions, or rearrangements of amino acids considered essential for tyrosine phosphatase activity. Expression studies indicated that most MdBV PTP genes are expressed in virus-infected host insects, with transcript abundance usually being highest in hemocytes. MdBV-infected hemocytes also exhibited higher levels of tyrosine phosphatase activity than noninfected hemocytes. We produced expression constructs for four of the most abundantly expressed PTP family members and conducted functional studies with hemocyte-like Drosophila S2 cells. These experiments suggested that recombinant PTP-H2 and PTP-H3 are functional tyrosine phosphatases whereas PTP-H1 and PTP-J1 are not. PTP-H2 and -H3 localized to focal adhesions in S2 cells, and coexpression with another MdBV gene product, Glc1.8, resulted in complete inhibition of phagocytosis.
Survival of pathogenic microorganisms and multicellular parasites often depends on their ability to impair the immune response of their hosts (10, 23, 36, 57). Viruses in the family Polydnaviridae are symbiotically associated with parasitoid wasps and are among the most virulent immunosuppressive pathogens of insects known (60). Polydnaviruses (PDVs) are divided into two genera, bracoviruses (BVs) and ichnoviruses, on the basis of their association with wasps in the families Braconidae and Ichneumonidae (61). PDVs in both genera persist as stably integrated proviruses in the genome of their associated wasp and replicate in the ovaries of females. Virus accumulates to high densities in the lumens of the lateral and common oviducts to form a suspension of virus and protein called calyx fluid. When a wasp lays an egg into its insect host, she injects a quantity of virus that infects immune cells and other host tissues. PDVs do not replicate in the parasitized host insect, but viral gene products prevent the host's immune system from killing the parasitoid's offspring and cause alterations in host growth that likely facilitate parasitoid development (42, 59, 62). Thus, a true mutualism exists between PDVs and wasps, as viral transmission depends on survival of the parasitoid and parasitoid survival depends on infection and expression of gene products encoded by the virus.
PDVs coexist with more than 30,000 species of parasitoids, and comparison of selected ichnovirus and BV genomes strongly suggests that each PDV carried by a given parasitoid species is genetically unique (29, 60, 62, 65). Correspondingly, the physiological effects PDVs have on parasitized hosts vary. Some PDVs only impair the ability of the host's immune system to eliminate the wasp's offspring, while others immunosuppress hosts more broadly (62). An example of the latter is Microplitis demolitor BV (MdBV), which is carried by the microgastrine braconid M. demolitor. This wasp parasitizes the larval stage of several moth species, including Pseudoplusia includens (53). The innate immune system of P. includens and other insects consists of a coordinated network of humoral and cellularly (hemocyte) mediated defense responses (25, 27, 33, 63). Individual hemocytes eliminate small foreign objects by phagocytosis, whereas large numbers of hemocytes cooperate to kill larger intruders, like parasitoids, by encapsulation (33). Both of these defense responses depend on the ability of hemocytes to bind to foreign targets and activate downstream signaling pathways that mobilize the actin cytoskeleton and effector functions like antimicrobial peptide expression (25, 30, 31). However, hosts parasitized by M. demolitor are unable to phagocytize or encapsulate any foreign target because MdBV-infected hemocytes lose the capacity to adhere to foreign surfaces and exhibit alterations in key immune signaling pathways (7, 42, 54, 62).
Complete sequencing of the MdBV genome identified 61 predicted genes encoding proteins of ≥100 amino acids (62). Most of these genes form families of related variants that have likely arisen from duplication events and subsequent divergence (60, 62). Transcriptome analysis also indicates that most gene family members are expressed in parasitized host insects (5, 55, 56, 58, 60). Previous studies have implicated two MdBV gene families in immunosuppression. The Glc genes encode cell surface mucins that disrupt adhesion and phagocytosis of hemocytes and hemocyte-like cell lines (5, 6, 50). The vankyrin (IκB-like) gene family encodes homologs related to insect and mammalian inhibitor κBs (IκBs) that function as negative regulators of nuclear factor κB transcription factors (NF-κBs). Insect NF-κBs mediate the expression of a diversity of immune factors, including antimicrobial peptide genes (15, 26). Two MdBV IκB-like family members, IκB-H4 and -N5, bind NF-κBs associated with the Toll and Imd signaling pathways and suppress expression of multiple antimicrobial peptide genes (56).
The largest MdBV gene family encodes predicted proteins related to protein tyrosine phosphatases (PTPs) (60, 62). Reversible tyrosine phosphorylation is a key factor in the regulation of immunity, as well as many other physiological processes (37-39). Notably, several other BVs also encode multiple predicted PTPs, suggesting that this gene family is both widespread and functionally important in parasitism (11, 13, 17). Here we report that most members of the MdBV PTP gene family are expressed in virus-infected hosts. We also present evidence that two family members are functional PTPs involved in disrupting adhesion and phagocytosis of insect immune cells.
MATERIALS AND METHODS
Insects and cell cultures.
M. demolitor was reared by parasitizing third-instar P. includens at 27 ± 1°C with a 16-h light, 8-h dark photoperiod as previously described (49). Drosophila S2 cells (American Type Culture Collection) were maintained in HyQ medium (HyClone) and passaged as adherent cells in Corning 75-cm2 tissue culture flasks. Most experiments were conducted with 12-well culture plates (Corning). Cells were seeded into wells at specific densities, cultured for 18 to 24 h, and then used in bioassays.
Sequence analysis.
PTP-related gene family members were aligned with selected insect and mammalian PTPs by using Lasergene software (DNASTAR, Inc.) and the CLUSTAL W method with gap creation penalties of 10.00 and gap extension penalties of 0.20. Motifs and other structural features were identified by using the PFAM (http://www.sanger.ac.uk/Software/Pfam/) or NCBI conserved-domain (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) database. Transmembrane domains were identified by using the “DAS” transmembrane prediction server (http://mendel.imp.univie.ac.at/sat/DAS/DAS.html). The sequences of selected MdBV PTP genes were reconfirmed by partial or full sequencing of genomic and/or cDNA clones by the chain termination method with an ABI Prism BigDye Termination Cycle kit (Applied Biosystems). Sequence reactions were run at the University of Wisconsin—Madison sequencing facility.
Calyx fluid collection and infection studies.
M. demolitor calyx fluid consists almost entirely of MdBV virions, which is the only factor in calyx fluid that is infectious and immunosuppressive (49, 53). During the present study, calyx fluid was collected from wasps and used to infect host cells as previously described (5-7). In accordance with the convention in the PDV literature, the amount of calyx fluid from a single wasp was defined as one wasp equivalent. M. demolitor normally injects 0.1 to 0.02 wasp equivalent of virus per host (7, 52, 59). To infect hosts or S2 cells, MdBV isolated from wasps was diluted in TC-100 medium (Sigma), sterilized with a 0.2-μm syringe Acrodisc filter (Pall). We then injected 0.05 wasp equivalent in 2 μl of medium into fifth-instar P. includens, which results in infection of greater than 95% of the circulating hemocytes (49). Mock-infected hosts were injected with 2 μl of TC-100 medium only. Hemocytes were collected for use in PTP activity assays (see below) 18 h later by bleeding larvae from a cut proleg into saline and pelleting the cells by gentle centrifugation (400 × g for 1 min).
Total-RNA isolation and real-time PCR.
For real-time PCR studies, third-instar P. includens larvae (6 to 12 h postecdysis) were singly parasitized by M. demolitor. Parasitized and nonparasitized larvae were then CO2 anesthetized and processed at 18 h postparasitism. For tissue samples, host larvae were dissected in physiological saline to collect the fat body, gut (digestive tract plus Malpighian tubules), salivary glands, and nervous system (brain plus ventral nerve cord) (7). Hemocytes were collected by bleeding larvae from a cut proleg and pelleting the cells by centrifugation as described above. Total RNA was extracted from each tissue sample with TRIzol Reagent (Invitrogen) according to the manufacturer's instructions with slight modifications. To remove contaminating genomic DNA, the isolated RNA was treated with DNase I (Ambion), extracted with phenol-chloroform, precipitated with isopropanol, and resuspended in RNase-free water. RNA amounts were quantified with RiboGreen (Molecular Probes) and a fluorescence microplate reader (FLUOstar Galaxy; BMG) (31, 41).
For first-strand cDNA synthesis, 500 ng of total RNA per sample was reverse transcribed with Superscript II reverse transcriptase (Invitrogen) and random hexamers in 20-μl reaction mixtures according to the manufacturer's recommendations. Relative quantitative real-time PCRs (rqRT-PCRs) were run on a Rotor-Gene 3000 Real-Time PCR Thermal Cycler (Corbett Research) in 10-μl reaction mixtures as described previously (7). Briefly, each reaction mixture contained 4 μl of template cDNA, 5 μl of iQ SYBR Green Supermix (Bio-Rad), and each of the corresponding gene-specific forward and reverse primers at 0.25 μM (Table 1). To normalize differences in total RNA amounts that were reverse transcribed in each reaction, an 18S ribosomal gene from P. includens (GenBank accession no. AY298945) was used as an endogenous control (31). Cycling conditions were the same for all primer combinations: 3 min of initial denaturation at 95°C, followed by 40 cycles of 20 s of denaturation at 95°C, 20 s of annealing at 50°C, and 20 s of extension at 72°C, except in the case of PTP-J2, for which the annealing temperature was 52°C. Data were acquired during the extension step and analyzed with the Rotor-Gene application software (version 6.0.27). Reaction mixtures for each amplicon were prepared in quadruplicate, and the relative abundance of each PTP in different host tissues was calculated by the comparative CT or 2−δ,δ CT method (7). Transcript abundance for each PTP was standardized to 1.0 in the host gut sample, and abundance in the other tissues was determined as the relative increase compared to the gut. For the visualization of PCR products on agarose gels, conditions were identical to those described above, with the exception that reactions were stopped at 25 cycles for each PTP and at 15 cycles for the 18S ribosomal control.
TABLE 1.
Gene-specific primers used for real-time PCRs
| Family member | Forward and reverse primer sequences (5′-3′)a | Expected product size (bp) |
|---|---|---|
| ptp-H1 | (F) TATATAATGCGTTATCACAAGTGCG | 201 |
| (R) ACCTTGTCTTGGAATGCAGAGAAT | ||
| ptp-H2 | (F) CGTTGCAGCAGACAGCCTAC | 187 |
| (R) CTTAAGTGAGTTTCTATTTGGGTGGT | ||
| ptp-H3 | (F) CAGGCTATTGTTTCGAAATCTTTAAC | 194 |
| (R) GGAGTTATCCCAACACGTTACATCA | ||
| ptp-H5 | (F) GATCCATCACATATAACCCTAACACC | 258 |
| (R) TCTTCCAAATATCTCCATACCTTCC | ||
| ptp-J1 | (F) CCAATTCGGAAGGGTCTCG | 198 |
| (R) GGGGTAGCACTTTTGTTTGTTATCT | ||
| ptp-J2 | (F) TATACGCTGATTTAAGAAAGCATAAGAC | 212 |
| (R) GACATAATCATTTAATCGGTTCATACAA | ||
| ptp-J3 | (F) TGGAACATCCGGTGTCATTATC | 236 |
| (R) TTTGGTGTAAATGTACTTCCGCTT | ||
| ptp-J4 | (F) ATAAGCTATCTGCACGAAACTCCC | 201 |
| (R) CATAATTCTCCGGCTGACTAATGG | ||
| ptp-N1 | (F) TGTGGTACATTGCAGTGACGG | 174 |
| (R) GTTGTAATATTAAGTAATCATCTGGCG | ||
| ptp-N2 | (F) AGAAGAGTTTCATTATATCGTGCTCC | 143 |
| (R) CGCCGTTTTTCGACTCTATCA | ||
| ptp-N3 | (F) CTACAACATATCTGCATCATCATCAA | 203 |
| (R) CCTCAAAGCTAAAGAAATCTAATTGC |
F, forward; R, reverse.
Plasmids and expression.
The coding sequences for ptp-H1, -H2, -H3, and -J1 were cloned into the expression vector pIZT/V5-His (Invitrogen), which uses the OpIE2 promoter from the Orgyia pseudotsugata baculovirus for constitutive expression of the gene of interest and the OpIE1 promoter to express a Zeocin-green fluorescent (GFP) gene fusion which can be used for selection and as a marker. This vector also produces recombinant proteins with a V5 epitope tag. Each PTP family member was PCR amplified with MdBV genomic DNA as the template and the primers ptp-H1 (5′-ATTCTAGAATGGGGCGTCATAGTTTC-3′; forward), ptp-H1 (5′-ATCCGCGGATACTTATTATTAAATGAATG-3′; reverse), ptp-H2 (5′-AGAATTCATAAAATGGGTCGATGCAAATT-3′; forward), ptp-H2 (5′-TATCCGCGGATAGTTATCTTTTAGATGAAG-3′; reverse), ptp-H3 (5′-CTAGTGTTTTCTAGAATGGCAGGCTATTGTTTCG-3′; forward), ptp-H3 (5′-GCCGCGGAAGCTGTGAAACCAGATAATAA-3′; reverse), ptp-J1 (5′-GCTCTAGAATGGGTTCACATTGTTCTAAAAAC-3′; forward), and ptp-J1 (5′-TCCCGCGGATAACAAGGCGTTGAC-3′; reverse). XbaI or EcoRI (forward) and SacII restriction sites (reverse) were incorporated into the primers (underlined) for directional cloning of each gene. As recommended by the manufacturer, we also modified the sequence around the start codon in some of the forward primers (bold) to create Kozak sequences for proper initiation of translation and mutated the stop codons in the reverse primers (bold) to express the protein product in frame with the vector-encoded V5 epitope tag and six-His tag. Standard 50-μl reaction mixtures contained 2 μl of Elongase Enzyme Mix (Invitrogen), 1.5 mM MgCl2, 250 μM deoxynucleoside triphosphate mix, each primer at 200 nM, and 100 ng of genomic MdBV DNA. After a 3-min initial denaturation at 94°C, the target sequence was amplified by 30 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 50°C, and a 4-min extension step at 68°C. PCR products were first cloned into pCRdual-TOPO (Invitrogen) and then cloned into pIZT/V5-His as previously described (5) to yield the expression constructs pIZT/PTP-H1, pIZT/PTP-H2, pIZT/PTP-H3, and pIZT/PTP-J1. In addition, ptp-H2 was also cloned into pHSP70polyA (16) under control of the Drosophila hsp70 promoter. Briefly, ptp-H2 plus a V5 epitope tag and a six-His tag was PCR amplified from pIZT/H2 with the primers 5′-AGATCTATGGGTCGATGCAAATTCAGG-3′ (forward) and 5′-AGATCTTCAATGGTGATGGTGATGATG-3′ (reverse), which each contained a BglII restriction site (underlined). The ptp-H2 stop codon was also reintroduced into the reverse primer so as not to express GFP. PCR products were again initially cloned into pCRdual-TOPO, followed by cloning into pHSP70polyA to produce pHSP70/PTP-H2. The structure and orientation of all constructs were confirmed by sequencing as described above. A previously made Glc1.8 expression construct (pIZT/Glc1.8) was also used in functional assays (5). All constructs were transiently expressed in S2 cells by cationic lipid-mediated transfection with 1 μg of plasmid per ml of medium (5, 50). Transfection efficiencies averaged 75 to 80% as measured by GFP expression.
PTP activity assays.
For each replicate, hemocytes from MdBV- or mock-infected hosts and transfected Drosophila S2 cells were counted with a hemocytometer. We then lysed 5 × 105 hemocytes and 1 × 106 S2 cells per replicate on ice in 250 μl of lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 1% [vol/vol] Nonidet P-40 [Pierce] plus protease inhibitor cocktail [Roche]). PTP activity in lysates was then measured in 96-well plates (Corning) with the tyrosine phosphatase assay system (Promega) according to the manufacturer's instructions. Briefly, lysates for each replicate were first precleared of free phosphate. Twenty microliters of the cleared lysate was then incubated with the tyrosine-containing phosphopeptide END(pY)INASL (0.13 mM) in the presence or absence of 1 mM sodium orthovanadate. The reaction buffer used was Tris-buffered saline containing 1 mM dithiothreitol and 1 mM phenylmethylsulfonyl fluoride (pH 5.5). After 30 min at 30°C, the reaction was terminated by addition of a stop buffer containing a molybdate dye for visualization of liberated phosphate at 600 nm with a plate reader (BMG). The number of picograms of phosphate released was then determined by comparison to a standard curve. Each treatment was replicated a minimum of four times with independently prepared and collected samples. The data were then analyzed by one-way analysis of variance and the Tukey-Kramer multiple-comparison procedure (α ≤ 0.05) with JMP 3.0 software (SAS Institute, Gary, NC) (47).
Western blotting.
Lysates prepared for PTP activity assays were also used in Western blotting experiments. Protein concentrations in lysates were determined by Bradford assay (Bio-Rad). Forty micrograms of protein per sample was then added to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer, boiled, and separated by SDS-PAGE. After transfer to nitrocellulose, membranes were probed with a mouse anti-V5 antibody (1:10,000; Invitrogen) that recognizes the V5 epitope tag on each recombinant protein. A goat anti-mouse horseradish peroxidase-conjugated secondary antibody (1:20,000; Jackson Labs) was then used, followed by visualization of bands by horseradish peroxidase-enhanced chemiluminescence (Amersham Biosciences).
Phagocytosis assays.
We assessed the ability of S2 cells to phagocytose rhodamine-labeled Escherichia coli (5) or inert polystyrene beads (0.5 μm; Polysciences). Cells were first transfected with different pIZT/PTP and/or Glc1.8 expression constructs, with the empty pIZT vector serving as the control. Forty-eight hours later, cells were collected and added to new 12-well culture plates in medium without serum at a density of 1 × 105 cells per well. After a 1-h preincubation period, bacteria or microspheres were added to each culture well at a particle-to-cell ratio of 15:1. Cells were allowed to phagocytose for 45 min at 27°C, followed by transfer of the culture plate to ice. We then scored the percentage of cells with one or more ingested particles (18) by counting 200 cells per well from four randomly selected fields of view with a Leica TCS inverted epifluorescence microscope. Particles were red, while cells expressing a gene of interest were green. Each treatment was replicated a minimum of five times with independently prepared samples. The proportional data were then arcsin transformed and analyzed by one-way analysis of variance and the Tukey-Kramer multiple-comparison procedure.
Immunofluorescence staining.
S2 cells transfected with pHSP70/PTP-H2 were processed for immunofluorescence microscopy as previously described (5). Briefly, cells at 48 h posttransfection (not heat shocked) were washed in phosphate-buffered saline (PBS) and then fixed for 20 min in 4% paraformaldehyde in PBS. Fixed cells were permeabilized with PBS-0.1% Triton X-100, blocked in 1% bovine serum albumin in PBS, and then incubated overnight at 4°C with mouse anti-V5 antiserum (1:1,000) that recognizes recombinant PTP-H2 and a rabbit anti-FakY397 antibody (1:100; Biosource) that recognizes Drosophila focal adhesion kinase (Fak56) (17, 19). After washing in PBS-0.1% Triton X-100, cells were incubated with Texas Red-conjugated goat anti-mouse (1:2,000) and fluorescein isothiocyanate-conjugated goat anti-rabbit (1:2,000; Jackson Labs) secondary antibodies. Incubation of cells in the secondary antibodies alone served as the negative control. Samples were examined on a Leica TCS scanning confocal microscope, and images were processed with Leica and Adobe Photoshop software.
RESULTS
MdBV encodes multiple PTP-related genes.
Sequencing of the MdBV genome and BlastX analysis (www.ncbi.nlm.nih.gov) previously identified 13 open reading frames (ORFs) with significant homology to known PTPs (60). Each MdBV PTP family member was named by its location in the MdBV genome with one ORF located on genomic segment D (ptp-D1), five on segment H (ptp-H1, -2, -3, -4, and -5), four on segment J (ptp-J1, -2, -3, and -4), and three on segment N (ptp-N1, -2, and -3) (Table 2). Further analysis during the present study indicated that all family members except ptp-D1 and ptp-N1 possessed 5′ TATA boxes that were located 7 to 192 bp upstream of the initiation methionine (Table 2). Each family member except ptp-D1 also possessed 3′ polyadenylation signal sequences (AATAA) that were 4 to 145 bp downstream of the stop codon (Table 2). Although several MdBV genes are spliced, none of the PTP-related ORFs contained introns (62). Predicted proteins encoded by each family member ranged from 141 (ptp-D1) to 337 (ptp-H1) amino acids and had molecular masses of 21.1 to 39.5 kDa (Table 2).
TABLE 2.
Predicted properties of M. demolitor PTP family members
| Family member | Genomic location (segment, position[s])a | No. of positions between:
|
Predicted protein length (amino acids) | Predicted protein mass (kDa) | |
|---|---|---|---|---|---|
| TATA and initiation codon | AATAAA and stop codon | ||||
| ptp-D1 | D, 7778-7823, 1-377 | None | None | 141 | 16.9 |
| ptp-H1 | H, 1250-2260 | −18 | 10 | 337 | 39.5 |
| ptp-H2 | H, 2846-3823 | −126 | 29 | 326 | 37.9 |
| ptp-H3 | H, 4941-5903 | −104 | 24 | 321 | 37.2 |
| ptp-H4 | H, 9043-9591 | −73 | 88 | 183 | 21.1 |
| ptp-H5 | H, 10409-11238, 1-129 | −70 | 63 | 319 | 36.9 |
| ptp-J1 | J, 1931-2833 | −82 | 126 | 301 | 34.7 |
| ptp-J2 | J, 6126-7019 | −7 | 78 | 298 | 34.5 |
| ptp-J3 | J, 10999-11754 | −13 | 20 | 251 | 29.7 |
| ptp-J4 | J, 12570-13469 | −60 | 4 | 299 | 35.0 |
| ptp-N1 | N, 4114-5070 | None | 46 | 319 | 37.1 |
| ptp-N2 | N, 5743-6512 | −78 | 136 | 256 | 30.3 |
| ptp-N3 | N, 14837-15796 | −29 | 37 | 320 | 38.0 |
PTPs form a large superfamily whose members are recognized by the presence of one or two centrally located catalytic domains (2, 39). Each catalytic domain contains a signature motif (catalytic site), HCSAGxGRxG, that includes an essential cysteine (underlined) required for PTP activity. The PTP superfamily is divided into three families: classical PTPs, dual-specificity phosphatases, and low-molecular-weight phosphatases (2). BlastX results indicated that all of the MdBV PTP family members encode predicted proteins in the classical family. Classical PTPs are further subdivided into 17 subtypes on the basis of sequence similarity of the catalytic domain(s) and other motifs with regulatory or targeting functions. These subtypes also divide themselves into nontransmembrane (NT) (i.e., cytosolic) and transmembrane forms. All predicted MdBV PTPs were of the NT form, with the highest similarity matches (1 e−81) being to PTPs encoded by BVs from the wasps Cotesia congregata (CcBV) and Cotesia plutellae (13, 44). Conserved-domain searches also identified high similarity matches to PTPs encoded by other organisms. For example, the catalytic domain of MdBV PTP-H2 had similarity matches of e−65 and e−56 to the catalytic domains of PTP1b from humans and Yop51 from Yersinia (Pasteurella) bacteria, respectively, which are both classical subtype NT1 members (2). Other MdBV PTPs were also most similar to the classical NT1 subtype.
The catalytic domains of classical PTPs contain 10 conserved motifs which include the previously mentioned signature motif (catalytic site, motif 9) plus nine other motifs with proposed roles in substrate recognition, secondary structure, or catalysis (2). Alignments with a predicted PTP from CcBV (designated PTPR) and with PTP1b indicated that seven MdBV PTP family members possess all 10 motifs: PTP-H1, -H2, -H3, -H5, -J1, -N1, and -N2 (Fig. 1). However, the invariant tryptophan in the WPD loop (motif 8) of PTP-H1 was replaced by a phenylalanine while the catalytic cysteine (Cys235) in motif 9 of PTP-J1 was replaced by a serine. PTP-H4 was almost identical to PTP-H3 but lacked motifs 1 to 3 because of truncation of its N terminus. The remaining family members (PTP-D1, -J3, -J4, and -N3) lacked five or more motifs because of more complex deletions and/or rearrangements (Fig. 1). Of these, only PTP-J4 had an intact catalytic site (Fig. 1). We also noted that PTP-D1 was nearly identical to PTP-J4, except for a small deletion in its N terminus and a large deletion in its C terminus that eliminated motifs 8 to 10 (Fig. 1). In summary, most MdBV PTP family members have features consistent with their being functional genes but only five, ptp-H2, -H3, -H5, -N1, and -N2, encode predicted proteins with fully intact catalytic domains. A severely degenerate catalytic domain combined with the absence of promoter elements or a polyadenylation signal strongly suggested that ptp-D1 is a pseudogene. Sequence similarity among ptp-H3 and -H4 and between ptp-J4 and -D1 also suggested that diversification of this gene family is due in part to recent duplication events within and between genomic segments.
FIG. 1.
Sequence analysis of MdBV PTP family members. The deduced amino acid sequences of MdBV PTP family members are aligned with the deduced sequences of PTPR from CcBV (EMBL accession number AJ632310) and PTP1b from humans (GenBank accession number M33689). Residues shared by a majority of the genes in the alignment are boxed in black. The consensus sequence of the 10 conserved motifs present in the catalytic domain of most classical PTPs is presented above the alignment (2).
Most PTP family members are expressed in MdBV-infected hosts.
Prior studies indicated that MdBV primarily infects the hemocytes, fat body, and nervous system of host insects, with expression of gene products beginning 2 h postinfection and continuing at near-steady-state levels for several days thereafter (7, 52). To assess whether MdBV PTPs were expressed in these or other host tissues, we conducted rqRT-PCR experiments with primer pairs specific for individual family members (Table 1). All family members were examined except ptp-D1 and ptp-H4, which are truncated but otherwise nearly identical to ptp-J4 and ptp-H3, respectively (see above). These experiments indicated that transcript abundance for ptp-H2, -J1, -J2, -J4, -N1, and -N3 was highest in hemocytes, while transcript abundance for ptp-H1, -H3, -H5, and -J3 was highest in the fat body and/or nervous system (Fig. 2). Transcript abundance for all family members was usually lower in the other tissues we sampled. As a control, PTP amplicons were not detected when the template from MdBV-infected samples was not reverse transcribed, indicating that our RNA samples were not contaminated with viral DNA. In addition, no amplicons were detected in samples from noninfected hosts (data not presented).
FIG. 2.
Relative rqRT-PCR analysis of MdBV PTP family members ptp-H1, -H2, -H3, -H5; -J1 to -J4; and -N1 to -N3 in gut (GT), fat body (FB), hemocytes (HE), nervous system (NS), and salivary glands (SG) from parasitized P. includens. All PTPs were expressed in one or more types of infected host tissue. Differences in total RNA amounts that were reverse transcribed and added to each rqRT-PCR mixture were normalized relative to an endogenous 18S rRNA control. The relative transcript abundance of each PTP is then reported in comparison to the gut sample, which is standardized to a value of 1.0 (see Materials and Methods). PCR products were run on 1% agarose gels to verify the size of each amplicon and to assure that no cross-amplification occurred. All treatments were replicated four times with tissues collected from different virus-injected larvae. Error bars for each sample equal 1 standard deviation.
MdBV-infected host hemocytes and transfected Drosophila S2 cells exhibit elevated levels of PTP activity.
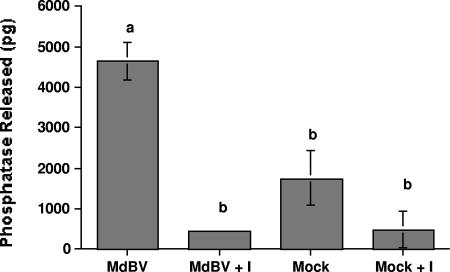
Given that transcript abundance was highest in hemocytes for several PTP family members, we next asked whether tyrosine phosphatase activity is higher in host hemocytes infected with a physiological dose of virus than in hemocytes from mock-infected hosts. We addressed this question with a commercially available kit optimized for studies on cell lysates. After preclearing lysates of free phosphate, we added a tyrosine-containing phosphopeptide [END(pY)INASL] recognized by most PTPs and then measured the free phosphate released after a 30-min incubation period. Addition of the PTP inhibitor sodium orthovanadate to each sample served as a negative control. Our results indicated that MdBV-infected hemocytes released significantly more free phosphate than did lysates from mock-infected hemocytes or lysates containing inhibitor, suggesting that one or more viral PTPs had tyrosine phosphatase activity (Fig. 3).
FIG. 3.
MdBV-infected host hemocytes exhibit elevated levels of tyrosine phosphatase activity. Hemocyte lysates from MdBV- or mock-infected P. includens larvae were precleared of free phosphate and incubated with a synthetic tyrosine phosphopeptide (Promega). Controls consisted of MdBV- or mock-infected lysates plus the inhibitor sodium orthovanadate (I). The number of picograms of free phosphate released ± the standard error after a 30-min incubation period was then measured at 600 nm (n = 5 per treatment). Means with the same letter are not significantly different (P < 0.05) by the Tukey-Kramer multiple-comparison procedure.
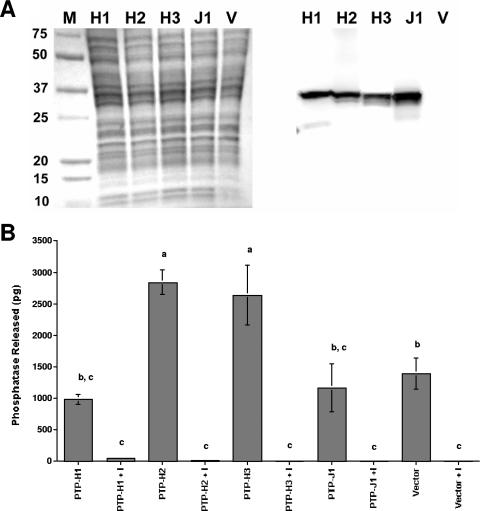
Sequence analysis suggested that ptp-H2, -H3, -H5, -N1, and -N2 were the strongest candidates for being functional PTPs since they were the only family members with fully intact catalytic domains (see above). Our rqRT-PCR studies, however, suggested that transcript abundance for ptp-H2 and -H3 was much higher in hemocytes than for ptp-N1, -H5, and -N2 (see Fig. 2). We therefore focused on ptp-H2 and -H3 by producing the expression constructs pIZT/H2 and pIZT/H3. Since the catalytic cysteine of ptp-J1 was replaced by a serine, we produced pIZT/J1 as a potential natural loss-of-function mutant. We also produced an expression construct for ptp-H1 (pIZT/H1) because it possessed a functional catalytic site but lacked an intact WPD loop (motif 8), which is also considered important for PTP activity (2). We then used hemocyte-like Drosophila S2 cells in bioassays because (i) these cells are excellent models for studies of insect cellular immune function, (ii) other MdBV virulence genes have immunosuppressive effects in S2 cells that are very similar to host hemocytes, (iii) more detailed knowledge of the pathway components regulating cellular immune responses is available for Drosophila than for other insects, and (iv) Drosophila is currently the only insect where homologues of focal adhesion components have been at least partially characterized (11, 19, 20, 22, 40). Following transfection of each construct into S2 cells, lysates were assayed for protein expression and PTP activity. An anti-V5 antibody detected a single protein band of the expected size (see Table 2) for each recombinant PTP family member on Western blots but did not detect any protein in lysates prepared from the empty-vector control (Fig. 4A). Enzymatic assays further indicated that cells transfected with pIZT/H2 and pIZT/H3 produced significantly more free phosphate than lysates from cells transfected with pIZT/H1, J1, or the empty vector (Fig. 4B). The presence of sodium orthovanadate, in contrast, reduced free-phosphate production in each treatment to almost undetectable levels (Fig. 4B).
FIG. 4.
Expression of PTP-H2 and -H3 significantly increases tyrosine phosphatase activity in Drosophila S2 cells. (A) Protein lysates from cells harvested 48 h posttransfection with different expression constructs were prepared and separated by SDS-PAGE on 8 to 16% gradient gels. A Coomassie blue-stained gel showing the molecular weights (103) of markers (M) and lysates from cells transfected with pIZT/H1 (H1), H2, H3, J1, or the empty vector (V) is shown on the left. The corresponding Western blot is shown on the right with recombinant PTP-H1, -H2, -H3, and -J1 detected with a mouse anti-V5 antibody. (B) Outcome of tyrosine phosphatase assays with lysates from S2 cells expressing PTP-H1, -H2, -H3, and -J1. Lysates from cells transfected with the empty vector or lysates containing sodium orthovanadate (I) served as controls. Assays were conducted as described in the legend to Fig. 3, and numbers of picograms of free phosphate released ± the standard error after a 30-min incubation period (n = 4 per treatment) are reported. Means with the same letter are not significantly different (P < 0.05) by the Tukey-Kramer multiple-comparison procedure.
PTP-H2 and -H3 reduce phagocytosis by S2 cells.
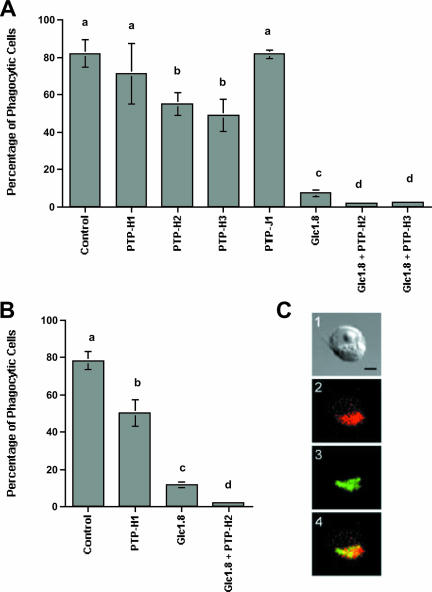
Phagocytosis and encapsulation are closely related defense responses that both involve receptor-mediated binding of a foreign target to the cell. Insect hemocytes remain bound to the surface of large foreign objects, whereas adhesion of smaller targets activates the formation of a phagosome and ingestion of the target via actin polymerization-dependent mechanisms (1, 9). Linkage to the cell surface and remodeling of the actin cytoskeleton in mammalian immune cells depend on the phosphorylation of several proteins localized in focal adhesions (1, 20, 40). As previously noted, MdBV infection disrupts adhesion and phagocytosis of insect hemocytes and hemocyte-like cell lines, including S2 cells (5, 6). One MdBV protein clearly involved in disrupting these defense responses is Glc1.8, which is expressed on the virus-infected cell surface (5, 6, 58). Given the importance of protein phosphorylation in these same processes, however, we assessed whether one or more MdBV PTPs also reduce phagocytosis by transfecting each PTP expression construct into S2 cells and conducting phagocytosis assays with bacteria and polystyrene beads that likely bind to cells via different surface receptors (15, 27, 45, 46). Our results indicated that a significantly smaller percentage of S2 cells expressing PTP-H2 or PTP-H3 phagocytized E. coli compared to the empty-vector control, whereas cells expressing PTP-H1 or -J1 did not (Fig. 5A). PTP-H2 expression also reduced the percentage of cells that phagocytized beads (Fig. 5B). However, cells expressing PTP-H2 or -H3 alone did not exhibit a reduction in phagocytosis as strong as that of cells expressing Glc1.8 which, as previously reported (6), reduced the percentage of phagocytic cells to less than 20% (Fig. 5A and B). However, cotransfection of cells with Glc1.8 and PTP-H2 or -H3 reduced phagocytosis even more strongly, with almost no cells being capable of internalizing bacteria or beads (Fig. 5A and B).
FIG. 5.
PTP-H2 and -H3 significantly reduce the phagocytosis of E. coli and polystyrene beads by S2 cells. Cells were transfected with pIZT/PTP-H1, -H2, -H3, -J1, or the empty vector (Vector) or cotransfected with PTP-H2 or PTP-H3 plus pIZT/Glc1.8. Forty-eight hours posttransfection, rhodamine-conjugated E. coli (A) or fluorescent polystyrene beads (B) were added to cells. Cells were examined 45 min later for phagocytosis by epifluorescence microscopy (n = 6 replicates per treatment). Means with the same letter in panel A or B are not significantly different (P < 0.05) by the Tukey-Kramer multiple-comparison procedure. (C) PTP-H2 colocalizes with Fak in focal adhesions. A light micrograph of a typical S2 cell expressing PTP-H2 is shown in image 1. The same cell after staining with anti-FakY397 (red) indicates that Fak56 localizes primarily to focal adhesions (2). Staining with anti-V5 (green) indicates that PTP-H2 localizes to the same region of the cell (3). Merging of images 2 and 3 results in an orange-yellow signal indicative of colocalization of Fak56 and PTP-H2. The scale bar in panel C1 equals 10 μm.
One component of focal adhesions in mammalian immune cells is focal adhesion kinase (Fak), which is a nonreceptor protein tyrosine kinase (PTK) (1, 20, 24, 40). The homologue of Fak in Drosophila (Fak56) also localizes to focal adhesions in adherent cells (19, 22). Since recombinant PTP-H2 exhibited phosphatase activity and reduced phagocytosis, we examined whether it localized to focal adhesions. Labeling with anti-FakY397 indicated that Fak56 localized primarily in focal adhesions of S2 cells similar to that shown in studies with other Drosophila cell types (Fig. 5C) (19). Visualization of PTP-H2 in transfected S2 cells with our anti-V5 antibody indicated that it too localized to focal adhesions in almost all of the cells we examined (Fig. 5C). Identical studies conducted with PTP-H3 yielded an identical result (data not presented), indicating that both family members predominantly localize in focal adhesions in S2 cells.
DISCUSSION
A key regulatory mechanism in immunity and other physiological processes is reversible tyrosine phosphorylation of essential proteins. In mammals, immune cells express more PTKs and PTP genes than any other cell type, with the possible exception of neurons (38). Far less is known about either the number or the function of PTKs and PTPs expressed in insect hemocytes. The Drosophila genome, for example, contains approximately 32 PTKs and 38 PTPs, but very few of these enzymes have been implicated in the regulation of any immune pathways or effector responses (3, 37). However, conservation between mammals and insects in immune signaling pathways such as Toll, JAK/STAT, and GATA, as well as in the protein complexes that regulate adhesion and phagocytosis, strongly suggests that specific PTKs and PTPs also regulate phagocytosis and other defense responses in insects (3, 22, 25, 33, 45, 46).
Several bacterial pathogens of mammals, including Yersinia spp., Salmonella enterica serovar Typhimurium, and Mycobacterium tuberculosis, encode phosphatases that disrupt phagocytosis, interfere with the activation of mitogen-activated protein kinases, or suppress other host defense responses (14, 23, 38, 48). In contrast, no experimental data prior to the present study indicate whether PTP-like genes encoded by PDVs function as virulence factors (12, 13, 44, 60, 62). Our previous studies indicate that MdBV immunosuppresses insects by inhibiting several effector functions, including adhesion and phagocytosis by immune cells. Here we report that most MdBV PTP family members are expressed in virus-infected hosts but only a subset of these genes likely encode catalytically functional PTPs. We also provide evidence that two of these putatively functional PTPs, PTP-H2 and -H3, are preferentially expressed in host hemocytes, localize to focal adhesions in S2 cells, and have antiadhesive or phagocytic activity.
Adhesion and phagocytosis of foreign targets involve multiple pathways in both mammals and insects (25, 27, 33). Opsonin-dependent pathways are regulated by binding of complement-like or other humoral pattern recognition molecules prior to uptake by hemocytes, while opsonin-independent pathways involve direct binding of the target to cell surface receptors like integrins and scavenger receptors (27, 34, 45, 46). Integrin expression by hemocytes is also implicated in the encapsulation of foreign targets, including parasitoids (27, 32, 35, 62). Integrin-mediated adhesion and signaling by mammalian immune cells require tyrosine phosphorylation of the PTK Fak or Pyk2, as well as other proteins, including p130Cas and paxillin (20, 24, 40). These proteins concentrate primarily in focal adhesions that link integrins and other components of the extracellular matrix to the actin cytoskeleton. Reciprocally, the antiphagocytic activity of PTP YopH from Yersinia sp. is associated with its ability to dephosphorylate Fak, paxillin, p130Cas, and several other proteins in mammalian macrophages, neutrophils, and lymphocytes (10, 14, 38). Far less is known about these processes in insect immune cells, but it is likely that adhesion and phagocytosis are regulated in part by similar protein complexes given that Drosophila encodes homologs of Fak (Fak56), Pyk2, and paxillin (DpaxA) that also localize to focal adhesions (11, 19, 22, 64). In turn, the antiphagocytic activity of recombinant MdBV PTP-H2 and -H3 combined with their localization in focal adhesions suggest Fak56 or related proteins are candidate substrates for these virus-encoded enzymes.
Some adhesion and phagocytic pathways do not require tyrosine phosphorylation for normal function (23). The ability of YopH to use multiple substrates has been suggested as one way by which Yersinia sp. simultaneously disables different phagocytic pathways. Another strategy is that pathogens can introduce multiple virulence factors into immune cells that disrupt phagocytosis and other effector functions by different mechanisms (10, 14, 38). Disrupting multiple immune pathways is clearly important to MdBV and other PDVs given that encapsulation and killing of the parasitoid, as well as potential clearance of the virus by the host immune system, likely involve multiple effector responses. Our previous studies indicated that MdBV encodes at least one other virulence gene (glc1.8) with antiadhesive and antiphagocytic activities that, unlike PTP-H2, targets the cell surface (5, 6). The present study indicates that PTP-H2 and -H3 complement the activity of Glc1.8 since expression of these PTPs and Glc1.8 together has a greater effect on adhesion and phagocytosis than expression of either factor alone. Combined with the expression of multiple IκBs that disrupt the Toll and Imd signaling pathways (56), the picture that collectively emerges is that MdBV encodes a suite of virulence factors that suppress multiple immune functions.
Although host hemocytes are the primary target of MdBV infection, other tissues are also infected (7, 52, 58, 62). Results of the present study further indicate that while transcript abundance of some PTP family members is highest in hemocytes, others are more abundant in the fat body and nervous system. This suggests that MdBV PTP-like gene products interact with other physiological processes of the host besides immunity. One potential target of interest is the ecdysone biosynthetic pathway, which involves the phosphorylation of several proteins in prothoracic glands (21). Hosts parasitized by many BV-carrying parasitoids exhibit reduced ecdysteroid titers that result in inhibition of molting and metamorphosis (8, 42), which has led to the suggestion that proteins in the ecdysone pathway are potential substrates for PTPs encoded by Toxoneuron nigriceps BV and CcBV (44). This could be the case for some MdBV PTP family members as well given that virus infection strongly suppresses host ecdysteroid titers (4, 51). A baculovirus-encoded PTP was also recently implicated in enhancing the locomotor activity of infected larvae (28). Lepidopteran larvae parasitized by BV-carrying braconid wasps exhibit reduced locomotor activity in concert with suppressed weight gain, but whether any viral gene product is responsible for these alterations is unknown.
Phylogenetic studies indicate that BV-carrying braconid wasps form a monophyletic lineage that now consists of approximately 17,000 species in four subfamilies (Microgastrinae, Miracinae, Cardiochilinae, and Cheloninae) (see references 42, 62, and 65 for a summary). Since BVs are only transmitted vertically, the presence of classical PTP genes in all of the BVs from microgastrine and cardiochiline wasps examined to date suggests that one or more PTP genes were acquired by a common ancestor relatively early in the evolution of the BV-braconid association. Subsequent diversification, however, has clearly resulted in pronounced differences among the PTP genes encoded by the thousands of BVs that exist today. The CcBV genome, for example, encodes 27 PTP family members, compared to only 13 in MdBV (17, 44, 60). Sequence comparisons among MdBV and CcBV PTP family members indicate that most differ considerably from one another. Within-species comparisons also reveal various degrees of similarity. For example, MdBV PTP-D1 and PTP-J4 are very similar and likely reflect a recent duplication event. In contrast, sequence variation is very high among other MdBV family members (Fig. 1), as well as PTP family members encoded by CcBV (44). The differences in the PTP-like genes encoded by BVs could reflect differences in the host insects their associated wasps parasitize since sequence variation in PTP catalytic domains greatly affects substrate specificities (2, 39). Most BV-carrying parasitoids also parasitize multiple host species, which further increases the diversity of protein targets with which PTP family members could interact (43, 60, 61, 63). While MdBV PTP-H2 and -H3 appear to be functional PTPs, sequence analysis suggests that most other family members likely are not. Analyses of PTP gene families in other organisms similarly reveal that catalytically inactive enzyme homologues commonly occur, and in some instances enzymatically inactive family members have assumed functions in other regulatory processes (43). Thus, some MdBV PTP family members could function as substrate traps or have other novel activities. Key challenges for the future are to determine what these other functions might be and what role they play in successful parasitism.
Acknowledgments
We thank M. H. Beck and R. Suderman for comments on the manuscript, J. Johnson for assistance in maintenance of the insects and cell lines used in this study, and J. Linder for assistance with the phagocytosis assays. We also thank L. Passarelli for sharing with us the pHSP70polyA expression vector.
This study was supported by grants from the U.S. Department of Agriculture NRI program to M.R.S.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Aderem, A., and D. M. Underhill. 1999. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 17:593-623. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J. N., O. H. Mortensen, G. H. Peters, P. G. Drake, L. F. Iversen, O. H. Olsen, P. G. Jansen, H. S. Andersen, N. K. Tonks, and N. P. Møller. 2001. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 21:7117-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baeg, G. H., R. Zhou, and N. Perrimon. 2005. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 19:1861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balgopal, M. M., B. A. Dover, W. G. Goodman, and M. R. Strand. 1996. Parasitism by Microplitis demolitor induces alterations in the juvenile hormone titers and juvenile hormone esterase activity of its host, Pseudoplusia includens. J. Insect Physiol. 42:337-345. [Google Scholar]
- 5.Beck, M., and M. R. Strand. 2005. Glc1.8 from Microplitis demolitor bracovirus induces a loss of adhesion and phagocytosis in insect High Five and S2 cells. J. Virol. 79:1861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck, M., and M. R. Strand. 2003. RNA interference silences Microplitis demolitor bracovirus genes and implicates glc1.8 in disruption of adhesion in infected host cells. Virology 314:521-535. [DOI] [PubMed] [Google Scholar]
- 7.Beck, M. H., R. Inman, and M. Strand. 9 October 2006. Microplitis demolitor bracovirus genome segments vary in abundance and are individually packaged in virions. Virology [Epub ahead of print.] doi: 10.1016/j.virol.2006.09.002. [DOI] [PubMed]
- 8.Beckage, N. E., and D. B. Gelman. 2004. Wasp parasitoid disruption of host development: implications for new biologically based strategies for insect control. Annu. Rev. Entomol. 49:299-330. [DOI] [PubMed] [Google Scholar]
- 9.Blander, J. M., and R. Medzhitov. 2004. Regulation of phagosome maturation by signals from Toll-like receptors. Science 304:1014-1018. [DOI] [PubMed] [Google Scholar]
- 10.Celli, J., and B. B. Finlay. 2002. Bacterial avoidance of phagocytosis. Trends Microbiol. 10:232-237. [DOI] [PubMed] [Google Scholar]
- 11.Chen, G.-C., B. Turano, P. J. Ruest, M. Hagel, J. Settleman, and S. M. Thomas. 2005. Regulation of Rho and Rac signaling to the actin cytoskeleton by paxillin during Drosophila development. Mol. Cell. Biol. 25:979-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, Y. P., P. B. Taylor, M. Shapiro, and D. E. Gundersen-Rindal. 2003. Quantitative expression analysis of a Glyptapanteles indiensis polydnavirus protein tyrosine phosphatase gene in its natural lepidopteran host, Lymantria dispar. Insect Mol. Biol. 12:271-280. [DOI] [PubMed] [Google Scholar]
- 13.Choi, J. Y., J. Y. Roh, J. N. Kang, H. J. Shim, S. D. Woo, B. R. Jin, M. S. Li, and Y. H. Je. 2005. Genomic segments cloning and analysis of Cotesia plutellae polydnavirus using plasmid capture system. Biochem. Biophys. Res. Commun. 332:487-493. [DOI] [PubMed] [Google Scholar]
- 14.Cornelis, G. R. 2002. Yersinia type III secretion: send in the effectors. J. Cell Biol. 158:401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Gregorio, E., P. T. Spellman, G. M. Rubin, and B. Lemaitre. 2001. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. USA 98:12590-12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Detvisitsakun, C., M. F. Berretta, C. Lehiy, and A. L. Passarelli. 2005. Stimulation of cell motility by a viral fibroblast growth factor homolog: proposal for a role in viral pathogenesis. Virology 336:308-317. [DOI] [PubMed] [Google Scholar]
- 17.Espagne, E., C. Dupuy, E. Huguet, L. Cattolico, B. Provost, N. Martins, M. Poirie, G. Periquet, and J. M. Drezen. 2004. Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science 306:286-289. [DOI] [PubMed] [Google Scholar]
- 18.Fällman, M., K. Andersson, S. Hakansson, K. E. Magnusson, O. Stendahl, and H. Wolf-Watz. 1995. Yersinia pseudotuberculosis inhibits Fc receptor-mediated phagocytosis in J774 cells. Infect. Immun. 63:3117-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimoto, J., K. Sawamoto, M. Okabe, Y. Takagi, T. Tezuka, S. Yoshikawa, H. Ryo, H. Okano, and T. Yamamoto. 1999. Cloning and characterization of Dfak56, a homolog of focal adhesion kinase, in Drosophila melanogaster. J. Biol. Chem. 274:29196-29201. [DOI] [PubMed] [Google Scholar]
- 20.Gelman, I. H. 2003. Pyk 2 FAKs, any two FAKs. Cell Biol. Int. 27:507-510. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert, L. I., R. Rybczynski, and J. T. Warren. 2002. Control and biochemical nature of the ecdysteroidogenic pathway. Annu. Rev. Entomol. 47:883-916. [DOI] [PubMed] [Google Scholar]
- 22.Grabbe, C., C. G. Zervas, T. Hunter, N. H. Brown, and R. H. Palmer. 2004. Focal adhesion kinase is not required for integrin function or viability in Drosophila. Development 131:5795-5805. [DOI] [PubMed] [Google Scholar]
- 23.Gruenheid, S., and B. B. Finlay. 2003. Microbial pathogenesis and cytoskeletal function. Nature 422:775-781. [DOI] [PubMed] [Google Scholar]
- 24.Hed, J., and O. Stendahl. 1982. Differences in the ingestion mechanisms of Igg and C3b particles in phagocytosis by neutrophils. Immunology 45:727-736. [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann, J. A., and J. M. Reichhart. 2002. Drosophila innate immunity: an evolutionary perspective. Nat. Immunol. 3:121-126. [DOI] [PubMed] [Google Scholar]
- 26.Imler, J.-L., and P. Bulet. 2005. Antimicrobial peptides in Drosophila: structures, activities and gene regulation, p. 1-21. In D. Kabelitz and J. M. Schröder (ed.), Mechanisms of epithelial defense, vol. 86. S. Karger AG, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 27.Irving, P., J. M. Ubeda, D. Doucet, L. Troxler, M. Lagueux, D. Zachary, J. A. Hoffmann, C. Hetru, and M. Meister. 2005. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell. Microbiol. 7:335-350. [DOI] [PubMed] [Google Scholar]
- 28.Kamita, S. G., K. Nagasaka, J. W. Chua, T. Shimada, K. Mita, M. Kobayashi, S. Maeda, and B. D. Hammock. 2005. A baculovirus-encoded protein tyrosine phosphatase gene induces enhanced locomotory activity in a lepidopteran host. Proc. Natl. Acad. Sci. USA 102:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroemer, J. A., and B. A. Webb. 2004. Polydnavirus genes and genomes: emerging gene families and new insights into polydnavirus replication. Annu. Rev. Entomol. 49:431-456. [DOI] [PubMed] [Google Scholar]
- 30.Lavine, M. D., and N. E. Beckage. 1996. Temporal pattern of parasitism-induced immunosuppression in Manduca sexta larvae parasitized by Cotesia congregata. J. Insect Physiol. 42:41-51. [Google Scholar]
- 31.Lavine, M. D., G. Chen, and M. R. Strand. 2005. Immune challenge differentially affects transcript abundance of three antimicrobial peptides in hemocytes from the moth Pseudoplusia includens. Insect Biochem. Mol. Biol. 35:1335-1346. [DOI] [PubMed] [Google Scholar]
- 32.Lavine, M. D., and M. R. Strand. 2003. Haemocytes from Pseudoplusia includens express multiple alpha and beta integrin subunits. Insect Mol. Biol. 12:441-452. [DOI] [PubMed] [Google Scholar]
- 33.Lavine, M. D., and M. R. Strand. 2002. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 32:1295-1309. [DOI] [PubMed] [Google Scholar]
- 34.Levashina, E. A., L. F. Moita, S. Blandin, G. Vriend, M. Lagueux, and F. C. Kafatos. 2001. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell 104:709-718. [DOI] [PubMed] [Google Scholar]
- 35.Levin, D. M., L. N. Breuer, S. Zhuang, S. A. Anderson, J. B. Nardi, and M. R. Kanost. 2005. A hemocyte-specific integrin required for hemocytic encapsulation in the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 35:369-380. [DOI] [PubMed] [Google Scholar]
- 36.Mason, N. J., D. Artis, and C. A. Hunter. 2004. New lessons from old pathogens: what parasitic infections have taught us about the role of nuclear factor-kappa B in the regulation of immunity. Immunol. Rev. 201:48-56. [DOI] [PubMed] [Google Scholar]
- 37.Morrison, D. K., M. S. Murakami, and V. Cleghon. 2000. Protein kinases and phosphatases in the Drosophila genome. J. Cell Biol. 150:F57-F62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mustelin, T., T. Vang, and N. Bottini. 2005. Protein tyrosine phosphatases and the immune response. Nat. Rev. Immunol. 5:43-57. [DOI] [PubMed] [Google Scholar]
- 39.Neel, B. G., and N. K. Tonks. 1997. Protein tyrosine phosphatases in signal transduction. Curr. Opin. Cell Biol. 9:193-204. [DOI] [PubMed] [Google Scholar]
- 40.Parsons, J. T. 2003. Focal adhesion kinase: the first ten years. J. Cell Sci. 116:1409-1416. [DOI] [PubMed] [Google Scholar]
- 41.Pech, L. L., and M. R. Strand. 1996. Granular cells are required for encapsulation of foreign targets by insect haemocytes. J. Cell Sci. 109:2053-2060. [DOI] [PubMed] [Google Scholar]
- 42.Pennacchio, F., and M. R. Strand. 2006. Evolution of developmental strategies in parasitic hymenoptera. Annu. Rev. Entomol. 51:233-258. [DOI] [PubMed] [Google Scholar]
- 43.Pils, B., and J. Schultz. 2004. Inactive enzyme-homologues find new function in regulatory processes. J. Mol. Biol. 340:399-404. [DOI] [PubMed] [Google Scholar]
- 44.Provost, B., P. Varricchio, E. Arana, E. Espagne, P. Falabella, E. Huguet, R. La Scaleia, L. Cattolico, M. Poirie, C. Malva, J. A. Olszewski, F. Pennacchio, and J. M. Drezen. 2004. Bracoviruses contain a large multigene family coding for protein tyrosine phosphatases. J. Virol. 78:13090-13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rämet, M., P. Manfruelli, A. Pearson, B. Mathey-Prevot, and R. A. B. Ezekowitz. 2002. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416:644-648. [DOI] [PubMed] [Google Scholar]
- 46.Rämet, M., A. Pearson, P. Manfruelli, X. H. Li, H. Koziel, V. Gobel, E. Chung, M. Krieger, and R. A. B. Ezekowitz. 2001. Drosophila scavenger receptor Cl is a pattern recognition receptor for bacteria. Immunity 15:1027-1038. [DOI] [PubMed] [Google Scholar]
- 47.Sall, J., and A. Lehman. 1996. JMP start statistics: a guide to statistics and data analysis using JMP and JMP IN software. Duxbury Press, New York, N.Y.
- 48.Singh, R., V. Rao, H. Shakila, R. Gupta, A. Khera, N. Dhar, A. Singh, A. Koul, Y. Singh, M. Naseema, P. R. Narayanan, C. N. Paramasivan, V. D. Ramanathan, and A. K. Tyagi. 2003. Disruption of mptpB impairs the ability of Mycobacterium tuberculosis to survive in guinea pigs. Mol. Microbiol. 50:751-762. [DOI] [PubMed] [Google Scholar]
- 49.Strand, M. R. 1994. Microplitis demolitor polydnavirus infects and expresses in specific morphotypes of Pseudoplusia includens hemocytes. J. Gen. Virol. 75:3007-3020. [DOI] [PubMed] [Google Scholar]
- 50.Strand, M. R., M. H. Beck, M. D. Lavine, and K. D. Clark. 2006. Microplitis demolitor bracovirus inhibits phagocytosis by hemocytes from Pseudoplusia includens. Arch. Insect Biochem. Physiol. 61:134-145. [DOI] [PubMed] [Google Scholar]
- 51.Strand, M. R., and B. A. Dover. 1991. Developmental disruption of Pseudoplusia includens and Heliothis virescens larvae by the calyx fluid and venom of Microplitis demolitor. Arch. Insect Biochem. Physiol. 18:131-145. [DOI] [PubMed] [Google Scholar]
- 52.Strand, M. R., D. I. McKenzie, V. Grassl, B. A. Dover, and J. M. Aiken. 1992. Persistence and expression of Microplitis demolitor polydnavirus in Pseudoplusia includens. J. Gen. Virol. 73:1627-1635. [DOI] [PubMed] [Google Scholar]
- 53.Strand, M. R., and T. Noda. 1991. Alterations in the hemocytes of Pseudoplusia includens after parasitism by Microplitis demolitor. J. Insect Physiol. 37:839-850. [Google Scholar]
- 54.Strand, M. R., and L. L. Pech. 1995. Immunological basis for compatibility in parasitoid host relationships. Annu. Rev. Entomol. 40:31-56. [DOI] [PubMed] [Google Scholar]
- 55.Strand, M. R., R. A. Witherell, and D. Trudeau. 1997. Two Microplitis demolitor polydnavirus mRNAs expressed in hemocytes of Pseudoplusia includens contain a common cysteine-rich domain. J. Virol. 71:2146-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thoetkiattikul, H., M. H. Beck, and M. R. Strand. 2005. Inhibitor κB-like proteins from a polydnavirus inhibit NF-κB activation and suppress the insect immune response. Proc. Natl. Acad. Sci. USA 102:11426-11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 58.Trudeau, D., R. A. Witherell, and M. R. Strand. 2000. Characterization of two novel Microplitis demolitor polydnavirus mRNAs expressed in Pseudoplusia includens haemocytes. J. Gen. Virol. 81:3049-3058. [DOI] [PubMed] [Google Scholar]
- 59.Turnbull, M., and B. Webb. 2002. Perspectives on polydnavirus origins and evolution. Adv. Virus Res. 58:203-254. [DOI] [PubMed] [Google Scholar]
- 60.Webb, B. A., M. R. Strand, S. E. Dickey, M. H. Beck, R. S. Hilgarth, W. E. Barney, K. Kadash, J. A. Kroemer, K. G. Lindstrom, W. Rattanadechakul, K. S. Shelby, H. Thoetkiattikul, M. W. Turnbull, and R. A. Witherell. 2006. Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology 347:160-174. [DOI] [PubMed] [Google Scholar]
- 61.Webb, B. A., N. E. Beckage, Y. Hayakawa, P. J. Krell, B. Lanzrein, M. R. Strand, D. B. Stoltz, and M. D. Summers. 2000. Polydnaviridae, p. 253-259. In M. H. van Regenmortel et al. (ed.), Virus taxonomy. Academic Press, San Diego, CA.
- 62.Webb, B. A., and M. R. Strand. 2005. The biology and genomics of polydnaviruses, p. 260-323. In L. I. Gilbert, K. Iatrou, and S. S. Gill (ed.), Comprehensive molecular insect science, vol. 5. Elsevier Press, San Diego, CA. [Google Scholar]
- 63.Wertheim, B., A. R. Kraaijeveld, E. Schuster, E. Blanc, M. Hopkins, S. D. Pletcher, M. R. Strand, L. Partridge, and H. C. J. Godfray. 2005. Genome-wide gene expression in response to parasitoid attack in Drosophila. Genome Biol. 6:R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wheeler, G. N., and R. O. Hynes. 2001. The cloning, genomic organization and expression of the focal contact protein paxillin in Drosophila. Gene 262:291-299. [DOI] [PubMed] [Google Scholar]
- 65.Whitfield, J. B. 2002. Estimating the age of the polydnavirus/braconid wasp symbiosis. Proc. Natl. Acad. Sci. USA 99:7508-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]