Abstract
Viruses are extremely abundant in seawater and are believed to be significant pathogens to photosynthetic protists (microalgae). Recently, several novel RNA viruses were found to infect marine photosynthetic protists; one of them is HcRNAV, which infects Heterocapsa circularisquama (Dinophyceae). There are two distinct ecotypes of HcRNAV with complementary intraspecies host ranges. Nucleotide sequence comparison between them revealed remarkable differences in the coat protein coding gene resulting in a high frequency of amino acid substitutions. However, the detailed mechanism supporting this intraspecies host specificity is still unknown. In this study, virus inoculation experiments were conducted with compatible and incompatible host-virus combinations to investigate the mechanism determining intraspecies host specificity. Cells were infected by adding a virus suspension directly to a host culture or by transfecting viral RNA into host cells by particle bombardment. Virus propagation was monitored by Northern blot analysis with a negative-strand-specific RNA probe, transmission electron microscopy, and a cell lysis assay. With compatible host-virus combinations, propagation of infectious progeny occurred regardless of the inoculation method used. When incompatible combinations were used, direct addition of a virus suspension did not even result in viral RNA replication, while in host cells transfected with viral RNA, infective progeny virus particles with a host range encoded by the imported viral RNA were propagated. This indicates that the intraspecies host specificity of HcRNAV is determined by the upstream events of virus infection. This is the first report describing the reproductive steps of an RNA virus infecting a photosynthetic protist at the molecular level.
Viruses are extremely abundant in seawater (29) and are considered to play a major role as pathogens of the primary marine producers photosynthetic protists (microalgae). Algal viruses were previously assumed to consist almost entirely of double-stranded DNA viruses that belong to the family Phycodnaviridae (28). However, recent investigations revealed that previously unknown RNA viruses also infect marine photosynthetic protists. HaRNAV (15, 30) and RsRNAV (25) are positive-sense single-stranded RNA viruses that infect the toxic-bloom-forming microalga Heterosigma akasiwo (Raphidophyceae) and the diatom Rhizosolenia setigera (Bacillariophyceae), respectively. Although the genomes of HaRNAV and RsRNAV have some characteristics similar to those of viruses in the proposed order Picornavirales, phylogenetic analysis based on the deduced amino acid sequence of the RNA-dependent RNA polymerase (RdRP) domain suggests that these two viruses have an independent lineage that evolved from a deep internal branch in Picornavirales (6). MpRNAV is a double-stranded RNA virus in the family Reoviridae that infects the cosmopolitan photosynthetic protist Micromonas pusilla (Prasinophyceae) (1).
HcRNAV infects the bloom-forming photosynthetic dinoflagellate Heterocapsa circularisquama (32). The genome of HcRNAV consists of 4.4 kb of undivided positive-sense single-stranded RNA with no cap structure at the 5′ end (H. Mizumoto and K. Nagasaki, unpublished data) and no poly(A) tail at the 3′ end (20). Instead, the 3′-terminal sequence of HcRNAV is predicted to construct a stable stem-loop structure (20), as observed in other nonpolyadenylated RNA viruses (19). The genome of HcRNAV contains two open reading frames (ORFs) (20). The deduced amino acid sequence suggests that ORF1 contains two conserved domains coding for essential replication enzymes: a serine protease domain and an RdRP domain (20). This implies that ORF1 codes for a 110-kDa polyprotein that is translated and cleaved into smaller functional proteins including the RdRP. Phylogenetic analysis of the RdRP amino acid sequence suggests that HcRNAV belongs to a previously unrecognized virus group (20). ORF2 codes for a 38-kDa viral coat protein (CP) (20).
Previously, we showed that HcRNAV infection is ecotype specific rather than species specific. Using a cross-reactivity test with 56 clonal H. circularisquama isolates and 107 clonal virus isolates, we found that HcRNAV is divided into two distinct ecotypes having complementary intraspecies host specificity, ecotypes UA and CY (32). Analyses of representative virus isolates belonging to each ecotype (HcRNAV34 and HcRNAV109; UA and CY, respectively) show that the viral genomes are about 97% identical to each other at the nucleotide sequence level (Fig. 1) (20, 32). The noticeable difference between these two viral genomes is four variable regions (I to IV) in ORF2 with a high frequency of amino acid substitutions (Fig. 1). We assume that this determines the intraspecies host specificity of HcRNAV, i.e., viral cell tropism (20); however, the details of the mechanism supporting the cell tropism of HcRNAV are not fully understood.
FIG. 1.
Schematic diagrams of the HcRNAV34 and HcRNAV109 genome structures (20). The HcRNAV genome is shown as a thick line with the protein coding regions depicted as boxes. The filled box indicate a nucleotide (nt) deletion region. Shaded boxes indicate high-frequency nucleotide substitution regions.
The major step determining tropism is the specific attachment of viruses to susceptible host cells; i.e., viral tropism can be determined by cellular receptor-viral ligand interactions. The most intensively studied example is poliovirus. The immunoglobulin superfamily molecule CD155 is regarded as the sole human cellular binding molecule conferring susceptibility to poliovirus (17). Although rodents ordinarily resist poliovirus infection because of the absence of CD155, transgenic mice expressing CD155 became susceptible to poliovirus infection (23). Another determinant of tropism is virus replication. Poxvirus tropism is considered to be regulated by intracellular events downstream of virus binding and entry, rather than at the level of specific host receptors (16).
The objective of this study was to determine whether the intraspecies host specificity of HcRNAV is determined at the entry process or the intracellular multiplication process. Instead of using the conventional virus-induced cell lysis assay, we examined viral RNA accumulation by using a highly sensitive strand-specific Northern blot analysis. In addition, direct transfection of viral RNA into host cells was performed to analyze viral RNA replication. Our results show that the intraspecies host specificity of HcRNAV is determined at early steps of virus infection. This is the first report showing the reproduction steps of RNA viruses infecting photosynthetic protists at the molecular level.
MATERIALS AND METHODS
Hosts and viruses.
The origins of the virus isolates (HcRNAV34 and HcRNAV109) and host photosynthetic protist isolates (H. circularisquama HU9433-P and HCLG-1) used in this study were previously reported (32). Cell cultures were grown in modified SWM3 medium enriched with 2 nM Na2SeO3 (4, 11, 13) and incubated under a 12-h light, 12-h dark cycle; light (130 to 150 μmol of photons m−2 s−1) was provided by cool white fluorescent illumination at 20°C. Because HcRNAV isolates have an ecotype-specific host range (32), HcRNAV34 and HcRNAV109 were maintained by using the suitable hosts HU9433-P and HCLG-1, respectively. The virus stock was inoculated into a fresh culture of H. circularisquama and incubated until host cell lysis was observed. The lysate was filtered through a 0.2-μm-pore-size polycarbonate membrane filter (Whatman, Middlesex, United Kingdom) to remove host cell debris. The viral titer of the filtrate was estimated by the extinction dilution method as described previously (32).
Virus inoculation.
An exponentially growing culture of H. circularisquama (25 ml) was inoculated with virus lysate at a multiplicity of infection of 10 and incubated. An aliquot of cell suspension (1.5 ml) was taken from the culture immediately after inoculation (0 h) and at 24, 48, and 72 h postinoculation (hpi). Host cells were centrifuged (13,000 × g for 3 min), and the pellets were stored at −80°C for RNA extraction.
Particle bombardment.
Viral RNA transfection of H. circularisquama cells was performed with the Bio-Rad Biolistic PDS-1000/He Particle Delivery System (Bio-Rad Laboratories, Hercules, CA). Gold particles (0.6 μm; Bio-Rad Laboratories) were coated with viral RNA as described below. HcRNAV virions were purified as described previously (32). Viral RNA was isolated and purified with an RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The RNA concentration was determined spectrophotometrically, and its integrity was verified by 1.5% denaturing agarose gel electrophoresis. Viral RNA (1.5 μg) was precipitated onto the gold particles (1.5 mg) by adding 1/10 volume of 5 M ammonium acetate and 2 volumes of 2-propanol. The suspension was gently mixed and chilled at −20°C for 1 h. After a quick centrifugation (2,000 × g for 3 s), the supernatant was discarded and the gold pellet was washed twice with 140 μl of 99.5% ethanol and resuspended in 24 μl of 99.5% ethanol. Six microliters was fixed onto a macrocarrier and dried.
Exponentially growing H. circularisquama cells (2.5 × 106) were collected for particle bombardment on no. 3 quantitative filter paper (47 mm in diameter; Advantec, Tokyo, Japan) by gravity filtration. The filter paper was set approximately 6 cm below the microcarrier launch assembly and bombarded with the viral RNA-coated gold particles at a rupture pressure of 1,350 lb/in2 in a vacuum of 28.5 in. Hg. After bombardment, the filter paper was placed in fresh SWM3 medium (50 ml) and gently shaken to release transfected cells. The culture was incubated as described above. A 1.5-ml aliquot was sampled from the cell suspension immediately after transfection (0 h) and at 24 and 48 h posttransfection. Cells were pelleted by centrifugation (13,000 × g for 3 min) and stored at −80°C prior to RNA extraction. The remaining 40 ml of culture was sampled at 48 h posttransfection and centrifuged at 860 × g for 10 min. The supernatant was filtered through a 0.2 μm-pore-size polycarbonate membrane filter (Whatman) and used for the cell lysis assay (see below). The remaining cell pellet was fixed with 1% glutaraldehyde at 4°C and processed for transmission electron microscopy (TEM). Preparation for TEM was performed as described previously (31, 32).
Northern blot analysis.
Frozen cells were homogenized with plastic pestles in 100 μl of RNase-free water and 40 μl of RNA extraction buffer (14). The aqueous phase was extracted with phenol prior to ethanol precipitation. Northern blot analysis of the purified RNA was performed as previously described (7). Strand-specific digoxigenin (DIG)-labeled RNA probes were transcribed from plasmid pBSSK+MCP. The plasmid was constructed as follows. A cDNA fragment of HcRNAV34 from nucleotides 3182 to 4261 (containing the full sequence of ORF2) was amplified by reverse transcription-PCR with primers 34MCPBam1 (5′-CGG GAT CCA TGA CCC GTC CCC TAG CTC TTA CC-3′; the BamHI site is underlined) and 34MCPEco1 (5′-CGG AAT TCT TAA GCA GCC ATC AAT GCT GGC ATA GC-3′; the EcoRI site is underlined). The amplified DNA fragment was digested with BamHI and EcoRI and ligated into the corresponding restriction enzyme sites of pBluescript II SK+ (Stratagene, La Jolla, CA). DIG-labeled RNA probes specific for the positive and negative strands of the HcRNAV genome were transcribed from BamHI-linearized pBSSK+MCP with T7 RNA polymerase and from EcoRI-linearized pBSSK+MCP with T3 RNA polymerase according to the manufacturer's (Roche, Basel, Switzerland) protocols. The RNA signals were detected with a luminescence image analyzer (LAS-3000 mini; Fuji Photo Film, Tokyo, Japan).
Cell lysis assay.
The cell lysis assay was performed as described previously (32). Briefly, an aliquot (100 μl) of the 0.2-μm-filtered lysate from the viral RNA-transfected H. circularisquama culture was added to 8 wells of a 96-well round-bottom cell culture plate (Becton Dickinson Labware, Franklin Lakes, NJ) containing 150 μl of an exponentially growing host culture (HU9433-P or HCLG-1) and incubated. Cell lysis was monitored for 10 to 14 days by optical microscopy. Cell lysis was defined as lysis of >90% of the host cells.
RESULTS
Viral replication in host cells inoculated with HcRNAV.
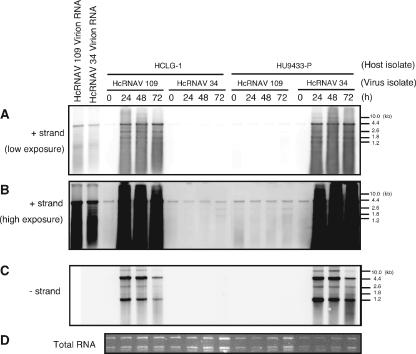
We used HCLG-1 and HU9433-P as representative host strains for HcRNAV109 (type CY) and HcRNAV34 (type UA), respectively (30). To determine whether viral genomic RNA can replicate in incompatible host-virus combinations, the accumulation of viral RNA was examined by Northern blot analysis with a positive-strand-specific RNA probe (Fig. 2A). In HCLG-1 and HU9433-P cultures inoculated with HcRNAV109 and HcRNAV34, respectively (the compatible combinations), accumulation of viral genomic RNA (4.4 kb) was detected at 24, 48, and 72 hpi (Fig. 2A). In contrast, in the HU9433-P culture inoculated with HcRNAV109 and in the HCLG-1 culture inoculated with HcRNAV34 (the incompatible combinations), no positive-strand RNA accumulation was detected (Fig. 2A). When the membrane was exposed for a longer time, a faint band of viral genomic RNA (4.4 kb) was detected in the lanes containing incompatible host-virus combinations. However, faint bands were also visible immediately after inoculation with either compatible or incompatible host-virus combinations (Fig. 2B). To determine if these faint signals were due to the initial virus inoculum, we analyzed the accumulation of the complementary negative-strand RNA that is specifically synthesized during the replication process of the positive-strand RNA virus (2) with the negative-strand-specific RNA probe (Fig. 2C). Genomic-length negative-strand RNAs (4.4 kb) were observed only in the compatible combinations but not in the incompatible combinations with intensive exposure. This suggests that the viral RNA was not replicated in incompatible host-virus combinations where viruses were inoculated by simply adding a virus suspension to the host culture.
FIG. 2.
Accumulation of positive (A and B)- and negative (C)-strand RNA of HcRNAV in H. circularisquama cells inoculated with HcRNAV34 and HcRNAV109 virions. Total RNA was extracted from H. circularisquama cells immediately after inoculation (0 h) and at 24, 48, and 72 hpi; separated by gel electrophoresis; and blotted onto membranes. The membranes were then probed with strand-specific, DIG-labeled RNA probes. The same amount of genomic RNA of HcRNAV34 or HcRNAV109 was used as a positive control. RNA length is indicated on the right. Total RNA stained with SYBR gold is shown in panel D.
Four additional RNA bands (10 kb, 2.6 kb, 1.8 kb, and 1.2 kb) other than the HcRNAV genomic RNA (4.4 kb) were observed in a Northern blot analysis (Fig. 2A and C).
Viral RNA replication in host cells transfected with HcRNAV genomic RNA.
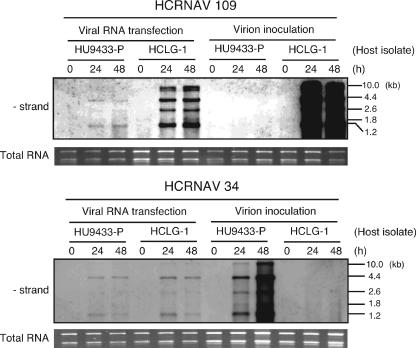
To determine whether incompatible H. circularisquama cells are permissive for HcRNAV replication, direct transfection of viral RNA into H. circularisquama cells was performed by particle bombardment (Fig. 3). For a negative control, we added viral RNA-coated gold particles and detected no RNA signal corresponding to the complementary strand of HcRNAV genomic RNA in the H. circularisquama cells (data not shown). This indicates that the purified viral RNA was not contaminated with infectious virions. In HU9433-P and HCLG-1 transfected, respectively, with purified genomic RNAs of HcRNAV34 and HcRNAV109 (the compatible combinations), RNA corresponding to the complementary strand of genomic RNA (4.4 kb) was observed (Fig. 3). We also observed the complementary strand of genomic RNA (4.4 kb) in incompatible host-virus combinations (i.e., HU9433-P transfected with HcRNAV109 RNA and HCLG-1 transfected with HcRNAV34 RNA) where host cells were transfected with viral RNA by particle bombardment (Fig. 3). This shows that H. circularisquama cells are permissive for HcRNAV replication in incompatible host-virus combinations, as well as in compatible combinations. Hence, it is therefore unlikely that the intraspecies host specificity of HcRNAV is determined during the viral RNA replication process. However, it should be noted that the RNA accumulation levels were different among host-virus combinations. Further quantitative analysis is required to determine whether the efficiency of viral replication differs among host-virus combinations.
FIG. 3.
Accumulation of negative-strand HcRNAV RNA in H. circularisquama cells transfected with HcRNAV34 or HcRNAV109 RNA. Total RNA was extracted from H. circularisquama cells immediately after inoculation (0 h) and 24 h and 48 hpi, separated by gel electrophoresis, and blotted onto membranes. The membranes were then probed with a negative-strand-specific, DIG-labeled RNA probe. RNA length is indicated on the right. Total RNA stained with SYBR gold is shown in panel D.
Properties of virions produced in incompatible host cells.
To determine whether progeny virions were formed in H. circularisquama cells that were directly transfected with purified HcRNAV RNA, intracellular accumulation of HcRNAV-like particles was tested for by TEM. Although no HcRNAV-like particle was observed in healthy cells (data not shown), crystalline arrays or unordered aggregations of HcRNAV-like particles were observed with both compatible (i.e., HU9433-P transfected with HcRNAV34 RNA and HCLG-1 transfected with HcRNAV109 RNA) and incompatible (i.e., HU9433-P transfected with HcRNAV109 RNA and HCLG-1 transfected with HcRNAV34 RNA) host-virus combinations (Fig. 4).
FIG. 4.
Transmission electron micrographs of H. circularisquama cells at 48 h posttransfection with HcRNAV34 or HcRNAV109 RNA. Note the propagation of HcRNAV-like particles forming crystalline arrays and/or unordered aggregation in the cytoplasm. Bars, 200 nm.
The 0.2-μm filtrate of cell cultures transfected with HcRNAV34 genomic RNA caused lysis of HU9433-P cells but not HCLG-1 cells (Table 1). Similarly, the 0.2-μm filtrate of cell cultures transfected with HcRNAV109 genomic RNA caused lysis of HCLG-1 cells but not HU9433-P cells (Table 1). This suggests that the progeny viruses produced in transfected H. circularisquama cells retained the original infection specificity encoded by the imported viral RNA.
TABLE 1.
Lytic activity of 0.2-μm filtrate of viral RNA-transfected H. circularisquama cells
| Combination of viral RNA and transfected host clone | Lytic activitya
|
|
|---|---|---|
| HU9433-P | HCLG-1 | |
| HcRNAV34 RNA → HU9433-P | + | − |
| HcRNAV34 RNA → HCLG-1 | + | − |
| HcRNAV109 RNA → HU9433-P | − | + |
| HcRNAV109 RNA → HCLG-1 | − | + |
+, Cell lysis occurred in all eight wells of a culture plate; −, no effect.
DISCUSSION
Infection steps determining intraspecies host specificity.
When HcRNAV virions were simply added to H. circularisquama cultures, viral RNA replication, propagation of progeny virions, and host cell lysis occurred with compatible host-virus combinations but not with incompatible combinations (Fig. 2) (32). When viral RNA was transfected into H. circularisquama cells by particle bombardment, viral RNA replication occurred with both compatible and incompatible host-virus combinations (Fig. 3). In addition, infectious progeny virus was produced in both incompatible host-virus combinations (Fig. 4 and Table 1). This indicates that the intraspecies host specificity of HcRNAV is not determined by viral RNA replication and the encapsidation process. Instead, it is determined by upstream events, most probably the specific binding of virus to the host cell surface receptor, entry into host cells, and disassembly of viral particles.
The upstream events of nonenveloped RNA viruses are not sufficiently understood, except for poliovirus and its relatives (8, 27). Poliovirus infection is initiated when virus particles attach to specific cell surface receptors (17). Virus-receptor interaction induces an irreversible conformational change in the virus capsid (33, 34) and produces a particle with altered sedimentation, antigenicity, and sensitivity to proteases. This metastable particle further undergoes a secondary conformational change in which the viral RNA is ejected into the cytoplasm (9). However, the factors triggering RNA release are unknown. This receptor-induced conformational change in the viral particle is also proposed for Flock house virus (genus Nodaviridae) (35). Our results suggest that specific binding of a virus particle to its corresponding host cell surface receptor may be the primary determinant of the intraspecies host specificity of HcRNAV.
Possible molecular mechanism for intraspecies host specificity of HcRNAV.
Two remarkable differences were observed between the HcRNAV34 and HcRNAV109 genomes at the nucleotide sequence level (20). One is the 15-nucleotide deletion in HcRNAV34 ORF1 (Fig. 1). However, no relationship between this deletion and intraspecies host specificity is predicted (Y. Tomaru, unpublished data). The other difference is the high-frequency nucleotide substitutions in ORF2 (variable regions I to IV) that cause amino acid substitutions in the CP (Fig. 1). A tertiary structure prediction of the CP shows that most of the substituted amino acid residues are located on the exterior surface of the virion (20). Therefore, the amino acid substitutions in the CP likely alter the host specificity of HcRNAV.
Similar phenomena have also been observed in other virus studies. For example, canine parvovirus is a host range variant of feline parvovirus that acquired the ability to infect dogs through changes in surface amino acid residues in its capsid protein, VP2 (3, 22). Furthermore, a single amino acid substitution in VP2 altered its host cell specificity (21) and its ability to bind the transferrin receptor specifically (10). With influenza virus, human viruses preferentially bind to cell surface oligosaccharides that contain the 5-N-acetylneuraminic acid-α-2,6-galactose (Neu5Acα2,6Gal) linkage, while avian and equine viruses bind to Neu5Acα2,3Gal (24). Ito et al. (12) found that if human influenza virus was passed in allantoic cavities of embryonated chicken eggs containing Neu5Acα2,3Gal but not Neu5Acα2,6Gal, progeny virions acquired Neu5Acα2,3Gal specificity. This change in receptor specificity was accompanied by amino acid substitutions in the receptor-binding site of the hemagglutinin molecule (12).
What is the virus receptor of H. circularisquama? In many cases, the external portions of membrane proteins have a complex of branched carbohydrate chains and such membrane glycoproteins are frequently used as viral receptors (26). Costas and Rodas (5) reported that the binding spectra of fluorescence-labeled lectins to marine dinoflagellates varied not only at the species level (i.e., between toxic Gymnodinium catenatum and nontoxic Gymnodinium sp.) but also at the clone level (e.g., Alexandrium minutum and A. excavatum). We speculate that the cell surface glycoprotein composition of H. circularisquama may vary between ecotypes, and this difference is implicated in the intraspecies host specificity of HcRNAV.
Characteristics of HcRNAV replication.
In this study, we analyzed the accumulation of negative-strand viral RNA as a marker of viral replication. In addition to the genomic-length RNA band (4.4 kb), an intense RNA band (1.2 kb) and three weak RNA bands (10 kb, 2.6 kb, and 1.8 kb) were also observed (Fig. 2C and 3), and correspondingly sized RNA species positively reacted with a positive-strand-specific probe (Fig. 2A). On the basis of the nucleotide sequence, the expected size of the ORF2 subgenomic RNA is >1.2 kb. In addition, we have preliminary data showing that the positive-strand 1.2-kb band was detected by an RNA probe containing the ORF2 sequence but not by a probe containing the ORF1 sequence (H. Mizumoto and K. Nagasaki, unpublished data). It is possible that the 1.2-kb-long RNA corresponds to subgenomic RNA for ORF2.
Transcription of the viral subgenomic RNAs is a strategy frequently used by eukaryotic positive-strand RNA viruses. Subgenomic RNA mediates the expression of the 3′-proximal ORF on multicistronic genomic RNA because only the first ORF on the eukaryotic mRNA is translated. Three general mechanisms of subgenomic RNA synthesis are proposed (18). In one of these mechanisms, premature termination occurs during negative-strand synthesis from the genomic RNA template. This results in the synthesis of a subgenomic-length negative-strand RNA (36), as observed with HcRNAV (Fig. 2C). In contrast to the accumulation of negative-strand RNAs (Fig. 2C), intense positive-strand RNA signals, except for genomic RNA, were not observed (Fig. 2A). It may be that subgenomic RNAs corresponding to the accumulated negative-strand RNAs were synthesized only at earlier stages during replication, as observed with Flock house virus (37). Further analysis will shed light on the replication and gene expression mechanisms of HcRNAV.
When viral RNA was transfected into H. circularisquama cells by particle bombardment, propagation of progeny virions occurred in both compatible and incompatible host-virus combinations (Fig. 3 and 4 and Table 1). This indicates that all of the cellular factors essential for HcRNAV replication, transcription, translation, and encapsidation are present in both H. circularisquama isolates. However, it should be noted that the RNA accumulation levels differed among host-virus combinations (Fig. 3). The positive-strand-specific RNA probe used here was synthesized with HcRNAV34 genomic RNA as a template, and it hybridized with similar intensities to both of the genomic RNAs of HcRNAV34 and HcRNAV109 (Fig. 2). This suggests that the cells permitting virus production may have different levels of permissiveness; i.e., some H. circularisquama isolates may produce large amounts of virus while others may permit only a low level of viral replication. Additionally, with Heterocapsa triquetra cells transfected with the HcRNAV RNA, accumulation of viral negative-stranded RNAs was not detected (H. Mizumoto, Y. Tomaru, and K. Nagasaki, unpublished data). This suggests that the intracellular condition of H. triquetra is not suitable for replication of HcRNAV. Therefore, the mechanisms determining the host specificity of dinoflagellate RNA viruses are considered to be different at the intraspecies and interspecies levels.
Acknowledgments
We are grateful to J. Lawrence (University of New Brunswick, Canada) for critical reading of this paper and to T. Okuno, K. Mise (Kyoto University, Japan), M. Yamaguchi, S. Sakamoto, S. Nagai, and K. Tarutani (National Research Institute of Fisheries and Environment of Inland Sea, Japan) for helpful advice. We also thank I. Imai (Kyoto University, Japan) for providing the HU9433-P culture.
This work was partially supported by funding from the Fisheries Agency of Japan; the Ministry of Agriculture, Forestry and Fisheries of Japan; and the Ministry of Education, Science, Sports and Culture (Grant-in-Aid for JSPS Fellows).
Footnotes
Published ahead of print on 15 November 2006.
REFERENCES
- 1.Brussaard, C. P., A. A. Noordeloos, R. A. Sandaa, M. Heldal, and G. Bratbak. 2004. Discovery of a dsRNA virus infecting the marine photosynthetic protist Micromonas pusilla. Virology 319:280-291. [DOI] [PubMed] [Google Scholar]
- 2.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, S. F., J. Y. Sgro, and C. R. Parrish. 1992. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 66:6858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, L. C. M., T. Edelstein, and J. McLachlan. 1969. Bonnemaisonia hamifera Hariot in nature and in culture. J. Phycol. 5:211-220. [DOI] [PubMed] [Google Scholar]
- 5.Costas, E., and V. L. Rodas. 1994. Identification of marine dinoflagellates using fluorescent lectins. J. Phycol. 30:987-990. [Google Scholar]
- 6.Culley, A. I., A. S. Lang, and C. A. Suttle. 2006. Metagenomic analysis of coastal RNA virus communities. Science 312:1795-1798. [DOI] [PubMed] [Google Scholar]
- 7.Damayanti, T. A., H. Nagano, K. Mise, I. Furusawa, and T. Okuno. 2002. Positional effect of deletions on viability, especially on encapsidation, of brome mosaic virus D-RNA in barley protoplasts. Virology 293:314-319. [DOI] [PubMed] [Google Scholar]
- 8.Hogle, J. M. 2002. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 56:677-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, Y., J. M. Hogle, and M. Chow. 2000. Is the 135S poliovirus particle an intermediate during cell entry? J. Virol. 74:8757-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hueffer, K., J. S. Parker, W. S. Weichert, R. E. Geisel, J. Y. Sgro, and C. R. Parrish. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 77:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai, I., S. Itakura, Y. Matsuyama, and M. Yamaguchi. 1996. Selenium requirement for growth of a novel red tide flagellate Chattonella verruculosa (Raphidophyceae) in culture. Fish. Sci. 62:834-835. [Google Scholar]
- 12.Ito, T., Y. Suzuki, A. Takada, A. Kawamoto, K. Otsuki, H. Masuda, M. Yamada, T. Suzuki, H. Kida, and Y. Kawaoka. 1997. Differences in sialic acid-galactose linkages in the chicken egg amnion and allantois influence human influenza virus receptor specificity and variant selection. J. Virol. 71:3357-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh, K., and I. Imai. 1987. Raphidophyceae, p. 122-130. In Japan Fisheries Resource Conservation Association (ed.), A guide for studies of red tide organisms. Shuwa, Tokyo, Japan.
- 14.Kroner, P., D. Richards, P. Traynor, and P. Ahlquist. 1989. Defined mutations in a small region of the brome mosaic virus 2a gene cause diverse temperature-sensitive RNA replication phenotypes. J. Virol. 63:5302-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang, A. S., A. I. Culley, and C. A. Suttle. 2004. Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo. Virology 320:206-217. [DOI] [PubMed] [Google Scholar]
- 16.McFadden, G. 2005. Poxvirus tropism. Nat. Rev. Microbiol. 3:201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendelsohn, C. L., E. Wimmer, and V. R. Racaniello. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855-865. [DOI] [PubMed] [Google Scholar]
- 18.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Mizumoto, H., Y. Hikichi, and T. Okuno. 2002. The 3′-untranslated region of RNA1 as a primary determinant of temperature sensitivity of red clover necrotic mosaic virus Canadian strain. Virology 293:320-327. [DOI] [PubMed] [Google Scholar]
- 20.Nagasaki, K., Y. Shirai, Y. Takao, H. Mizumoto, K. Nishida, and Y. Tomaru. 2005. Comparison of genome sequences of single-stranded RNA viruses infecting the bivalve-killing dinoflagellate Heterocapsa circularisquama. Appl. Environ. Microbiol. 71:8888-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker, J. S., and C. R. Parrish. 1997. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J. Virol. 71:9214-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parrish, C. R., C. F. Aquadro, M. L. Strassheim, J. F. Evermann, J. Y. Sgro, and H. O. Mohammed. 1991. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 65:6544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren, R. B., F. Costantini, E. J. Gorgacz, J. J. Lee, and V. R. Racaniello. 1990. Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell 63:353-362. [DOI] [PubMed] [Google Scholar]
- 24.Rogers, G. N., and J. C. Paulson. 1983. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127:361-373. [DOI] [PubMed] [Google Scholar]
- 25.Shirai, Y., Y. Takao, H. Mizumoto, Y. Tomaru, D. Honda, and K. Nagasaki. 2006. Genomic and phylogenetic analysis of a single-stranded RNA virus infecting Rhizosolenia setigera (Stramenopiles: Bacillariophyceae). J. Mar. Biol. Assoc. U. K. 86:475-483. [Google Scholar]
- 26.Smith, A. E., and A. Helenius. 2004. How viruses enter animal cells. Science 304:237-242. [DOI] [PubMed] [Google Scholar]
- 27.Smyth, M. S., and J. H. Martin. 2002. Picornavirus uncoating. Mol. Pathol. 55:214-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suttle, C. A. 2000. Viral infection of cyanobacteria and eukaryotic algae, p. 248-286. In C. J. Hurst (ed.), Viral ecology. Academic Press, San Diego, CA.
- 29.Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 30.Tai, V., J. E. Lawrence, A. S. Lang, A. M. Chan, A. I. Culley, and C. A. Suttle. 2003. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J. Phycol. 39:343-352. [Google Scholar]
- 31.Tarutani, K., K. Nagasaki, S. Itakura, and M. Yamaguchi. 2001. Isolation of a virus infecting the novel shellfish-killing dinoflagellate Heterocapsa circularisquama. Aquat. Microb. Ecol. 23:103-111. [Google Scholar]
- 32.Tomaru, Y., N. Katanozaka, K. Nishida, Y. Shirai, K. Tarutani, M. Yamaguchi, and K. Nagasaki. 2004. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat. Microb. Ecol. 34:207-218. [Google Scholar]
- 33.Tsang, S. K., B. M. McDermott, V. R. Racaniello, and J. M. Hogle. 2001. Kinetic analysis of the effect of poliovirus receptor on viral uncoating: the receptor as a catalyst. J. Virol. 75:4984-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuthill, T. J., D. Bubeck, D. J. Rowlands, and J. M. Hogle. 2006. Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles. J. Virol. 80:172-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walukiewicz, H. E., J. E. Johnson, and A. Schneemann. 2006. Morphological changes in the T=3 capsid of Flock House virus during cell entry. J. Virol. 80:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White, K. A. 2002. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology 304:147-154. [DOI] [PubMed] [Google Scholar]
- 37.Zhong, W., and R. R. Rueckert. 1993. Flock house virus: down-regulation of subgenomic RNA3 synthesis does not involve coat protein and is targeted to synthesis of its positive strand. J. Virol. 67:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]