Abstract
The oncogenic herpesvirus, Kaposi's sarcoma-associated herpesvirus, also identified as human herpesvirus 8, contains genes producing proteins that control transcription and influence cell signaling. Open reading frame 36 (ORF36) of this virus encodes a serine/threonine protein kinase, which is designated the viral protein kinase (vPK). Our recent efforts to elucidate the role of vPK in the viral life cycle have focused on identifying viral protein substrates and determining the effects of vPK-mediated phosphorylation on specific steps in viral replication. The vPK gene was transcribed into 4.2-kb and 3.6-kb mRNAs during the early and late phases of viral reactivation. vPK is colocalized with viral DNA replication/transcription compartments as marked by a polymerase processivity factor, and K-bZIP, a protein known to bind the viral DNA replication origin (Ori-Lyt) and to regulate viral transcription. The vPK physically associated with and strongly phosphorylated K-bZIP at threonine 111, a site also recognized by the cyclin-dependent kinase Cdk2. Both K-bZIP and vPK were corecruited to viral promoters targeted by K-bZIP as well as to the Ori-Lyt region. Phosphorylation of K-bZIP by vPK had a negative impact on K-bZIP transcription repression activity. The extent of posttranslational modification of K-bZIP by sumoylation, a process that influences its repression function, was decreased by vPK phosphorylation at threonine 111. Our data thus identify a new role of vPK as a modulator of viral transcription.
Kaposi's sarcoma-associated herpesvirus (KSHV), also designated human herpesvirus 8, has been linked to several malignancies, including Kaposi's sarcoma, B-cell lymphomas, primary effusion lymphomas, and multicentric Castleman's disease in immunocompromised individuals (reviewed in reference 46). Kaposi's sarcoma is the most common malignancy associated with AIDS (45). The viral genome is double-stranded DNA, approximately 165 kbp, and encodes over 81 open reading frames (ORFs) (43). The majority of the ORFs are essential for viral replication and include genes necessary for viral DNA replication, transcription, and assembly of infectious particles (51). In addition, the KSHV genome contains a large number of ORFs with homology to known cellular genes. Several of these viral ORFs are implicated in modulating host immune responses, promoting angiogenesis, and dysregulating cell growth (14). A model for regulated expression of KSHV genes has been deduced from numerous genetic and biochemical studies of viral RNA patterns and activities of viral transactivators (22, 31, 39, 49). As for other herpesviruses, KSHV genes in productive infection have been classified into the following temporally distinct classes: immediate-early, early, and late. This virus can also establish a latent state that is characterized by presence of a multicopy circular episome of viral DNA, which expresses a small subset of viral proteins. Productive (lytic) viral replication can be induced by treatment of latently infected cell lines with butyrate, phorbol esters, or hypoxia (13). After induction, the transcriptional transactivator K-Rta (ORF50) is activated, and this protein then induces the K-bZIP (also called K8 or RAP) transcriptional regulator and ORF57 (posttranscriptional transactivator). These latter viral regulators activate other early and late phase genes, and thus ensues the complete viral life cycle.
Alpha-, beta-, and gamma-herpesviruses encode phosphotransferases that phosphorylate proteins and nucleosides (reviewed in reference 24). One group of viral protein kinases is conserved among all alpha-, beta-, and gamma-herpesviruses. The conserved protein kinases are UL13 of herpes simplex virus (HSV), ORF47 of varicella-zoster virus, UL97 of cytomegalovirus (CMV), BGLF4 of Epstein-Barr virus (EBV), and ORF36 of KSHV. Located within the catalytic region of these kinases are 11 conserved subdomains that are common to cellular serine/threonine protein kinases. Experimental studies on members of each of the three major groups of herpesviruses have identified a wide array of viral and cellular protein substrates of the conserved viral protein kinases. Structural analysis of purified virions of several herpesviruses, including HSV, CMV, and EBV, indicate that these protein kinases are associated with virus particles (2, 12, 37, 50, 52); thus, the kinases are in a position to influence virion assembly and events that take place after entry of the virion into the cell. A number of studies using kinase-null viral mutants demonstrated the importance of the kinase for regulating viral gene expression, replication, tissue tropism, or infection in animal models (7, 11, 26, 35, 41, 48). Various studies have implicated the viral protein kinase in influencing viral gene expression (6, 58), viral DNA replication (27, 52, 53), or nucleocapsid egress from the nucleus during virus assembly (26, 34, 53). The importance of the viral protein kinase (vPK) for HSV and CMV replication is supported by studies showing that changes in this gene can confer resistance to certain antiviral agents (e.g., ganciclovir) (7, 26). Taken together, these studies demonstrate that the conserved herpesvirus protein kinases impact multiple steps in viral replication.
Orf-36 protein (hereafter designated vPK) of KSHV is a serine protein kinase, which is localized in the nucleus (38). In vitro protein kinase assays indicated that this viral protein was autophosphorylated and that the lysine residue in the catalytic kinase subdomain II was essential for enzymatic activity (38). Previous analysis on levels of KSHV transcripts in productive infection indicated that vPK RNA was accumulated in the late phase of viral replication (22, 39). However, a recent study detected vPK RNA at early time points and in the presence of an inhibitor of viral DNA synthesis (31). Detailed analysis of KSHV RNA revealed two polycistronic transcripts encoding vPK that are initiated from promoters that are active in the early stage of the viral life cycle and inducible by hypoxic conditions (18). With respect to potential therapeutic significance, vPK was shown to phosphorylate the antiherpesvirus drug ganciclovir (10). In a recent study that explored a potential role for vPK on cell signaling, we established that vPK phosphorylated components of the Jun N-terminal protein kinase signal transduction pathway, which in turn activated c-Jun in the activating protein 1 transcription complex (17).
We have continued to study the vPK of KSHV by focusing on viral targets of this protein kinase. The main finding is that vPK interacts with the transcriptional regulator K-bZIP in productively infected cells. We demonstrated that a specific residue of K-bZIP, threonine 111, was phosphorylated by vPK and that this phosphorylation modulated the transcription function of K-bZIP. Interestingly, this threonine residue of K-bZIP is also the target of the cyclin-dependent kinase Cdk2. Chromatin immunoprecipitation experiments showed that vPK and K-bZIP were recruited to selected viral promoters as well as the Ori-Lyt region. High-resolution microscopy revealed that vPK was localized to replication/transcription complexes in infected cells. Interestingly, vPK is packaged in mature virions; this finding raises the possibility that vPK may influence events at viral entry into host cells, including at a very early stage of viral gene expression. We discuss a model whereby vPK exerts pleiotropic effects by modulating several steps in the lytic replication cycle of KSHV, in part by interacting with and influencing K-bZIP function on viral gene expression.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney epithelial 293 cells and 293T cells were grown in monolayer culture in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS) in the presence of 5% CO2. The TREx-K-Rta BCBL-1 cell line, generated by Nakamura et al. (36), was cultured in RPMI 1640 medium supplemented with 15% FBS, 100 μg/ml of blasticidin (Invitrogen), and 100 μg/ml of hygromycin (Invitrogen).
Plasmids.
Plasmids encoding the full-length K-bZIP and K-Rta genes were described previously (20). This cloning introduced a CpoI site and a Flag tag or T7 tag at the N terminus of each protein as described previously (20). The resulting plasmids were designated pFlag-K-bZIP or pT7-K-bZIP. Plasmids encoding the full-length vPK wild type and kinase-dead mutant vPK-K108Q were described previously (17). Deletion fragments of K-bZIP were amplified by PCR using Pfu Turbo (Stratagene) and cloned into the CpoI site of pGEX-modified vector (21) using cDNA encoding K-bZIP wild type or phosphor-mutant K-bZIP as a template. Newly synthesized primers for preparing deletion DNA fragments are listed in Table 1. Plasmids encoding phosphor-mutant K-bZIP-T111D and K-bZIP-T111A were prepared by site-directed mutagenesis (Strategene). Full-length wild-type vPK, kinase-dead mutant vPK-K108Q, ORF6, ORF9, ORF44, ORF56, and ORF59 were amplified by PCR and cloned into pVL1392 vector (Invitrogen), which introduced a Flag tag and CpoI site (pVL-Flag). Full-length primase-associated factor (ORF40 to ORF41) (5) was amplified from cDNA of tetradecanoyl phorbol acetate (TPA)-treated BCBL-1 and cloned into the CpoI site of pVL-Flag vector. The CpoI DNA fragments of K-Rta and K-bZIP were also cloned in the pVL-Flag vector. Primers used in this study are listed in Table 1. Cloned DNA sequences were confirmed by sequencing.
TABLE 1.
Primers prepared in this study
| Primer | Sequence (5′→3′)a |
|---|---|
| ORF6 CpoI-F | aaaCGGTCCGACCATGGCGCTAAAGGGACCA |
| ORF6 CpoI-R | aaaCGGACCGCTACAAATCCAGGTCAGAGAG |
| ORF9 CpoI-F | aaaCGGTCCGATCATGGATTTT TCAATCCA |
| ORF9 CpoI-R | aaaCGGACCGCTAGGGCGTGGGAAAAGTCAC |
| ORF40 CpoI-F | aaaCGGTCCGTCAATGGCAACGAGCGAAGAA |
| ORF41 CpoI-R | aaaCGGACCGTCAAAATAAAGATAAAAGCCT |
| ORF44 CpoI-F | aaaCGGTCCGACCATGGACAGCTCGGAAGGGTGC |
| ORF44 CpoI-R | aaaCGGACCGTCAGTAGATCAGAGTAGTCTT |
| ORF56 CpoI-F | aaaCGGTCCGACCATGGAGACGACATACCGC |
| ORF56 CpoI-R | aaaCGGACCGTTAACTGGCCAGTCCCACTGG |
| ORF59 CpoI-F | aaaCGGTCCGATATGCCTGTGGATTTTCAC |
| ORF59 CpoI-R | aaaCGGACCGCACATGGTGTCAAATCAGG |
| vPK CpoI-F | aaaCGGTCCGATGCGCTGGAAGAGAATGGAG |
| vPK CpoI-R | aaaCGGACCGTCAGAAAACAAGTCCGCGGGT |
| K-bZIP aa75 CpoI-R | aaaCGGACCGAAGGTCAATGACCGTTTCACA |
| K-bZIP aa76 CpoI-F | aaaCGGTCCGACAGCGCCTTCCCAAAGTGGC |
| K-bZIP aa150 CpoI-R | aaaCGGACCGGCGACCTGCGCCCTGTTTGGC |
| K-bZIP aa151 CpoI-F | aaaCGGTCCGCCCGTGCCTGCGTCTGTAGTT |
| K-bZIP aa72 CpoI-F | aaaCGGTCCGGTCATTGACCTTACAGCGCCTTC |
| K-bZIP aa87 CpoI-F | aaaCGGTCCGGAACATCTGCCATGCTCACTGAA |
| K-bZIP aa113 CpoI-F | aaaCGGTCCGCCAAGAGGACCACACATTTCGCA |
| K-bZIP aa128 CpoI-F | aaaCGGTCCGAAGAGGCGACTACATAGAAAGTT |
| K-bZIP aa102 CpoI-R | aaaCGGACCGTTAGGGGATGTGGAATTTAGTTT |
| K-bZIP aa123 CpoI-R | aaaCGGACCGTTATGGAAGCTGTTGCGAAATGT |
| K-bZIP aa154 CpoI-R | aaaCGGACCGTTACGCAGGCACGGGGCGACCTG |
| K-bZIP T111A-F | TCTCACGCACCACCAAGAGGACCACACATT |
| K-bZIP T111A-R | TGGTGGTGCGTGAGAGAGCGTCCAGGATC |
| K-bZIP T111D-F | CTCTCTCACGACCCACCAAGAGGACCACAC |
| K-bZIP T111D-R | TCTTGGTGGGTCGTGAGAGAGCGTCCAGGA |
| ORF34 probe-F | aaaGAATTCGTGGGCTTTGACTCTGAATAAG |
| ORF34 probe-R | aaaGGATCCGCCGCCCATGTCCCGGAAAAA |
| ORF35 probe-F | aaaGAATTCGGCTCTGGAGGCCAACATCAA |
| ORF35 probe-R | aaaGGATCCCTTTTCAGCGCCTCAAACCTCT |
| ORF36 probe-F | AGCCTTGCATACCCTGTAGATCGA |
| ORF36 probe-R | GGATAGGTAACACTGGAAGAGGAC |
| ORF37 probe-F | GTACACCTTTGCGAAAATGGAGTG |
| ORF37 probe-R | AGTAGCTGTGACGGACGTTCACGA |
| ORF38 probe-F | ATGGGATTTCTCCTATCTATCTGCAA |
| ORF38 probe-R | CTTTTTTAATGCGGGATATGGGGATTG |
Boldfaced nucleotides represent restriction enzyme sites used for cloning the PCR products; underlining indicates an initiation codon, stop codon, or mutated codon; lowercase letters indicate nucleotides added to enhance recognition by the restriction enzyme.
Purification of KSHV virions.
Purification of KSHV virions was performed essentially with the method previously described by Zhu et al. (59). One litter of TREx-K-Rta BCBL-1 was cultured in a roller bottle with 15% FBS. For virus production, KSHV reactivation was induced by adding a final concentration of 1 μg/ml of doxycycline (DOX). After a 96-h induction, the medium was collected and cleared by centrifugation at 4,000 × g for 30 min and then at 8,000 × g for 15 min to remove cells and cell debris. Virions were pelleted at 27,000 rpm for 1 h through a 5% sucrose cushion (5 ml) in a Beckman SW28 rotor and resuspended in 1× phosphate-buffered saline (PBS) in 1/100 of the original volume. The concentrated virus particles were centrifuged through a 10 to 50% sucrose gradient at 27,000 rpm for 2 h in a Beckman SW55Ti rotor. The virus band at the gradient junction was collected. This virus band was diluted with 1× PBS and pelleted at 27,000 rpm for 1 h. Pellets were resuspended in 1× PBS and again purified through a 10 to 50% continuous sucrose gradient at 27,000 rpm overnight with a Beckman SW55Ti rotor. Virions were diluted with 1× PBS and pelleted at 27,000 rpm for 1 h. The pellet was resuspended in 1 ml of 1× PBS. Purified virions were digested with trypsin for 30 min at 37°C in either the absence or presence of 1% Triton-X and subjected to immunoblotting analysis with either anti-K8.1 mouse monoclonal antibody (Advanced Biotechnology Inc.) or anti-vPK rabbit immunoglobulin G (IgG).
Northern blotting.
Total RNA from TREx-K-Rta BCBL-1 cells was prepared by ISOGEN (Nippon Gene, Tokyo, Japan) as recommended by the manufacturer. Where indicated, cells were incubated with 300 μg/ml phosphonoacetic acid (PAA) during reactivation to inhibit viral DNA replication. Total RNA (10 μg/lane) was separated on a 1.0% agarose-formaldehyde gel and transferred to a nylon membrane (Biodyne; Pall BioSuport, New York). The RNA was immobilized on the membrane by drying at room temperature (RT) for 1 h and then cross-linked by UV light. DNA probes were prepared from total genomic DNA of BCBL-1 cells by PCR amplification with specific primer sets (Table 1). DNA fragments purified from agarose gels were radiolabeled with [α32-P]dATP using a Strip-EZ DNA kit (Ambion) as recommended by the supplier. To detect low-abundance ORF34 and ORF35 transcripts, RNA probes were generated by T7 RNA polymerase with Strip-EZ RNA kit (Ambion) and incorporated with [α32-P]UTP. Hybridization was performed at 42°C for DNA probes and at 64°C for RNA probes as described previously (20).
Immunoprecipitation and immunoblot analysis.
TREx-K-Rta BCBL-1 cells were rinsed in ice-cold PBS, and 1 × 107 cells were lysed in EBC lysis buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 0.5% NP-40, 50 mM NaF, 200 μM Na2VO4, 1 mM phenylmethylsulfonyl fluoride) supplemented with a protease inhibitor cocktail (Roche). After centrifugation (15,000 × g for 10 min at 4°C), 20 μl of protein A and protein G Sepharose beads (Upstate) was added to the supernatants and preincubated overnight at 4°C. Five hundred micrograms of each of the cleared supernatants was reacted with 3 μg of anti-vPK or preinoculated rabbit IgG for 3 h at 4°C with gentle rotation. The immune complex was then captured by the addition of 20 μl of a protein A and protein G Sepharose bead mixture and rocked for an additional 2 h at 4°C. Beads were washed four times with EBC buffer and boiled for 5 min in 20 μl of 2× sodium dodecyl sulfate (SDS) sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.6% bromophenol blue). Protein samples from total cell lysates (50 μg/lane) or immunoprecipitates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene difluoride membrane (Millipore) using a semidry transfer apparatus (Amersham Pharmacia).
For detecting a molecular weight shift by phosphorylation, cells were lysed with phosphoprotein-specific lysis buffer (50 mM Tris-HCl [pH 7.4], 1% Triton-X, 10% glycerol, 50 mM KCl, 50 mM β-glycerophosphate, 50 mM NaF, 2 mM Na2VO4, 5 mM EDTA, 2 mM benzamide) supplemented with a protease inhibitor cocktail (Roche). 293T cells were cotransfected with 2 μg of pT7-K-bZIP and 2 μg of pFlag-vPK or pFlag-empty expression plasmids using Fugene 6 (Roche) according to the supplier's recommendations. The cells were harvested 48 h after transfection and lysed in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.5], 1.0% NP-40, 0.1% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 10 μg/ml of aprotinin). Five hundred micrograms of cell lysate was immunoprecipitated with the addition of 25 μl of anti-Flag antibody-conjugated agarose (Sigma). Beads were washed four times with RIPA buffer and then boiled for 5 min in 20 μl of 2× SDS sample buffer. Protein samples from total cell lysates (50 μg/lane) or immunoprecipitates were subjected to SDS-PAGE and then transferred to polyvinylidene difluoride membrane as described above and previously (20). For detecting small ubiquitin-like modifier protein (SUMO)-modified K-bZIP, cells were washed twice with PBS and lysed with SUMO lysis buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 20 μM N-ethylmalemide [Sigma]) supplemented with a protease inhibitor cocktail (Roche). Cell lysates were briefly sonicated and cleared by centrifugation (15,000 × g for 10 min at 4°C), and the supernatant was used for immunoblot analysis. For immunoprecipitation of proteins modified by sumoylation, cell lysates were boiled for 10 min at 95°C and diluted 10-fold with either phosphate buffer (for immunoprecipitation with His tag) or RIPA buffer (for immunoprecipitation with anti-Flag antibody). Final dilutions of the primary antibodies for immunoblotting were 1 μg/ml of anti-K-bZIP, 1 μg/ml of anti-vPK, 1 μg/ml of anti-Flag M2 (Sigma), and 1 μg/ml of anti-SUMO-2/3 (Zymed Laboratories Inc).
ChIP assay.
After 48 h with or without induction of viral reactivation with DOX (1 μg/ml), 107 TREx-K-Rta BCBL-1 cells were fixed with 1% formaldehyde at RT for 10 min and washed with ice-cold PBS. Cells were washed in buffer I (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5). Cell pellets were collected by centrifugation and washed in buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5). A total of 200 μl of cell pellets was resuspended in 1 ml of lysis buffer (0.5% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 1× protease inhibitor cocktail [Roche]) and sonicated four times for 30 s with 0.5-s pulses (Fisher 550 Sonic Dismembrator). Cell debris was removed by centrifugation, and the chromatin solutions were diluted five times in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8.1], 1× protease inhibitor cocktail). A sample of total chromatin was collected to serve as a total input DNA control. Chromatin fragments were immunoprecipitated with 5 μg of anti-K-bZIP IgG, preinoculated rabbit IgG, or anti-vPK rabbit IgG overnight at 4°C with gentle rotation. Immunocomplexes were recovered and eluted as described before (19). After reverse cross-linking at 65°C overnight, the DNA fragments were purified with a QIAquick PCR Purification Kit (QIAGEN) after pH was adjusted with 3 M sodium acetate (pH 7.0) and eluted with 100 μl of 1× TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]). The chromatin-immunoprecipitation (ChIP) DNA was analyzed by PCR for Fig. 5 with primer sets described previously (19).
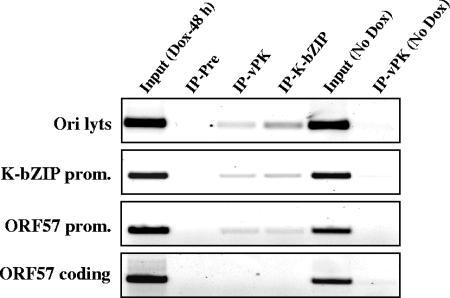
FIG. 5.
Recruitment of vPK to the KSHV genome. ChIP assay was performed on cell lysates of the TREx-K-Rta BCBL-1 cell line by using the indicated rabbit IgG. Cell lysates were prepared before (No Dox) and after 48 h of reactivation (Dox-48 h). For each primer set, a PCR amplification with the total input DNA (Input) before immunoprecipitation was carried out. ChIP fragments of preimmune IgG (IP-Pre), anti-vPK IgG (IP-vPK), or anti-K-bZIP IgG (IP-K-bZIP) were subjected to PCR amplification analysis. Prom, promoter.
Immunofluorescence assay.
BCBL-1 cells were treated with TPA for 48 h, collected by low-speed centrifugation, fixed with methanol-acetone (1:1) for 15 min at RT, and then washed three times with PBS. After treatment with blocking solution, 2% bovine serum albumin (BSA; Fisher) in PBS, cells on the coverslip were incubated with anti-vPK rabbit IgG (1:1,000) in PBS-2% BSA for 1 h at RT. For the colocalization study, the coverslip was incubated with anti-ORF59 mouse monoclonal antibody (1:3,000) (Advanced Biotechnology Inc.) together with anti-vPK rabbit IgG. After four washes with PBS, Alexa Fluor 555-conjugated goat anti-mouse IgG F(ab′)2 (1:5,000) (Molecular Probes) and/or Alexa Fluor 488-conjugated goat anti-rabbit IgG F(ab′)2 (1:5,000) (Molecular Probes) in blocking solution was applied and allowed to react for 1 h. Purified anti-vPK IgG and anti-K-bZIP IgG were labeled with either Alexa Fluor 488 or Alexa Fluor 647 with a protein labeling kit (Molecular Probes) and were applied and allowed to react for 1 h at concentration of 1 μg/ml each. After four washes with PBS, coverslips were mounted on glass slides. Imaging was performed by using a confocal microscope (LSM 510-MicroSystem; Carl Zeiss Co., Ltd).
Generation of recombinant baculoviruses and protein purification.
Spodoptera fugiperda Sf9 cells were maintained in EX-CELL 420 medium (JRH Biosciences). The pVL Flag-ORF36 wild type, pVL Flag-ORF36 K108Q, pVL Flag-ORF6, pVL Flag-ORF9, pVL Flag-ORF40-41, pVL Flag-ORF44, pVL Flag-K-Rta, pVL Flag-K-bZIP, pVL Flag-ORF56, or pVL Flag-ORF59 was cotransfected with linearized BaculoGold viral DNA (Pharmingen) into Sf9 cells by using Fugene 6 (Roche), and recombinant viruses were subsequently amplified. Expression of the proteins in Sf9 cells was confirmed by immunoblotting with anti-Flag monoclonal antibody (Sigma). A large-scale culture of Sf9 cells (100 ml) was infected with recombinant baculovirus, and cells were harvested 48 h postinfection. Cells were lysed with Flag lysis buffer (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 1 mM EDTA, 1% Triton X-100) supplemented with a protease inhibitor cocktail (Roche). Cell lysates were cleared by centrifugation and immunoprecipitated with the addition of 100 μl of anti-Flag antibody-conjugated agarose (Sigma). Beads were washed three times with Flag lysis buffer and twice with TBS buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl). Flag-tagged protein was eluted with 100 μg/ml of 3× Flag peptide (Sigma) in TBS buffer. Purification of protein was confirmed by immunoblotting with anti-Flag antibody, and the amount of purified protein was measured by SDS-PAGE with BSA as a standard.
Preparation and purification of GST fusion proteins.
Glutathione transferase (GST) fusion proteins were expressed in Escherichia coli strain BL21 transformed with pGEX-ORF36 N-151, pGEX-K-bZIP N-75, pGEX-K-bZIP 76-150 pGEX-K-bZIP 151-C, pGEX-K-bZIP 72-102, pGEX-K-bZIP 87-123, pGEX-K-bZIP 87-123 (T111A), pGEX-K-bZIP 113-154, pGEX-K-bZIP 128-154, or pGEX4T-2 (N and C correspond to termini; number represents amino acid position). The procedure of purification of GST fusion proteins was described previously (20).
In vitro kinase assay.
Purified proteins were dialyzed to Tris buffer (50 mM Tris-HCl [pH 7.5], 1 mM dithiothreitol). Purified wild-type kinase or kinase-dead mutant (100 ng) was incubated with 2 μg of purified substrates in kinase buffer (20 mM HEPES [pH 7.5], 5 mM MnCl2, 10 mM β-mercaptoethanol) supplemented with 10 μCi [γ-32P]ATP at 37°C for 30 min. The reaction was stopped by adding 2× sample buffer, protein was separated by 10% SDS-PAGE, and the gel was subjected to autoradiography.
Reporter assays in transient transfection experiments.
Reporter plasmid was constructed by inserting the promoter region (29) upstream of the firefly luciferase coding region (Luc) in the pGL3-Basic vector (Promega). 293 cells were seeded in 12-well plates at 1 × 105 cells/well in 1.0 ml of Dulbecco's modified Eagle medium supplemented with 10% FBS and incubated at 37°C with 5% CO2. For each well, an equal amount of plasmid DNA, including the reporter and the control or expression plasmid, was transfected using the Fugene 6 reagent following the manufacturer's protocol (Roche). Cell lysates were prepared 48 h after transfection with 1× passive lysis buffer (Promega). A luciferase assay was performed according to the manufacturer's protocol using a Lumat LB 9501 Luminometer (Wallac Inc., California). At least three independent determinations were performed at each setting.
RESULTS
vPK transcription during lytic replication.
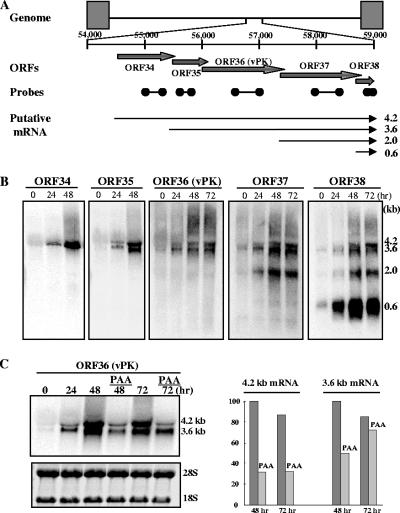
In previous reports, the vPK gene was shown to be expressed as either an early or a late gene during lytic replication, depending on the conditions used to reactivate virus in latently infected B-lymphoid tumor cells (22, 31, 38, 39). Because the mode of action of the inducers (e.g., phorbol ester or butyrate) is complex and the fraction of cells affected is relatively small, we chose to measure the expression kinetics of vPK by K-Rta-mediated reactivation. In the TREx-K-Rta BCBL-1 cell line (36), a derivative of the BCBL cell line, the expression of K-Rta is induced by DOX, resulting in a well-ordered KSHV gene expression program and a relatively synchronized cycle of viral replication in nearly all cells (19, 36). RNA from induced cells was analyzed by Northern blot analysis, which showed that vPK is encoded in two transcripts (4.2 and 3.6 kb), together with ORF37 and ORF38. These transcripts were detected as early as 24 h after induction and maintained essentially the same intensity thereafter. ORF34 probe detected the 4.2-kb transcript but not the 3.6-kb transcript; this result indicates that the 4.2-kb mRNA initiated from ORF34 and that the 3.6-kb mRNA is transcribed from ORF35. In addition to the 4.2- and 3.6-kb transcripts, ORF37 probe detected a 2.0-kb transcript, and ORF38 probe detected 2.0- and 0.6-kb transcripts. The transcript data are consistent with a transcriptional map of these ORFs shown in Fig. 1A, where all transcripts are coterminal at the strong poly(A) site at nucleotide (nt) positions 58852 to 58858 (accession no. U75698).
FIG. 1.
Transcription of vPK (ORF36). (A) Schematic diagram of the KSHV genome structure and gene organization of ORF36 region. Positions of probes for Northern blotting are indicated by ORF number: 34 (nt 55042 to 55346), 35 (nt 55639 to 55872), 36 (nt 56415 to 56812), 37 (nt 58022 to 58392), and 38 (nt 58688 to 58843). The nucleotide number corresponds to the sequence of accession no. U75698. Putative direction and coding regions are shown as arrows. The direction of transcripts and their coding regions were determined from the size and position of the polyadenylation site. (B) Northern blot analysis. After induction of K-Rta expression, total RNA was prepared at the indicated time points. Ten micrograms was loaded per lane, electrophoresed, and transferred to nylon membranes. The results of Northern hybridization with the indicated probes are shown. (C) Classification of ORF36 transcripts. The TREx-K-Rta BCBL-1 cell line was induced with DOX in the presence of 300 μg/ml of PAA. Ten micrograms of total RNA was loaded per lane. The results of Northern hybridization with the ORF36 probe are shown (left). Transcripts were quantified (right) by using Quantity-One (Bio-Rad). The highest transcript level was normalized to a value of 100%.
The kinetics of vPK expression suggests that it is an early or early-late gene. To define this further, the expression of vPK was examined by using the DNA polymerase inhibitor, PAA, a treatment for temporal classification of viral genes. The TREx-K-Rta BCBL-1 cells were induced with DOX in the presence of PAA. Expression of the 4.2-kb mRNA was reduced to 31% in the presence of PAA, but the 3.6-kb mRNA was relatively resistant to PAA treatment, and, in fact, the level of the 3.6-kb transcript increased from 48 h to 72 h in the presence of PAA (Fig. 1C). These results indicated that vPK was expressed as both an early (3.6 kb) and an early-late (4.2 kb) gene.
Identification of vPK protein in naturally infected cells.
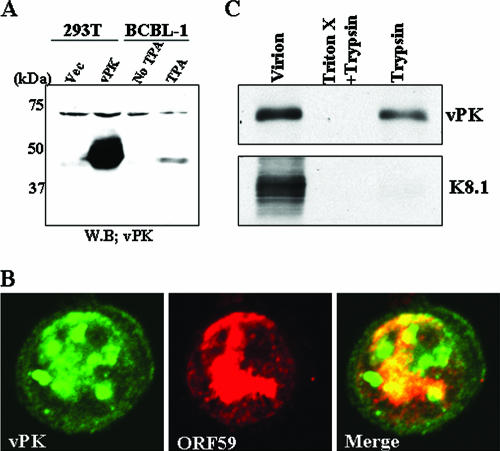
Previously, we showed that vPK activates the Jun N-terminal protein kinase cellular signal transduction pathway (17). Other investigators reported the localization of vPK in the nucleus of transiently transfected cells (38). These studies, while informative, were carried out with ectopically expressed, epitope-tagged vPK in transfected cells. To characterize vPK during lytic replication, we generated rabbit serum, using a GST-vPK (residues 1 to 150) fusion protein as an antigen. The anti-vPK antibody reacted with the cell lysates from KSHV-reactivated BCBL-1 (Fig. 2A, lane TPA, showing the lytic phase) but not with nonreactivated BCBL-1 (Fig. 2A, lane No TPA, showing the latent phase); the size, 48 kDa, corresponds well with the size calculated from the amino acid sequence of ORF36. Furthermore, this antibody interacts with Flag-tagged vPK protein expressed in transiently transfected 293T cells (Fig. 2A, lane vPK) but not with proteins from vector-transfected cells (lane Vec), attesting to the specificity of the antibody.
FIG. 2.
Characterization of vPK in infected cells. (A) Immunoblot analyses of vPK. Cell lysates were prepared from 293T cells transfected with the vPK expression plasmid or control vector (Vec) or from TPA-treated or nontreated (No TPA) BCBL-1 cells. Total cell lysate (50 μg/lane) from each cell culture was subjected to immunoblot analysis. (B) Colocalization of vPK with Orf59 protein. Confocal analysis was performed by using affinity-purified anti-vPK IgG and anti-ORF59 mouse monoclonal antibody. vPK (green) and ORF59 (red) were detected with Alexa Fluor 488-conjugated anti-rabbit IgG and Alexa Fluor 555-conjugated anti-mouse IgG. (C) vPK localization inside virions. KSHV virions were prepared as described in Materials and Methods. Virion protein was digested with trypsin in either the presence or absence of 1% Triton-X. The indicated protein was probed with specific antibodies.
With the specific antibody in hand, we examined the localization of vPK in BCBL-1 cells 48 h after induction by TPA. As shown in Fig. 2B, during the replicative phase of the lytic cycle, vPK is localized primarily in the nucleus, exhibiting a punctate pattern in the midst of the diffuse staining; this pattern resembles KSHV DNA replication/transcription compartments (54). Antibody to polymerase processivity factor (PPF)/ORF59, known to be part of the replication/transcriptional complex (54), was used to “mark” such a structure, and, indeed, the merged picture showed colocalization of vPK and PPF (Fig. 2B).
vPK is packaged in the virion.
After establishing the expression kinetics and subcellular localization of vPK in BCBL-1 cells, we determined whether vPK is packaged in the virion. Viruses released from BCBL-1 cells at 96 h after K-Rta induction were purified through a sucrose gradient, and the presence of vPK protein was probed with anti-vPK antibody with or without trypsin digestion. The K8.1 virion membrane associated glycoprotein was used as a control. As shown in Fig. 2C, vPK but not K8.1 was protected from trypsin digestion, suggesting that vPK is primarily packaged inside the viral membrane. Addition of the detergent Triton X, which solubilized the membrane, rendered vPK susceptible to trypsin digestion. These results demonstrate that a fraction of vPK is packaged in the virion and suggest that vPK function may not be limited to just the early/late phase. Through phosphorylation, vPK may regulate the functions of virion proteins, as well as viral regulatory proteins, upon entry into the host cell.
vPK phosphorylates K-bZIP in vitro.
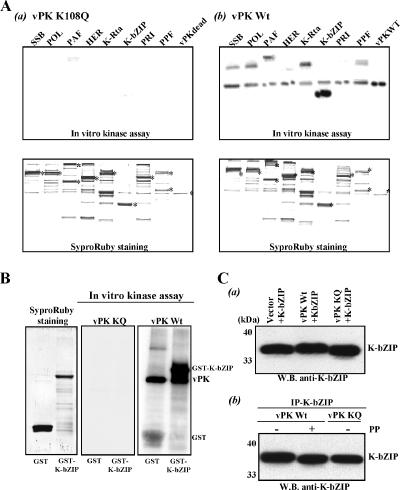
Given the close proximity of vPK with the KSHV replication complex, we determined whether the replication proteins of this virus are potential substrates for vPK. Eight KSHV gene products, shown to be essential for viral DNA replication and localized in the replication complex (3, 30, 54), were tagged with Flag epitope and expressed by baculovirus vectors. These tagged proteins were purified with Flag antibody conjugated to agarose beads and subsequently eluted with Flag peptide. The eight viral proteins are single-strand DNA binding protein (SSB), DNA polymerase (POL), primase-associated factor (PAF) (5), helicase (HER), primase (PRI), PPF, K-bZIP, and K-Rta. The vPK wild-type and kinase-dead mutant proteins were similarly prepared. An in vitro protein kinase assay was performed by incubating vPK and the individual replication-associated proteins in the presence of ATP and 5 mM MnCl2 (38). The wild-type vPK exhibited significant autokinase activity compared to the kinase-dead (K108Q) mutant (Fig. 3A). Among the eight proteins tested, K-bZIP was the most efficiently phosphorylated by vPK. In addition, SSB, POL, PAF, K-Rta, and PPF were phosphorylated but to a much smaller extent. Phosphorylation of K-bZIP by vPK was further verified by using GST-K-bZIP as a substrate (Fig. 3B). Strong phosphorylation of this K-bZIP by vPK wild type but not the vPK-K108Q mutant strongly suggests that the observed phosphorylation was due to intrinsic kinase activity of vPK.
FIG. 3.
In vitro protein kinase assay. (A) Viral substrates. Substrates and the protein kinases (wild-type vPK and kinase-dead vPK) were expressed by using the baculovirus vector and purified with Flag-agarose. SypoRuby-stained gel is shown at the bottom of the panel, and autoradiography of the same gel after drying is shown at top. Proteins and the associated ORFs are the following: SSB (ORF6), POL (ORF9), PAF (ORF40 to ORF41), HER (ORF44), K-Rta (ORF50), K-bZIP (K8), PRI (ORF56), PPF (ORF59), and vPK (ORF36). (B) vPK wild type phosphorylates GST-K-bZIP. The in vitro kinase assay was performed by using either GST-K-bZIP or GST as a substrate. Purified GST and GST-K-bZIP are shown at the far left. The vPK wild type but not kinase-dead mutant significantly phosphorylates GST-K-bZIP. (C) In vivo phosphorylation. The K-bZIP expression plasmid was cotransfected with indicated plasmids into 293 cells. K-bZIP was probed with anti-K-bZIP antibody. (a) Molecular mass of K-bZIP was increased when K-bZIP was cotransfected with vPK wild-type plasmid but not mutant vPK plasmid or vector control. (b) K-bZIP was treated with 100 U of lambda phosphate (PP) for 30 min at 37°C or carrier (−) after immunoprecipitation (IP). After phosphate treatment, K-bZIP was separated by PAGE, and probed with anti-K-bZIP. WB, Western blotting; vPK KQ, vPK with the mutation K108Q (kinase dead).
In vivo phosphorylation was examined by mobility shift of K-bZIP in transfected cells. K-bZIP was coexpressed with either wild-type vPK or the kinase-dead K108Q mutant in 293 cells. Cell lysates were subjected to SDS-PAGE, and K-bZIP protein was probed with anti-K-bZIP antibody. We observed a mobility shift of K-bZIP only in cells transfected with wild-type vPK and not with the K108Q kinase-dead mutant (Fig. 3C upper blot), suggesting that K-bZIP is phosphorylated by vPK in vivo. To show that this mobility shift was indeed due to phosphorylation, the K-bZIP immunoprecipitates were treated with lambda protein phosphatase before Western blotting, and as shown in Fig. 3C (lower blot), K-bZIP mobility was restored to the fast-migrating form. This finding is consistent with K-bZIP phosphorylation by vPK in vivo.
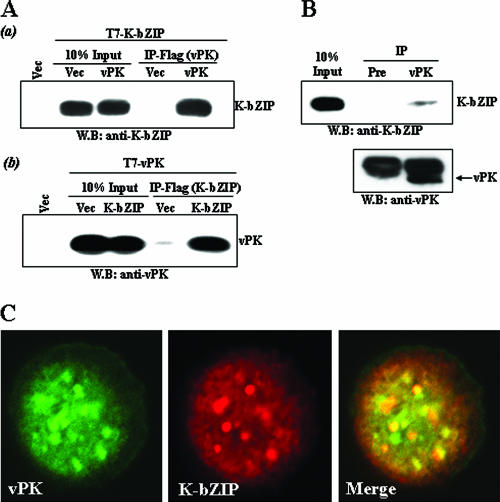
vPK physically associates with K-bZIP in vivo.
The data shown above suggest that vPK phosphorylates and colocalizes with K-bZIP. We determined whether vPK and K-bZIP physically associate with each other in vivo. This was first done by a coimmunoprecipitation assay in cotransfected 293T cells. Flag-vPK was cotransfected with T7-K-bZIP. Flag-empty vector served as a negative control in a separate transfection. Twenty-four hours after transfection, cells were lysed with RIPA buffer and immunoprecipitated with anti-Flag mouse monoclonal antibody. To avoid detecting mouse IgG heavy chain, we performed immunoblotting with rabbit IgG against either vPK or K-bZIP to detect coprecipitants. As shown in Fig. 4A (upper blot), K-bZIP was efficiently coprecipitated with vPK. The reciprocal experiment, shown in the lower blot in Fig. 4A, confirmed the association of these two viral proteins. We further extended the studies to the naturally infected BCBL-1 cell line. Forty-eight hours after activation of KSHV lytic replication by TPA, vPK was immunoprecipitated with affinity-purified anti-vPK antibody. As a negative control, preinoculated rabbit IgG was used. This experiment showed that K-bZIP was coprecipitated with vPK (Fig. 4B), demonstrating the association of these two viral proteins in the natural setting of viral reactivation. This association was verified by additional microscopy studies revealing colocalization of K-bZIP and vPK after anti-K-bZIP IgG and anti-vPK IgG were conjugated with Alexa Fluor 647 and Alexa Fluor 488, respectively. In addition to PPF, K-bZIP showed colocalization with vPK (Fig. 4C). It is noteworthy that the vPK-K-bZIP colocalization pattern is not restricted to KSHV DNA replication/transcription foci, suggesting a complex association between vPK and K-bZIP, involved in both replication and transcriptional regulation. The data also revealed that the subcellular locations of vPK are not static, perhaps depending on the different stages of replication and different viral proteins associated with vPK.
FIG. 4.
Association between K-bZIP and vPK. (A) Association between K-bZIP and vPK in cotransfected 293T cells. 293T cells were cotransfected with the indicated plasmids. Cell lysates were precipitated with Flag-antibody conjugated to agarose, and coimmunoprecipitation of K-bZIP (a) or vPK (b) was detected by using anti-K-bZIP rabbit IgG or anti-vPK rabbit IgG. The expression of K-bZIP or vPK in total cell lysates is shown in the same blot as a control. (B) Coimmunoprecipitation assay with KSHV-positive BCBL-1. BCBL-1 cells induced for KSHV lytic replication for 48 h with TPA were harvested, and 500 μg of cell lysate was immunoprecipitated with either preimmune rabbit IgG or anti-vPK rabbit IgG. The same membrane was stripped and reprobed with anti-vPK rabbit IgG. (C) Colocalization of vPK with K-bZIP. vPK (green) and K-bZIP (red) were detected with Alexa Fluor 488-conjugated anti-vPK rabbit IgG (green) and Alexa Fluor 647-conjugated anti-K-bZIP rabbit IgG (red). IP, immunoprecipitation; WB, Western blotting; Vec, vector.
vPK and K-bZIP are corecruited to viral promoters.
Previously, we showed that K-bZIP is a strong transcriptional repressor, which modulates the transcription of K-bZIP and ORF57 promoters mediated by K-Rta (20, 29). Based on ChIP assays, K-bZIP is recruited to these two promoters and Ori-Lyt DNA during the early phase of KSHV lytic replication (19). Given the close association between vPK and K-bZIP, we determined whether vPK is also recruited to these sites on the viral genome by the ChIP assay on lysates of reactivated TREx-K-Rta-BCBL-1 cells. As shown in Fig. 5, vPK and K-bZIP are corecruited to both the K-bZIP and ORF57 promoter regions as well as Ori-Lyt DNA but not to the ORF57 coding region. These data, together with results described in the previous section, indicate that vPK and K-bZIP are likely to be functional partners in vivo.
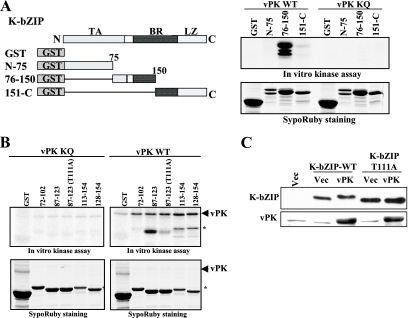
The major vPK phosphorylation site of K-bZIP.
To map the phosphorylation site(s) of K-bZIP by vPK, we first generated a series of GST-K-bZIP deletion mutants, roughly dividing the protein into three sections. Bacterially expressed proteins, purified as above, were incubated with either wild-type or kinase-dead vPK. As shown in Fig. 6A, strong phosphorylation was detected on the middle segment encoding amino acid residues 76 to 150 of K-bZIP. Additional deletion constructs spanning this region were made for additional tests of phosphorylation by vPK (Fig. 6B). This approach narrowed the region of major phosphorylation to residues 87 to 123. Threonine 111 within this region was determined to be a major phosphorylation site, because its mutation to alanine significantly diminished the phosphorylation level of K-bZIP (Fig. 6B). The fact that the K-bZIP-T111A mutant still retains some phosphorylation suggests that there are additional vPK phosphorylation site(s) yet to be identified. K-bZIP-T111A was cotransfected with vPK in 293 cells, and the proteins were separated in SDS-PAGE and probed with anti-K-bZIP antibody; K-bZIP-T111A, unlike the wild type, showed little mobility shift, reinforcing the notion that the mobility shift of the wild-type K-bZIP is due to phosphorylation by vPK (Fig. 6C). We thus identified K-bZIP threonine 111 as the major phosphorylation target of vPK.
FIG. 6.
Mapping of vPK phosphorylation site of K-bZIP. (A) The domains of K-bZIP and the GST-K-bZIP mutants are indicated in the left panel. TA, transactivation domain; BR, basic domain; LZ, leucine zipper domain. Results of the in vitro protein kinase assay and SyproRuby staining of the same gel are shown at right. (B) The coding region of recombinant K-bZIP is indicated at the top of the panels. Results of the in vitro protein kinase assay and SyproRuby staining of the same gels are shown. Asterisks show phosphorylated protein originated from E. coli. (C) In vivo phosphorylation. The K-bZIP-wild type (WT) or K-bZIP-T111A expression plasmid was cotransfected with the indicated plasmids into 293 cells. K-bZIP or vPK was probed with specific antibody. The molecular weight of K-bZIP wild type but not K-bZIP-T111A was increased when K-bZIP was cotransfected with vPK. WB, Western blotting; Vec, vector; vPK KQ, vPK with the mutation K108Q.
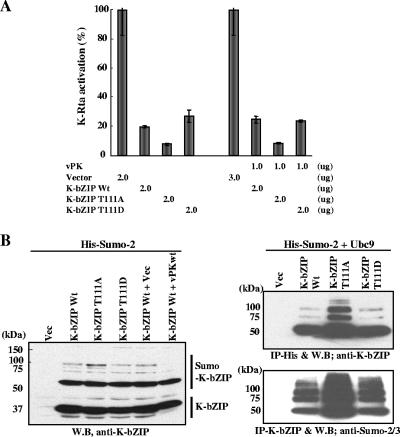
Phosphorylation modulates K-bZIP repression function.
To study the functional significance of vPK phosphorylation on K-bZIP, in addition to the mutant T111A, we mutated threonine 111 to aspartic acid, thereby mimicking the phosphorylated form. Our previous work showed that one of the K-bZIP functions is to serve as a transcriptional repressor of K-Rta (20, 29); this function depends on the sumoylation of lysine 158 of K-bZIP (19). To determine whether phosphorylation of K-bZIP at threonine 111 impacts the repression function, wild-type K-bZIP or its phosphor-acceptor mutants were cotransfected with K-Rta and the reporter plasmid carrying the K-bZIP promoter, which directs luciferase expression. As expected, wild type K-bZIP strongly represses K-Rta-mediated activation of the K-bZIP promoter to about the 30% level (Fig. 7A). The K-bZIP-T111A mutant was even more potent in its repression function, whereas K-bZIP-T111D displayed a repression level similar to wild-type K-bZIP (Fig. 7A). These data suggest that phosphorylation of threonine 111 has a negative impact on the repression activity of K-bZIP. Because K-bZIP repression activity largely depends on its sumoylation, we compared the sumoylation patterns of the wild-type K-bZIP and K-bZIP-T111A mutant. Consistent with its higher potency of repression activity, K-bZIP-T111A is more heavily sumoylated compared to K-bZIP wild type and K-bZIP-T111D, particularly in the formation of SUMO multimers. Inclusion of vPK in the transfection mix significantly reduced the sumoylation of K-bZIP wild type (Fig. 7B, left). The differential sumoylation of the wild type and T111A is even more pronounced when the conjugation enzyme Ubc9 for SUMO was cotransfected to further catalyze the reactions (Fig. 7B, right). To demonstrate that the high-molecular-weight species are sumoylated K-bZIP, His-tagged SUMO-2 and Ubc9 were cotransfected with K-bZIP, and SUMO-modified proteins were affinity purified with nickel-charged resin. K-bZIP was probed with anti-K-bZIP antibody. Monomer, dimer, and trimer forms of sumoylated K-bZIP were detected (Fig. 7B, right). Consistent with immunoblotting results (Fig. 7B, left), K-bZIP-T111A was more heavily sumoylated. The reciprocal experiment further confirmed this result (Fig. 7B, right). These data again demonstrated the potential of K-bZIP to be sumoylated and suggest that phosphorylation by vPK may modulate the ability of K-bZIP to be modified by sumoylation.
FIG. 7.
Phosphorylation and sumoylation of K-bZIP. (A) Phosphorylation alters K-bZIP transcription function. Transient reporter assays were performed with 293 cells cotransfected with reporter plasmid containing K-bZIP promoter. The amount of K-Rta expression vector was kept at 0.5 μg. The amounts of K-bZIP wild type, phosphor mutants, or vPK expression plasmid are shown at the bottom of the panel. Luciferase activity of K-Rta alone is normalized to a value of 100%. (B) Sumoylation of the wild type and phosphoacceptor mutant of K-bZIP. Indicated expression plasmids were cotransfected with SUMO-2 expression vector into 293 cells. (b) Identification of sumoylation of K-bZIP. 293 cells were cotransfected with the indicated plasmids. After isolation of SUMO-2-modified protein (IP-His) or K-bZIP (IP-Flag), immunoblotting was performed with the indicated antibody. IP, immunoprecipitation; WB, Western blotting; Vec; vector control.
DISCUSSION
This report focuses on the characterization of vPK of KSHV, with the goal of building knowledge of the role of this protein in the viral life cycle. Basic features of vPK were examined including the transcriptional pattern of the vPK gene, incorporation of vPK protein into virions, and localization within infected cells. Importantly, viral proteins that interact with vPK, and viral targets of its phosphorylation activity, were identified. The results of these studies were used to propose a model of vPK function in KSHV replication.
The transcriptional pattern of the vPK gene was defined in the BCBL-1 cell line harboring latent virus. During reactivation, two vPK transcripts, 4.2 and 3.6 kb, are produced with different kinetics. The result of kinetics study indicated that vPK was expressed as both an early (3.6 kb) and an early-late (4.2 kb) gene, and maximum expression required DNA replication. This kinetics of expression was closely related to EBV BGLF4, a homolog of vPK (15). Other investigators using microarray analysis, coupled with the use of an inhibitor of viral DNA synthesis, also showed that the vPK gene was expressed early in the viral lytic cycle (31). In K-Rta-inducible BCBL-1 cells, the 4.2-kb transcript, encoding ORF34 to ORF38, was sensitive to PAA; this finding differs from an earlier report (18). We also detected two additional transcripts (2.0 kb and 0.6 kb) that had not previously been reported (18). These differences in transcription patterns might be caused by the different method for reactivation, i.e., overexpression of the transactivator, K-Rta. The method used for viral reactivation may partially overcome the defect in late gene expression. In addition, we cannot rule out that the 0.6-kb transcript is derived from the opposite strand, as double-stranded DNA probe was used in that experiment. The two vPK encoding transcripts were coterminal with transcripts encoding ORF37 and ORF38 (see also reference 18). Precise transcription initiation sites were recently reported at position 54566 for the 4.2-kb transcript and at position 55567 for the 3.6 kb transcript (18). Potential functional implications of such multicistronic viral transcripts remain to be determined.
To aid in the study of vPK, a highly specific antibody was produced by immunizing rabbits with purified recombinant vPK protein. This monospecific antibody identified vPK as a 48-kDa protein in cells producing virus (i.e., reactivated BCBL-1 cells). Interestingly, the antibody readily detected vPK in purified virions. Structural analysis of purified virions of other herpesviruses, including HSV, CMV, EBV, and rhesus rhadinovirus, indicate that the conserved protein kinases of these viruses are also incorporated into mature virus particles (2, 12, 37, 50, 52). This finding suggests that this kinase may play a role in phosphorylating viral proteins during virion assembly and/or regulate the functions of viral and cellular proteins upon entry into host cells and during the early stage of de novo infection (47).
Analysis of subcellular localization by immunofluorescence experiments in induced BCBL-1 cells revealed that vPK is primarily accumulated in the nucleus (Fig. 2C), consistent with an earlier study based on transient expression of genetically engineered vPK in transfected 293 cells (38). Interestingly, we also found that the subcellular localization pattern of vPK, in the context of viral infection, was more varied than shown in this previous report. In some cells, staining for vPK displayed a discrete dot-like pattern. This pattern of vPK compartmentalization coincided with viral replication foci marked by the KSHV DNA PPF (ORF59) (54). In other cells, staining for vPK exhibited a more diffuse arrangement (Fig. 4C). These patterns could reflect different phases of viral replication, presumably where vPK is assembled into different protein complexes. This diverse pattern, coupled with the presence of vPK transcripts both early and late, implies that vPK could influence multiple steps in viral replication. The pattern of vPK localization is similar to the EBV vPK homolog BGLF4, which is found in the viral replication compartment, with variation in subcellular localization at different times after reactivation of virus (2, 52). Likewise, CMV UL97, the vPK homolog, is also colocalized with UL44 (the PPF homolog) in the viral DNA replication compartment; UL97 phosphorylates UL44 (33). Additionally, the viral protein kinase of EBV, BGLF4, was shown to phosphorylate the processivity factor, EA-D, of this virus (16). Thus, herpesviruses encode conserved protein kinases that appear to have a common role in viral DNA replication.
The subcellular localization studies, suggesting that vPK is associated with the DNA replication apparatus, prompted further investigation on a role for vPK in the KSHV life cycle. Accordingly, several recombinant viral proteins involved in viral DNA replication were tested as substrates of vPK activity. This included a group of six proteins that directly participate in viral DNA synthesis (SSB, POL, PAF, HER, PRI, and PPF) as well as two regulatory proteins, K-Rta and K-bZIP, which have been shown to physically associate with the lytic origin of DNA replication (Ori-Lyt) (3, 30). In the cell-free system with purified vPK and purified substrates, K-bZIP was the most highly phosphorylated target. Although the vPK kinase-dead mutant failed to phosphorylate these substrates, it is still possible that the phosphorylation observed by wild-type vPK could be due to an associated protein kinase that was activated by wild-type vPK. Either way, it is clear that vPK alone, or a vPK complex with an additional protein(s), strongly phosphorylates K-bZIP in vitro. Additionally, ChIP analysis of infected cells showed that both vPK and K-bZIP were corecruited to Ori-Lyt DNA. The potential role of vPK in viral DNA synthesis can be investigated in the future in an in vitro replication system, which is built from the aforementioned viral DNA proteins, as described for several herpesviruses including KSHV (3, 4).
Additional studies were done to define the site of phosphorylation of K-bZIP and assess the effects of this posttranslational modification on K-bZIP function in regulating viral gene expression. The threonine residue at position 111 was determined to be the major site of vPK phosphorylation. Mutation of this residue to alanine (T111A) largely diminished but did not completely eliminate the phosphorylation by vPK. Our previous studies showed that K-bZIP strongly repressed the transcriptional activation of K-Rta and that this repression depended to a large extent on sumoylation of K-bZIP (19). The K-bZIP-T111A mutant exhibited an even stronger repression function that correlated with its ability to be significantly sumoylated. In contrast, a phosphor-mimetic mutant, K-bZIP-T111D, retained the repression activity like wild-type K-bZIP. Taken together, these data suggest that phosphorylation of threonine 111 of K-bZIP has a negative effect on its repression activity and a positive effect on its transactivation function (data not shown). Thus, K-bZIP joins a growing list of transcriptional factors whose activities are modulated by sumoylation and phosphorylation in an antagonistic manner (8, 56). Accordingly, we propose a model whereby vPK switches K-bZIP from being a strong repressor of K-Rta, which targets immediate-early genes, to a transactivator that synergizes with K-Rta to activate early and late viral gene expression (data not shown). A key feature of this model is that vPK phosphorylation may play a role as the molecular switch for regulating (i.e., balancing) these opposing activities of K-bZIP. Interestingly, Zta of EBV, the homologue of K-bZIP, is also modified by sumoylation (1); however, the significance of sumoylation of Zta on EBV transcription has not been reported.
The studies in this report also revealed functional similarity of KSHV vPK with the homologous protein kinases of other herpesviruses that modulates the transactivation function of viral genes; for example EBV BGLF4 modulates EBNA2 (58), and HSV UL13 modulates ICP22 (42). Going beyond these previous studies, the present report utilized ChIP analysis to show that vPK of KSHV was corecruited with K-bZIP to viral promoters.
The phosphorylation target of the conserved protein kinases of herpesviruses resembles that of the cyclin dependent protein kinase. Intriguingly, threonine 111 [H-T(111)-P-P-R] was also identified as a major site of phosphorylation when K-bZIP was tested as a substrate for the Cdk2 and Cdc2 (40). Furthermore, a test of vPK phosphorylation on a library of synthetic peptide substrates implicated the consensus motif T-P-X-X-R, R-R-Ψ-S-P or R-R-P-T-Ψ (where Ψ indicates a hydrophobic residue), which resembles the consensus motif of the cyclin dependent protein kinase (data not shown). Interestingly, the EBV BGLF4 and HSV UL13 protein kinases were shown to phosphorylate some of the Cdk2 target motifs (23, 25, 57, 58). Taken together, these findings suggest that the conserved protein kinases of herpesviruses in some cases can substitute for cellular protein kinases, perhaps to advance the cell cycle in situations where cells are growth arrested. It has been suggested that during the early stage of herpesvirus infection, cells are often blocked at G1 stage to allow viral mRNA and proteins to be synthesized before the onset of cellular DNA replication. In previous studies, K-bZIP was shown to be a key cell cycle regulator that binds Cdk2 and activates p21 to slow down the cell cycle at the immediate-early stage of lytic infection (21, 55). Accordingly, we suggest that, at the later stages of viral replication, vPK restores the Cdk2 function by phosphorylating Cdk2 targets, and vPK may compete with Cdk2, thereby releasing Cdk2 from its complex with K-bZIP. Cdk2 plays an important role in DNA replication by phosphorylating the licensing factor MCM (32). Additionally, Cdk2 was shown to be required for herpesvirus replication (9, 28, 44). Thus, it is conceivable that vPK may serve a similar role, i.e., as a surrogate for Cdk2, during the replication of the KSHV genome.
In summary, our model shows that vPK switches K-bZIP from being a strong repressor of K-Rta transactivation of immediate-early genes to a protein that synergizes with K-Rta to activate early and late viral gene expression. Additionally, vPK may modulate the activity of K-bZIP and perhaps other viral proteins that play a role in replication of the KSHV genome. Interestingly, the incorporation of vPK into virions and the diverse patterns of vPK subcellular localization are findings which indicate that the kinase is assembled into different viral protein complexes; this differential compartmentalization implies that vPK functions at multiple steps in viral replication. The information presented here sets the stage for future studies to fully define the role of vPK in the KSHV life cycle and also to explore the effects of vPK on the host cell. Detailed knowledge of vPK targets and function(s) in the virion and infected cell will establish a basis for novel therapies against KSHV and the pathology associated with infection with this virus.
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA111185 to H.J.K.) and the U.S. Department of Agriculture (2004-35204-14207 to H.J.K. and Y.I.). Y.I. was supported by the California university-wide AIDS Research Program (F03-D-206). Additional funding was provided by the University of California-Davis Cancer Center.
Footnotes
Published ahead of print on 15 November 2006.
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, R., A. Kato, K. Kato, M. Kanamori-Koyama, K. Sugimoto, T. Sairenji, Y. Nishiyama, and Y. Kawaguchi. 2006. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J. Virol. 80:5125-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. [DOI] [PubMed] [Google Scholar]
- 4.AuCoin, D. P., K. S. Colletti, Y. Xu, S. A. Cei, and G. S. Pari. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 76:7890-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AuCoin, D. P., and G. S. Pari. 2002. The human herpesvirus-8 (Kaposi's sarcoma-associated herpesvirus) ORF 40/41 region encodes two distinct transcripts. J. Gen. Virol. 83:189-193. [DOI] [PubMed] [Google Scholar]
- 6.Baek, M. C., P. M. Krosky, A. Pearson, and D. M. Coen. 2004. Phosphorylation of the RNA polymerase II carboxyl-terminal domain in human cytomegalovirus-infected cells and in vitro by the viral UL97 protein kinase. Virology 324:184-193. [DOI] [PubMed] [Google Scholar]
- 7.Biron, K. K., R. J. Harvey, S. C. Chamberlain, S. S. Good, A. A. Smith, 3rd, M. G. Davis, C. L. Talarico, W. H. Miller, R. Ferris, R. E. Dornsife, S. C. Stanat, J. C. Drach, L. B. Townsend, and G. W. Koszalka. 2002. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole l-riboside with a unique mode of action. Antimicrob. Agents Chemother. 46:2365-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossis, G., C. E. Malnou, R. Farras, E. Andermarcher, R. Hipskind, M. Rodriguez, D. Schmidt, S. Muller, I. Jariel-Encontre, and M. Piechaczyk. 2005. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol. Cell. Biol. 25:6964-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bresnahan, W. A., I. Boldogh, P. Chi, E. A. Thompson, and T. Albrecht. 1997. Inhibition of cellular Cdk2 activity blocks human cytomegalovirus replication. Virology 231:239-247. [DOI] [PubMed] [Google Scholar]
- 10.Cannon, J. S., F. Hamzeh, S. Moore, J. Nicholas, and R. F. Ambinder. 1999. Human herpesvirus 8-encoded thymidine kinase and phosphotransferase homologues confer sensitivity to ganciclovir. J. Virol. 73:4786-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulter, L. J., H. W. Moss, J. Lang, and D. J. McGeoch. 1993. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J. Gen. Virol. 74:387-395. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham, C., A. J. Davison, A. Dolan, M. C. Frame, D. J. McGeoch, D. M. Meredith, H. W. Moss, and A. C. Orr. 1992. The UL13 virion protein of herpes simplex virus type 1 is phosphorylated by a novel virus-induced protein kinase. J. Gen. Virol. 73:303-311. [DOI] [PubMed] [Google Scholar]
- 13.Davis, D. A., A. S. Rinderknecht, J. P. Zoeteweij, Y. Aoki, E. L. Read-Connole, G. Tosato, A. Blauvelt, and R. Yarchoan. 2001. Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 97:3244-3250. [DOI] [PubMed] [Google Scholar]
- 14.Ensoli, B., C. Sgadari, G. Barillari, M. C. Sirianni, M. Sturzl, and P. Monini. 2001. Biology of Kaposi's sarcoma. Eur. J. Cancer. 37:1251-1269. [DOI] [PubMed] [Google Scholar]
- 15.Gershburg, E., M. Marschall, K. Hong, and J. S. Pagano. 2004. Expression and localization of the Epstein-Barr virus-encoded protein kinase. J. Virol. 78:12140-12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershburg, E., and J. S. Pagano. 2002. Phosphorylation of the Epstein-Barr virus (EBV) DNA polymerase processivity factor EA-D by the EBV-encoded protein kinase and effects of the L-riboside benzimidazole 1263W94. J. Virol. 76:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamza, M. S., R. A. Reyes, Y. Izumiya, R. Wisdom, H. J. Kung, and P. A. Luciw. 2004. ORF36 protein kinase of Kaposi's sarcoma herpesvirus activates the c-Jun N-terminal kinase signaling pathway. J. Biol. Chem. 279:38325-38330. [DOI] [PubMed] [Google Scholar]
- 18.Haque, M., V. Wang, D. A. Davis, Z. M. Zheng, and R. Yarchoan. 2006. Genetic organization and hypoxic activation of the Kaposi's sarcoma-associated Herpesvirus ORF34-37 gene cluster. J. Virol. 80:7037-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izumiya, Y., T. J. Ellison, E. T. Yeh, J. U. Jung, P. A. Luciw, and H. J. Kung. 2005. Kaposi's sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification. J. Virol. 79:9912-9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izumiya, Y., S. F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H. J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izumiya, Y., S. F. Lin, T. J. Ellison, A. M. Levy, G. L. Mayeur, C. Izumiya, and H. J. Kung. 2003. Cell cycle regulation by Kaposi's sarcoma-associated herpesvirus K-bZIP: direct interaction with cyclin-CDK2 and induction of G1 growth arrest. J. Virol. 77:9652-9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, K., A. Yokoyama, Y. Tohya, H. Akashi, Y. Nishiyama, and Y. Kawaguchi. 2003. Identification of protein kinases responsible for phosphorylation of Epstein-Barr virus nuclear antigen leader protein at serine-35, which regulates its coactivator function. J. Gen. Virol. 84:3381-3392. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi, Y., and K. Kato. 2003. Protein kinases conserved in herpesviruses potentially share a function mimicking the cellular protein kinase cdc2. Rev. Med. Virol. 13:331-340. [DOI] [PubMed] [Google Scholar]
- 25.Kawaguchi, Y., K. Kato, M. Tanaka, M. Kanamori, Y. Nishiyama, and Y. Yamanashi. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J. Virol. 77:2359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krosky, P. M., M. C. Baek, and D. M. Coen. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krosky, P. M., M. C. Baek, W. J. Jahng, I. Barrera, R. J. Harvey, K. K. Biron, D. M. Coen, and P. B. Sethna. 2003. The human cytomegalovirus UL44 protein is a substrate for the UL97 protein kinase. J. Virol. 77:7720-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kudoh, A., T. Daikoku, Y. Sugaya, H. Isomura, M. Fujita, T. Kiyono, Y. Nishiyama, and T. Tsurumi. 2004. Inhibition of S-phase cyclin-dependent kinase activity blocks expression of Epstein-Barr virus immediate-early and early genes, preventing viral lytic replication. J. Virol. 78:104-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao, W., Y. Tang, S. F. Lin, H. J. Kung, and C. Z. Giam. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 77:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, C. L., H. Li, Y. Wang, F. X. Zhu, S. Kudchodkar, and Y. Yuan. 2003. Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin. J. Virol. 77:5578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, M., J. Suen, C. Frias, R. Pfeiffer, M. H. Tsai, E. Chuang, and S. L. Zeichner. 2004. Dissection of the Kaposi's sarcoma-associated herpesvirus gene expression program by using the viral DNA replication inhibitor cidofovir. J. Virol. 78:13637-13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mailand, N., and J. F. Diffley. 2005. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 122:915-926. [DOI] [PubMed] [Google Scholar]
- 33.Marschall, M., M. Freitag, P. Suchy, D. Romaker, R. Kupfer, M. Hanke, and T. Stamminger. 2003. The protein kinase pUL97 of human cytomegalovirus interacts with and phosphorylates the DNA polymerase processivity factor pUL44. Virology 311:60-71. [DOI] [PubMed] [Google Scholar]
- 34.Marschall, M., A. Marzi, P. aus dem Siepen, R. Jochmann, M. Kalmer, S. Auerochs, P. Lischka, M. Leis, and T. Stamminger. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280:33357-33367. [DOI] [PubMed] [Google Scholar]
- 35.Moffat, J. F., L. Zerboni, M. H. Sommer, T. C. Heineman, J. I. Cohen, H. Kaneshima, and A. M. Arvin. 1998. The ORF47 and ORF66 putative protein kinases of varicella-zoster virus determine tropism for human T cells and skin in the SCID-hu mouse. Proc. Natl. Acad. Sci. USA 95:11969-11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Connor, C. M., and D. H. Kedes. 2006. Mass spectrometric analyses of purified rhesus monkey rhadinovirus reveal 33 virion-associated proteins. J. Virol. 80:1574-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, J., D. Lee, T. Seo, J. Chung, and J. Choe. 2000. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) open reading frame 36 protein is a serine protein kinase. J. Gen. Virol. 81:1067-1071. [DOI] [PubMed] [Google Scholar]
- 39.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polson, A. G., L. Huang, D. M. Lukac, J. D. Blethrow, D. O. Morgan, A. L. Burlingame, and D. Ganem. 2001. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 75:3175-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prichard, M. N., N. Gao, S. Jairath, G. Mulamba, P. Krosky, D. M. Coen, B. O. Parker, and G. S. Pari. 1999. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J. Virol. 73:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purves, F. C., and B. Roizman. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein alpha 22. Proc. Natl. Acad. Sci. USA 89:7310-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez, V., A. K. McElroy, J. Yen, S. Tamrakar, C. L. Clark, R. A. Schwartz, and D. H. Spector. 2004. Cyclin-dependent kinase activity is required at early times for accurate processing and accumulation of the human cytomegalovirus UL122-123 and UL37 immediate-early transcripts and at later times for virus production. J. Virol. 78:11219-11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scadden, D. T. 2003. AIDS-related malignancies. Annu. Rev. Med. 54:285-303. [DOI] [PubMed] [Google Scholar]
- 46.Schulz, T. F. 2006. The pleiotropic effects of Kaposi's sarcoma herpesvirus. J. Pathol. 208:187-198. [DOI] [PubMed] [Google Scholar]
- 47.Sharma-Walia, N., H. H. Krishnan, P. P. Naranatt, L. Zeng, M. S. Smith, and B. Chandran. 2005. ERK1/2 and MEK1/2 induced by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection. J. Virol. 79:10308-10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soong, W., J. C. Schultz, A. C. Patera, M. H. Sommer, and J. I. Cohen. 2000. Infection of human T lymphocytes with varicella-zoster virus: an analysis with viral mutants and clinical isolates. J. Virol. 74:1864-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Zeijl, M., J. Fairhurst, E. Z. Baum, L. Sun, and T. R. Jones. 1997. The human cytomegalovirus UL97 protein is phosphorylated and a component of virions. Virology 231:72-80. [DOI] [PubMed] [Google Scholar]
- 51.Verma, S. C., and E. S. Robertson. 2003. Molecular biology and pathogenesis of Kaposi sarcoma-associated herpesvirus. FEMS Microbiol. Lett. 222:155-163. [DOI] [PubMed] [Google Scholar]
- 52.Wang, J. T., P. W. Yang, C. P. Lee, C. H. Han, C. H. Tsai, and M. R. Chen. 2005. Detection of Epstein-Barr virus BGLF4 protein kinase in virus replication compartments and virus particles. J. Gen. Virol. 86:3215-3225. [DOI] [PubMed] [Google Scholar]
- 53.Wolf, D. G., C. T. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc. Natl. Acad. Sci. USA 98:1895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, F. Y., J. H. Ahn, D. J. Alcendor, W. J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, F. Y., Q. Q. Tang, H. Chen, C. ApRhys, C. Farrell, J. Chen, M. Fujimuro, M. D. Lane, and G. S. Hayward. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc. Natl. Acad. Sci. USA 99:10683-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, S. H., E. Jaffray, R. T. Hay, and A. D. Sharrocks. 2003. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell 12:63-74. [DOI] [PubMed] [Google Scholar]
- 57.Yokoyama, A., M. Tanaka, G. Matsuda, K. Kato, M. Kanamori, H. Kawasaki, H. Hirano, I. Kitabayashi, M. Ohki, K. Hirai, and Y. Kawaguchi. 2001. Identification of major phosphorylation sites of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP): ability of EBNA-LP to induce latent membrane protein 1 cooperatively with EBNA-2 is regulated by phosphorylation. J. Virol. 75:5119-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yue, W., E. Gershburg, and J. S. Pagano. 2005. Hyperphosphorylation of EBNA2 by Epstein-Barr virus protein kinase suppresses transactivation of the LMP1 promoter. J. Virol. 79:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, F. X., J. M. Chong, L. Wu, and Y. Yuan. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:800-811. [DOI] [PMC free article] [PubMed] [Google Scholar]