Abstract
One or more of the unique 3′-proximal open reading frames (ORFs) of the severe acute respiratory syndrome (SARS) coronavirus may encode determinants of virus virulence. A prime candidate is ORF6, which encodes a 63-amino-acid membrane-associated peptide that can dramatically increase the lethality of an otherwise attenuated JHM strain of murine coronavirus (L. Pewe, H. Zhou, J. Netland, C. Tangudu, H. Olivares, L. Shi, D. Look, T. Gallagher, and S. Perlman, J. Virol. 79:11335-11342, 2005). To discern virulence mechanisms, we compared the in vitro growth properties of rJ.6, a recombinant JHM expressing the SARS peptide, with isogenic rJ.6-KO, which has an inactive ORF containing a mutated initiation codon and a termination codon at internal position 27. The rJ.6 infections proceeded rapidly, secreting progeny about 1.5 h earlier than rJ.6-KO infections did. The rJ.6 infections were also set apart by early viral protein accumulation and by robust expansion via syncytia, a characteristic feature of JHM virus dissemination. We found no evidence for protein 6 operating at the virus entry or assembly stage, as virions from either infection were indistinguishable. Rather, protein 6 appeared to operate by fostering viral RNA and protein synthesis, as RNA quantifications by reverse transcription-quantitative PCR revealed viral RNA levels in the rJ.6 cultures that were five to eight times higher than those lacking protein 6. Furthermore, protein 6 coimmunoprecipitated with viral RNAs and colocalized on cytoplasmic vesicles with replicating viral RNAs. The SARS coronavirus encodes a novel membrane protein 6 that can accelerate replication of a related mouse virus, a property that may explain its ability to increase in vivo virus virulence.
The coronaviruses (CoVs) include many strains that collectively infect a variety of mammalian and avian hosts, causing respiratory, enteric, and neurologic diseases, often with severe clinical sequelae. Most notable are those causing human severe acute respiratory syndrome (SARS), an acute and frequently fatal epidemic pneumonia acquired from infection by animal SARS CoV strains (43). More common human coronaviruses include CoVs 229E, OC43, HKU-1 (47), and NL63 (38), which are frequent causes of upper respiratory tract infections, pneumonia, and croup (39, 48). The prevalence of the coronaviruses, their predilection for interspecies transfer to humans (1), and their potential for aggressive pathogenesis have all brought increased attention to these infectious agents.
Infectious coronavirus virions are enveloped particles with interiors harboring remarkably large ∼30-kb monopartite plus-strand RNA genomes. Translation of the 5′ ∼20 kb generates the so-called nonstructural proteins (nsp's) that largely function in viral RNA replication and transcription, while the remaining 3′ ∼10-kb template generates a nested set of subgenomic RNAs that are then translated to form the “structural” virion proteins S (spike), E (envelope), M (membrane), and N (nucleocapsid). Individual coronavirus types are set apart by additional viral genes, which are often designated as group-specific or “accessory” open reading frames (ORFs) to signify their dispensability for virus growth (49). SARS CoV genomes are replete with eight accessory ORFs, compacted into the 3′ region encoding the essential virion proteins (45). Notably, SARS CoVs containing these “extra” ORFs have been isolated from humans, terrestrial mammals, and bats (19), lending credence to the hypothesis that these accessory ORFs have been maintained in evolution for virus maintenance in changing in vivo environments.
Most of the SARS CoV accessory ORFs do not have any easily recognized homologs in other coronaviruses, and the functions of the ORF products remain unknown. Therefore, to begin to gain some understanding of these proteins and their relevance to virus infection, Pewe et al. (30) used reverse genetics (21) to transfer each SARS CoV ORF into an accommodating portion of the related mouse hepatitis virus (MHV) strain JHM-2.2v1 (42), thereby creating a set of recombinant JHM (rJ) viruses, each encoding their typical spectrum of JHM-specific products in conjunction with one of the SARS CoV ORF products. Each rJ virus was evaluated for growth in murine fibroblast tissue culture and in a well-established mouse model for JHM pathogenesis (8). One recombinant, rJ.6, exhibited a remarkably unique, hypervirulent character. These findings revealed that the SARS CoV protein functioned in a heterologous murine coronavirus, somehow operating to increase viral pathogenicity.
In SARS CoV genomes, the ORF6 coding sequence is within the 3′ regions encoding virion proteins, but interestingly, current evidence suggests that the translation product is “nonstructural” (30). Protein 6 is 63 amino acids in length, with ∼43 N-terminal residues being predominantly hydrophobic; indeed, the protein localizes exclusively to intracellular membranes in rJ.6-infected cells (30). Protein 6 has also been documented in SARS CoV infections using immunofluorescence assays (IFAs) (2), and its C-terminal, largely charged ∼23 residues were weakly immunogenic in some SARS CoV-infected patients (4). These findings indicate that ORF6 is normally expressed in SARS CoV infections and likely operates during the membrane-associated events of coronavirus growth, events that include viral RNA synthesis (12, 40), viral membrane protein synthesis, and virus assembly and secretion (5).
Our preliminary data suggested that protein 6 acted on the virus infection itself, independent of any in vivo effects, including the host immune response, because rJ.6 grew to a higher titer than isogenic recombinants in which the ORF6 sequence was mutated to prevent translation (30). We set out to explore this finding in mechanistic detail. Our results clarify how protein 6 operates during coronavirus infection and offer the intriguing insight that this small hydrophobic peptide may function to augment coronavirus RNA-dependent RNA synthesis.
MATERIALS AND METHODS
Cells.
17cl1 fibroblasts were grown in Dulbecco's modified Eagle medium (DMEM) containing 5% tryptose phosphate broth (Difco Laboratories) and 5% heat-inactivated fetal bovine serum (FBS). HeLa-MHVR5 cells (33), which stably synthesize MHV receptor carcinoembryonic antigen cell adhesion molecule isoform 1a (CEACAM1a), were grown in DMEM supplemented with 10% FBS (DMEM-10% FBS). All growth media included 0.01 M sodium HEPES (pH 7.4). Cell lines were propagated as adherent monolayer cultures.
Recombinant viruses.
Recombinant viruses were constructed from cDNAs of MHV JHM strain 2.2-v1 (42), a virus derived from neurovirulent MHV4 (44) but containing the neuroattenuating S mutation Leu1114Phe. The “parental” recombinant lacking SARS genes was designated rJ.2.2. Within rJ.2.2, dispensable gene 4 was replaced with SARS ORF3a or ORF6 to create rJ.6 or rJ.3a, respectively. An additional recombinant, isogenic to rJ.6 but lacking the SARS ORF6 initiation codon and containing a premature termination codon at ORF6 position 27, was also constructed and designated the knockout virus rJ.6-KO. All SARS ORFs included sequences at their 3′ ends encoding an influenza virus hemagglutinin (HA) monoclonal antibody (MAb) epitope. Further details on recombinant virus constructions are in reference 30. All viruses were cultured and evaluated for infectivity by titration on 17cl1 cells. Typical output titers ranged from 0.1 × 106 to 1 × 106 PFU per ml.
Infectious center assays.
17cl1 cell monolayers were infected at 0.1 PFU/cell, rinsed extensively with isotonic saline at 1 h postinfection (hpi), then trypsinized until the cells were suspended in a solution, and immediately diluted serially (1/2 log10 dilutions) with DMEM-10% FBS. Cell dilutions were then seeded onto fresh 17c1l cell monolayers and incubated for 3 days under agar plugs for development of infectious centers.
Cell-cell fusion assays.
HeLa-MHVR5 cells were treated with Lipofectin with pEMC-T7-luc (25), which encodes firefly luciferase under the transcriptional control of a bacteriophage T7 promoter. Twelve hours later, the cells were infected with indicated recombinant JHM viruses at 0.1 PFU/cell. Separate monolayers of HeLa-MHVR5 cells were simultaneously infected with vTF7.3 (10), which constitutively produces T7 RNA polymerase. At 8 h postinfection, the vTF7.3-infected cells were trypsinized, pelleted, resuspended in DMEM-10% FCS, and overlaid onto recombinant JHM (rJHM)-infected cells at 1:1 vTF7.3:rJHM coculture ratios. At various times after coculturing, the cells were dissolved and luciferase activities were quantified using commercial reagents (luciferase assay system; Promega Corp., Madison, WI) and a Turner Designs 20/20 bioluminometer.
Pulse-chase analyses.
HeLa-MHVR5 cells, infected with recombinant JHM viruses at 0.01 PFU/cell, were starved at 9 h postinfection for 30 min in cysteine- and methionine-free DMEM, replenished with the same medium containing 35S-labeled amino acids (Amersham, Inc.) for 30 min, and then rinsed extensively with isotonic saline before incubating in DMEM-10% FCS containing 2 mM methionine and 2 mM cysteine. At defined intervals thereafter, cells were placed on ice, rinsed with saline, and dissolved in NP-40-DOC buffer (25 mM sodium HEPES [pH 7.2], 100 mM NaCl, 0.01% bovine serum albumin [BSA], 0.5% NP-40, 0.5% sodium deoxycholate [DOC], 0.1% Sigma P2714 protease inhibitor cocktail). To capture 35S-labeled S proteins from lysates, N-domain (N)-CEACAM-human IgG1 Fc domain (Fc) (11) and protein A magnetic beads (New England Biolabs) were added for 2 h at 4°C, beads were collected by magnets and washed by sequential cycles of resuspension in NP-40-DOC buffer and collection by using magnets. Beads were then suspended in endoglycosidase H buffer (0.1 M sodium citrate [pH 5.5], 0.02% sodium dodecyl sulfate [SDS], 0.1 mM 2-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride), aliquoted, and incubated for 18 h at 37°C with or without 1 U of endoglycosidase H (Roche, Inc.). The 35S-labeled protein samples were then prepared for electrophoresis, subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by autoradiography as previously described (11).
Western blot analyses.
Recombinant JHM-infected HeLa-MHVR5 cells were dissolved at indicated times postinfection using NP-40-DOC buffer, and 105 cell equivalents were subjected to SDS-PAGE. Separated proteins were immunoblotted with anti-S murine monoclonal antibody 10G, kindly provided by Fumihiro Taguchi, anti-M MAb J.3.1 (9), and anti-N MAb J.3.3 (9). Bound antibodies were detected with horseradish peroxidase-conjugated secondary antibodies and Western Lightning chemiluminescence reagents (both from Perkin-Elmer, Inc.) and Kodak XAR film.
Reverse transcription and real-time PCR.
Total RNAs were isolated from rJHM-infected HeLa-MHVR5 cells at various times postinfection using RNeasy mini kit reagents (QIAGEN) and by following the manufacturer's instructions. Complementary DNAs were then synthesized using random primers as described previously (29). For PCR, 2-μl cDNA aliquots were added to 23-μl PCR cocktail containing 2× SYBR green master mix (ABI, Foster City, CA) and 0.2 μM concentrations of each sense and antisense primer (IDT DNA, Coralville, IA). Amplifications were performed in an ABI Prism 7700 thermocycler. The specificity of the amplification was confirmed using melting curve analysis. Data were collected and recorded by ABI Prism 7700 software and expressed as a function of the cycle threshold (CT), the cycle at which the fluorescence intensity in a given reaction tube rises above background (calculated as 10 times the mean standard deviation of fluorescence in all wells over the baseline cycles). Specific primer sets used for MHV-JHM and the murine housekeeping gene were as follows (5′ to 3′): JHM nucleocapsid sense, GACACAACCGACGTTCCTTT; JHM nucleocapsid antisense, and HPRT antisense, CAGATTCAACTTGCGCTCATC. Quantitative JHM nucleocapsid abundance was calculated as follows: for each cDNA sample assayed, CT values for reactions amplifying JHM nucleocapsid and HPRT were determined. JHM N CT values for each sample were corrected by subtracting the CT for HPRT. The ratio of JHM N RNA to HPRT RNA was calculated by the formula 2CT. The validity of this approach was confirmed by using serial 10-fold dilutions of cDNA. The amplification efficiencies for JHM nucleocapsid antisense and murine HPRT amplimers were found to be identical.
To determine relative virion RNA levels, media from 17cl1 cells infected with various rJ viruses were collected at 18 h postinfection and clarified by centrifugation. To 0.5-ml samples of clarified media, 105 PFU of vesicular stomatitis virus (VSV) was added to provide internal RNA controls. RNAs were extracted using MagMax viral RNA isolation reagents (catalog no. 1929; Ambion, Inc.), and cDNAs were prepared using RETRO Script (catalog no. 1710; Ambion, Inc.). Complementary DNAs were then quantitated by PCR as described above, using primer sets for JHM gene 1 (sense, ACGTCGATCGCAAGAGCCT; antisense, CTCTCGTCCGTAACCTCAACG) and for VSV nucleocapsid (sense, AGAGCAAGGAATGCCCGAC; antisense, GACCACATCTCTGCCTTGTGG). The amplification efficiencies for VSV nucleocapsid, which were similar between samples, were used to normalize for modest variations in RNA extraction. The CT values for the JHM gene 1 amplifications were then related to the respective rJ virus infectivity (plaque-forming units [PFU] per milliliter) to obtain relative specific infectivities of the different recombinant viruses.
RNA immunoprecipitations.
HeLa-MHVR5 cells (107) infected with the various JHM recombinants (0.01 PFU/cell) were rinsed extensively at 12 h postinfection with phosphate-buffered saline (PBS) and then suspended in 1-ml volumes of buffer K (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.2 mM EDTA, 0.1% protease inhibitor cocktail, 40 U/ml recombinant RNasin from Promega, Inc.). After incubation on ice for 30 min, cell suspensions were extruded five times through 20-gauge needles, followed by another five extrusions through 27-gauge needles. Cell lysates were then clarified by centrifugation at 2,000 × g for 5 min. Supernatants were incubated with nonspecific murine immunoglobulin G (IgG) and protein A magnetic beads for 1 h at 4°C, and then the beads were collected and removed before incubating for 2 h at 4°C with specific antibodies J.3.3 (anti-N) or 12CA5 (anti-HA epitope) along with protein A magnetic beads. Beads were washed extensively with ice-cold buffer K by five cycles of suspension and collection by using magnets, then suspended in RNA elution buffer (300 mM sodium acetate [pH 5.2], 0.2% SDS, 1 mM EDTA, 10 μg/ml proteinase K) and incubated for 10 min at 65°C. Eluted RNAs were further extracted with phenol and chloroform, precipitated with ethanol, and dissolved in 30 μl water. Five-microliter aliquots were then subjected to reverse transcription, and the cDNAs were used for PCR amplifications. Specific primer sets were for JHM gene 1a and HPRT as noted above.
Dual immunofluorescence and confocal microscopy.
HeLa-MHVR5 cells were grown on glass coverslips and infected with recombinant JHM viruses. At 11 h postinfection, actinomycin D (5 μg/ml) was added to cultures, and 1 h later, 5-bromouridine-5′-triphosphate (BrUTP) (Sigma, Inc.) was introduced into the cells using FuGENE-6 reagent (Roche, Inc.). Briefly, the FuGENE reagent was diluted to 10% (vol/vol) with DMEM, incubated for 5 min at 22°C, and then BrUTP (from 100 mM stock) and actinomycin D (from 5-mg/ml stock) were added to final concentrations of 3 mM and 5 μg/ml, respectively. Cells on coverslips were then incubated with 20 μl of the BrUTP mixture for 15 min at 4°C, and the coverslips were then placed in 0.5-ml DMEM-10% FBS and incubated for 15 min at 37°C. Cells were rinsed with PBS, fixed with 4% formaldehyde in PBS, and permeabilized with digitonin (5 μg/ml digitonin in 300 mM sucrose, 100 mM KCl, 2.5 mM MgCl2,1 mM EDTA,10 mM HEPES [pH 6.9]). Cells were then blocked with PBS containing 2% bovine serum albumin. BrUTP-labeled viral RNA was detected using mouse monoclonal anti-BrdU (Roche, Inc.) and anti-mouse IgG Alexa Fluor 568-conjugated anti-mouse IgG (Molecular Probes, Inc.). HA-tagged protein 6 (protein 6-HA) was directly detected using fluorescein isothiocyanate-conjugated rabbit anti-HA antibody (Immunology Consultancy Laboratories).
Immunofluorescence assays to detect MHV proteins 6 and nsp3 were performed on HeLa-MHVR5 cells, fixed, and permeabilized with digitonin at 12 h postinfection. Mouse monoclonal antibody 12CA5 (Roche, Inc.) was used to detect protein 6-HA, and rabbit anti-D3 serum (35), generously provided by Susan Baker, was used to detect nsp3. Secondary antibodies were Alexa Fluor 448-conjugated anti-mouse IgG (for protein 6-HA) and Alexa Fluor 568-conjugated anti-rabbit IgG (for nsp3). For detection of proteins 6 and M, infected HeLa-MHVR5 cells were permeabilized with methanol. Biotinylated mouse monoclonal anti-HA antibody 3F10 (Roche, Inc.) was used to detect protein 6-HA, and mouse monoclonal antibody 5B11.5, a gift from Michael Buchmeier, was used to detect M proteins. Secondary detection reagents were streptavidin-horseradish peroxidase-tyramide signal amplification-cyanine 3 (PerkinElmer) for HA-tagged protein 6 and fluorescein isothiocyanate-conjugated donkey anti-mouse sera for M.
After all immunostainings, coverslips were washed with PBS and mounted with ProLong Gold antifade reagent (Invitrogen). Epifluorescent images were obtained using a Leica DM-IRB microscope equipped with an Optronics charge-coupled device camera. One-micron confocal sections were visualized using a Carl Zeiss model 510 laser-scanning confocal microscope.
RESULTS
Effects of protein 6 on rJHM infection kinetics.
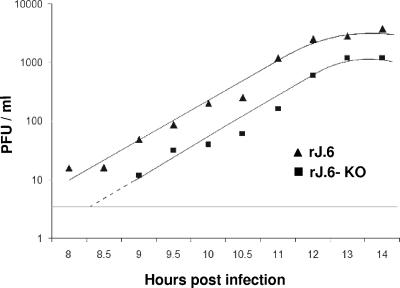
We recently reported that recombinant rJ.6, which encodes protein 6, yields infectious titers that are ∼0.5 to 1 log10 unit higher than parallel infections by rJ.2.2, the parent recombinant JHM, or by rJ.6-KO, an isogenic recombinant in which ORF6 is rendered incompetent for translation via mutation of the initiation codon and insertion of a premature termination codon (30). These results were obtained from infected L929 murine cells, with data points obtained at 6- to 12-h intervals. We extended these investigations with infected HeLa-MHVR cells and with special attention to the early kinetics of infective virion secretion. Time course experiments (Fig. 1) revealed rates of infective virus accumulation that were similar for both viruses throughout a ∼8- to 14-h postinfection period. The clear distinguishing feature of the rJ.6 infection was that virion secretion began ∼1 to ∼1.5 h earlier than for rJ.6-KO infection. Indeed, this was a modest kinetic difference, but it could be significant in accelerating multicycle in vivo infections. As these observations were reproduced in murine 17cl1 and L929 cells, at both 0.01 and 0.1 PFU/cell input multiplicities (data not shown), we suggest that host cell type and inoculum dose play little role in protein 6-mediated acceleration of infection.
FIG. 1.
Time course analysis of secreted virion infectivities. HeLa-MHVR5 cells were infected for 1 h at 0.01 PFU/cell with rJ.6#1 or rJ.6.KO#1, then rinsed, and replaced with fresh growth media. Infectious virus in the media collected at the indicated time points were quantified by plaque assay on 17cl1 indicator cells. The horizontal line indicates the sensitivity of the plaque assays (4 PFU/ml). Similar results were obtained in three independent experiments.
Effects of protein 6 on secreted virions.
The early secretion of rJ.6 might be attributed to acceleration of virus entry, eclipse phase events, and/or virion assembly and secretion through exocytic pathways. We first considered whether rJ.6 virions were set apart from rJ.6-KO and thus potentially capable of entering more rapidly into host cells. To this end, we incubated infected cells with 35S-labeled amino acids, collected and purified secreted virions by two cycles of sucrose gradient sedimentation, and then inspected virion proteins by SDS-PAGE and fluorography. We found the 35S-labeled virion protein profiles for both viruses to be essentially indistinguishable, with no evidence for protein 6 among the rJ.6 virion proteins (data not shown). This was in concert with our earlier discovery that protein 6 is not detectable in purified virions as judged by Western immunoblot analysis (30). Furthermore, from plaque assays we discovered that the infectivities of the two virion preparations, in terms of PFUs per 35S cpm, differed by less than twofold. We further compared the specific infectivities of rJ.6 and rJ.6-KO preparations by quantitating their genomic RNA content via reverse transcription-quantitative PCR (RT-qPCR). Our results (Table 1) confirmed that the specific infectivities of the two virion preparations, in terms of PFU per unit of genomic RNA, were essentially identical. These findings suggested that protein 6 did not change the structure or infectivity of secreted JHM virions, making it unlikely that the early secretion of rJ.6 could be explained by special virions with unusually rapid entry kinetics.
TABLE 1.
Relative specific infectivities of rJ.6 and rJ.6-KO preparations
| Expt | Virusa | Infectivity (PFU/ml)b | CT MHVc | CT VSVd | Relative MHV RNA contente | Relative specific infectivityf |
|---|---|---|---|---|---|---|
| 1 | rJ.6 | 1.1 × 106 | 24 (1.62-fold) | 16.1 (0.87-fold) | 1.86 | 1.18 |
| rJ.6-KO | 5 × 105 | 24.7 | 15.9 | 1 | 1 | |
| 2 | rJ.6 | 2 × 105 | 24 (7.46-fold) | 18.1 (1.62-fold) | 4.6 | 0.87 |
| rJ.6-KO | 5 × 104 | 26.9 | 18.8 | 1 | 1 |
In two independent experiments, rJ.6 and rJ.6-KO were grown in parallel and collected from infected 17cl1 cell supernatants at 18 h postinfection.
Infectivities were determined by plaque assays on 17cl1 indicator cells.
CT values for each virus pair were determined by RT-qPCR, and the differences in the CT values were converted to the changes in the differences (n-fold), using 2ΔCT = change in difference (n-fold).
CT values for the VSV RNA extracted from the VSV-spiked samples were determined by parallel RT-qPCR, and the CT differences were converted to the changes in the differences (n-fold).
Relative MHV RNA contents were determined by dividing the change in MHV RNA levels by the change in control VSV RNA levels. MHV RNA contents were normalized relative to rJ.6-KO.
Relative specific infectivity values (PFU/unit of genomic RNA) were determined by dividing the PFU ratios by the RNA ratios for each virus pair.
Effects of ORF6 on events during the eclipse phase.
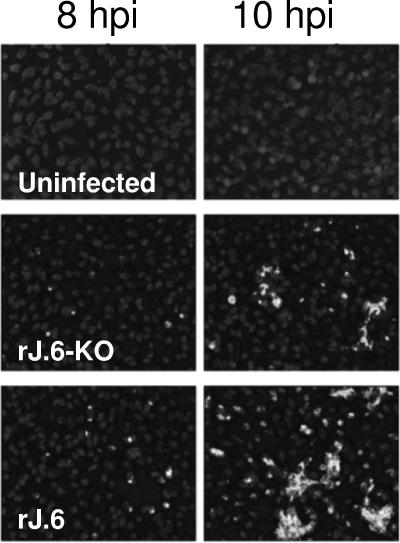
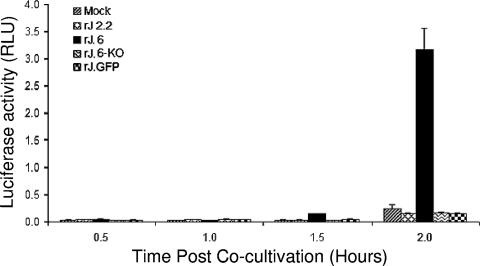
We turned our attention to comparing the virus infections during the eclipse phase. In our first approach, we determined the infection times required to accumulate sufficient viral proteins for detection by immunofluorescence assays. To this end, parallel HeLa-MHVR cultures were inoculated with rJHMs at identical, low input multiplicities. At 2 h postinfection, test cultures were evaluated by an infectious center assay, as described in Materials and Methods, with the results revealing the expected identical percentages of infected cells at this early time period. At later times postinfection, IFAs employing MAbs to M and N proteins were used to reveal infected cells. By this method, we found only 0.25% and 1% of cells infected by rJ.6-KO at 6 and 8 hpi, respectively, while 1% and 7% levels were observed for rJ.6 at these times. By 10 hpi, the rJ.6 infections were expanded throughout the monolayers, an effect of neighboring cells being recruited into syncytia (Fig. 2). These differing rates of syncytial expansion in the three cultures were further quantitated by an intercellular fusion-dependent reporter gene activation assay (24), and the results (Fig. 3) confirmed that the rJ.6 infections disseminated via syncytia significantly earlier and more robustly than infections by viruses lacking protein 6. These findings suggested that protein 6 operates very early after infection to accelerate the growth of JHM viruses.
FIG. 2.
Immunofluorescence detection of infected cells at 8 and 10 h postinfection. HeLa-MHVR5 cells were infected with rJ.6-KO or rJ.6 at 0.01 PFU/cell, fixed with paraformaldehyde at the indicated times, and then permabilized with methanol. Cells were immunostained for M protein (MAb J.1.3), and nuclei were recognized by incubation with Hoescht 33258. The panels in this figure are representative of ∼20 panels from each culture that were evaluated to obtain the percentage of IFA-positive cells stated in the text.
FIG. 3.
Quantification of syncytial expansions in rJ.6- and rJ.6-KO-infected cultures. vTF7.3-infected HeLa-MHVR cells were cocultivated with pEMC-T7-luc-transfected HeLa-MHVR cells that had been infected 8 h earlier with the indicated recombinant viruses. At the indicated times after cocultivation, cells were dissolved, mixed with luciferin substrate, and evaluated in a luminometer for light emissions. Data are plotted in relative light units (RLU).
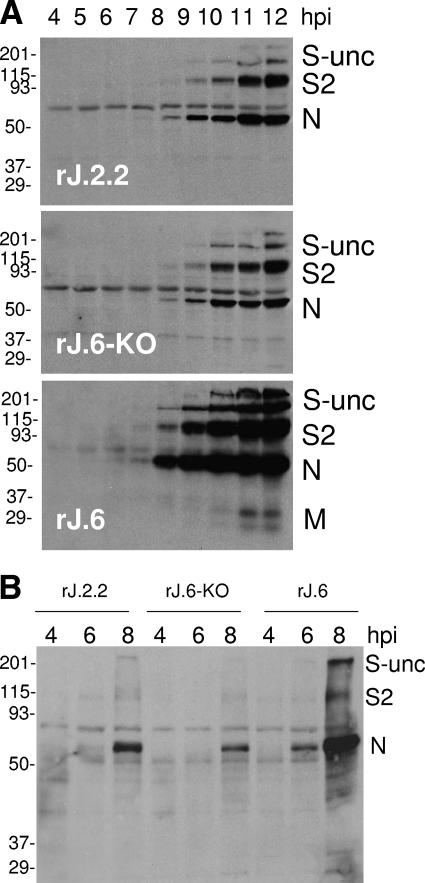
We next determined the time course of viral protein accumulation in the infected cultures by Western immunoblotting, and the findings (Fig. 4A) revealed that viral N proteins were detectable as early as 6 hpi, 2 h earlier than the times required to detect the same proteins in the other two cultures lacking ORF6. The striking accumulation of viral S, M, and N proteins in the rJ.6 cells at the later 10- to 12-hpi time points was not simply an outcome of cellular recruitment into growing syncytia, because even when syncytia were blocked with an anti-CEACAM antibody, viral proteins still accumulated earlier and to higher levels in the rJ.6 infections (Fig. 4B). Indeed, the syncytium-inducing S proteins did not appear to be preferential or direct targets of protein 6 activity, because exocytic transport rates of S proteins were identical in all infected cultures, as measured by pulse-chase labeling with 35S-labeled amino acids (data not shown). Indeed, S proteins in all cultures adopted the conformations required for precipitation by N-CEACAM-Fc (18) within ∼30 min (data not shown), suggesting that protein 6 did not grossly alter the protein folding environment in the endoplasmic reticulum (ER). We considered these findings to be relevant because protein 6 localizes to intracellular vesicles (30) and might alter the function of the exocytic pathway in some fashion if it accumulated to high densities on the ER or Golgi apparatus.
FIG. 4.
Time course analysis of virion protein accumulations. (A) HeLa-MHVR5 cells were infected with the indicated viruses at 0.01 PFU/cell, and individual cultures were dissolved at hourly intervals. A total of 105 cell equivalents were electrophoresed, and virion proteins S, N, and M were detected by Western blotting. All Western blots were incubated together in the same antibody solutions and then exposed equally via chemiluminescence detection methods. The positions of molecular mass standards (in kilodaltons) are listed to the left of the gels. S-unc, uncleaved S; S2, C-terminal S posttranslation product. (B) HeLa-MHVR cells were infected with the indicated viruses at 0.01 PFU/cell. At 3 hpi, rabbit anti-MHVR antiserum (1:50) was added to infected cultures at 3 hpi to block syncytial developments. Cell lysates were then collected at the indicated times postinfection and evaluated by Western blotting for S, N, and M proteins. The positions of molecular mass standards (in kilodaltons) are listed to the left of the gel.
Relationships between protein 6 and viral RNA synthesis.
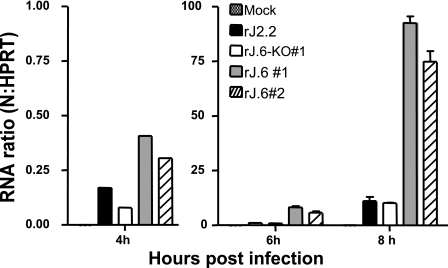
The early accumulations of viral proteins in the rJ.6-infected cultures were clearly observed before the detection of secreted virions (Fig. 1) and also before the development of syncytia (Fig. 2), pointing the evidence toward protein 6-mediated kinetic effects during the eclipse phase of infection. The effects could be at the level of viral replication, transcription, and/or translation. To investigate the relationships between protein 6 and viral RNA synthesis, we first measured total viral RNA accumulations at 4, 6, and 8 hpi by RT-qPCR. We found that rJ.6 viral RNAs accumulated to levels that were five- to eightfold higher than the rJ.6-KO levels (Fig. 5). These results, as expected, paralleled our findings on viral protein accumulations (Fig. 4). Notably, these higher viral RNA levels were observed as early as 4 hpi, a time when we were unable to detect protein 6 by either immunofluorescence or Western blot assays, suggesting that the very early synthesis of minute quantities of protein 6 can quickly act to augment either viral translation or replication.
FIG. 5.
Time course analysis of viral RNA accumulations. HeLa-MHVR cells were infected with the indicated recombinant viruses at a multiplicity of infection of 0.01 PFU/cell, and total cellular RNAs were harvested from individual cultures at 4, 6 and 8 h postinfection. MHV N gene-specific RNAs were then quantified by RT-qPCR, normalizing the level of N gene amplicons to that of HeLa cell HPRT amplicons, as described in Materials and Methods.
To further consider whether the effect of protein 6 might be at the level of viral RNA synthesis, we investigated possible linkages with viral RNAs. In addressing biochemical associations, a coimmunoprecipitation approach was employed (3). The procedure involved disrupting infected cells by extrusion through 27-gauge needles, a method that preserves intracellular vesicles, then immunoprecipitating protein 6-HA and associated lipid organelle fragments onto HA-specific antibody-coated beads. The adsorbed subcellular components were digested with SDS and proteinase K, and any coprecipitated RNAs were purified by phenol extraction. The RNAs captured in this way were then recognized by their ability to act as a template for RT-PCR amplicons.
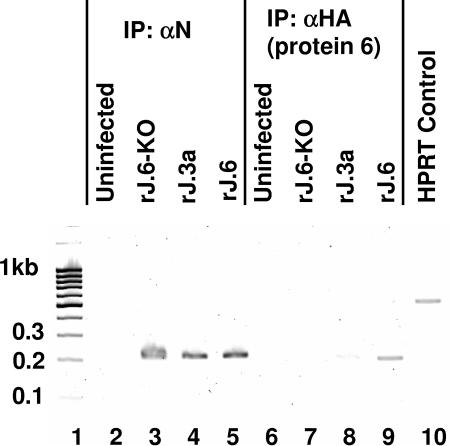
Results obtained from this approach revealed that JHM gene 1 sequences did indeed coprecipitate with protein 6-HA (Fig. 6, lane 9). RNAs collected in parallel from rJ.6KO-infected cells did not contain the gene 1 sequences (lane 7), and the RNAs isolated in complex with ORF3a-HA contained minimal gene 1 levels (lane 8). The ORF3a protein is a known virion-associated SARS accessory protein (16). Controls employing antibodies binding to N ribonucleoproteins revealed that all infected-cell lysates contained immunoprecipitable viral RNAs (lanes 2 to 4). Notably, all PCRs in these approaches included primers specific for cellular HPRT RNA, and in no case were HPRT DNA sequences amplified from the immunoprecipitated nucleic acids (Fig. 6, note control lane 10). These findings indicated that protein 6-HA specifically associated with viral RNAs, either directly or indirectly through proteins or lipid membranes.
FIG. 6.
Coimmunoprecipitation of protein 6 with JHM-specific RNAs. Cytoplasmic extracts were prepared by needle extrusion of rJHM-infected HeLa-MHVR5 cells and then incubated with the indicated antibody-coated magnetic beads. Coimmunoprecipitating RNAs were eluted from the harvested beads and used to act as templates in RT-PCR mixtures designed to amplify both viral nucleocapsid and cellular HPRT sequences. PCR amplification products were separated by electrophoresis and imaged by ethidium bromide staining. Molecular size standards in kilobase pairs are illustrated in lane 1. HPRT amplicons templated by total uninfected cell RNAs are revealed in lane 10. IP: αN, immunoprecipitation with anti-N.
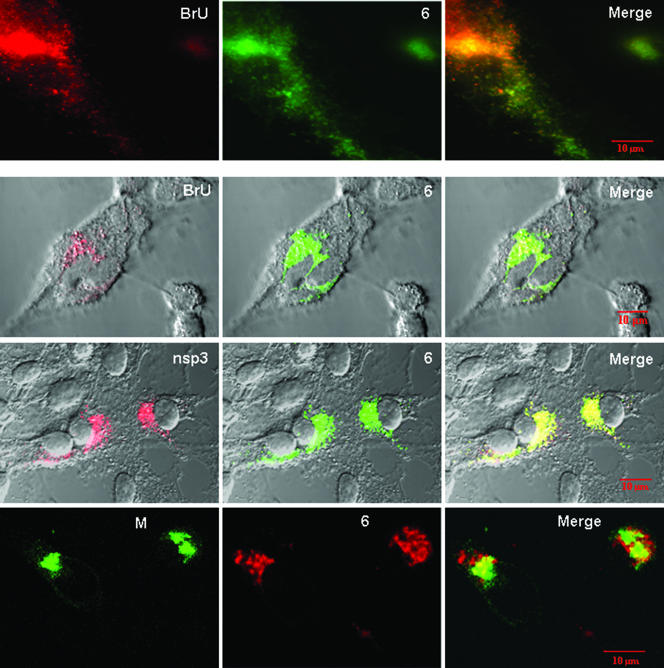
Although the biochemical approach yielded convincing data, the findings did not provide information on the in situ locations of HA-tagged protein 6-RNA complexes, nor did they reveal whether protein 6-HA localized with actively replicating viral RNAs. Therefore, a complementary cell biological approach was employed to determine whether protein 6 colocalized with intracellular sites of viral RNA synthesis (6, 12). Briefly, viral RNA synthesis was localized by inhibiting cellular transcription with actinomycin D, pulsing with bromouridine (BrU), and then applying anti-BrU antibodies. Our results (Fig. 7) revealed punctate patterns of BrU deposition that were similar to those observed previously in MHV-infected cells (12, 31). We presume that these sites of BrU accumulation are the membrane vesicles known to act as platforms for viral RNA synthesis. These vesicular structures were also sites of anti-HA deposition (Fig. 7), indicating significant colocalization of protein 6 with replicating viral RNAs.
FIG. 7.
Intracellular localizations of protein 6 relative to replicating viral RNAs and nsp3 and M proteins. HeLa-MHVR cells infected with rJ.6 viruses were pulse-labeled with BrUTP in the presence of actinomycin D (top two rows of panels) or incubated without BrUTP labeling (bottom two rows). All cells were then fixed, permeabilized, and incubated with fluorescent antibodies recognizing incorporated BrU (top two rows), MHV nsp3 (third row), MHV M (bottom row), and the C-terminal HA epitopes were appended to protein 6 (all panels). The images in the top row were taken with a Leica DM-IRB epifluorescence microscope; all remaining images were taken with a Zeiss model 510 laser-scanning confocal microscope. Images depicting the subcellular locations of BrUTP-labeled RNAs, nsp3, and M are shown in the leftmost column, images of ORF6-HA are shown in the middle column, and superimposed images (Merge) are shown in the rightmost column.
To continue localizing protein 6 in infected cells, additional assays were performed using rabbit antiserum D3 (35), specific for MHV nsp3, a gene 1 posttranslation product that has been carefully localized to sites of viral RNA replication (12). In double-label immunofluorescence assays with D3 and anti-HA antibodies, we found striking colocalizations (Fig. 7). These similar positions of protein 6 and nsp3 contrasted with those obtained in immunofluorescence assays for proteins 6 and M, a central virion assembly subunit (Fig. 7). Together our findings argue that protein 6 operates during eclipse-phase MHV RNA replicative events, not during concomitant and subsequent MHV assembly processes.
DISCUSSION
The fundamental events of coronavirus infection take place in association with cellular membranes. Entry into cells involves fusion of virion and endosome membranes (14), newly synthesized viral RNA replicases function on intracellular membranes during the eclipse phase (51), nascent virion proteins incorporate into ER membranes, and ER-Golgi intermediate compartment membranes are then acquired to form new virion coats (5). Therefore, it is not surprising that several coronavirus accessory proteins are associated with membranes, including SARS CoV proteins 3a (50), 6 (30), 7a (23), 7b (27), and 9b (22). How these accessory membrane proteins function at each infection stage depends, in part, on whether they are virion components. Recent findings indicate that proteins 3a (16) and 7a (15) are virion components, while our current evidence reveals that protein 6 is not. In murine fibroblast cell cultures infected with rJ.6, we have never observed any significant incorporation of protein 6 into infective virions (30). Indeed, in this study we found that protein 6 effected no significant changes in the infectivity of secreted recombinant JHM viruses (Table 1), nor did protein 6 change the rate of JHM S protein folding and transport through the Golgi apparatus. These findings suggested that the accumulating protein 6 did not compromise the integrity of the exocytic pathway, at least not enough to impede the progress of virion-associated S proteins, nor did protein 6 change the character of secreted virions such that subsequent entry into neighboring cells would be impacted. The findings lead us to the view that protein 6 accelerates rJHM infection kinetics at stages distinct from virus entry into cells or virus assembly and exit out of cells.
Thus, we considered how protein 6 might function during the eclipse phase of rJHM infection, and in doing so, we speculated on its relationships to other viral peptides. Protein 6 is largely hydrophobic at its amino terminus and charge-rich and relatively basic at its carboxy terminus, an overall architecture that bears some similarity to other “nonstructural” proteins that are translated from the subgenomic RNAs of other coronaviruses. For example, the 7-kDa ORF3a protein encoded by the group 3 coronavirus infectious bronchitis virus, is not present in virions but is abundant on the ER membranes of infected cells (28). Similar to SARS CoV protein 6, the infectious bronchitis virus 3a protein is dispensable for virus growth in immortalized cell cultures, its functional relevance being discerned only in cultures mimicking the in vivo environment (13). The 15-kDa ORF4 protein of MHV, the very ORF replaced by the SARS ORF6 in rJ.6, is similarly hydrophobic at its amino terminus and basic at its carboxy end and is also dispensable for MHV growth in tissue culture (26). The ORF7 protein encoded by the group 1 coronavirus transmissible gastroenteritis virus is a hydrophobic 9-kDa membrane-associated peptide. Remarkably, the transmissible gastroenteritis virus ORF7 product has been suggested to act in tethering viral replicases to membranes (37), a function that, on the basis of the findings in this report, we might suggest for SARS CoV protein 6. With the development of complementation assays in which accessory genes are expressed in trans during infection, we can evaluate whether protein 6 or any of the other small hydrophobic proteins foster viral RNA replicative events. These investigations are in progress.
While the infection-accelerating function for SARS CoV protein 6 was revealed in the context of a recombinant MHV infection, it is not yet clear how readily this same function might be evident in authentic SARS CoV infections. Recombinant SARS CoVs lacking the ORF6 gene have been constructed and evaluated relative to complete SARS CoV, with findings of little, if any, effect of ORF6 deletion on in vitro or in vivo (lung) virus growth (49). It is possible that the choices of different cell lines and mouse strains accounted for the absence of an overt protein 6 phenotype in this study. Additionally, the relatively complex SARS CoV genome may encode functions rendering its protein 6 more redundant than that observed in the heterologous rJHM infections. Regardless, it is clear that advancing our studies will require careful comparisons of recombinant SARS CoVs with or without ORF6 deletions, as performed in this study with recombinant JHMs, for viral RNA synthesis rates and protein 6 localizations.
Our evidence linking protein 6 to coronavirus RNA synthesis includes its striking positive effect on JHM RNA accumulation (Fig. 5), its coimmunoprecipitation with viral RNAs (Fig. 6), and its subcellular vesicular colocalization with both recently synthesized virus-specific RNAs and replication component nsp3 (Fig. 7). While none of these findings can resolve possible direct linkages between protein 6 and viral RNA replication machinery, they do suggest that protein 6 might assist in the development and/or maintenance of these subcellular sites of viral RNA synthesis. How might this be accomplished? One possibility is that the N-terminal portion of protein 6 might adopt an amphipathic alpha-helical configuration in complex with membranes, perhaps with the putative helix parallel to the bilayer in analogy to the amphipathic residues of the alphavirus nsp1 (17), a well-known plus-strand RNA virus replicase component (34). Notably, protein 6 has several arginines within its N-terminal region, spaced roughly seven residues apart, making it possible that intercalation on membranes creates a platform of positive charges that are set apart from the negatively charged phospholipid surface. It will be interesting to determine whether these charged residues within the largely hydrophobic N-terminal region are essential for protein 6 to foster viral RNA amplification.
Lipid bilayer architectures surrounding sites of viral RNA-dependent RNA synthesis are relatively diverse and are therefore variably designated as convoluted membranes, spherules, vesicle packets, and double-membrane vesicles (20, 46). Coronavirus-induced structures appear to be double-membrane vesicles (12). The organelles contributing membrane to these coronavirus-induced structures are not yet certain, with perhaps both endosomes and ER participating (35, 36, 40). Additional contributors may include membranous components of the autophagic pathway, as double-membrane vesicles with colocalizing viral nsp's and autophagic marker LC3 have been identified in coronavirus-infected cells (31). Protein 6 may mobilize or remodel these intracellular membranes, perhaps affecting the rate at which double-membrane vesicles are formed, as this function would likely enhance RNA synthesis. That protein 6 may act to alter membrane shape is suggested by parallels to the reticulons, a class of membrane morphogenetic proteins stabilizing highly curved ER membranes (41). The long amphipathic N-terminal portion of protein 6 can theoretically intercalate into membranes in a hairpin configuration, similar to that proposed for reticulons Rtn4a/NogoA (41), thereby distorting and perhaps vesiculating intracellular membranous organelles.
SARS CoV protein 6 also has remarkable similarities with the coronavirus E proteins, in that both are small hydrophobic peptides with membrane-intercalating N-terminal regions. The E proteins are known to induce highly curved “tubular bodies,” an outcome of their accumulation on ER-Golgi intermediate compartment membranes (32). E proteins thus operate in virion morphogenesis, perhaps creating appropriate particle curvatures (7). We have no evidence for SARS CoV protein 6 assisting in virion production and are thus intrigued by the possibility that SARS CoV in particular encodes separate membrane remodeling proteins, for example, protein 6 to create vesicles fostering viral RNA synthesis and E protein to generate secreted virion vesicles.
We believe that the ability of protein 6 to enhance virus replication is linked to the separate finding that rJ.6 is highly neurovirulent. Consistent with this, we observed preferential growth of rJ.6 in the murine central nervous system 6 to 9 days postinfection (30). Additionally, protein 6, perhaps by deforming cytoplasmic membranes, may also accelerate virus-induced cell death, thereby contributing to virulence. To further appreciate how protein 6 functions in viral RNA metabolism and in in vivo virulence, we are working toward a structural resolution of this 63-residue peptide in membranes.
Acknowledgments
We thank Joseph Boscarino, Jason Lacny, and Lecia Pewe for technical assistance. We are grateful to Susan Baker (Loyola University), John Fleming (University of Wisconsin), Michael Buchmeier (Scripps Research Institute), and Fumihiro Taguchi (National Institute of Infectious Diseases, Tokyo, Japan) for generously providing antibodies used in this study.
This work was supported by NIH grant P01 AI060699.
Footnotes
Published ahead of print on 15 November 2006.
REFERENCES
- 1.Baric, R. S., B. Yount, L. Hensley, S. A. Peel, and W. Chen. 1997. Episodic evolution mediates interspecies transfer of a murine coronavirus. J. Virol. 71:1946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, W. S., C. Wu, S. C. Chow, T. Cheung, K. F. To, W. K. Leung, P. K. Chan, K. C. Lee, H. K. Ng, D. M. Au, and A. W. Lo. 2005. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS). Mod. Pathol. 18:1432-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi, K. S., P. Huang, and M. M. Lai. 2002. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology 303:58-68. [DOI] [PubMed] [Google Scholar]
- 4.Chow, S. C., C. Y. Ho, T. T. Tam, C. Wu, T. Cheung, P. K. Chan, M. H. Ng, P. K. Hui, H. K. Ng, D. M. Au, and A. W. Lo. 2006. Specific epitopes of the structural and hypothetical proteins elicit variable humoral responses in SARS patients. J. Clin. Pathol. 59:468-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Haan, C. A., and P. J. Rottier. 2005. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 64:165-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Hage, N., and G. Luo. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84:2761-2769. [DOI] [PubMed] [Google Scholar]
- 7.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleming, J. O. 1995. Coronaviruses. J. Neurovirol. 1:323-325. [DOI] [PubMed] [Google Scholar]
- 9.Fleming, J. O., S. A. Stohlman, R. C. Harmon, M. M. C. Lai, J. A. Frelinger, and L. P. Weiner. 1983. Antigenic relationships of murine coronaviruses: analysis using monoclonal antibodies to JHM (MHV-4) virus. Virology 131:296-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallagher, T. M. 1997. A role for naturally occurring variation of the murine coronavirus spike protein in stabilizing association with the cellular receptor. J. Virol. 71:3129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodgson, T., P. Britton, and D. Cavanagh. 2006. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 80:296-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann, H., and S. Pohlmann. 2004. Cellular entry of the SARS coronavirus. Trends Microbiol. 12:466-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, C., N. Ito, C. T. Tseng, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 80:7287-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, N., E. C. Mossel, K. Narayanan, V. L. Popov, C. Huang, T. Inoue, C. J. Peters, and S. Makino. 2005. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 79:3182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lampio, A., I. Kilpelainen, S. Pesonen, K. Karhi, P. Auvinen, P. Somerharju, and L. Kaariainen. 2000. Membrane binding mechanism of an RNA virus-capping enzyme. J. Biol. Chem. 275:37853-37859. [DOI] [PubMed] [Google Scholar]
- 18.Lewicki, D. N., and T. M. Gallagher. 2002. Quaternary structure of coronavirus spikes in complex with carcinoembryonic antigen-related cell adhesion molecule cellular receptors. J. Biol. Chem. 277:19727-19734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676-679. [DOI] [PubMed] [Google Scholar]
- 20.Mackenzie, J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masters, P. S., and P. J. Rottier. 2005. Coronavirus reverse genetics by targeted RNA recombination. Curr. Top. Microbiol. Immunol. 287:133-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier, C., A. R. Aricescu, R. Assenberg, R. T. Aplin, R. J. Gilbert, J. M. Grimes, and D. I. Stuart. 2006. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure 14:1157-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson, C. A., A. Pekosz, C. A. Lee, M. S. Diamond, and D. H. Fremont. 2005. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure 13:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okuma, K., M. Nakamura, S. Nakano, Y. Niho, and Y. Matsuura. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235-244. [DOI] [PubMed] [Google Scholar]
- 26.Ontiveros, E., L. Kuo, P. S. Masters, and S. Perlman. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 289:230-238. [DOI] [PubMed] [Google Scholar]
- 27.Pekosz, A., S. R. Schaecher, M. S. Diamond, D. H. Fremont, A. C. Sims, and R. S. Baric. 2006. Structure, expression, and intracellular localization of the SARS-CoV accessory proteins 7a and 7b. Adv. Exp. Med. Biol. 581:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pendleton, A. R., and C. E. Machamer. 2005. Infectious bronchitis virus 3a protein localizes to a novel domain of the smooth endoplasmic reticulum. J. Virol. 79:6142-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pewe, L., G. F. Wu, E. M. Barnett, R. F. Castro, and S. Perlman. 1996. Cytotoxic T cell-resistant variants are selected in a virus-induced demyelinating disease. Immunity 5:253-262. [DOI] [PubMed] [Google Scholar]
- 30.Pewe, L., H. Zhou, J. Netland, C. Tangudu, H. Olivares, L. Shi, D. Look, T. Gallagher, and S. Perlman. 2005. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 79:11335-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raamsman, M. J., J. K. Locker, A. de Hooge, A. A. de Vries, G. Griffiths, H. Vennema, and P. J. Rottier. 2000. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 74:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao, P. V., and T. M. Gallagher. 1998. Intracellular complexes of viral spike and cellular receptor accumulate during cytopathic murine coronavirus infections. J. Virol. 72:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. C. Baker, J. W. Oh, and M. M. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J. Virol. 73:5957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tung, F. Y., S. Abraham, M. Sethna, S. L. Hung, P. Sethna, B. G. Hogue, and D. A. Brian. 1992. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology 186:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Hoek, L., K. Sure, G. Ihorst, A. Stang, K. Pyrc, M. F. Jebbink, G. Petersen, J. Forster, B. Berkhout, and K. Uberla. 2005. Croup is associated with the novel coronavirus NL63. PLoS Med. 2:e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voeltz, G. K., W. A. Prinz, Y. Shibata, J. M. Rist, and T. A. Rapoport. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124:573-586. [DOI] [PubMed] [Google Scholar]
- 42.Wang, F. I., J. O. Fleming, and M. M. C. Lai. 1992. Sequence analysis of the spike protein gene of murine coronavirus variants: study of genetic sites affecting neuropathogenicity. Virology 186:742-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, M., M. Yan, H. Xu, W. Liang, B. Kan, B. Zheng, H. Chen, H. Zheng, Y. Xu, E. Zhang, H. Wang, J. Ye, G. Li, M. Li, Z. Cui, Y. F. Liu, R. T. Guo, X. N. Liu, L. H. Zhan, D. H. Zhou, A. Zhao, R. Hai, D. Yu, Y. Guan, and J. Xu. 2005. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 11:1860-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiner, L. P. 1973. Pathogenesis of demyelination induced by a mouse hepatitis virus (JHM virus). Arch. Neurol. 28:298-303. [DOI] [PubMed] [Google Scholar]
- 45.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wileman, T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312:875-878. [DOI] [PubMed] [Google Scholar]
- 47.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo, P. C., S. K. Lau, H. W. Tsoi, Y. Huang, R. W. Poon, C. M. Chu, R. A. Lee, W. K. Luk, G. K. Wong, B. H. Wong, V. C. Cheng, B. S. Tang, A. K. Wu, R. W. Yung, H. Chen, Y. Guan, K. H. Chan, and K. Y. Yuen. 2005. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J. Infect. Dis. 192:1898-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yount, B., R. S. Roberts, A. C. Sims, D. Deming, M. B. Frieman, J. Sparks, M. R. Denison, N. Davis, and R. S. Baric. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79:14909-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan, X., J. Li, Y. Shan, Z. Yang, Z. Zhao, B. Chen, Z. Yao, B. Dong, S. Wang, J. Chen, and Y. Cong. 2005. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 109:191-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 287:57-94. [DOI] [PMC free article] [PubMed] [Google Scholar]