Abstract
One of the critical steps in the progression to cervical cancer appears to be the establishment of persistent human papillomavirus (HPV) infection. We have demonstrated that the lack of cytotoxic T-lymphocyte response to HPV type 16 (HPV 16) E6 protein was associated with persistence and that the potential presence of dominant CD8 T-cell epitopes was most frequently found (n = 4 of 23) in the E6 16-40 region by examining the pattern of CD8 T-cell epitopes within the E6 protein in women who had cleared their HPV 16 infections. The goal of this study was to define the minimal/optimal amino acid sequences and the HLA restricting molecules of these dominant CD8 T-cell epitopes as well as those of subdominant ones if present. Three dominant epitopes, E6 29-38 (TIHDIILECV; restricted by the HLA-A0201 molecule), E6 29-37 (TIHDIILEC; restricted by B48), and E6 31-38 (HDIILECV; restricted by B4002), and one subdominant epitope, E6 52-61 (FAFRDLCIVY; restricted by B35) were characterized. Taken together with a previously described dominant epitope, E6 52-61 (FAFRDLCIVY; restricted by B57), the CD8 T-cell epitopes demonstrated striking HLA class I binding promiscuity. All of these epitopes were endogenously processed, but the presence of only two of the five epitopes could have been predicted based on the known binding motifs. The HLA class I promiscuity which has been described for human immunodeficiency virus may be more common than previously recognized.
Cervical cancer is the second most common cancer among women worldwide (CANCERMondial [http://www-dep.iarc.fr/], 2005). The association between human papillomavirus (HPV) infections, most commonly HPV type 16 (HPV 16), and the development of cervical cancer has been well described (46). However, the exact mechanisms leading from HPV infection to malignancy are not well understood. One of the critical steps in the progression to cervical cancer appears to be the establishment of persistent infection. Epidemiologic evidence suggests that over 90% of women are able to clear their HPV infections, including the oncogenic HPV 16 type. The remaining women whose HPV infections persist are at risk for the development of invasive cancer. The E6 and E7 proteins of high-risk HPV types are expressed early after infection and are known to mediate cell transformation by interacting with products of cellular human tumor suppressor genes. The E6 protein can bind and promote degradation of cell-encoded p53, while the E7 protein interacts with the retinoblastoma susceptibility gene product (7, 18, 32, 36, 37, 41). In a longitudinal study using multiple chromium release assays, we found that detectable responses to HPV 16 E6 were more common in those women who cleared their HPV 16 infections than in women who had viral persistence. Interestingly, no association was found for E7 (29). Furthermore, we examined the pattern of HPV 16 E6 CD8 T-cell epitopes recognized by women who had evidence of HPV 16 clearance (27). Among 23 women with evidence of HPV 16 clearance, we found evidence of potential antigenic CD8 T-cell epitopes in 8 women, and there was a propensity of finding these epitopes in the N-terminal half of the E6 protein. Among these women, the presence of dominant epitopes was found most commonly (n = 4) in the E6 16-40 region. The goal of this study was to characterize the dominant T-cell epitopes in this region by defining their minimal and optimal amino acid sequences and their restriction HLA molecules. Subdominant epitopes from the same subjects, if present, were also described. A method previously used to describe the E6 52-61 epitope (FAFRDLCIVY) restricted by the HLA-B57 molecule from one of the subjects was used (26). Here, we describe three new endogenously processed T-cell epitopes of the HPV 16 E6 protein, E6 29-37 (TIHDIILEC; restricted by HLA-B48), E6 31-38 (HDIILECV; restricted by HLA-B4002), and E6 52-61 (FAFRDLCIVY; restricted by HLA-B35). In addition, E6 29-38 (TIHDIILECV), restricted by HLA-A0201, was described, but this epitope has been previously reported by other investigators (19, 33). These five CD8 T-cell epitopes identified in the HPV 16 E6 protein demonstrate striking HLA class I binding promiscuity and are found in either the E6 29-38 region (HLA-A0201, -B48, and -B4002 restricted) or the E6 52-61 region (HLA-B35 and -B57 restricted).
MATERIALS AND METHODS
Subjects.
The generation of CD8 T-cell lines and enzyme-linked immunospot (ELISPOT) assays to examine the pattern of CD8 T-cell epitopes, in women who were able to clear their HPV 16 infections, have been described previously (27). The CD8 T cells from subjects who demonstrated the potential presence of dominant epitopes in the HPV 16 E6 16-40 region were characterized further. The study protocol was approved by the University of California at San Francisco Committee on Human Research and by the Institutional Review Board of the University of Arkansas for Medical Sciences.
HLA typing.
HLA class I typing was performed at the University of California at San Francisco Immunogenetics Laboratory or at the University of Arkansas for Medical Sciences HLA Laboratory using peripheral blood lymphocytes (PBL) or an Epstein-Barr virus (EBV)-transformed B-lymphoblastoid cell line (LCL). A serological method or PCR sequence-specific amplification (PCR-SSP) methods was used. High-resolution HLA class I A, B, and/or C locus typing, by sequencing or by PCR-SSP, was also performed whenever necessary.
Peptides.
Overlapping 9-mer peptides (overlapping by 8 amino acids) and 15-mer peptides (overlapping by 10 amino acids) covering the entire HPV 16 E6 protein were described previously (26). Additional HPV 16 E6 peptides (7-mers, 8-mers, 10-mers, and 11-mers) were synthesized as necessary to define the minimal and optimal amino acid sequences of the CD8 T-cell epitopes. The purity of peptides ranged from 70% to 99%. The amino acid sequences of all peptides were derived from the HPV 16 German prototype (38).
Magnetic selection of IFN-γ-secreting cells to isolate T-cell clones.
To isolate T-cell clones recognizing the E6 antigenic epitope from subjects 5, 7, and 15, their CD8 T-cell lines were stimulated as described previously (27) for two additional 7-day cycles, so that the frequency of targeted T-cell clones would be above the threshold of selection (approximately 0.1%). The peptide-specific T cells were positively selected using the gamma interferon (IFN-γ) secretion assay enrichment kit, according to the manufacturer's instructions (Miltenyi Biotec), after stimulating cells with 10 μM of each peptide contained in the positive peptide pools (Table 1). Selected cells were plated at a concentration of 0.5 cells per well in the presence of a 0.5× feeder cell mixture (Yssel's medium containing 1% pooled human serum, penicillin G [100 units/ml], streptomycin [100 μg/ml], 5 × 105/ml irradiated allogeneic PBL, 5 × 104/ml irradiated JY cells, and 0.1 μg/ml phytohemagglutinin [PHA]). Control wells for growth contained 1 to 1,000 cells per well. On day 5, 100 μl of Yssel's medium containing 20 U/ml of recombinant interleukin-2 was added to each well. Growing microcultures were transferred to 24-well plates containing 1 ml of a 1× feeder cell mixture per well (Yssel's medium containing 1% pooled human serum, penicillin G [100 units/ml], streptomycin [100 μg/ml], 1 × 106/ml irradiated allogeneic PBL, 1 × 105/ml irradiated JY cells, and 0.1 μg/ml PHA). Although the limiting-dilution technique described here is a standard method of isolating T-cell clones, a formal analysis of clonality of these T-cell clones using a molecular method was not performed.
TABLE 1.
Positive peptide pools for CD8 T-cell responses to the HPV16 E6 protein in subjects with dominant peaks in the E6 16-40 region
| Subject | T-cell responsea for HPV 16 E6 regionb:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1-25 | 16-40 | 31-55 | 46-70 | 61-85 | 76-100 | 91-115 | 106-130 | 121-145 | 136-158 | |
| 5 | X | x | ||||||||
| 7 | X | x | ||||||||
| 15 | X | |||||||||
| 20 | X | x | ||||||||
X, strongest T-cell response for a given subject; x, subdominant T-cell response for a given subject.
Each region contains three 15-mer peptides overlapping by the central 10 amino acids.
ELISPOT assays for screening of T-cell clones.
The ELISPOT assay method described by Larsson and colleagues was used with minor modifications (23). Briefly, a 96-well plate (Millititer; Millipore, Bedford, MA) was coated with 5 μg/ml primary anti-IFN-γ monoclonal antibody (Mabtech, Stockholm, Sweden) and stored at 4°C overnight. The plate was then washed four times with phosphate-buffered saline (PBS) and blocked using RPMI 1640 plus 5% pooled human serum for 1 h at 37°C. Fifty to 100 μl of culture medium containing the T-cell clones was collected, and the cells were washed, resuspended with RPMI 1640 with 5% pooled human serum, and divided into a single well at the same position in replicate plates. One hundred thousand autologous LCL cells were also added to all wells. PHA at 10 μg/ml was added as positive control in one well, and no T-cell clone cells were added to another well (negative control). One 15-mer peptide (10 μM) of the positive peptide pools (three peptides in each pool) was added to each plate along with the last plate, to which only medium was added (no-peptide negative control). For some subjects with multiple positive peptide pools, the pools were used per plate to screen the T-cell clones in order to reduce the number of ELISPOT plates. After a 20-h incubation at 37°C, the plate was washed four times with PBS plus 0.05% Tween 20. A total of 50 μl of secondary antibody (1 μg/ml biotin-conjugated anti-IFN-γ monoclonal antibody; Mabtech) was added, and the plate was incubated for 2 h at 37°C. The plate was then washed four times with PBS plus 0.1% Tween 20. Avidin-bound biotinylated horseradish peroxidase H (Vectastain Elite kit; Vector laboratories, Inc., Burlingame, CA) was added for 1 h at 37°C. After four washings with PBS with 0.1% Tween 20, stable diaminobenzene (Research Genetics, Huntsville, AL) was added to develop the reaction. The plates were washed with deionized water three times and air dried overnight. Spot-forming units were counted using an automated ELISPOT analyzer (Cell Technology, Inc., Jessup, MD).
ELISPOT assays to characterize the CD8 T-cell epitopes.
To assess whether the T-cell clones recognize endogenously processed antigen, autologous LCLs infected with recombinant vaccinia virus expressing HPV 16 E6 (E6-vac) at a multiplicity of infection (MOI) of 10 were used as antigen-presenting cells (APC) in ELISPOT assays. In later experiments, an MOI of 5 was used since other experiments have shown little difference in the numbers of spot-forming units with an MOI of 5 or 10 (K. H. Kim, W. Greenfield, E. Shotts, and M. Nakagawa, submitted for publication). The wild-type virus, Western Reserve (WR), and recombinant vaccinia virus expressing HPV 16 E7 (E7-vac), if available, were used as the negative controls. Autologous LCL cells were washed twice using RPMI 1640 with 1% pooled human serum, and appropriate amounts of virus were added to the respective tubes. The tubes were incubated at 37°C for 1 h with mixing every 20 min. One thousand cells from each T-cell clone were plated per well along with 1 × 105 infected autologous LCL cells in duplicates or triplicates. Otherwise, the ELISPOT assays were carried out as described above.
The standard approach used to define the minimal and optimal peptide antigen epitope was to determine which of the three 15-mers was positive in each positive peptide pool and then to test seven overlapping 9-mer peptides contained within the positive 15-mer peptide. Typically only one of the seven 9-mer peptides happened to be positive, and the testing of two 10-mers containing the 9-mer and two 8-mers within the 9-mer followed. All peptides were used at a concentration of 10 μM, and 1 × 103 cells from each T-cell clone along with 1 × 105 autologous LCL cells were plated to each well. The minimal and optimal peptide was defined as the shortest peptide which was able to elicit the highest number of spot-forming units. When there was an uncertainty as to which peptide may be minimal and optimal, the candidate peptides were serially diluted (10−5 M to 10−10 M) and the shortest peptide with more spot-forming units at the lower concentrations of the peptide was determined to be minimal and optimal. If the antigenicity was determined to be equivalent among multiple peptides with the limiting-dilution experiments, the shortest peptide was designated as “the minimal peptide among equally effective peptides”.
ELISPOT assay and chromium release assay for identifying the restricting HLA class I molecules.
The putative restricting HLA class I molecule was identified using allogeneic LCLs sharing one or a few class I molecules by an ELISPOT assay in such a manner that all of the subjects' HLA class I molecules (A, B, and C) were examined. An autologous LCL was used as a positive control. One thousand T-cell clone cells, 1 × 105 allogeneic LCL cells, and the antigenic peptide were plated in triplicates, and the assay was otherwise performed as described above. High background spot-forming units were previously observed (26), likely due to HLA molecules on the surface of the T-cell clones being able to present the peptides to each other. Therefore, the chromium release assays were performed to confirm the ELISPOT results using multiple allogeneic LCLs expressing the putative restricting HLA class I molecule. The LCLs were pulsed with 10 μM of a given peptide antigen. The cells were radiolabeled using 100 μCi sodium chromate (Na251CrO4) and incubated with the peptide. After a washing, the cells were plated in triplicates in 96-well plates at 3 × 103 cells per well. Effector cells were added at multiple effector/target cell ratios. The plated cells were pelleted by centrifugation and then incubated for 5 h at 37°C in a humidified 5% CO2 incubator. The supernatants were harvested using a Skatron harvesting press, and the chromium-51 was counted using a gamma counter (Packard Instruments, Meriden, CT). Percent specific lysis was calculated as described previously (28).
Fluorescence-activated cell sorter analysis.
The T-cell clones were stained for surface markers with anti-CD4-anti-CD8 and anti-CD3-anti-CD16 (Caltag, Burlingame, CA) and analyzed using FACSCalibur (Becton Dickinson Immunocytometry Systems, San Jose, CA) or Coulter EPICS XL-MLC flow cytometer (Beckman Coulter, Fullerton, CA).
RESULTS
Screening of CD8 T-cell clones.
Table 1 shows the patterns of possible HPV 16 E6 CD8 T-cell epitopes in the four subjects being studied. The frequencies of T cells specific for the potential dominant epitopes were 0.05%, 0.02%, 0.04%, and 0.05% of CD8 T cells for subjects 5, 7, 15, and 20, respectively (27). Cells of the CD8 T-cell lines (2 × 106 to 7.1 × 106 cells per subject) were used for magnetic selection, and the yields of IFN-γ-positive cells were 0.21%, 2.7%, 0.46%, and 0.52%, respectively. One-half or all of the IFN-γ-positive cells were used for limiting dilution, and 95 to 400 T-cell clones per subject that grew well were expanded in 24-well plates 2 weeks after limiting dilution. Ninety-four to 188 T-cell clones were screened using the IFN-γ ELISPOT assay, and most or all of T-cell clones that demonstrated higher numbers of spot-forming units with a peptide or a peptide pool compared with the no-peptide control were retested with individual 15-mer peptides (data not shown) and with the E6-vac-infected autologous LCL (Fig. 1).
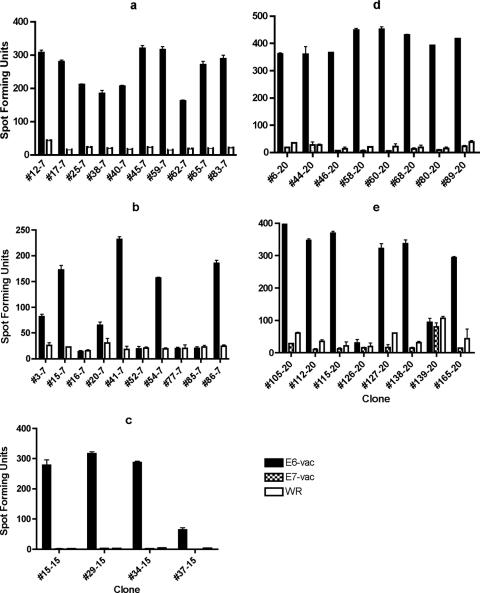
FIG. 1.
ELISPOT assays performed using E6-vac-, E7-vac-, and/or Western Reserve (WR)-infected autologous EBV LCLs as antigen-presenting cells revealed that the T-cell clones from subjects 7, 15, and 20 recognize endogenously processed E6 epitopes. The number after the hyphen indicates subject of origin. The bars represent standard errors of the means. (a) Twenty of 20 T-cell clones from the subject 7 screen positive for the E6 16-40 region were positive for an endogenously processed E6 epitope. Ten representative clones are shown. The experiment was done with an MOI of 5 in triplicates, and E6-vac and WR were tested. (b) Six of 10 T-cell clones from the subject 7 screen positive for the E6 46-70 region were positive for a endogenously processed E6 epitope. The experiment was done with an MOI of 5 in triplicates, and E6-vac and WR were tested. (c) Ten of 14 T-cell clones from the subject 15 screen positive for the E6 16-40 region were positive for a endogenously processed E6 epitope. Four representative clones are shown. The experiment was done with an MOI of 10 in duplicates, and E6-vac, E7-vac, and WR were tested. (d) Eight of eight T-cell clones from the subject 20 screen positive for the E6 16-40 region were positive for an endogenously processed E6 epitope. The experiment was done with an MOI of 5 in duplicates, and E6-vac, E7-vac, and WR were tested. (e) Six of eight T-cell clones from the subject 20 screen positive for the E6 31-55 region were positive for an endogenously processed E6 epitope. The experiment was done with an MOI of 5 in duplicates, and E6-vac, E7-vac, and WR were tested.
For subject 7, 10 (#12-7, #17-7, #25-7, #38-7, #40-7, #45-7, #59-7, #62-7, #65-7, and #83-7) of 10 T-cell clones positive for the E6 16-40 region in the screening ELISPOT assay were positive for the E6 26-40 15-mer peptide (data not shown) and 20 of 20 T cell clones were positive with the E6-vac-infected autologous LCL (Fig. 1a). For the E6 46-70 region, 6 (#3-7, #15-7, #20-7, #41-7, #54-7, and #86-7) of 6 T-cell clones were positive for the E6 51-65 15-mer peptide (data not shown) and 6 of 10 T-cell clones were positive for the E6-vac-infected autologous LCL (Fig. 1b). For subject 15, 10 (#1-15, #15-15, #27-15, #29-15, #34-15, #37-15, #68-15, #77-15, #92-15, and #93-15) of 14 CD8 T-cell clones positive for the E6 16-40 region were positive for the E6 26-40 15-mer peptide (data not shown) and 10 of 14 T-cell clones were positive with the E6-vac-infected autologous LCL (Fig. 1c). For subject 20, eight (#6-20, #44-20, #46-20, #58-20, #60-20, #68-20, #80-20, and #89-20) of eight CD8 T-cell clones positive for the E6 16-40 region were positive for the E6 26-40 15-mer peptide (data not shown) and eight of eight T-cell clones were also positive with the E6-vac-infected autologous LCL (Fig. 1d). For the E6 31-55 region, four (#105-20, #115-20, #127-20, and #138-20) of four positive CD8 T-cell clones were positive for the E6 31-45 15-mer peptide (data not shown) and six of eight positive clones were positive with the E6-vac-infected autologous LCL (Fig. 1e). For subject 5, none of the five T-cell clones positive for the E6 26-40 region was positive upon retesting with the three 15-mer peptides or with the E6-vac-infected autologous LCL. Two (clones #1-5 and #17-5) of 10 T-cell clones positive for the E6 136-158 region were positive with the E6 141-155 (15-mer) peptide (data not shown) but not with the E6-vac-infected autologous LCL. We concluded that this epitope was not endogenously processed.
Minimal and optimal amino acid sequences of the CD8 T cell epitope.
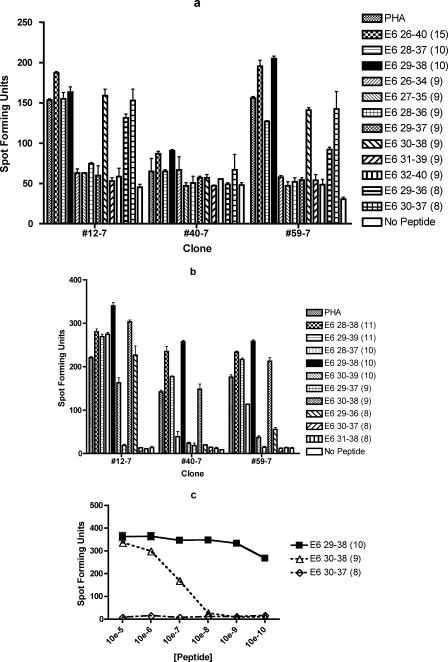
To characterize subject 7's dominant epitope within the E6 26-40 region, seven 9-mers, two 8-mers, two 10-mers were tested (Fig. 2a). Of the 9-mers, only the E6 30-38 peptide was positive for clones #12-7 and #59-7. The two 8-mers tested, E6 29-36 and E6 30-37, also demonstrated recognition by clones #12-7 and #59-7. For clone #40-7, only a small difference was detected between the PHA-positive control and the no-peptide negative control. Since this is likely to be due to the poor condition of the T-cell clone on the particular day, the experiment was repeated for clone #40-7 (data not shown). The highest number of spot-forming units were detected significantly above the no-peptide control with the E6 29-38 10-mer, followed in order by E6 26-40 15-mer, E6 30-38 9-mer, E6 30-37 8-mer, E6 29-36 8-mer, E6 28-37 10-mer, and PHA. In order to define the minimal and optimal peptide, the ELISPOT assay was repeated using the 11-mers surrounding the E6 29-38 10-mer (Fig. 2b). The highest number of spot forming units were detected with the E6 29-38 10-mer with all three clones tested in Fig. 2b. To further support the conclusion that the minimal and optimal epitope is the E6 29-38 10-mer peptide, serial dilutions of the E6 29-38 10-mer, E6 30-38 9-mer, and E6 30-37 8-mer were performed with clones #40-7 and # 59-7 (Fig. 2c). The results were consistent with the notion that the E6 29-38 10-mer is the minimal and optimal peptide.
FIG.2.
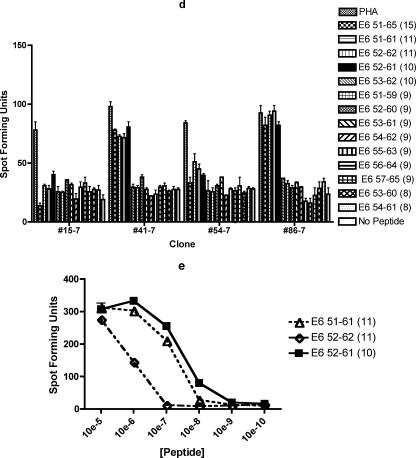
ELISPOT assays demonstrating that the shortest and optimal peptide for subject 7's dominant epitope is E6 29-38 and that the miminal among equally effective peptides of her subdominant epitope is E6 52-61. The numbers in parentheses indicate peptide lengths in amino acids. The bars represent standard errors of the means. (a) Two of three T-cell clones tested demonstrated the highest number of spot-forming units with E6 29-38 among a number of peptides tested including two 8-mers (E6 29-36 and E6 30-37), seven overlapping 9-mers covering the E6 26-40 region, two 10-mers (E6 28-37 and E6 29-38), and one 15-mer (E6 26-40). The experiment was done in duplicates. (b) Comparison of the E6 28-38 11-mer, E6 29-39 11-mer, E6 28-37 10-mer, E6 29-38 10-mer, E6 30-39 10-mer, E6 29-37 9-mer, E6 30-38 9-mer, E6 29-36 8-mer, E6 30-37 8-mer, and E6 31-38 8-mer revealed that the optimal peptide of minimum length is likely to be the E6 29-38 10-mer. The experiment was done in triplicates. (c) Comparison of the E6 29-38 10-mer, E6 30-38 9-mer, and E6 30-37 8-mer, ranging from 10−5 M to 10−10 M, confirmed the optimal peptide of minimum length to be the E6 29-38 10-mer peptide. The experiment was done in duplicates, and the results of one representative clone (#59-7) out of two clones tested are shown. (d) Two of four T-cell clones tested demonstrated the most number of spot-forming units with E6 52-61 among a number of peptides tested including two 8-mers (E6 53-60 and E6 54-61), seven overlapping 9-mers covering the E6 51-65 region, two 10-mers (E6 52-61 and E6 53-62), two 11-mers (E6 51-61 and E6 52-62), and one 15-mer (E6 51-65). The experiment was done in duplicates. (e) Comparison of the E6 51-61 11-mer, E6 52-62 11-mer, and E6 52-61 10-mer, ranging from 10−5 M to 10−10 M, confirmed that the minimal peptide between the two optimal peptides is the E6 52-61 10-mer peptide. The experiment was done in triplicates, and the graph of one representative clone (#86-7) out of two clones tested is shown.
The subdominant epitope from subject 7 was also characterized. In addition to seven 9-mers, two 8-mers, two 10-mers, and two 11-mers were examined (Fig. 2d). None of the 9-mers was positive, and E6 52-61 had the largest number of spot-forming units for clones #15-7 and #41-7. For clones #54-7 and #86-7, a larger number of spot-forming units was demonstrated with the E6 51-61 11-mer peptide among peptides examined. The dilutional analysis of the peptides has shown that the E6 52-61 10-mer peptide and the E6 51-61 11-mer peptide have similar patterns (#41-7 and #86-7). The representative graph for clone #86-7 is shown in Fig. 2e. Therefore, the minimal peptide between the two similarly optimal peptides for subject 7's subdominant epitope appears to be the E6 52-61 10-mer.
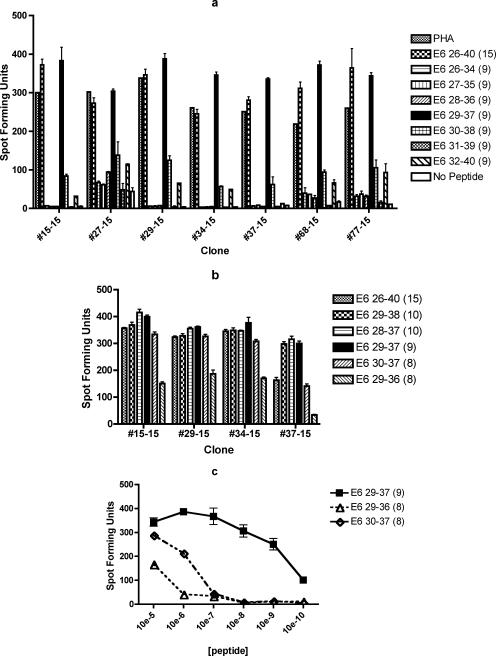
For subject 15, seven clones (#15-15, #27-15, #29-15, #34-15, #37-15, #68-15, and #77-15) were tested using seven 9-mer peptides (Fig. 3a). All clones were most strongly positive with E6 29-37 among the 9-mer peptides. To define the shortest and optimal sequence of this epitope, two 8-mer peptides within E6 29-37 and two 10-mer peptides surrounding E6 29-37 were tested (#15-15, #29-15, #34-15, and #37-15; Fig. 3b). The response to the E6 29-37 9-mer was stronger in all clones tested compared to either 8-mer. However, the difference between the E6 30-37 8-mer and E6 29-37 9-mer was >100 spot-forming units only for clone #37-15. The responses to the E6 26-40 15-mer, E6 29-38 10-mer, E6 28-37 10-mer, and E6 29-37 9-mer were similar. To clarify whether the E6 30-37 8-mer or the E6 29-37 9-mer was the optimal peptide, these two peptides and the E6 29-36 8-mer peptide were serially diluted and retested. The E6 29-37 9-mer was positive over a wider range of dilutions compared to either one of the 8-mers for four clones tested (#15-15, #29-15, #34-15, and #37-15). A representative graph for clone #15-15 is shown in Fig. 3c. These results suggest that the shortest and optimal peptide is the E6 29-37 9-mer.
FIG. 3.
ELISPOT assays demonstrating that the shortest and optimal peptide for subject 15's dominant epitope is E6 29-37. The bars represent standard errors of the means. (a) Seven of seven T-cell clones tested demonstrated the highest number of spot-forming units with E6 29-38 among the seven overlapping 9-mers covering the E6 26-40 region and the 15-mer. The experiment was done in duplicates. (b) Comparison of the E6 26-40 15-mer, E6 29-38 10-mer, E6 28-37 10-mer, E6 29-37 9-mer, E6 30-37 8-mer, and E6 29-36 8-mer revealed that the optimal peptide of minimum length is likely to be the E6 29-37 9-mer. The experiment was done in triplicates. (c) Comparison of the E6 29-37 9-mer, E6 30-37 8-mer, and E6 29-36 8-mer, ranging from 10−5 M to 10−10 M, confirmed the optimal peptide of minimum length to be the E6 29-37 9-mer peptide. The experiment was done in triplicates, and the graph of one representative clone (#15-15) out of four clones tested is shown.
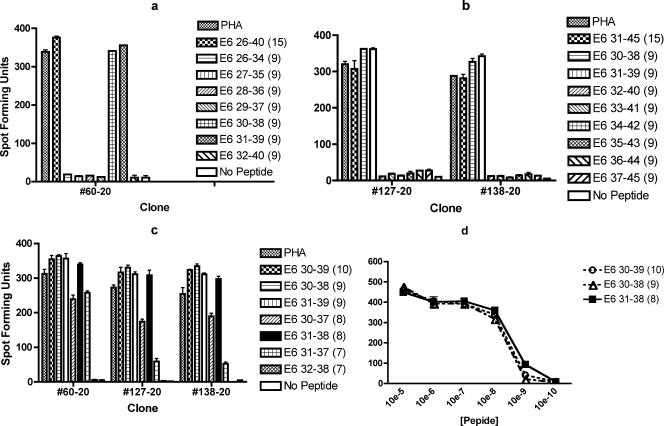
For subject 20, one clone (#60-20) positive for the E6 26-40 region and two clones (#127-20 and #138-20) positive for the E6 31-45 region were tested with separate sets of 9-mer peptides. All three clones (#60-20, #127-20, and #138-20) were positive for the E6 30-38 and E6 31-39 peptides (Fig. 4a and b). Therefore, a single epitope appeared to be present in an area of overlap of the E6 26-40 and E6 31-45 regions. The three clones were also tested with two 7-mers, two 8-mers, two 9-mers, and one 10-mer (Fig. 4c). Less spot-forming units were seen with the E6 31-37 7-mer, E6 32-38 7-mer, and E6 30-37 8-mer, but those with the E6 31-38 8-mer, E6 30-38 9-mer, E6 31-39 9-mer, and E6 30-39 10-mer were similar. Serial dilutions of the peptides were performed to compare the E6 30-39 10-mer, E6 30-38 9-mer, and E6 31-39 9-mer for clone #138-20 (data not shown) and to compare the E6 30-39 10-mer, E6 30-38 9-mer, and E6 31-38 8-mer for clones #60-20 and #138-20 (Fig. 4d). The E6 30-38 9-mer peptide seems to have better affinity compared to the E6 31-39 9-mer (data not shown). Those of the E6 31-38 (8-mer), E6 30-38 (9-mer), and E6 30-39 (10-mer) appear to be similar (Fig. 4d). Therefore, the shortest of these three peptides, E6 31-38 (8-mer), appears to be the minimal peptide among equally optimal peptides for subject 20's dominant epitope.
FIG. 4.
ELISPOT assays demonstrating that the shortest among equally optimal peptides for subject 20's dominant epitope present within the overlapping amino acids in the E6 26-40 and E6 31-45 regions is the E6 31-38 peptide. The bars represent standard errors of the means. (a) Clone #60-20 demonstrated large numbers of spot-forming units with E6 30-38 and E6 31-39 among the seven overlapping 9-mers covering the E6 26-40 region. The experiment was done in duplicates. (b) Clones #127-20 and #138-20 demonstrated large numbers of spot-forming units with E6 30-38 and E6 31-39 among the seven overlapping 9-mers covering the E6 31-45 region and the 15-mer. The experiment was done in duplicates. (c) Comparison of the E6 30-39 10-mer, E6 30-38 9-mer, E6 31-39 9-mer, E6 30-37 8-mer, E6 31-38 8-mer, E6 31-37 7-mer, and E6 32-38 7-mer revealed that the optimal peptide of minimum length is likely to be the E6 31-38 8-mer. The experiment was done in triplicates. (d) Comparison of the E6 30-39 10-mer, E6 30-38 9-mer, and E6 31-38 8-mer, ranging from 10−5 M to 10−10 M, showed that the minimal among three equally optimal peptides is the E6 31-38 8-mer peptide. The experiment was done in duplicates, and results with one representative clone (#60-20) out of two clones tested are shown.
The two T-cell clones from subject 5 (#1-5 and #17-5) which were positive with the E6 141-155 15-mer peptide were tested with seven overlapping 9-mers within this peptide along with three 11-mers (E6 142-152, E6 143-153, and E6 144-154). None of the 9-mers or 11-mers was positive by ELISPOT assay although the positivity to the E6 141-155 peptide was confirmed by ELISPOT assay with both clones (data not shown). This epitope was not characterized any further since it does not appear to be endogenously processed.
Restriction element of the CD8 T-cell epitopes.
Allogeneic LCLs sharing one or a few HLA class I molecules with each subject were used to determine the restriction element using ELISPOT assays (data not shown). None of the allogeneic LCLs appear to be positive for the E6 29-38 epitope (#12-7, #40-7, #45-7, and #59-7) (data not shown), an HLA-B35 matched allogeneic LCL appeared to be positive for the E6 52-61 epitope (#15-7, #41-7, #54-7, and #86-7) (data not shown), an HLA-B48 matched allogeneic LCL appeared to be positive for the E6 29-37 epitope (#15-15, #27-15, #29-15, #34-15, and #37-15) (data not shown), and an HLA-B4002 matched allogeneic LCL appeared to be positive for the E6 31-38 epitope (#60-20 and #138-20) (data not shown).
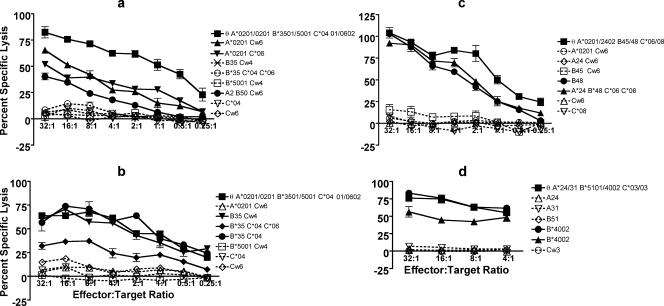
For subject 7's E6 29-38 epitope, HLA-A0201 matched LCLs demonstrated cytotoxicity but the level of lysis was noticeably less than that with the HLA-A0201-positive autologous LCL (clones #40-7 and #59-7; Fig. 5a). This does not seem to be due to the difference in the lysability of these EBV LCLs since an EBV LCL positive for HLA-A2, -B35, and -B57 also showed less cytotoxicity with the E6 29-37 (A0201-restricted) T-cell clone (#59-7) compared to that with the E6 52-61 (B35-restricted) T-cell clone (#86-7) and E6 52-61 (B57-restricted) T-cell clone (26) (data not shown). For subject 7's E6 52-61 epitope, the chromium release showed cytotoxicity with three HLA-B35 matched allogeneic LCLs (clones #41-7 and #86-7) but the level of lysis was lower for one of the allogeneic LCLs compared to other LCLs and the autologous LCL (Fig. 5b). It is possible that this allogeneic LCL may express a different allele of B35 compared to the autologous LCL. Similarly, the chromium release assay was able to confirm that HLA-B48 was the restriction molecule for the E6 29-37 epitope (#34-15 and #37-15; Fig. 5c) and that HLA-B4002 was the restriction molecule for the E6 31-38 epitope (#60-20, #127-20, and #138-20; Fig. 5d). The clones #34-15 and #37-15, specific for the E6 29-37 epitope, were also tested with another B48-restricted HPV epitope (E7 7-15 [26]) using the autologous LCL and two allogeneic LCLs expressing B48. No cytotoxicity was observed, underscoring the specificity of the T-cell clones (data not shown).
FIG. 5.
Identifying the restricting HLA class I molecule for the CD8 T-cell epitope using chromium release assays. θ, autologous LCL; *, HLA type determined using one of the molecular methods. The bars represent standard errors of means. (a) The E6 29-38 epitope appears to be restricted by the A0201 molecule. A representative (#59-7) of two clones is shown. (b) The E6 52-61 epitope appears to be restricted by the B35 molecule. A representative (#86-7) of two clones is shown. (c) The E6 29-37 epitope appears to be restricted by the B48 molecule. A representative (#34-15) of the two clones is shown. (d) The E6 31-38 epitope appears to be restricted by the B4002 molecule. A representative (#127-20) of three clones is shown.
The surface phenotype of all but one of the T-cell clones was CD3+ CD4− CD8+ CD16− (#12-7, #17-7, #25-7, #38-7, #40-7, #45-7, #59-7, #62-7, #65-7, #83-7, #3-7, #15-7, #20-7, #41-7, #54-7, #86-7, #15-15, #29-15, #34-15, #37-15, #6-20, #60-20, #89-20, #105-20, #115-20, #127-20, #138-20, and #165-20; data not shown). Clone #27-15 appeared to consist of a mixed population of T cells with a CD4+ subset and a CD8+ subset (data not shown).
DISCUSSION
In this study, we described three dominant epitopes (E6 29-38, restricted by the HLA-A0201 molecule, E6 29-37, restricted by B48, and E6 31-38, restricted by B4002) in the E6 16-40 region and one subdominant epitope (E6 52-61, restricted by B35) in women who cleared their HPV 16 infections. These data complement our prior work which had demonstrated the importance of the T-cell responses to the HPV 16 E6 protein in viral clearance (29). Taken together with a previously described dominant epitope by our group (E6 52-61, restricted by B57) (26), the CD8 T-cell epitopes demonstrate a striking HLA class I binding promiscuity (Fig. 6). We also identified another example of the HLA class I binding promiscuity in the HPV 16 E7 protein among previously published works (19, 26, 31) (Fig. 6). Although this kind of promiscuity has been previously described for human immunodeficiency virus (6, 14, 24, 44), these are the first such examples reported for HPV to our knowledge. It may have been underrecognized for local infections such as those caused by HPV due to technical difficulties in performing fine epitope mapping because of low frequencies of circulating T cells. The finding of HLA class I promiscuity may have significant implications for the development of vaccine and immunotherapies, since the same peptides derived from viral sequences may be used for people with different HLA types.
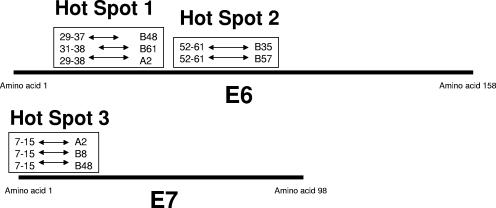
FIG. 6.
Diagram of the “epitope hot spots” within the HPV 16 E6 and E7 proteins, showing the clustering of the CD8 T-cell epitopes restricted by different HLA molecules.
Why would the immune system utilize multiple HLA molecules to present the same viral peptide antigen? After all, it is believed that an extensively polymorphic major histocompatibility complex (MHC) has evolved due to the selective advantages of being able to present epitopes by rare MHC types in instances in which pathogens have adapted to common MHC types (39). It may be that presenting the same viral peptide by multiple HLA types has similar evolutionary advantages to presenting multiple different viral peptides by multiple HLA types with the benefit of superior efficiency in terms of the work required by the host cells to process the peptide epitopes. It is, however, surprising that the same viral peptide such as the HPV 16 E6 29-38 peptide can be bound by the HLA-A0201, -B48, and -B61 molecules since they do not belong to a known HLA supertype.
Considerable effort has been made to identify antigenic epitopes of HPV using both mouse model systems and human systems (11, 12, 33-35). Kast and coauthors (19) identified potential cytotoxic T-lymphocyte (CTL) epitopes of HPV 16 E6 and E7 proteins for five common HLA types by measuring the binding of each of 150 nonamer peptides using purified HLA molecules and radiolabeled peptides. The immunogenicity of nine of these potential antigenic epitopes for HLA-A0201 was tested. Using HLA-A0201 transgenic mice in vivo, Ressing et al. identified four immunogenic peptides: E6 29-38, E7 11-20 (YMLDLQPETT), E7 82-90 (LLMGTLGIV), and E7 86-93 (TLGIVCPI). In vitro CTL induction of peripheral blood mononuclear cells (PBMC) from humans confirmed the immunogenicity of three of the four peptides (E7 11-20, E7 82-90, and E7 86-93) (33). CTLs to one of these peptides, E7 11-20, have been demonstrated in patients with squamous intraepithelial lesions (SIL) (45) and in cancer patients (2, 10, 45). One of the CD8 T-cell epitopes we described is the E6 29-38 epitope, restricted by the HLA-A0201 molecule. Although this epitope was described previously, we believe this is the first time the dominant presence of CD8 T cells with the specificity to this epitope was demonstrated in a woman whose HPV 16 infection has become undetectable. A different approach for identifying antigenic epitopes of HPV 16 E6 and E7 proteins was taken by Bourgault Villada and colleagues (4). Overlapping peptides of these proteins to stimulate PBMC from a healthy donor and binding assays to find candidate epitopes were used to identify two HLA-B18 epitopes, E6 80-88 (ISEYRHYCY) and E7 44-52 (QAEPDRAHY). E6 80-88 was also shown to be an endogenously processed epitope, which can be recognized by T cells from a patient with high-grade SIL. An attractive approach for defining new CD8 T-cell epitopes is to predict their presence based on motifs known to be involved in HLA class I binding, TAP transport, and/or proteasomal cleavage using computer programs, with hopes of minimizing experimental efforts (22, 30, 40). However, it is possible to miss some epitopes using this approach since the presence of only two of five HPV 16 E6 epitopes we described (Table 2) was predicted on the basis of binding motifs (HLA binding motif scanner, Los Alamos National Laboratory]http://hiv.lanl.gov/content/immunology/motif_scan/motif_scan[). A key feature of our approach is the identification of potential CD8 T-cell epitopes based on the magnitude of the T-cell response, with hopes of characterizing those with the best level of protection against the pathogen (16). We were successful in isolating T-cell clones of six of the seven positive peptide pools on the basis of IFN-γ secretion using a commercially available magnetic bead system. Furthermore, most of the CD8 T-cell epitopes were shown to be endogenously processed. However, the T-cell clones isolated from subject 5 (#1-5 and #17-5) recognized only the E6 141-155 15-mer peptide-pulsed target and not the E6-vac-infected ones. Given that none of the 9-mers and the 11-mers within the E6 141-155 region was recognized, these T-cell clones may be detecting a three-diminsional structure formed by the E6 141-155 15-mer peptide which mimics some other antigen (i.e., mimotope).
TABLE 2.
Summary of HPV 16 E6 CD8 T-cell epitopes described on the basis of strong T-cell response
| Epitope | Length (amino acids) | Subject | Sequence | Endogenously processed | Able to kill target | Binding motifs | Restriction element |
|---|---|---|---|---|---|---|---|
| E6 29-37 | 9 | 15 | TIHDIILEC | Yes | Yes | No | B48 |
| E6 29-38 | 10 | 7 | TIHDIILECV | Yes | Yes | No | A0201 |
| E6 31-38 | 8 | 20 | HDIILECV | Yes | Yes | No | B4002 |
| E6 52-61 | 10 | 1 | FAFRDLCIVY | Yes | Yes | Yes | B57 |
| E6 52-61 | 10 | 7 | FAFRDLCIVY | Yes | Yes | Yes | B35 |
We have demonstrated the endogenous processing of the newly described epitopes using autologous EBV LCLs infected with a recombinant vaccinia virus expressing the HPV 16 E6 protein. It has also been shown that the E6 52-61 epitope restricted by the HLA-B57 molecule is expressed by a primary tumor cell line derived from an HPV 16-positive, HLA-B57-positive cervical cancer patient using a chromium release assay and IFN-γ ELISPOT assay (21). Similarly, the expression of the E6 29-38 epitope restricted by the HLA-A0201 molecule and the E6 52-61 epitope restricted by the HLA-B35 molecule by the same primary tumor cell line has also be shown (the patient also happened to be HLA-A0201 and -B35 positive [K. H. Kim and M. Nakagawa, unpublished data]). Although these T-cell epitopes appear to be expressed by tumor cells, it is not clear whether they can be used as targets of cancer immunotherapy because of the small magnitude of recognition (21).
Since the T-cell clones isolated in our study came from women who were able to clear their HPV 16 infections, it is tempting to speculate that they had a role in viral clearance. However, the women were studied only at one time point (i.e., after clearance), and T-cell responses in women who were not able to clear their HPV 16 infections were not studied at the same time. A comparison of responses between women who were able to clear their HPV 16 infections and whose HPV 16 infections persisted may be able to elucidate the significant role of the CD8 T-cell epitopes.
During the past decade, numerous HPV prophylactic and therapeutic vaccine candidates have been evaluated in preclinical and clinical trials. Prophylactic vaccines based on HPV virus-like particles (VLP) have shown remarkable efficacy. The combined incidence of persistent infection or diseases (associated with HPV 6, 11, 16, and 18) was reduced by 90% (P < 0.0001) in the vaccinated group compared to placebo by the quadrivalent vaccine developed by Merck (43). However, such protection has been shown only for HPV-naive individuals, and therapeutic vaccines are needed for those who have already been exposed to HPV. A number of different types of therapeutic vaccines are currently in development. They include vaccines based on peptides (25), recombinant fusion proteins (8, 15), recombinant vaccinia viruses (1, 3, 5, 20), DNA (42), and dendritic cells (13). A spectrum of end points may be used to evaluate the efficacy of therapeutic vaccines, including eradication of HPV infection, prevention of SIL in HPV-infected individuals, regression of SIL, and prevention of recurrence of SIL and cervical cancer after conventional treatments. Many phase I or phase II clinical trials have used regression of SIL as end points while concurrently monitoring safety and immunogenicity (1, 3, 8, 15, 17, 20, 25). While virtually all clinical trials have demonstrated candidate vaccine safety, immunogenicity and clinical responses have been less consistent. Two HPV 16 E7 peptides (E7 12-20 and E7 86-93) were administered with incomplete Freund's adjuvant to patients with high-grade SIL or vulvar intraepithelial neoplasia (VIN) (25). Safety was demonstrated, and 10 of 16 (63%) subjects tested showed an increase in HPV E7-specific IFN-γ secretion, but only 3 of 18 (17%) subjects showed regression of their dysplasia after vaccination. TA-HPV, developed by Xenova Research, is a recombinant vaccinia virus expressing modified HPV 16/18 E6 and E7 proteins (5). Safety was demonstrated; however, measurable CTL responses were shown in 1 of 3 (33%) evaluable patients with advanced cervical cancer, 3 of 12 (25%) cervical intraepithelial neoplasia 3 (CIN 3) subjects, and 4 of 29 (14%) patients with early invasive cervical cancer (1, 3, 20). TA-CIN is a recombinant HPV 16 L2E6E7 fusion protein. Safety was demonstrated in healthy volunteers (9) and in patients with VIN who were previously vaccinated with TA-HPV (8). However, induction of HPV-specific cell-mediated immunity showed mixed results: T-cell proliferative responses to the fusion protein were demonstrated in all VIN patients tested, but IFN-γ ELISPOT assays using E7 peptides were negative for all subjects tested (8). Another version of fusion protein, HPV 16 E6E7, was administered with ISCOMATRIX adjuvant for patients with CIN (15). Safety was demonstrated, and antigen-specific IFN-γ secretion was shown in 10 of 24 (42%) patients tested after vaccination. A recently published clinical trial has shown promising clinical responses: MVA E2 recombinant vaccinia virus was evaluated in a phase II clinical trial and was used to treat high-grade lesions (CIN 2 and CIN 3) (17). Fifty-four participants were treated with this vaccine candidate or with conization. Among 34 patients who received MVA E2, complete responses were shown in the majority (19 patients [56%] by colposcopy and 20 patients [59%] by histology) and the remaining demonstrated partial regression. However, most of the therapeutic vaccine candidates evaluated to date appear to be less efficacious than the VLP-based prophylactic vaccine. In this paper, we have described what we believe to be the first examples of HLA class I binding promiscuity in HPV. The knowledge of epitope “hot spots” may offer a new alternative therapeutic vaccine (perhaps in a form of peptide vaccine or DNA vaccine with an appropriate adjuvant), particularly if more examples of such promiscuity are to be described in the future.
Acknowledgments
This work was supported by the National Institutes of Health (NCI CA051323, NCI K07 CA075974, and M01 RR01271) and the Arkansas Biosciences Institute, the major component of the Tobacco Settlement Proceeds Act of 2000.
Footnotes
Published ahead of print on 15 November 2006.
REFERENCES
- 1.Adams, M., L. Borysiewicz, A. Fiander, S. Man, B. Jasani, H. Navabi, C. Lipetz, A. S. Evans, and M. Mason. 2001. Clinical studies of human papilloma vaccines in pre-invasive and invasive cancer. Vaccine 19:2549-2556. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, M., M. L. Salgaller, E. Celis, A. Sette, W. A. Barnes, S. A. Rosenberg, and M. A. Steller. 1996. Generation of tumor-specific cytolytic T lymphocytes from peripheral blood of cervical cancer patients by in vitro stimulation with a synthetic human papillomavirus type 16 E7 epitope. Am. J. Obstet. Gynecol. 175:1586-1593. [DOI] [PubMed] [Google Scholar]
- 3.Borysiewicz, L. K., A. Fiander, M. Nimako, S. Man, G. W. Wilkinson, D. Westmoreland, A. S. Evans, M. Adams, S. N. Stacey, M. E. Boursnell, E. Rutherford, J. K. Hickling, and S. C. Inglis. 1996. A recombinant vaccinia virus encoding human papillomavirus types 16 and 18, E6 and E7 proteins as immunotherapy for cervical cancer. Lancet 347:1523-1527. [DOI] [PubMed] [Google Scholar]
- 4.Bourgault Villada, I., N. Beneton, C. Bony, F. Connan, J. Monsonego, A. Bianchi, P. Saiag, J. P. Levy, J. G. Guillet, and J. Choppin. 2000. Identification in humans of HPV-16 E6 and E7 protein epitopes recognized by cytolytic T lymphocytes in association with HLA-B18 and determination of the HLA-B18-specific binding motif. Eur. J. Immunol. 30:2281-2289. [DOI] [PubMed] [Google Scholar]
- 5.Boursnell, M. E., E. Rutherford, J. K. Hickling, E. A. Rollinson, A. J. Munro, N. Rolley, C. S. McLean, L. K. Borysiewicz, K. Vousden, and S. C. Inglis. 1996. Construction and characterisation of a recombinant vaccinia virus expressing human papillomavirus proteins for immunotherapy of cervical cancer. Vaccine 14:1485-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, S. A., T. D. Lockey, C. Slaughter, K. S. Slobod, S. Surman, A. Zirkel, A. Mishra, V. R. Pagala, C. Coleclough, P. C. Doherty, and J. L. Hurwitz. 2005. T cell epitope “hotspots” on the HIV type 1 gp120 envelope protein overlap with tryptic fragments displayed by mass spectrometry. AIDS Res. Hum. Retroviruses 21:165-170. [DOI] [PubMed] [Google Scholar]
- 7.Crook, T., J. A. Tidy, and K. H. Vousden. 1991. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell 67:547-556. [DOI] [PubMed] [Google Scholar]
- 8.Davidson, E. J., R. L. Faulkner, P. Sehr, M. Pawlita, L. J. Smyth, D. J. Burt, A. E. Tomlinson, J. Hickling, H. C. Kitchener, and P. L. Stern. 2004. Effect of TA-CIN (HPV 16 L2E6E7) booster immunisation in vulval intraepithelial neoplasia patients previously vaccinated with TA-HPV (vaccinia virus encoding HPV 16/18 E6E7). Vaccine 22:2722-2729. [DOI] [PubMed] [Google Scholar]
- 9.de Jong, A., T. O'Neill, A. Y. Khan, K. M. Kwappenberg, S. E. Chisholm, N. R. Whittle, J. A. Dobson, L. C. Jack, J. A. St Clair Roberts, R. Offringa, S. H. van der Burg, and J. K. Hickling. 2002. Enhancement of human papillomavirus (HPV) type 16 E6 and E7-specific T-cell immunity in healthy volunteers through vaccination with TA-CIN, an HPV16 L2E7E6 fusion protein vaccine. Vaccine 20:3456-3464. [DOI] [PubMed] [Google Scholar]
- 10.Evans, E. M., S. Man, A. S. Evans, and L. K. Borysiewicz. 1997. Infiltration of cervical cancer tissue with human papillomavirus-specific cytotoxic T-lymphocytes. Cancer Res. 57:2943-2950. [PubMed] [Google Scholar]
- 11.Feltkamp, M. C., H. L. Smits, M. P. Vierboom, R. P. Minnaar, B. M. de Jongh, J. W. Drijfhout, J. ter Schegget, C. J. Melief, and W. M. Kast. 1993. Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. Eur. J. Immunol. 23:2242-2249. [DOI] [PubMed] [Google Scholar]
- 12.Feltkamp, M. C., G. R. Vreugdenhil, M. P. Vierboom, E. Ras, S. H. van der Burg, J. ter Schegget, C. J. Melief, and W. M. Kast. 1995. Cytotoxic T lymphocytes raised against a subdominant epitope offered as a synthetic peptide eradicate human papillomavirus type 16-induced tumors. Eur. J. Immunol. 25:2638-2642. [DOI] [PubMed] [Google Scholar]
- 13.Ferrara, A., M. Nonn, P. Sehr, C. Schreckenberger, M. Pawlita, M. Durst, A. Schneider, and A. M. Kaufmann. 2003. Dendritic cell-based tumor vaccine for cervical cancer II: results of a clinical pilot study in 15 individual patients. J. Cancer Res. Clin. Oncol. 129:521-530. [DOI] [PubMed] [Google Scholar]
- 14.Frahm, N., P. Goulder, and C. Brander. 2003. Broad HIV-1 specific CTL responses reveal extensive HLA class I binding promiscuity of HIV-derived, optimally defined CTL epitopes, p. 3-24. In B. Korber, C. Brander, B. Haynes, R. Koup, J. Moore, B. Walker, and D. Watkins (ed.), HIV immunology and HIV/SIV vaccine databases 2003. Los Alamos National Laboratory, Los Alamos, NM.
- 15.Frazer, I. H., M. Quinn, J. L. Nicklin, J. Tan, L. C. Perrin, P. Ng, V. M. O'Connor, O. White, N. Wendt, J. Martin, J. M. Crowley, S. J. Edwards, A. W. McKenzie, S. V. Mitchell, D. W. Maher, M. J. Pearse, and R. L. Basser. 2004. Phase 1 study of HPV16-specific immunotherapy with E6E7 fusion protein and ISCOMATRIX adjuvant in women with cervical intraepithelial neoplasia. Vaccine 23:172-181. [DOI] [PubMed] [Google Scholar]
- 16.Gallimore, A., H. Hengartner, and R. Zinkernagel. 1998. Hierarchies of antigen-specific cytotoxic T-cell responses. Immunol. Rev. 164:29-36. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Hernandez, E., J. L. Gonzalez-Sanchez, A. Andrade-Manzano, M. L. Contreras, S. Padilla, C. C. Guzman, R. Jimenez, L. Reyes, G. Morosoli, M. L. Verde, and R. Rosales. 2006. Regression of papilloma high-grade lesions (CIN 2 and CIN 3) is stimulated by therapeutic vaccination with MVA E2 recombinant vaccine. Cancer Gene Ther. 13:592-597. [DOI] [PubMed] [Google Scholar]
- 18.Heck, D. V., C. L. Yee, P. M. Howley, and K. Munger. 1992. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc. Natl. Acad. Sci. USA 89:4442-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kast, W. M., R. M. Brandt, J. Sidney, J. W. Drijfhout, R. T. Kubo, H. M. Grey, C. J. Melief, and A. Sette. 1994. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J. Immunol. 152:3904-3912. [PubMed] [Google Scholar]
- 20.Kaufmann, A. M., P. L. Stern, E. M. Rankin, H. Sommer, V. Nuessler, A. Schneider, M. Adams, T. S. Onon, T. Bauknecht, U. Wagner, K. Kroon, J. Hickling, C. M. Boswell, S. N. Stacey, H. C. Kitchener, J. Gillard, J. Wanders, J. S. Roberts, and H. Zwierzina. 2002. Safety and immunogenicity of TA-HPV, a recombinant vaccinia virus expressing modified human papillomavirus (HPV)-16 and HPV-18 E6 and E7 genes, in women with progressive cervical cancer. Clin. Cancer Res. 8:3676-3685. [PubMed] [Google Scholar]
- 21.Kim, K. H., R. Dishongh, A. D. Santin, M. J. Cannon, S. Bellone, and M. Nakagawa. 2006. Recognition of a cervical cancer derived tumor cell line by a human papillomavirus type 16 E6 52-61-specific CD8 T cell clone. Cancer Immun. 6:9. [PubMed] [Google Scholar]
- 22.Larsen, M. V., C. Lundegaard, K. Lamberth, S. Buus, S. Brunak, O. Lund, and M. Nielsen. 2005. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur. J. Immunol. 35:2295-2303. [DOI] [PubMed] [Google Scholar]
- 23.Larsson, M., D. T. Wilkens, J. F. Fonteneau, T. J. Beadle, M. J. Merritt, R. G. Kost, P. A. Haslett, S. Cu-Uvin, N. Bhardwaj, D. F. Nixon, and B. L. Shacklett. 2002. Amplification of low-frequency antiviral CD8 T cell responses using autologous dendritic cells. AIDS 16:171-180. [DOI] [PubMed] [Google Scholar]
- 24.Masemola, A. M., T. N. Mashishi, G. Khoury, H. Bredell, M. Paximadis, T. Mathebula, D. Barkhan, A. Puren, E. Vardas, M. Colvin, L. Zijenah, D. Katzenstein, R. Musonda, S. Allen, N. Kumwenda, T. Taha, G. Gray, J. McIntyre, S. A. Karim, H. W. Sheppard, and C. M. Gray. 2004. Novel and promiscuous CTL epitopes in conserved regions of Gag targeted by individuals with early subtype C HIV type 1 infection from southern Africa. J. Immunol. 173:4607-4617. [DOI] [PubMed] [Google Scholar]
- 25.Muderspach, L., S. Wilczynski, L. Roman, L. Bade, J. Felix, L. A. Small, W. M. Kast, G. Fascio, V. Marty, and J. Weber. 2000. A phase I trial of a human papillomavirus (HPV) peptide vaccine for women with high-grade cervical and vulvar intraepithelial neoplasia who are HPV 16 positive. Clin. Cancer Res. 6:3406-3416. [PubMed] [Google Scholar]
- 26.Nakagawa, M., K. Kim, and A.-B. Moscicki. 2004. Different methods of identifying new antigenic epitopes of human papillomavirus type 16 E6 and E7 proteins. Clin. Diagn. Lab. Immunol. 11:889-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa, M., K. H. Kim, and A. B. Moscicki. 2005. Patterns of CD8 T-cell epitopes within the human papillomavirus type 16 (HPV 16) E6 protein among young women whose HPV 16 infection has become undetectable. Clin. Diagn. Lab. Immunol. 12:1003-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa, M., D. P. Stites, S. Farhat, J. R. Sisler, B. Moss, F. Kong, A. B. Moscicki, and J. M. Palefsky. 1997. Cytotoxic T lymphocyte responses to E6 and E7 proteins of human papillomavirus type 16: relationship to cervical intraepithelial neoplasia. J. Infect. Dis. 175:927-931. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa, M., D. P. Stites, S. Patel, S. Farhat, M. Scott, N. K. Hills, J. M. Palefsky, and A. B. Moscicki. 2000. Persistence of human papillomavirus type 16 infection is associated with lack of cytotoxic T lymphocyte response to the E6 antigens. J. Infect. Dis. 182:595-598. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen, M., C. Lundegaard, O. Lund, and C. Kesmir. 2005. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57:33-41. [DOI] [PubMed] [Google Scholar]
- 31.Oerke, S., H. Hohn, I. Zehbe, H. Pilch, K. H. Schicketanz, W. E. Hitzler, C. Neukirch, K. Freitag, and M. J. Maeurer. 2005. Naturally processed and HLA-B8-presented HPV16 E7 epitope recognized by T cells from patients with cervical cancer. Int. J. Cancer 114:766-778. [DOI] [PubMed] [Google Scholar]
- 32.Pirisi, L., S. Yasumoto, M. Feller, J. Doniger, and J. DiPaolo. l987. Transformation of human fibroblasts and keratinocytes with human papillomavirus type 16 DNA. J. Virol. 61:1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ressing, M. E., A. Sette, R. M. Brandt, J. Ruppert, P. A. Wentworth, M. Hartman, C. Oseroff, H. M. Grey, C. J. Melief, and W. M. Kast. 1995. Human CTL epitopes encoded by human papillomavirus type 16 E6 and E7 identified through in vivo and in vitro immunogenicity studies of HLA-A*0201-binding peptides. J. Immunol. 154:5934-5943. [PubMed] [Google Scholar]
- 34.Sadovnikova, E., X. Zhu, S. M. Collins, J. Zhou, K. Vousden, L. Crawford, P. Beverley, and H. J. Stauss. 1994. Limitations of predictive motifs revealed by cytotoxic T lymphocyte epitope mapping of the human papilloma virus E7 protein. Int. Immunol. 6:289-296. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar, A. K., G. Tortolero-Luna, P. N. Nehete, R. B. Arlinghaus, M. F. Mitchell, and K. J. Sastry. 1995. Studies on in vivo induction of cytotoxic T lymphocyte responses by synthetic peptides from E6 and E7 oncoproteins of human papillomavirus type 16. Viral Immunol. 8:165-174. [DOI] [PubMed] [Google Scholar]
- 36.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 37.Schlegel, R., W. C. Phelps, Y. L. Zhang, and M. Barbosa. 1988. Quantitative keratinocyte assay detects two biological activities of human papillomavirus DNA and identifies viral types associated with cervical carcinoma. EMBO J. 7:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seedorf, K., G. Krammer, M. Durst, S. Suhai, and W. G. Rowekamp. 1985. Human papillomavirus type 16 DNA sequence. Virology 145:181-185. [DOI] [PubMed] [Google Scholar]
- 39.Serjeantson, S. W. 1989. The reasons for MHC polymorphism in man. Transplant Proc. 21:598-601. [PubMed] [Google Scholar]
- 40.Stevanovic, S. 2005. Antigen processing is predictable: from genes to T cell epitopes. Transpl. Immunol. 14:171-174. [DOI] [PubMed] [Google Scholar]
- 41.Storey, A., D. Pim, A. Murray, K. Osborn, L. Banks, and L. Crawford. 1988. Comparison of the in vitro transforming activities of human papillomavirus types. EMBO J. 7:1815-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trimble, C. L. 2005. Prospects for therapeutic HPV vaccines. Gynecol. Oncol. 99:S249-S250. [DOI] [PubMed] [Google Scholar]
- 43.Villa, L. L., R. L. Costa, C. A. Petta, R. P. Andrade, K. A. Ault, A. R. Giuliano, C. M. Wheeler, L. A. Koutsky, C. Malm, M. Lehtinen, F. E. Skjeldestad, S. E. Olsson, M. Steinwall, D. R. Brown, R. J. Kurman, B. M. Ronnett, M. H. Stoler, A. Ferenczy, D. M. Harper, G. M. Tamms, J. Yu, L. Lupinacci, R. Railkar, F. J. Taddeo, K. U. Jansen, M. T. Esser, H. L. Sings, A. J. Saah, and E. Barr. 2005. Prophylactic quadrivalent human papillomavirus (types 6, 11, 16, and 18) L1 virus-like particle vaccine in young women: a randomised double-blind placebo-controlled multicentre phase II efficacy trial. Lancet Oncol. 6:271-278. [DOI] [PubMed] [Google Scholar]
- 44.Walker, B. D., and B. T. Korber. 2001. Immune control of HIV: the obstacles of HLA and viral diversity. Nat. Immunol. 2:473-475. [DOI] [PubMed] [Google Scholar]
- 45.Youde, S. J., P. R. Dunbar, E. M. Evans, A. N. Fiander, L. K. Borysiewicz, V. Cerundolo, and S. Man. 2000. Use of fluorogenic histocompatibility leukocyte antigen-A*0201/HPV 16 E7 peptide complexes to isolate rare human cytotoxic T-lymphocyte-recognizing endogenous human papillomavirus antigens. Cancer Res. 60:365-371. [PubMed] [Google Scholar]
- 46.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]