Abstract
Feline coronavirus (FCoV), porcine transmissible gastroenteritis coronavirus (TGEV), canine coronavirus (CCoV), and human coronavirus HCoV-229E, which belong to the group 1 coronavirus, use aminopeptidase N (APN) of their natural host and feline APN (fAPN) as receptors. Using mouse-feline APN chimeras, we identified three small, discontinuous regions, amino acids (aa) 288 to 290, aa 732 to 746 (called R1), and aa 764 to 788 (called R2) in fAPN that determined the host ranges of these coronaviruses. Blockade of infection with anti-fAPN monoclonal antibody RG4 suggested that these three regions lie close together on the fAPN surface. Different residues in fAPN were required for infection with each coronavirus. HCoV-229E infection was blocked by an N-glycosylation sequon present between aa 288 to 290 in murine APN. TGEV required R1 of fAPN, while FCoV and CCoV required both R1 and R2 for entry. N740 and T742 in fAPN and the homologous R741 in human APN (hAPN) were key determinants of host range for FCoV, TGEV, and CCoV. Residue N740 in fAPN was essential only for CCoV receptor activity. A conservative T742V substitution or a T742R substitution in fAPN destroyed receptor activity for the pig, dog, and cat coronaviruses, while a T742S substitution retained these receptor activities. Thus, the hydroxyl on T742 is required for the coronavirus receptor activity of fAPN. In hAPN an R741T substitution caused a gain of receptor activity for TGEV but not for FCoV or CCoV. Therefore, entry and host range of these group 1 coronaviruses depend on the ability of the viral spike glycoproteins to recognize small, species-specific amino acid differences in the APN proteins of different species.
Coronaviruses are important respiratory and enteric pathogens of humans and many animal species (32, 40, 58). Coronavirus phylogenetic groups 1, 2a, 2b, and 3 differ in host range and pathogenicity (17, 32, 40, 58). Group 1 contains two human coronaviruses, HCoV-229E and HCoV-NL63, that cause acute respiratory tract infections (4, 6, 56, 60) and several important veterinary viruses: in pigs, porcine epidemic diarrhea virus and transmissible gastroenteritis coronavirus (TGEV) cause enteric disease, and porcine respiratory coronavirus causes respiratory disease (25, 26); feline coronavirus (FCoV) causes enteric or systemic disease in cats (10, 27, 42, 57); and canine coronavirus (CCoV) causes enteric disease in dogs (44).
Human, feline, canine, and porcine group 1 coronaviruses cause transmissible disease within a single host species. However, experimental inoculation of several other species with these coronaviruses can result in viral replication, seroconversion, and, in some cases, nontransmissible disease (2, 3, 63, 64). For serial transmission to occur in a new host species, the spike glycoproteins of group 1 coronaviruses need to adapt to their receptor in the new host species by mutation or recombination with another coronavirus that naturally infects the new host species.
An important determinant of coronavirus host range is the interaction of the ∼200-kDa viral spike (S) glycoprotein with a receptor glycoprotein on the surface of susceptible cells (18, 30, 41, 49). Several coronavirus receptors have been identified. Mouse hepatitis virus, in phylogenetic group 2a, uses murine carcinoembryonic cell adhesion molecule 1a as a receptor (14, 15, 62). Human angiotensin-converting enzyme 2 (hACE-2) is a receptor for severe acute respiratory syndrome (SARS)-CoV in phylogenetic group 2b and HCoV-NL63 in group 1 (21, 36, 52). HCoV-229E, TGEV, FCoV, and CCoV in group 1 use aminopeptidase N (APN) of their natural host species to enter cells (12, 28, 54, 65). In cell culture, human APN (hAPN) is a receptor for only HCoV-229E, and porcine APN (pAPN) is a receptor for only TGEV (12, 65). However, feline APN (fAPN) is a receptor for not only FCoV but also HCoV-229E, TGEV, and CCoV (54). The purpose of this study was to identify key regions and residues in fAPN that determine the host range of these group 1 coronaviruses.
APN (CD13) is a 150- to 160-kDa type II transmembrane glycoprotein expressed as a homodimer on the apical membranes of epithelial cells in the respiratory and enteric tracts, endothelial cells, and kidney cells; at synaptic junctions; and on cells of the immune system (monocytes, dendritic cells, and granulocytes) (34). APN is a zinc-dependent protease that cleaves N-terminal amino acids from biologically active peptides (34). The APN proteins from human, mouse, rat, rabbit, pig, cow, cat, dog, and chicken are highly conserved at the amino acid level (70 to 80% amino acid identity). Secondary structure predictions and biochemical studies suggest that APN consists of seven domains (51). Domain I, at the N terminus, is a short cytoplasmic tail; domain II is the transmembrane domain; and domain III (amino acids [aa] 40 to 70 of hAPN) is the “stalk region.” In hAPN, domain IV includes aa 70 to 252. Domains V and VI (aa 253 to 580 of hAPN) contain the active site of the enzyme and a conserved zinc-binding motif (HELAH). Domain VII (aa 581 to 967) at the C terminus of hAPN is predicted to be mainly α-helical (51).
Until the crystal structures for APN and group 1 coronavirus S glycoproteins are determined, the identification of receptor determinants on APN that affect coronavirus host range depends on the use of mutant and chimeric APN proteins. Several regions of APN that are important for entry of group 1 coronaviruses were previously identified using chimeras between APN proteins of different species (13, 28). Human-pig APN chimeras showed that aa 717 to 813 in domain VII of pAPN are essential for TGEV receptor activity (11), while aa 288 to 295 in domain V of hAPN are necessary for HCoV-229E receptor activity (29). The introduction into hAPN of a sequon encoding a potential N-glycosylation sequon at aa 291, as found in pAPN, abrogates HCoV-229E receptor activity (59). Regions in fAPN that are important for HCoV-229E, TGEV, and FCoV receptor activity were identified using pig-feline and human-feline APN chimeras. Residues 670 to 840 in domain VII of fAPN are required for TGEV and FCoV receptor activity, and aa 135 to 297 in domain V of fAPN are required for HCoV-229E receptor activity (19, 29). In addition, aa 643 to 841 in domain VII of canine APN in an hAPN backbone can mediate entry of CCoV, TGEV, and FCoV (5).
In this study, chimeras between fAPN and mouse APN (mAPN), which lacks coronavirus receptor activity, were used to identify three small, discontinuous regions in fAPN between aa 288 to 290 in domain V and aa 732 to 746 (called R1) and aa 764 to 788 (called R2) in domain VII that were critical determinants of coronavirus entry and host range. Using mutant APN proteins, we identified single residues in fAPN and hAPN that are critical determinants of group 1 coronavirus host range and infection in vitro. These results provide a model for the evolution and emergence of coronaviruses as they adapt to recognize species-specific differences in the APN proteins of different host species.
MATERIALS AND METHODS
Cell lines and viruses.
Felis catus whole fetus (Fcwf) cells (provided by Niels Pedersen, University of California at Davis, Davis, CA), a hamster kidney (BHK) fibroblast cell line, a swine testicle cell line (provided by David Brian, University of Tennessee, Knoxville, TN), canine tumor cell line A-72 (provided by Leonard Binn, Walter Reed Army Institute for Research, Silver Spring, MD), and human lung fibroblast cell line MRC5 (ATCC CCL-171, Manassas, VA) were grown as previously described (54, 59). FCoV genotype II strain 79-1146, CCoV genotype II strain 1-71 (provided by Leonard Binn, Walter Reed Army Institute for Research), TGEV clone E (provided by David Brian, University of Tennessee), and HCoV-229E (ATCC VR-740) were propagated, and titers of infectious viruses were determined by plaque assay as previously described (54, 59).
Chimeric and mutant APN plasmids.
All mouse-feline APN (m/fAPN) chimeric and mutant APN cDNA sequences were constructed by standard fusion PCR techniques (24) or by site-directed mutagenesis as described previously (59) using Pfu Turbo polymerase (Stratagene, La Jolla, CA). All cDNAs encoding chimeric m/fAPN, mutant, and wild-type fAPN (accession number NM 001009252) (54) and mAPN (accession number NM 008486; provided by Linda Shapiro, University of Connecticut Health Center, Farmington, CT) were cloned into the pcDNA3.1D/V5-His-TOPO mammalian expression vector (Invitrogen, Carlsbad, CA) in frame with the C terminal V5 and six-His tags, according to the manufacturer's instructions. The expression plasmid containing hAPN cDNA (59) was used to introduce mutations into the hAPN DNA sequence. All DNA constructs were sequenced by the University of Colorado Cancer Center DNA Sequencing and Analysis Core Facility.
Transient transfections.
BHK cells were transfected with plasmids containing cDNA encoding fAPN, mAPN, hAPN, and mutant or chimeric APN proteins using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Twenty-four hours after transfection, cells were seeded on glass coverslips, and 48 h after transfection, they were used for virus inoculation or for detection of APN protein expression by immunofluorescence or flow cytometry as described below.
Generation of cells stably expressing fAPN.
A plasmid construct containing the fAPN cDNA in the pCiNeo mammalian expression vector (Promega Corp., Madison, WI), fAPN-pCiNeo, was generated by subcloning the fAPN cDNA fragment from the pCR3 expression plasmid (Invitrogen) described by Tresnan et al. (54) into the pCiNeo expression vector using EcoRI and NotI. BHK cells were transfected with the fAPN-pCiNeo construct using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, and 48 h after transfection cells were placed under G418 selection (GIBCO BRL, Grand Island, NY). Two weeks later, cells expressing fAPN were selected by fluorescence-activated cell sorting with a mouse anti-fAPN RG4 monoclonal antibody (MAb) from a hybridoma cell line (23) kindly provided by Tsutomu Hohdatsu (Kitasato University, Japan). Cell sorting was done at the University of Colorado Cancer Center Flow Cytometry Core Facility.
Virus inoculation.
BHK cells transfected with cDNAs encoding wild-type, chimeric, or mutant APN proteins were inoculated 48 h after transfection with HCoV-229E, FCoV, TGEV, or CCoV diluted in Dulbecco's modified Eagle's minimal essential medium (GIBCO BRL) supplemented with heat-inactivated 10% fetal bovine serum (HyClone, Logan, UT) and 2% antibiotic-antimycotic (GIBCO BLR) at a multiplicity of infection (MOI) of 0.3 to 0.8. After 1 h of adsorption, the inocula were removed and replaced with fresh medium. Inoculated cells were fixed with methanol:acetic acid (3:1) for detection of viral antigens as described below.
Immunofluorescence assay to detect expression of APN proteins or viral antigens.
To detect APN expression, transfected cells on coverslips were fixed with 1.6% paraformaldehyde (Ted Pella, Inc., Redding, CA), and an immunofluorescence assay (IFA) was performed with purified, fluorescein-conjugated rat anti-mAPN R3-242 MAb (BD Biosciences Pharmingen, San Jose, CA), purified RG4 MAb, or anti-hAPN DW1 MAb. Incubation with DW1 or RG4 MAbs was followed by incubation with fluorescein-conjugated goat anti-mouse immunoglobulin G (IgG; Jackson Immunoresearch, West Grove, PA). FCoV, TGEV, and CCoV antigens were detected with a polyclonal fluorescein-conjugated feline anti-FCoV serum (FITC-FIP; VMRD, Inc., Pullman, WA) that cross-reacts with TGEV and CCoV. HCoV-229E antigens were detected with a polyclonal goat anti-HCoV-229E serum (59). Immunolabeled cells were analyzed using a Zeiss Axioplan 2 microscope (Carl Zeiss, Inc., New York, NY).
Flow cytometry.
Surface expression of APN protein was detected with anti-fAPN RG4 MAb, anti-mAPN R3-242 MAb, or anti-hAPN DW1 MAb. Cells were washed and incubated with phycoerythrin-conjugated goat anti-mouse IgG (Jackson Immunoresearch) or fluorescein-conjugated goat anti-rat IgG (Jackson Immunoresearch) and fixed with 1.6% paraformaldehyde. Cells were analyzed at the University of Colorado Cancer Center Flow Cytometry Core Facility.
Blockade of coronavirus infection with anti-fAPN RG4 MAb.
BHK cells stably expressing fAPN were preincubated with 4.8 μg of total protein of purified RG4 MAb or a control mouse MAb (control MAb) against an irrelevant antigen in medium for 45 min at 4°C and then inoculated with FCoV, TGEV, CCoV, or HCoV-229E at an MOI of 0.1. After virus adsorption for 1 h, cells were washed, and fresh medium was added containing 16 ng/μl of RG4 or control MAb. Cells were fixed with methanol:acetic acid (3:1) 16 to 18 h after inoculation, and viral antigens were detected by IFA. In two experiments, BHK cells transiently transfected with plasmids encoding fAPN or m/fAPN chimeric proteins were preincubated with 14.4 μg or 4.8 μg of total protein of RG4 MAb or control MAb and then inoculated with FCoV or HCoV-229E at an MOI of 0.6 or 0.3. Viral antigens were detected as described above. Cells expressing viral antigens were counted in five fields at a magnification of ×40 for all APN constructs except for m/fAPN containing aa 582 to 967 of fAPN in the mAPN backbone (m/fAPN582-967), for which positive cells were counted in five fields at a magnification of ×10. For each wild-type or chimeric APN protein, the number of cells positive for viral antigens in samples treated with the control MAb was set as 100%. Samples treated with the RG4 MAb were scored as positive for receptor blockade if the percentage of infected cells was <5% of control MAb-treated samples and scored as negative for receptor blockade if the percentage of infected cells was ≥90% of control MAb-treated samples. The Fab fragments of purified RG4 MAb and a control MAb were prepared using the ImmunoPure Fab preparation kit (Pierce, Rockford, IL) according to the manufacturer's instructions. BHK cells transiently expressing wild-type fAPN were preincubated with 20 μg of total protein of RG4 Fab or control Fab, inoculated with HCoV-229E or FCoV, and then processed as described above.
RESULTS
HCoV-229E and the animal coronaviruses FCoV, CCoV, and TGEV require different regions of fAPN for entry.
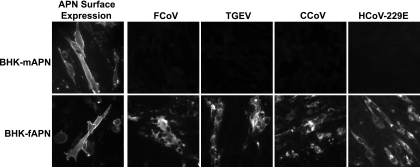
BHK cells transfected with plasmids encoding fAPN or mAPN expressed the APN protein on the cell surface 48 h after transfection (Fig. 1). Cells expressing fAPN were susceptible to infection with HCoV-229E, FCoV, TGEV, and CCoV as previously reported (54), while cells expressing mAPN were resistant to infection with all four of these group 1 coronaviruses (Fig. 1).
FIG. 1.
BHK cells transfected with fAPN but not mAPN are susceptible to infection with FCoV, TGEV, CCoV, and HCoV-229E. Surface expression of fAPN or mAPN proteins on paraformaldehyde-fixed BHK cells transiently transfected with plasmids encoding fAPN or mAPN was detected by immunolabeling with anti-fAPN RG4 or anti-mAPN R3-242 MAb, respectively. Viral antigens in cells inoculated with FCoV, TGEV, CCoV, or HCoV-229E were detected after 24 h by immunolabeling with antiviral antibodies.
To identify fAPN regions required for coronavirus receptor activity, BHK cells were transfected with chimeric m/fAPN proteins containing different regions of fAPN in an mAPN backbone. All chimeric m/fAPN proteins were expressed at the cell surface 48 h after transfection (Fig. 2). Two m/fAPN chimeras, m/fAPN1-582 and m/fAPN251-582, had receptor activity for HCoV-229E but not for FCoV or TGEV (Fig. 2). In contrast, m/fAPN582-967, m/fAPN704-967, and m/fAPN704-831 had receptor activity for FCoV and TGEV but not for HCoV-229E (Fig. 2). Chimera m/fAPN704-831 also had receptor activity for CCoV (data not shown). In contrast, m/fAPN831-967 had no receptor activity for the human or animal group 1 coronaviruses (Fig. 2). Thus, the region between aa 251 to 582 of fAPN was required for HCoV-229E entry, while the region between aa 704 to 831 of fAPN was required for entry of the animal coronaviruses tested.
FIG. 2.
HCoV-229E and the animal coronaviruses FCoV and TGEV require different regions of fAPN for receptor activity. BHK cells were transiently transfected with plasmids encoding m/fAPN chimeric proteins. The numbers in parentheses indicate fAPN residues present in the m/fAPN protein. At 48 h posttransfection, the surface expression of the chimeric m/fAPN1-582 and m/fAPN251-582 proteins was detected with anti-fAPN RG4 MAb, and the surface expression of all other m/fAPN chimeric proteins was detected with anti-mAPN R3-242 MAb. At 24 h after inoculation, viral antigens were detected with antiviral antibodies.
The anti-fAPN RG4 MAb binds to an epitope between aa 251 to 582 of fAPN and blocks fAPN-mediated infection of HCoV-229E, FCoV, CCoV, and TGEV.
Immunolabeling and flow cytometry were used to test the ability of the anti-fAPN RG4 and anti-mAPN R3-242 MAbs to recognize fAPN, mAPN, and chimeric m/fAPN proteins expressed on BHK cells. The RG4 MAb recognized fAPN, as previously reported (23), but not mAPN. The anti-mAPN MAb recognized mAPN but not fAPN (Table 1 and Fig. 1). Two chimeras, m/fAPN251-967 and m/fAPN251-582, were recognized by both MAbs. In contrast, m/fAPN582-967 was recognized by only the anti-mAPN MAb, and m/fAPN1-582 was recognized by only the anti-fAPN MAb (Table 1 and Fig. 2). Thus, the anti-mAPN MAb R3-242 bound to an epitope on mAPN between aa 1 to 251. The anti-fAPN RG4 MAb bound to an epitope on fAPN that lies between aa 251 to 582 and includes domain V that is required for HCoV-229E entry, but anti-fAPN RG4 did not bind to aa 582 to 967 in domain VII of fAPN that is required for entry of FCoV, CCoV, and TGEV.
TABLE 1.
Mapping the binding epitopes of anti-fAPN RG4 and anti-mAPN R3-242 MAbsa
| APN protein | RG4 MAb binding
|
R3-242 MAb binding
|
||
|---|---|---|---|---|
| IFA | FC | IFA | FC | |
| Mock transfected | − | − | − | − |
| fAPN | + | + | − | − |
| mAPN | − | − | + | + |
| m/fAPN251-967 | + | + | + | + |
| m/fAPN251-582 | + | + | + | + |
| m/fAPN582-967 | − | − | + | + |
| m/fAPN1-582 | + | + | − | − |
BHK cells transiently transfected with plasmids encoding fAPN, mAPN, or chimeric m/fAPN proteins were tested for reactivity with anti-fAPN RG4 or anti-mAPN R3-242 MAbs by IFA and flow cytometry (FC).
Anti-fAPN RG4 MAb was previously shown to block FCoV, TGEV, and CCoV infection of feline cells expressing fAPN (23). Infection of BHK cells stably expressing fAPN by these three animal coronaviruses and by HCoV-299E was blocked by treatment of the cells with RG4 MAb, but virus infection was not blocked in cells treated with a control MAb (data not shown). Thus, the RG4 MAb blocked infection by the three animal coronaviruses and HCoV-299E even though these viruses required different domains of fAPN for entry. Treatment of cells transiently expressing fAPN or m/fAPN chimeric proteins with a control MAb did not block FCoV or HCoV-229E infection, but treatment with anti-fAPN RG4 MAb blocked HCoV-229E infection of BHK cells expressing fAPN, m/fAPN251-967, or m/fAPN251-582 (Table 2). RG4 MAb also blocked FCoV infection of cells expressing fAPN, or m/fAPN251-967, but RG4 MAb did not block FCoV infection of cells expressing m/fAPN582-967 (Table 2). In addition, the small (∼50 kDa) Fab protein of RG4 MAb, but not the Fab fragment of a control MAb, blocked both FCoV and HCoV-229E infection of cells expressing fAPN (data not shown).
TABLE 2.
Blockade of FCoV and HCoV-229E infection with the anti-fAPN RG4 MAb in cells expressing fAPN or chimeric m/fAPN proteinsa
| APN protein | HCoV-229E infection
|
FCoV infection
|
||
|---|---|---|---|---|
| Control MAb | RG4 MAb | Control MAb | RG4 MAb | |
| fAPN | − | + | − | + |
| m/fAPN251-967 | − | + | − | + |
| m/fAPN251-582 | − | + | NA | NA |
| m/fAPN582-967 | NA | NA | − | −b |
| m/fAPN1-582 | − | + | NA | NA |
Infection with FCoV or HCoV-229E was detected by immunofluorescence with antiviral antibodies 16 to 18 h after inoculation. +, antibody blockade of virus infection (<5% of cells infected); −, no antibody blockade of virus infection (≥90% of cells infected). NA, not applicable; the m/fAPN chimeric protein has no receptor activity for the virus.
This antibody does not bind to this m/fAPN chimera.
The recognition of m/fAPN proteins (Table 1) and the inhibition of coronavirus infection (Table 2) by the RG4 MAb showed that this MAb bound to an epitope within aa 251 to 582 of fAPN, which was necessary for HCoV-229E infection, and not to aa 582 to 967 of fAPN that contained domain VII, which is necessary for FCoV, CCoV, and TGEV receptor activity. The blockade of fAPN-mediated FCoV and HCoV-229E infection by the Fab fragment of RG4 MAb suggested that the discontinuous regions in fAPN required for infection by these two viruses may lie close enough on the surface of the APN protein for binding of the small RG4 Fab fragment to block infection by both of these coronaviruses.
The N-glycosylation sequon at aa 288 to 290 in mAPN is an important determinant of HCoV-229E host range.
Using m/fAPN chimeric proteins we showed that aa 251 to 582 of fAPN are important for HCoV-229E receptor activity, in agreement with previous reports that showed that aa 135 to 297 of fAPN were required for HCoV-229E receptor activity (19, 29). To determine if the overlapping residues within these two regions were sufficient for HCoV-229E entry, we tested the coronavirus receptor activities of chimeric m/fAPN251-296 and m/fAPN287-296 proteins (data not shown). Figure 7 summarizes the data showing that aa 287 to 296 of fAPN in the mAPN backbone conferred receptor activity for HCoV-229E but not for TGEV, FCoV, or CCoV. We compared aa 287 to 296 of fAPN with APN proteins of other species and identified an N-linked glycosylation sequon, NIS, between aa 288 to 290 in mAPN and one, NET, between aa 286 to 288 in pAPN that are not present in the corresponding regions of hAPN or fAPN proteins. We previously showed that introduction of a sequon encoding an N-glycosylation sequon at aa 291 in hAPN abrogated HCoV-229E receptor activity (59). To determine if a potential N-linked glycan at N288 on the mAPN protein was responsible for the lack of HCoV-229E receptor activity of mAPN, we eliminated the glycosylation sequon in mAPN by substituting N288 and S290 for the corresponding fAPN residues, Y and E, respectively (mAPNN288Y,S290E). The mutant mAPNN288Y,S290E, lacking the NIS motif had weak receptor activity for HCoV-229E but no receptor activity for TGEV, FCoV, or CCoV (see Fig. 7; also data not shown), suggesting that mAPN contains all the residues sufficient for HCoV-229E receptor activity but that an N-linked glycan at N288 in mAPN may block HCoV-229E entry.
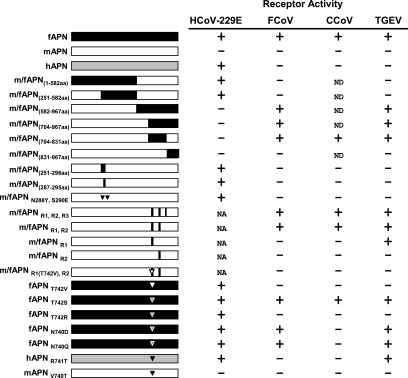
FIG. 7.
Summary of the receptor activities of wild-type, chimeric, and mutant APN proteins. The receptor activities for the APN proteins are summarized for each group 1 coronavirus. For m/fAPN chimeras, amino acids in parenthesis are the fAPN amino acids present in the chimeric protein (see text for and R1, R2, and R3). This figure includes some data presented by Tusell and Holmes (55). Triangles indicate single amino acid substitutions in the APN with amino acids present in fAPN (black), mAPN (white), or hAPN (gray); white-outlined triangles represent a single amino acid substitution that is not present in fAPN, mAPN, or hAPN. NA, not applicable; ND, not done.
Two small regions of fAPN, called R1 and R2, are required for FCoV and CCoV receptor activity, but only R1 of fAPN is required for TGEV receptor activity.
To further characterize the region in fAPN between aa 704 to 831 that was required for group 1 animal coronavirus receptor activity (Fig. 2), we aligned the amino acid sequences of APN proteins from different species and identified residues in this region that were unique to fAPN and might therefore contribute to its receptor activity for the animal coronaviruses (Fig. 3). By comparing the amino acid sequences of APN proteins that have coronavirus receptor activity—feline, canine, porcine, and human APN proteins—with sequences of APN proteins that have no known coronavirus receptor activity—mouse, rat, rabbit, and bovine APN proteins—we identified two areas of high amino acid sequence variation, aa 732 to 746 and aa 764 to 788, which we called R1 and R2, respectively (Fig. 3). Secondary structure predictions indicated that R1 is a loop or unstructured region containing several surface-exposed residues: N732, K739, N740, T742, and D743. The R2 region is predicted to be mainly α-helical with several surface-exposed residues (Fig. 3). We identified a region called R3 (aa 819 to 821) of fAPN, which contained a potential N-glycosylation sequon in canine, mouse, rabbit, and human APN proteins but not in feline, porcine, rat or bovine APN proteins (Fig. 3).
FIG. 3.
Alignment of aa 701 to 821 of APN proteins of different species. Align Plus 5 in the Clone Manager 7 software suite (Scientific and Educational Software) was used to align amino acid sequences of APN proteins from different species using a BLOSUM 62 scoring matrix. Accession numbers for feline, canine, porcine, human, bovine, mouse, rat, and rabbit APN are P79171, XP536190, P15145, P15144, P79098, P97449, P15684, and P15541, respectively. APN proteins that have coronavirus receptor activity are indicated in black, and APN proteins with no known coronavirus receptor activity are in gray. Two regions of high variability between APN sequences of different species, R1 and R2, are underlined. The R3 region corresponds to an N-glycosylation sequon that is absent in fAPN but present in APN sequences of several other species. The PROFsec and PROFacc software programs (47) were used to predict the secondary structure and solvent accessibility, respectively, of aa 701 to 821 of fAPN. The predicted secondary structure of this region of fAPN is shown above the sequence alignment. Predicted helical regions are shown as thick lines, and predicted loops or unstructured regions are thin lines. Gray dots indicate residues in R1, R2, and R3 that are predicted to be surface exposed. Arrowheads indicate residues N740 and T742 of fAPN that are required for FCoV, TGEV, and/or CCoV receptor activity.
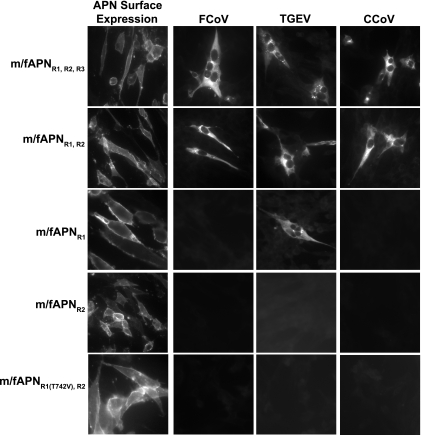
Since the amino acid sequences of the R1 and R2 regions are highly variable among APN proteins of different species and are predicted to contain surface-exposed residues, we analyzed the roles of these regions in coronavirus receptor activity. We also tested whether a potential N-linked glycan at amino acid residue N819 in the R3 of mAPN could block entry of FCoV, TGEV, or CCoV. The R1, R2, and R3 regions of mAPN were replaced with the corresponding fAPN regions. All m/fAPN chimeric proteins were expressed on the cell surface of transfected cells (Fig. 4). Cells expressing the chimeric m/fAPN proteins carrying the R1, R2, and R3 regions of fAPN (m/fAPNR1,R2,R3) or m/fAPNR1,R2 were susceptible to infection with FCoV, TGEV, and CCoV (Fig. 4). Thus, the R3 region of fAPN was not necessary for infection with these animal coronaviruses.
FIG. 4.
Two small discontinuous regions, R1 and R2, of fAPN are required for FCoV and CCoV receptor activity, but only R1 of fAPN is required for TGEV receptor activity. The mutant m/fAPNR1(T742V),R2 protein is an m/fAPNR1,R2 protein with a T742V substitution. Surface expression of the m/fAPN proteins 48 h after transfection was detected with anti-mAPN R3-242 MAb. Viral antigens were detected with antiviral antibody 10 to 12 h after inoculation.
The m/fAPNR1 chimeric protein had receptor activity for TGEV but not for FCoV or CCoV, although this protein had less TGEV receptor activity than m/fAPNR1,R2 (Fig. 4 and data not shown). Therefore, while R1 is sufficient for TGEV infection, interactions with R2 may increase the efficiency of TGEV infection. However, the R2 region of fAPN alone in an mAPN backbone was not sufficient for entry of these pig, cat, and dog coronaviruses (Fig. 4). These experiments identified two small regions of 15 aa (R1) and 25 aa (R2) of fAPN in an mAPN backbone that were necessary and sufficient for FCoV and CCoV infection, whereas only R1 of fAPN was sufficient for TGEV infection.
Asparagine 740 and threonine 742 in R1 of fAPN are important determinants of host range for FCoV, TGEV, and CCoV but not for HCoV-229E.
To identify specific residues in APN that determine the host ranges of FCoV, TGEV and CCoV we substituted single amino acids in the R1 region of the chimeric m/fAPNR1,R2 protein with the corresponding residues from mAPN. The mutant m/fAPNR1,R2 proteins with the single amino acid substitution K739N, D743N, H744R, or Q746P all had receptor activity for FCoV, TGEV, and CCoV (data not shown). In contrast, the mutant m/fAPNR1,R2 with a conservative T742V substitution (m/fAPNR1(T742V),R2) completely lacked receptor activity for FCoV, TGEV, and CCoV (Fig. 4).
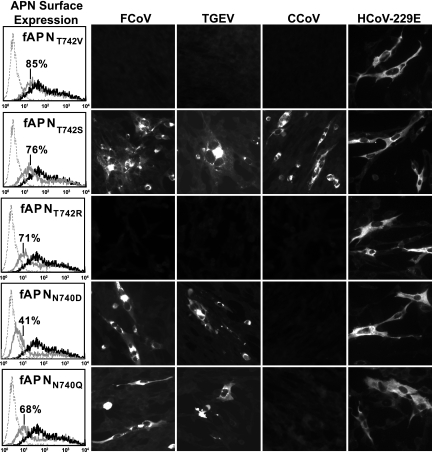
The effects of a single T742V substitution on the coronavirus receptor activity of the wild-type fAPN protein were also analyzed. Although the mutant fAPNT742V protein was expressed on the cell surface at nearly wild-type levels, it completely lacked receptor activity for FCoV, TGEV, or CCoV (Fig. 5). T742 in R1 of fAPN is part of an N-glycosylation sequon (NWT) between aa 740 to 742 (Fig. 3). To test whether this N-glycosylation sequon in fAPN affected the entry of FCoV, TGEV, and/or CCoV, we made single amino acid substitutions at N740 or T742 of fAPN that either removed or retained the glycosylation sequon. All of the N740 and T742 fAPN mutant proteins were expressed on the surface of the transfected cells and retained HCoV-229E receptor activity (Fig. 5). The fAPNT742S mutant that retained the N-glycosylation sequon had receptor activity for FCoV, TGEV, and CCoV (Fig. 5). In contrast, the fAPNT742R mutant that lacked the glycosylation sequon had no receptor activity for FCoV, TGEV, or CCoV. However, both fAPNN740Q and fAPNN740D mutant proteins that lacked the glycosylation sequon had receptor activity for FCoV and TGEV but not for CCoV (Fig. 5).
FIG. 5.
Amino acid residues T742 and N740 in fAPN are key determinants of host range for FCoV, TGEV, and CCoV but not for HCoV-229E. The surface expression of mutant fAPN proteins with the single amino acid substitution N740D, N740Q, T742V, T742S, or T742R at 48 h after transfection was analyzed by flow cytometry with anti-fAPN RG4 MAb. In the flow cytometry panels, the dotted line indicates mock-transfected cells, the gray line indicates cells expressing the mutant fAPN protein, and the black line shows cells expressing the wild-type fAPN protein. Ninety-two percent of transfected cells expressed wild-type fAPN. The percentage of cells expressing the mutant fAPN protein is shown for each overlay. Viral antigens were detected with antiviral antibody 10 to 12 h after inoculation.
In summary, T742 of fAPN was a critical determinant of host range for FCoV, TGEV, and CCoV but not for HCoV-229E, and N740 of fAPN was required only for CCoV receptor activity. Thus, even though FCoV, TGEV, and CCoV all require R1 of fAPN to infect cells, different key residues in this region are critical for entry of each virus, suggesting that the spike proteins of these coronaviruses have distinctive interactions with the R1 region of fAPN.
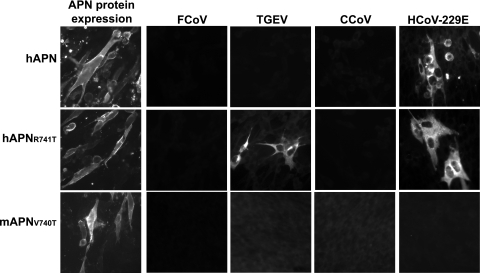
A single amino acid substitution R741T in hAPN confers TGEV receptor activity.
Since the single amino acid substitutions T742R and T742V (corresponding to R741 in hAPN or to V740 in mAPN) abrogated the FCoV, TGEV, and CCoV receptor activities of fAPN (Fig. 5), we tested the effects on coronavirus receptor activity of an R741T substitution in hAPN (hAPNR741T) or a V740T substitution in mAPN (mAPNV740T). All of the APN proteins were expressed on the surface of transfected cells (Fig. 6). As expected (65), wild-type hAPN had receptor activity for HCoV-229E but not for FCoV, TGEV, and CCoV (Fig. 6). Cells expressing mAPNV740T were resistant to infection with all four coronaviruses (Fig. 6), indicating that a single V740T substitution in mAPN was not sufficient to make this murine protein a receptor for these group 1 coronaviruses. In contrast, the mutant hAPNR741T protein retained receptor activity for HCoV-229E and gained receptor activity for TGEV but not for FCoV or CCoV (Fig. 6).
FIG. 6.
The single amino acid substitution R741T in hAPN mediates receptor activity for TGEV but not for FCoV, TGEV, or CCoV. The surface expression of wild-type hAPN and the hAPNR741T proteins was detected with anti-hAPN DW1 MAb, and the expression of wild-type mAPN and mAPNV740T proteins was detected with anti-mAPN R3-242 MAb. Viral antigens were detected with antiviral antibody 10 to 12 h after inoculation.
Additional amino acid substitutions in the R2 region of the hAPNR741T mutant protein were introduced to determine if they would confer FCoV or CCoV receptor activity. Mutant hAPN proteins with double or triple substitutions, hAPNR741T,N763Y, hAPNR741T,K777A, and hAPNR741T,M780K,E781K, were tested for HCoV-229E, FCoV, TGEV, and CCoV receptor activity. Flow cytometry showed that all of these mutant hAPN proteins were expressed on the cell surface (data not shown). Each of these mutant hAPN proteins, like the hAPNR741T protein, had receptor activity for HCoV-229E and TGEV but no receptor activity for FCoV or CCoV (data not shown). In summary, amino acid residue R741 in hAPN is responsible for the lack of TGEV receptor activity in this human protein, and additional residues, probably in R1 or R2, contribute to the lack of FCoV and CCoV receptor activity of hAPN. The V740T substitution in mAPN had no effect on coronavirus receptor activity (Fig. 6), and additional amino acids in R1 and R2 of fAPN are required for this murine protein to gain coronavirus receptor activity (Fig. 4).
DISCUSSION
The first steps in virus infection depend upon the interactions of an attachment protein on the surface of a virion with a specific receptor on the cell surface. For many coronaviruses, specific interactions between the viral spike glycoprotein and a receptor glycoprotein are important determinants of viral host range, tissue tropism, and virulence (30, 41, 48-50, 53). Adaptation of a coronavirus to a new host species may require mutations and/or recombination in the spike glycoprotein gene to accommodate differences in amino acids on the homologous receptors of the new host species (20, 30, 39, 53). To understand the role of receptor specificity in coronavirus adaptation to a new host, it is important to identify domains and residues in the receptor and spike glycoproteins that interact to mediate virus entry and contribute to virus host range. Several group 1 coronaviruses that cause disease in humans, pigs, cats, or dogs (HCoV-229E, TGEV, FCoV, and CCoV, respectively) use the APN protein of their natural host as their receptor as well as fAPN as a receptor for entry in cell culture (12, 54, 65). To identify regions and residues in the receptor protein that determine the species specificity of infection by these coronaviruses, we analyzed their interactions with their common receptor, fAPN, with human and murine APN and with chimeric mouse-feline APN proteins (Table 3). In these experiments, receptor activity was assayed by detection of viral antigens in the cytoplasm after virus entry, which requires not only binding of the spike glycoproteins to APN, but also spike-mediated fusion of the viral envelope with host cell membranes and synthesis of viral RNA and proteins. Therefore, the receptor determinants identified in this study are biologically relevant to the host range of these coronaviruses.
TABLE 3.
Key residues on APN that determine the species specificity of group 1 coronavirus infection
| APN residue or region(s) (aa)a | Group 1 coronavirus(es) | Effect on coronavirus entry |
|---|---|---|
| 288-290 | HCoV-229E | Entry is blocked by the presence of an N-glycosylation sequon in mAPN and pAPN that is absent in fAPN and hAPN |
| T742 | TGEV, FCoV, CCoV | Entry via fAPN is blocked by substitution with V from mAPN or R from hAPN |
| R741 | TGEV | Entry via hAPN requires substitution with T from fAPN or pAPN |
| R1 (732-746) | TGEV | In an mAPN backbone, entry requires substitution of the R1 region from fAPN |
| R1 (732-746) and R2 (764-788) | FCoV, CCoV | In an mAPN backbone, entry requires the substitution of both the R1 and R2 regions from fAPN |
| N740 | CCoV | Entry requires the presence of this residue that is part of an N-glycosylation sequon and is conserved in most APN proteins |
All amino acid numbers correspond to fAPN, except for R741, the residue in hAPN homologous to T742 in fAPN.
Previous studies on chimeric APN proteins identified two discontinuous regions that are required for entry of different group 1 coronaviruses (19, 29). Residues 670 to 840 in domain VII of fAPN are required for FCoV and TGEV receptor activity (19), while aa 135 to 297 in domain V of fAPN are required for HCoV-229E receptor activity (29). We have identified three smaller regions in fAPN (aa 288 to 290 in domain V and R1 and R2 in domain VII) that are critical determinants of the species specificity of coronavirus entry (Table 3). Although infection with HCoV-229E and FCoV require different regions of fAPN, the anti-fAPN RG4 MAb and its ∼50-kDa Fab fragment blocked fAPN-mediated infection by both viruses (Table 2 and Fig. 2; also data not shown). Therefore, the three discontinuous regions of fAPN (aa 288 to 290, R1, and R2) that are key determinants of the host range of the human, feline, canine, and porcine coronaviruses probably lie close together on the surface of the fAPN protein, forming part of a large interface that contains the binding sites for RG4 MAb and for the spike glycoproteins of these four group 1 coronaviruses. Cocrystals between the receptor-binding domains (RBDs) of the viral attachment proteins for herpes simplex virus, human immunodeficiency virus, and SARS-CoV with their cellular receptors HveA, CD4, and ACE2, respectively, identified numerous contact residues in the large (∼1,500 to 1,700 Å2) binding interfaces between the viral RBDs and receptor proteins (8, 9, 31, 35). The cocrystal of the SARS-CoV RBD with its hACE-2 receptor showed that 18 residues in the receptor contact 14 residues in the spike (35). Mutational analyses of the SARS-CoV RBD and ACE-2 proteins from different species showed that only a few of these contact residues are essential determinants of receptor specificity and virus entry (38, 39). Like these reports, we showed that within the large, discontinuous receptor/spike interface only a few residues on fAPN are critical determinants of coronavirus entry and host range.
We identified small, species-specific differences between APN proteins of different mammalian species that determine the host range of HCoV-229E, FCoV, TGEV, and CCoV infection (Fig. 7 and Table 3). For human coronavirus HCoV-229E, the presence or absence of a potential N-glycosylation sequon at or near aa 288 to 290 in APN is a critical determinant of host range (Fig. 7). Human and feline APN proteins that lack a glycosylation sequon at this position have receptor activity for the human coronavirus, while the porcine (59) and murine APN proteins which have an N-linked glycosylation sequon at or near this position lack HCoV-229E receptor activity. Removal of the N-glycosylation sequon at aa 288 to 290 made the mutant mAPN a receptor for HCoV-229E. These results suggest that a potential glycan at this position in mAPN and pAPN blocks entry of HCoV-229E and that residues conserved between mAPN, fAPN, and hAPN are sufficient for HCoV-229E receptor activity. Similarly, a potential N-linked glycan at the spike-receptor interface may block entry of SARS-CoV pseudotyped viruses and is a critical determinant of SARS-CoV host range (39). Rat ACE-2, which lacks receptor activity for SARS-CoV, has an N-glycosylation sequon, NFS, between aa 82 to 83, while human ACE-2 has no glycosylation sequon at this position. Substitution of this glycosylation sequon in rat ACE-2 with the homologous residues from hACE-2 and an additional H353K substitution were required for receptor activity for SARS-CoV pseudotypes (39). Thus, N-linked glycans in the virus-receptor interface may be key determinants of the receptor specificity and host range of coronaviruses.
Key determinants of the host range for FCoV, CCoV, and TGEV were identified in the R1 region (aa 732 to 746) of fAPN. Residue T742 was critical for the receptor activity for all three animal coronaviruses (Table 3). The feline, canine, and porcine APN proteins all have a T at this position that is not found in APN proteins of species that lack receptor activity for these viruses, including hAPN and mAPN, which have R or V residues at this position, respectively (Fig. 3). A T742V or T742R substitution in fAPN destroyed its receptor activity for all three animal coronaviruses, but a T742S substitution in fAPN retained receptor activity for these viruses (Fig. 5). These data suggest that a hydroxyl group from T or S at aa 742 in fAPN may be essential for its animal coronavirus receptor activity. Substitution of the large, positively charged R741 residue in hAPN with T from fAPN made the mutant hAPN a receptor for TGEV but not for FCoV and CCoV. This R741T mutation in hAPN retained HCoV-229E receptor activity (Fig. 6). T742 in the feline, canine, and porcine APN proteins is part of an N-glycosylation sequon, NWT, at or near aa 740 to 742, that is not found in mAPN and hAPN, which lack receptor activity for the three animal coronaviruses. Residue N740 of this glycosylation sequon is conserved in APN proteins of most species (Fig. 3). Although N740 of this glycosylation sequon is conserved in most APN proteins, substitution of this residue in fAPN for D or Q abrogated receptor activity for only CCoV but not HCoV-229E, FCoV, or TGEV (Fig. 5). Thus, either the N740 residue itself or a potential N-linked glycan at this position in fAPN is required for CCoV receptor activity (Fig. 5). In summary, residues N740 and T742 in fAPN and the homologous R741 residue in hAPN are critical determinants of the species specificity of group 1 animal coronavirus entry and receptor specificity.
Like these results, species-specific amino acid differences between ACE-2 proteins of human, palm civet, mouse, and rat affect the binding and entry of different SARS-CoV isolates (38, 39). Mouse ACE-2 has poor SARS-CoV receptor activity, but a single substitution of H353K, as found in the hACE-2, makes it as efficient a receptor for SARS-CoV as hACE-2 (38). However, multiple substitutions in rat ACE2 were required to confer receptor activity for SARS-CoV pseudotypes (39). In the hAPN protein, which is a receptor for HCoV-229E, a single R741T substitution makes this protein a receptor for TGEV. In contrast, a single V740T substitution in mAPN did not make this protein a receptor for any of the coronaviruses tested, and additional fAPN substitutions in R1 or R2, and/or removal of the N-linked glycan at aa 288 of the mAPN protein were required for coronavirus receptor activity (Table 3 and Fig. 7). We identified these three regions of fAPN as both sufficient and necessary for coronavirus receptor activity. However, the observation that the efficiency of virus entry decreased as smaller regions of fAPN were substituted into the mAPN backbone (data not shown) suggested that other residues of fAPN outside these regions may also contribute to efficient coronavirus entry.
Previous studies using different APN chimeras suggest that the TGEV spike protein interacts with homologous regions in the feline and porcine APN proteins. In an hAPN backbone, TGEV entry requires aa 717 to 813 in domain VII of pAPN (11), which contains the homologous region to R1 of fAPN that is sufficient for TGEV entry in an mAPN backbone. Similar findings were obtained for HCoV-229E and its feline and human APN receptors (19, 29). Although these group 1 coronaviruses interact with homologous regions on their APN receptor proteins and they can all use fAPN for entry, their RBDs are in nonhomologous regions of the viral spike glycoproteins. Enzyme-linked immunosorbent assays showed that aa 407 to 547 of the HCoV-299E spike glycoprotein were sufficient to bind soluble hAPN (7). Flow cytometry showed that retrovirus pseudotypes with chimeric HCoV-NL63/HCoV-229E spikes or truncated HCoV-229E spikes bound to hAPN if they contained both HCoV-229E aa 278 to 329 and a large downstream domain that includes aa 407 to 547 (22). Perhaps these two discontinuous regions in the HCoV-229E spike protein form the hAPN-binding domain. The domain of the TGEV spike glycoprotein that binds pAPN was mapped to aa 506 to 728 in coimmunoprecipitation assays of truncated spike proteins coexpressed with pAPN in insect cells (16). Similarly, although SARS-CoV and HCoV-NL63 both use hACE-2 as a receptor, their RBDs are not homologous, and amino acid substitutions in hACE-2 that reduce entry of SARS-CoV pseudotypes do not affect entry of HCoV-NL63 pseudotypes (1, 22, 39). In this paper we identified specific residues and regions in fAPN that determine the host range of HCoV-229E, FCoV, TGEV, and CCoV, but the specific fAPN-binding residues in the RBDs of the viral spike proteins that bind to these sites on fAPN and affect viral host range and receptor specificity remain to be identified. For SARS-CoV reports show that only two residues in the RBD determine the efficiency of binding to human or palm civet ACE-2 and, thus, determine the host range and efficiency of transmission of this virus in different host species (38, 39). Therefore, small amino acid differences in the RBDs of group 1 coronaviruses that use APN proteins as receptors may also determine their receptor specificity and host range. Palm civet ACE-2 is a receptor for SARS-CoV strains isolated from both humans and palm civets. The spike glycoproteins of these SARS-CoV strains must accommodate the species-specific amino acid differences between palm civet and human ACE-2 proteins. The fAPN protein, like palm civet ACE-2, is a receptor for several related coronaviruses that cause disease in different mammalian species. The spikes of the group 1 coronaviruses must also accommodate to species-specific differences in fAPN and the APN proteins of their natural hosts. We showed that group 1 coronaviruses HCoV-229E, FCoV, TGEV, and CCoV interact with fAPN in different ways, requiring different regions and residues in the receptor protein for virus entry. The differences in interaction of the virus spike proteins with fAPN correlate well with the phylogenetic relationships of the spike glycoproteins of the viruses. HCoV-229E spike is more divergent at the amino acid level than FCoV, TGEV, and CCoV spikes, and HCoV-229E requires a different region of fAPN than the animal coronaviruses do (Fig. 2). The spike glycoproteins of FCoV and CCoV are more closely related to each other than to TGEV spike, and FCoV and CCoV require both R1 and R2 of fAPN, while TGEV requires only the R1 region of fAPN for virus entry (Fig. 4). Furthermore, within the R1 domain of fAPN, we identified several specific residues that affect receptor specificity for FCoV and CCoV (Fig. 5).
HCoV-229E, FCoV, TGEV, and CCoV likely originated from a common ancestral group 1 coronavirus that may have infected cats using fAPN as its receptor. As HCoV-229E, TGEV, and CCoV evolved from this common ancestral virus, viruses with mutant spike proteins may have been selected for the ability to efficiently infect other host species via their APN proteins. As these group 1 coronaviruses were further selected for efficient serial transmission within the new host species, they may have lost their ability to be efficiently transmitted from cat to cat using fAPN, although they can all still use fAPN as a receptor for cell entry in vitro. In cats coinfected with FCoV and CCoV, recombination between the spike genes has been detected that gives rise to different FCoV and CCoV genotypes (20, 45, 46, 57). Spike genes of some CCoV strains have higher nucleotide sequence identity to TGEV than to other CCoV strains (61). Viral genome sequences of several bat coronaviruses closely related to human SARS-CoV and to group 1 coronaviruses were sequenced from different Asian bat species, suggesting that known human viruses, such as SARS-CoV, may have emerged from a bat coronavirus (33, 37, 43). Coronavirus evolution and emergence into new hosts is likely an ongoing process depending in part on accumulation of amino acid substitutions in the RBDs of the viral spike proteins and their receptor proteins. We identified several small differences in amino acid sequences of the feline, human, and murine APN proteins that determine their coronavirus receptor specificity and affect the host ranges of several group 1 coronaviruses. These studies contribute to an understanding of coronavirus evolution and suggest that coronaviruses may adapt to new host species by acquiring mutations in their spike proteins that enable them to recognize small, species-specific differences in the receptor protein of a new host species.
Acknowledgments
This work was supported by NIH grants AI 26075 and AI 59576. S.T. was partially supported by NIH training grant T32 AI 52066.
We thank Tanya Miura, Emily Travanty, Scott Jeffers, M. K. Smith, Susanna McReynolds, Samuel Dominguez, and Megan Howard for helpful discussions and reviews of the manuscript.
Footnotes
Published ahead of print on 8 November 2006.
REFERENCES
- 1.Babcock, G. J., D. J. Esshaki, W. D. Thomas, Jr., and D. M. Ambrosino. 2004. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 78:4552-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlough, J. E., C. M. Johnson-Lussenburg, C. A. Stoddart, R. H. Jacobson, and F. W. Scott. 1985. Experimental inoculation of cats with human coronavirus 229E and subsequent challenge with feline infectious peritonitis virus. Can. J. Comp. Med. 49:303-307. [PMC free article] [PubMed] [Google Scholar]
- 3.Barlough, J. E., C. A. Stoddart, G. P. Sorresso, R. H. Jacobson, and F. W. Scott. 1984. Experimental inoculation of cats with canine coronavirus and subsequent challenge with feline infectious peritonitis virus. Lab. Anim. Sci. 34:592-597. [PubMed] [Google Scholar]
- 4.Bastien, N., J. L. Robinson, A. Tse, B. E. Lee, L. Hart, and Y. Li. 2005. Human coronavirus NL-63 infections in children: a 1-year study. J. Clin. Microbiol. 43:4567-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benbacer, L., E. Kut, L. Besnardeau, H. Laude, and B. Delmas. 1997. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J. Virol. 71:734-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradburne, A. F., and B. A. Somerset. 1972. Coronavirus antibody titers in sera of healthy adults and experimentally infected volunteers. J. Hyg. 70:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breslin, J., I. Mork, M. K. Smith, L. K. Vogel, E. M. Hemmila, A. Bonavia, P. J. Talbot, H. Sjostrom, O. Noren, and K. V. Holmes. 2003. Human coronavirus 229E: receptor binding domain and neutralization by soluble receptor at 37C. J. Virol. 77:4435-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 9.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Groot-Mijnes, J. D., J. M. van Dun, R. G. van der Most, and R. J. de Groot. 2005. Natural history of a recurrent feline coronavirus infection and the role of cellular immunity in survival and disease. J. Virol. 79:1036-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmas, B., J. Gelfi, E. Kut, H. Sjostrom, O. Noren, and H. Laude. 1994. Determinants essential for the transmissible gastroenteritis virus-receptor interaction reside within a domain of aminopeptidase N that is distinct from the enzymatic site. J. Virol. 68:5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmas, B., J. Gelfi, R. L'Haridon, L. K. Vogel, H. Sjostrom, O. Noren, and H. Laude. 1992. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 357:417-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delmas, B., J. Gelfi, H. Sjostrom, O. Noren, and H. Laude. 1993. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 342:293-298. [DOI] [PubMed] [Google Scholar]
- 14.Dveksler, G. S., C. W. Dieffenbach, C. B. Cardellichio, K. McCuaig, M. N. Pensiero, G. S. Jiang, N. Beauchemin, and K. V. Holmes. 1993. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus-A59. J. Virol. 67:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dveksler, G. S., M. N. Pensiero, C. B. Cardellichio, R. K. Williams, G. S. Jiang, K. V. Holmes, and C. W. Dieffenbach. 1991. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 65:6881-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godet, M., J. Grosclaude, B. Delmas, and H. Laude. 1994. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 68:8008-8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorbalenya, A. E., E. J. Snijder, and W. J. Spaan. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 78:7863-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haijema, B. J., H. Volders, and P. J. Rottier. 2003. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 77:4528-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hegyi, A., and A. F. Kolb. 1998. Characterization of determinants involved in the feline infectious peritonitis virus receptor function of feline aminopeptidase N. J. Gen. Virol. 79:1387-1391. [DOI] [PubMed] [Google Scholar]
- 20.Herrewegh, A. A., I. Smeenk, M. C. Horzinek, P. J. Rottier, and R. J. de Groot. 1998. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 72:4508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann, H., K. Pyrc, L. van der Hoek, M. Geier, B. Berkhout, and S. Pohlmann. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 102:7988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann, H., G. Simmons, A. J. Rennekamp, C. Chaipan, T. Gramberg, E. Heck, M. Geier, A. Wegele, A. Marzi, P. Bates, and S. Pohlmann. 2006. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 80:8639-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hohdatsu, T., Y. Izumiya, Y. Yokoyama, K. Kida, and H. Koyama. 1998. Differences in virus receptor for type I and type II feline infectious peritonitis virus. Arch. Virol. 143:839-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton, R. M. 1993. SOEing together tailor-made genes, p. 251-261. In B. A. White (ed.), PCR protocols: current methods and applications, vol. 15. Humana Press Inc., Totowa, NJ. [Google Scholar]
- 25.Kim, L., J. Hayes, P. Lewis, A. V. Parwani, K. O. Chang, and L. J. Saif. 2000. Molecular characterization and pathogenesis of transmissible gastroenteritis coronavirus (TGEV) and porcine respiratory coronavirus (PRCV) field isolates co-circulating in a swine herd. Arch. Virol. 145:1133-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, O., and C. Chae. 2003. Experimental infection of piglets with a Korean strain of porcine epidemic diarrhoea virus. J. Comp. Pathol. 129:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss, I., A. M. Poland, and N. C. Pedersen. 2004. Disease outcome and cytokine responses in cats immunized with an avirulent feline infectious peritonitis virus (FIPV)-UCD1 and challenge-exposed with virulent FIPV-UCD8. J. Feline Med. Surg. 6:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolb, A. F., A. Hegyi, J. Maile, A. Heister, M. Hagemann, and S. G. Siddell. 1998. Molecular analysis of the coronavirus-receptor function of aminopeptidase N. Adv. Exp. Med. Biol. 440:61-67. [DOI] [PubMed] [Google Scholar]
- 29.Kolb, A. F., A. Hegyi, and S. G. Siddell. 1997. Identification of residues critical for the human coronavirus 229E receptor function of human aminopeptidase N. J. Gen. Virol. 78:2795-2802. [DOI] [PubMed] [Google Scholar]
- 30.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae: the viruses and their replication, p. 1163-1185. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 33.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lendeckel, U., T. Kahne, D. Riemann, K. Neubert, M. Arndt, and D. Reinhold. 2000. Review: the role of membrane peptidases in immune functions. Adv. Exp. Med. Biol. 477:1-24. [DOI] [PubMed] [Google Scholar]
- 35.Li, F., W. Li, M. Farzan, and S. C. Harrison. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864-1868. [DOI] [PubMed] [Google Scholar]
- 36.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676-679. [DOI] [PubMed] [Google Scholar]
- 38.Li, W., S. K. Wong, F. Li, J. H. Kuhn, I. C. Huang, H. Choe, and M. Farzan. 2006. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J. Virol. 80:4211-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, W., C. Zhang, J. Sui, J. H. Kuhn, M. J. Moore, S. Luo, S. K. Wong, I. C. Huang, K. Xu, N. Vasilieva, A. Murakami, Y. He, W. A. Marasco, Y. Guan, H. Choe, and M. Farzan. 2005. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24:1634-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perlman, S., and A. A. Dandekar. 2005. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 5:917-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillips, J. J., M. M. Chua, E. Lavi, and S. R. Weiss. 1999. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 73:7752-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poland, A. M., H. Vennema, J. E. Foley, and N. C. Pedersen. 1996. Two related strains of feline infectious peritonitis virus isolated from immunocompromised cats infected with a feline enteric coronavirus. J. Clin. Microbiol. 34:3180-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poon, L. L., D. K. Chu, K. H. Chan, O. K. Wong, T. M. Ellis, Y. H. Leung, S. K. Lau, P. C. Woo, K. Y. Suen, K. Y. Yuen, Y. Guan, and J. S. Peiris. 2005. Identification of a novel coronavirus in bats. J. Virol. 79:2001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pratelli, A. 2006. Genetic evolution of canine coronavirus and recent advances in prophylaxis. Vet. Res. 37:191-200. [DOI] [PubMed] [Google Scholar]
- 45.Pratelli, A., N. Decaro, A. Tinelli, V. Martella, G. Elia, M. Tempesta, F. Cirone, and C. Buonavoglia. 2004. Two genotypes of canine coronavirus simultaneously detected in the fecal samples of dogs with diarrhea. J. Clin. Microbiol. 42:1797-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pratelli, A., V. Martella, M. Pistello, G. Elia, N. Decaro, D. Buonavoglia, M. Camero, M. Tempesta, and C. Buonavoglia. 2003. Identification of coronaviruses in dogs that segregate separately from the canine coronavirus genotype. J. Virol. Methods 107:213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rost, B., G. Yachdav, and J. Liu. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321-W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rottier, P. J., K. Nakamura, P. Schellen, H. Volders, and B. J. Haijema. 2005. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J. Virol. 79:14122-14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez, C. M., A. Izeta, J. M. Sanchez-Morgado, S. Alonso, I. Sola, M. Balasch, J. Plana-Duran, and L. Enjuanes. 1999. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 73:7607-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schickli, J. H., L. B. Thackray, S. G. Sawicki, and K. V. Holmes. 2004. The N-terminal region of the murine coronavirus spike glycoprotein is associated with the extended host range of viruses from persistently infected murine cells. J. Virol. 78:9073-9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sjostrom, H., O. Noren, and J. Olsen. 2000. Structure and function of aminopeptidase N. Adv. Exp. Med. Biol. 477:25-34. [DOI] [PubMed] [Google Scholar]
- 52.Smith, M. K., S. Tusell, E. A. Travanty, B. Berkhout, L. van der Hoek, and K. V. Holmes. 2006. Human angiotensin-converting enzyme 2 (ACE2) is a receptor for human respiratory coronavirus NL63. Adv. Exp. Med. Biol. 581:285-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thackray, L. B., and K. V. Holmes. 2004. Amino acid substitutions and an insertion in the spike glycoprotein extend the host range of the murine coronavirus MHV-A59. Virology 324:510-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tresnan, D. B., R. Levis, and K. V. Holmes. 1996. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 70:8669-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tusell, S. M., and K. V. Holmes. 2006. Molecular interactions of group 1 coronaviruses with feline APN. Adv. Exp. Med. Biol. 581:289-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Hoek, L., K. Sure, G. Ihorst, A. Stang, K. Pyrc, M. F. Jebbink, G. Petersen, J. Forster, B. Berkhout, and K. Uberla. 2005. Croup is associated with the novel coronavirus NL63. PLOS Med. 2:e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vennema, H., A. Poland, J. Foley, and N. C. Pedersen. 1998. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology 243:150-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wentworth, D. E., and K. V. Holmes. 2001. Molecular determinants of species specificity in coronavirus receptor aminopeptidase N (CD13): influence of N-linked glycosylation. J. Virol. 75:9741-9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wenzel, R. P., J. O. Hendley, J. A. Davies, and J. M. Gwaltney, Jr. 1974. Coronavirus infections in military recruits. Three-year study with coronavirus strains OC43 and 229E. Am. Rev. Respir. Dis. 109:621-624. [DOI] [PubMed] [Google Scholar]
- 61.Wesley, R. D. 1999. The S gene of canine coronavirus, strain UCD-1, is more closely related to the S gene of transmissible gastroenteritis virus than to that of feline infectious peritonitis virus. Virus Res. 61:145-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams, R. K., G. S. Jiang, and K. V. Holmes. 1991. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. USA 88:5533-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woods, R. D., N. F. Cheville, and J. E. Gallagher. 1981. Lesions in the small intestine of newborn pigs inoculated with porcine, feline, and canine coronaviruses. Am. J. Vet. Res. 42:1163-1169. [PubMed] [Google Scholar]
- 64.Woods, R. D., and R. D. Wesley. 1992. Seroconversion of pigs in contact with dogs exposed to canine coronavirus. Can. J. Vet. Res. 56:78-80. [PMC free article] [PubMed] [Google Scholar]
- 65.Yeager, C. L., R. A. Ashmun, R. K. Williams, C. B. Cardellichio, L. H. Shapiro, A. T. Look, and K. V. Holmes. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]