Abstract
Adeno-associated virus (AAV) integrates site specifically into the AAVS1 locus on human chromosome 19. Although recruitment of the AAV nonstructural protein Rep78/68 to the Rep binding site (RBS) on AAVS1 is thought to be an essential step, the mechanism of the site-specific integration, particularly, how the site of integration is determined, remains largely unknown. Here we describe the identification and characterization of a new cellular regulator of AAV site-specific integration. TAR RNA loop binding protein 185 (TRP-185), previously reported to associate with human immunodeficiency virus type 1 TAR RNA, binds to AAVS1 DNA. Our data suggest that TRP-185 suppresses AAV integration at the AAVS1 RBS and enhances AAV integration into a region downstream of the RBS. TRP-185 bound to Rep68 directly, changing the Rep68 DNA binding property and stimulating Rep68 helicase activity. We present a model in which TRP-185 changes the specificity of the AAV integration site from the RBS to a downstream region by acting as a molecular chaperone that promotes Rep68 complex formation competent for 3′→5′ DNA helicase activity.
Adeno-associated virus (AAV) is a nonpathogenic human parvovirus that contains a linear single-stranded DNA genome of approximately 4.7 kb, carrying palindromic inverted terminal repeats (ITRs) at both ends that serve as the viral origin of replication. The AAV genome consists of two major open reading frames, rep and cap. The cap gene encodes three structural proteins, VP1, VP2, and VP3. The rep gene encodes four nonstructural proteins, Rep78, Rep68, Rep52, and Rep40. The nonstructural proteins have multiple activities, such as DNA binding, ATPase, helicase, and endonuclease activities, and play pivotal roles in various stages of the viral life cycle, including integration, replication, and regulation of viral gene expression. For efficient AAV replication and gene expression, coinfection by a helper virus, such as adenovirus or herpesvirus, is required. In the absence of a helper virus, AAV infection results in stable integration of the viral DNA genome into a specific locus on chromosome 19, called AAVS1 (17, 25). This property of AAV is unique among eukaryotic DNA viruses.
In the current model of AAV site-specific integration, following infection, the single-stranded AAV genome is converted to duplex DNA. Next, the viral p5 promoter is activated and directs the synthesis of Rep78 and Rep68. These proteins bind to the 16-bp Rep binding site (RBS) present in the AAV ITRs and the p5 promoter. Rep78/68 also binds to an RBS in the AAVS1 region of chromosome 19 and recruits the AAV genome to AAVS1 (8, 31). Rep78/68 then creates a nick at the 6-bp terminal resolution site (trs) flanking the RBS on AAVS1, and its helicase activity unwinds the duplex trs in the 3′→5′ direction to facilitate DNA replication (8, 29). Subsequently, the AAV genome is integrated into the AAVS1 site within ca. 1 kb downstream of the RBS (8, 21). Currently, there are two models of the formation of the AAV-AAVS1 junction. One model assumes that there is Rep78/68-mediated strand switching during DNA replication (21); the other model assumes the involvement of Rep in the ligation of AAV and AAVS1 sequences (27).
AAV site-specific integration requires three components: (i) the RBS on the ITRs and the p5 promoter of the AAV genome, (ii) a 33-bp sequence containing the trs and the RBS in AAVS1 of chromosome 19, and (iii) Rep78 or Rep68 (12, 24). Rep78 and Rep68 function in the same way in AAV site-specific integration, and either protein is sufficient for integration in vitro. However, as the levels of Rep proteins are extremely low in latently infected cells (36), additional cellular factors that help Rep function may exist and contribute to AAV site-specific integration. Indeed, it has been reported that the high-mobility-group protein 1 (HMG-1) binds to and promotes the DNA binding and endonuclease activities of Rep68 in vitro, thereby stimulating junction formation at the RBS (3, 6). Nevertheless, the complicated mechanism of AAV site-specific integration, particularly, the mechanism of integration of the AAV genome within ca. 1 kb downstream of the RBS in AAVS1 in vivo (8, 21), has not yet been fully elucidated. Although Rep helicase activity is thought to be involved in AAV integration into a region downstream of the RBS (21), it is unclear how the helicase activity is regulated during latent infection by AAV.
We have been interested in identifying cellular regulators of AAV site-specific integration, which we assume interact with central components of the integration machinery, such as Rep68 and AAVS1 DNA. In a previous paper (9), we described searching for Rep68-binding proteins and identifying two members of the 14-3-3 protein family. Subsequent analysis suggested that 14-3-3 is involved in AAV DNA replication but not in integration (reference 9 and data not shown). In the current study, we focused on AAVS1 and searched for proteins that bind to the core elements of AAVS1. Towards this end, we carried out affinity chromatography using latex beads to which one can conjugate various biologically active components, such as chemical compounds (26), nucleic acids (30), and proteins (9). In a one-step affinity chromatography procedure, we purified TRP-185, a 185-kDa protein previously implicated in the activation of human immunodeficiency virus type 1 gene expression (33), from HeLa cell nuclear extracts (NE). We present several lines of evidence suggesting that TRP-185 controls selection of the AAV integration site through its interaction with the AAVS1 RBS and Rep68.
MATERIALS AND METHODS
Plasmids.
An approximately 1.6-kb fragment of AAVS1 was excised from pUC18 (HindIII) AAVS1 (a gift from Tadahito Kanda) and cloned into EcoRI and BamHI sites of pBluescript SK+ to create pBSAAVS1. The EcoRI and BamHI sites of pBSAAVS1 were defined as nucleotide numbers 0 and 1612, respectively (see Fig. 2A). The RBS of pBSAAVS1 (GCTCGCTCGCTCGCTG) was mutated to CCTCCCTCCCTCCCTG by the PCR-based overlap extension method. Mutated nucleotides in the above sequence are underlined. To construct pFASTRep68, a DNA fragment containing Rep68 was excised from pBS-Rep68 (9) and cloned into pFastBac1 (Invitrogen). A Rep68 K340H mutant was generated by PCR-based mutagenesis. The TRP-185 cDNA, modified to express a C-terminal His or Flag tag, was cloned into pFastBac1 or pcDNA3.1(+) (Invitrogen), respectively, thus generating pFASTTRP-185-His and pcTRP-185-Flag. For the expression of a short hairpin RNA (shRNA) that targeted nucleotides 1414 to 1434 of the TRP-185 mRNA, the oligonucleotides 5′-ACTCAGTATATAGCGGAAAGTTCAAGAGACTTTCCGCTATACTGAGTCTTTTT-3′ and 5′-GATCAAAAAGACTCAGTATAGCGGAAAGTCTCTTGAACTTTCCGCTATACTGAGTCA-3′ were annealed and inserted into pBS-U6 (35), which contains the mouse U6 promoter. Next, a DNA fragment containing the U6 promoter and the oligonucleotides was inserted into the BanIII and SacII sites of pLenti6/V5-GW/lacZ (Invitrogen).
FIG. 2.
Rep68 mediates AAV-AAVS1 junction formation specifically at the RBS in vitro. (A) Schematic diagrams of the AAV genome and the plasmid pBSAAVS1 containing a 1.6-kb AAVS1 sequence (left) and primer sets (right) used for in vitro junction formation assays. The positions of the primers used are indicated in the diagrams. (B) rRep68 used in this study was visualized by silver staining. The positions of molecular mass markers are shown on the right. (C) In vitro junction formation assays were performed with or without 1.5 pmol of rRep68. The recombination products and the indicated amounts of PCR standards were subjected to PCR analysis with various primer sets as shown in panel A. The PCR products were analyzed by 2% agarose gel electrophoresis and stained with ethidium bromide. The positions of size markers are shown on the right. (D) Junction formation assays were performed with the indicated amounts of rRep68 and wild-type (w) or mutant (mt) pBSAAVS1. The recombination products were examined by PCR with primer set 1 (A). The positions of size markers are shown on the right. Quantitation of the results from three independent experiments is shown below. Data represent the means ± standard errors of the mean. (E) AAV-AAVS1 junction sequences from eight clones. The RBS-flanking regions of the two substrates are shown above. In the junction sequences, overlapping sequences between AAV and AAVS1 are indicated by open boxes, and respective junction points are indicated by numbers according to the published numbering system (16). The RBS sequences are indicated by letters. nt, nucleotide.
Preparation of recombinant proteins.
Recombinant hemagglutinin (rHA)- and Flag-tagged Rep68 (rRep68) and His-tagged TRP-185 (rTRP-185) proteins were expressed in insect cells (sf9) as described in reference 9. rRep68 was purified by affinity chromatography using anti-Flag M2 agarose (Sigma) followed by elution with a Flag peptide. rTRP-185 was purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography according to the manufacturer's instructions (QIAGEN).
Affinity purification by DNA-immobilized latex particles.
AAVS1 DNA binding proteins were affinity purified from HeLa cell NE that had been prepared by Dignam's method (4). The following oligonucleotides were annealed and covalently conjugated to latex particles as described in reference 14: 5′-AATTCGGCGGTTGGGGCTCGGC(GCTC)3GCTGGGCGGGCGG-3′ and 5′-GATCCCGCCCGCCCAGC(GAGC)3GCCGAGCCCCAACCGCCG-3′ for wild-type minimal AAVS1 sequence (mnAAVS1) and 5′-AATTCGGCGGTTGGGGCTCGGC(CCTC)3CCTGGGCGGGCGG-3′ and 5′-GATCCCGCCCGCCCAGG(GAGG)3GCCGAGCCCCAACCGCCG-3′ for mutant mnAAVS1. Mutated nucleotides in the above sequences are underlined. Latex particles (0.43 mg) carrying 1 μg of DNA were equilibrated with HGEDN (10 mM HEPES [pH 7.9], 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.1% NP-40) containing 0.1 M KCl (0.1 HGEDN; the number preceding HGEDN denotes the molar concentration of KCl) and incubated with HeLa NE (800 μg), salmon sperm DNA (50 μg), and poly(dI-dC) (2 μg) at 4°C for 1 h. The latex particles were washed three times with 0.2 HGEDN, and bound proteins were eluted with 0.3 HGEDN.
Formaldehyde cross-linking and ChIP analysis.
HeLa cells (7 × 106 cells on a 15-cm dish) were transfected with 6 μg of pcTRP-185-Flag using Lipofectamine 2000 (Invitrogen). Two days posttransfection, the cells were harvested for chromatin immunoprecipitation (ChIP) analysis as described previously (7). Genomic DNA fragments in the input and immunoprecipitated samples were purified and subjected to real-time PCR analysis using iQ SYBR green supermix (Bio-Rad). The following primers were used: 5′-ATCCGTGACGTCAGCAAGC-3′ and 5′-CATCCTCTCCGGACATCG-3′ for the AAVS1 RBS region and 5′-GCCTTAAGGTTTATACCAAAATCA-3′ and 5′-GGAAGGCACTGTTAAAGTTGAG-3′ for a chromosome 2q34 region. Amplification conditions consisted of 95°C for 3 min, followed by 41 cycles of 95°C for 15 s, 56°C for 15 s, and 72°C for 15 s.
In vitro junction formation.
In vitro junction formation assays were performed as described previously (6), except that the reactions were carried out in a 15-μl solution containing 40 mM HEPES (pH 7.9), 7 mM MgCl2, 4 mM ATP, 2 mM DTT, 15 fmol of the AAV genome substrate (19), 30 fmol of wild-type or mutant pBSAAVS1, and various amounts of rRep68. HeLa NE, rTRP-185, and bovine serum albumin (BSA) were included in the reaction mixture where indicated. DNA was purified and subjected to real-time PCR analysis using iQ SYBR green supermix and the following primers: V-4526/PITR (5′-TTAACTACAAGGAACCCCTA-3′; AAV positions 4526 to 4545), PITR left/V-149 (5′-CTCCAGGAACCCCTAGT-3′; AAV positions 133 to 149), S1-447/H4d1 (5′-GGCAAGCTTCCATCCTCTCCGGACATCGCAC-3′; pBSAAVS1 positions 426 to 447), S1-1562 (5′-GCAACACAGCAGAGAGCAAG-3′; pBSAAVS1 positions 1543 to 1562), S1-2513 (5′-GCCTACATACCTCGCTCTGC-3′; pBSAAVS1 positions 2494 to 2513), and S1-3693 (5′-TTTGCCTTCCTGTTTTTGCT-3′; pBSAAVS1 positions 3674 to 3693) (6, 10, 11). Amplification conditions consisted of 95°C for 3 min, followed by 30 cycles of 95°C for 15 s, 56°C for 15 s, and 72°C for 1 min. For each primer set, 500-bp linear DNA containing primer-binding sites at both ends was made by PCR and used to generate a standard curve. PCR products were analyzed by 2% agarose gel electrophoresis or cloned into pGEM-T Easy (Promega) for sequencing.
Immunodepletion of TRP-185.
Immunodepletion of TRP-185 from HeLa NE was carried out essentially as described previously (7). NE (100 μl) was incubated three times for 2 h each at 4°C with protein A-Sepharose onto which 160 μg of anti-TRP-185 antiserum (raised against TRP-185 amino acids 1411 to 1621) or preimmune serum was absorbed.
TRP-185 knockdown.
For knockdown experiments, a recombinant lentivirus expressing shRNA against TRP-185 was prepared using a ViraPower lentiviral expression system (Invitrogen) and pLentisiTRP-185. Quantification of the TRP-185 mRNA level was carried out using QuantiTect SYBR green reverse transcription-PCR (RT-PCR) master mix (QIAGEN) and the TRP-185-specific primers 5′-CAGGTGACTGGTCTCAGCAA-3′ and 5′-CTGAAGCCCCAAATACCTCA-3′.
In vivo AAV integration assay.
AAV preparation and infection were performed as previously described (11). Sixteen hours postinfection, genomic DNA was prepared using a DNeasy tissue kit (QIAGEN). PCR was carried out using iQ supermix (Bio-Rad), 200 ng of genomic DNA, and the following primers: V-3569 (5′-ACGCAGTCAAGGCTTCAGTT-3′; AAV positions 3569 to 3588) and S1-447/H4d1 for the RBS, V-3569 and S1-1615 (5′-ATCCGCTCAGAGGACATCAC-3′; AAVS1 positions 1596 to 1615) for the RBS downstream region, and V-4526/PITR and the Alu element primer (5′-GCCTCCCAAAGTGCTGGGATTACAG-3′) for flanking regions of Alu elements. Amplification conditions consisted of 95°C for 3 min, followed by 35 cycles of 95°C for 1 min, 56°C for 1 min, and 72°C for 3 min. Amplified products were analyzed by Southern hybridization using a 32P-labeled AAV-specific probe (AAV positions 4526 to 4679) or cloned into pGEM-T Easy (Promega) for sequencing.
Helicase assays.
Circular M13 substrate was prepared as described previously (13). Linear AAVS1 substrates were prepared by annealing the complementary oligonucleotides 5′-TGGGGCTCGGCGCTCGCTCGCTCGCTGGG-3′ and 5′-GCCCGCCCAGCGAGCGAGCGAGCGCCGAGCCCCAACCGCCGCCACCACCCGCCCGCCCGC-3′ (for wild-type AAVS1) or 5′-TGGGGCTCGGCcCTCcCTCcCTCcCTGGG-3′ and 5′-GCCCGCCCAGgCAGgCAGgGAGgG CCGAGCCCCAACCGCCGCCACCACCCGCCCGCCCGC-3′ (for mutant AAVS1), followed by labeling of the 3′ ends of the sense strands with Klenow fragment and [α-32P]dCTP. The RBS sequence in each oligonucleotide is underlined, and mutated positions are shown in lowercase. Wild-type or mutant AAVS1 substrate (12.5 fmol) or M13 substrate (10 fmol) was incubated with the indicated amounts of rRep68 and rTRP-185 in a 20-μl reaction mixture containing 25 mM HEPES (pH 7.9), 0.5 mM ATP, 5 mM MgCl2, 1 mM DTT, and 200 ng of BSA for 30 min at 37°C, and the reactions were terminated by the addition of 10 μl of stop buffer (0.5% sodium dodecyl sulfate [SDS], 50 mM EDTA). The mixtures were then electrophoresed on nondenaturing 10% (AAVS1) or 6% (M13) polyacrylamide gels and visualized by autoradiography.
Gel-shift assays.
Gel-shift probe was made by end-labeling of double-stranded wild-type mnAAVS1 oligonucleotides (the same as the one used for affinity purification) with Klenow fragment. Reaction mixtures (10 μl) containing 5 μg of BSA, 10 ng of poly(dI-dC), 0.01 pmol of probe, and various amounts of rRep68 and rTRP-185 in 0.1 HGEDN were incubated on ice for 30 min and separated by 4% polyacrylamide gel electrophoresis in 0.5× Tris-borate-EDTA at 4°C.
Gel filtration.
Reaction mixtures (50 μl) containing BSA (25 μg), rTRP-185 (7.5 pmol), rRep68 (0.75 pmol), and mnAAVS1 DNA (2.5 pmol) in 0.1 HGEDN were incubated on ice for 30 min and then applied to a Superose 6 gel filtration column (Amersham Pharmacia) equilibrated with 0.1 HGEDN. The fractions were analyzed by immunoblotting for rRep68 and rTRP-185 or Southern blotting for mnAAVS1 DNA.
His-tag pull-down assays.
The Ni-NTA resin was equilibrated with 0.1 HGEDN containing 20 mM imidazole and incubated with 1.2 pmol of rTRP-185, 3.5 pmol of rRep68, and 50 μg of BSA in the presence or absence of 2.5 pmol of wild-type or mutant mnAAVS1 DNA in a 100-μl reaction mixture for 30 min at 4°C. The resin was washed three times with the imidazole-containing buffer. Bound proteins were eluted with SDS sample buffer (30), separated by 7.5% SDS-polyacrylamide gel electrophoresis (PAGE), and immunoblotted.
RESULTS
Identification of TRP-185 as an AAVS1-binding factor.
To purify cellular factors that interacted with AAVS1, we performed affinity chromatography using an approximately 50-bp wild-type mnAAVS1, containing the trs and the RBS, or its mutant, immobilized on latex beads (Fig. 1A). The latex beads were incubated with HeLa NE, and proteins that bound to the DNA-conjugated beads were eluted with high-salt buffer and analyzed by SDS-PAGE. As shown in Fig. 1B, a protein of approximately 185 kDa in size was specifically eluted from the mnAAVS1-conjugated beads (Fig. 1B, compare lanes 3 and 4). On the silver-stained gel, proteins of 80, 60, and 50 kDa (indicated by dots) also seemed to be recovered selectively from the wild-type mnAAVS1-conjugated beads; however, the observation was not reproducible (data not shown). The 185-kDa protein band was therefore subjected to in-gel digestion with lysyl-C endopeptidase, and the resulting peptides were subjected to sequence analysis by tandem mass spectrometry. We obtained four sequences, GRPAGGPDPSLQP, LLPVLVQCGGAALR, LLDKDELVSK, and QQLLSHGDTKP, that were identical to the protein sequence of TRP-185. We confirmed the identity of the purified protein using immunoblot analysis. As shown in Fig. 1B, lower panel, the 185-kDa protein was recognized by anti-TRP-185 monoclonal antibody.
FIG. 1.
Identification of TRP-185 as an AAVS1-binding factor. (A) Wild-type and mutant mnAAVS1 DNA sequences and schematic representation of DNA-immobilized latex beads. (B) Input NE (lane 1), eluate from control beads (lane 2), and eluate from wild-type (lane 3) or mutant (lane 4) mnAAVS1-immobilized beads were separated on a 5 to 20% SDS-polyacrylamide gel, and proteins were visualized by silver staining. The protein bands that were differentially purified by wild-type mnAAVS1-conjugated beads are marked by dots. In the lower panel, the same samples were immunoblotted using anti-TRP-185 monoclonal antibody (NK5.18, a gift of Richard B. Gaynor). The positions of molecular mass markers are shown on the left. (C) HeLa cells were transiently transfected with pcTRP-185-Flag or pcDNA3.1(+) (mock) and then subjected to ChIP analysis using anti-Flag antibody. Input and immunoprecipitated (Ppt) DNAs were subjected to real-time PCR analysis using primers that specifically amplified the AAVS1 RBS region or a control region on chromosome 2q34. PCR products were analyzed by 2% agarose gel electrophoresis. Data represent the means ± standard errors of the mean from six independent experiments, and statistical significance, indicated by a bracket, was determined by Student's t test (P < 0.05).
To assess whether TRP-185 interacted with AAVS1 in cells, we performed ChIP analysis of HeLa cells expressing or not expressing Flag-tagged TRP-185. After formaldehyde cross-linking, immunoprecipitation was carried out with anti-Flag antibody, and coprecipitated DNA and input DNA were subjected to real-time PCR analysis with primers that specifically amplified the AAVS1 RBS region or a control region on chromosome 2q34. As shown in Fig. 1C, TRP-185-Flag associated with the AAVS1 RBS region, whereas its association with the control region on chromosome 2q34 was comparable to background levels. These results together indicated that TRP-185 binds to the AAVS1 RBS region in vitro and in vivo.
Rep68 mediates AAV-AAVS1 junction formation specifically at the RBS in vitro.
Prior to further characterization of TRP-185, we carried out a reevaluation of previously developed in vitro junction formation assays. In a previous study (6), it was shown that junctions between AAV and AAVS1 were formed almost precisely at the RBS of AAV and AAVS1 in the presence of Rep68 and ATP, and that the junction formation was enhanced by components of HeLa cell extracts. In our hands, however, integration products were obtained in amounts sufficient for reliable quantification in the absence of crude cell extracts (Fig. 2), and in fact, HeLa cell NE showed an inhibitory, rather than a stimulatory, effect on integration (see Fig. 3 and Discussion). Hence, our standard reaction mixtures contained two DNA substrates (Fig. 2A), ATP, and purified rRep68 (Fig. 2B) as the only source of protein factor. Since the previous study used only PCR primers that would detect AAV-AAVS1 junctions at or near the RBS (6), we first used various AAVS1 primers at different distances away from the RBS in combination with AAV primers at both ends of the AAV genome in order to detect various potential integration products (Fig. 2A). The results were that integration events were efficiently observed only at the RBS region of AAVS1 in the presence of rRep68, although some weak signals were also seen in other regions in a Rep68-independent manner (Fig. 2C). In addition, integration at the AAVS1 RBS was dependent on intact RBS and the concentrations of rRep68 indicated in the legend to Fig. 2D. Furthermore, sequence analysis of eight cloned products revealed that all the junctions were formed precisely at the RBS of AAV and AAVS1 (Fig. 2E). These results lead us to conclude that our in vitro integration assay is specific to the AAVS1 RBS, Rep68 dependent, and orientation independent, which is consistent with the previous report (6). In a subsequent study, we therefore used only primer set 1, specified in Fig. 2A.
FIG. 3.
TRP-185 inhibits Rep68-mediated AAV-AAVS1 junction formation at the RBS in vitro. (A) rTRP-185 used in this study was visualized by silver staining. The positions of molecular mass markers are shown on the right. (B) Junction formation assays were performed with the indicated amounts of rRep68, rTRP-185, heat-denatured rTRP-185 (h.d. rTRP-185), and BSA. The positions of size markers are shown on the right. Quantitation of the results from three independent experiments is shown below. Data represent the means ± standard errors of the mean. (C) Immunoblot analysis of HeLa NE depleted of TRP-185. HeLa NE was repeatedly (one to three times) passed over protein A-Sepharose to which either anti-TRP-185 or preimmune serum (ctrl) was absorbed. The supernatants were analyzed for the presence of TRP-185 and HMG-1 by immunoblotting. Quantitation of each protein level is shown below. (D) AAV-AAVS1 junction formation reactions were performed in the presence or absence of control (ctrl) or TRP-185-depleted (deltaTRP) NE (8 μg) and the indicated amounts of rRep68 and rTRP-185. The positions of size markers are shown on the right. Quantitation of the results from three independent experiments is shown below. Data represent the means ± standard errors of the mean.
TRP-185 inhibits Rep68-mediated AAV-AAVS1 junction formation at the RBS in vitro.
To determine whether TRP-185 plays a role in AAV-AAVS1 junction formation, we performed in vitro junction formation assays using histidine-tagged rTRP-185 that was expressed in insect cells and affinity-purified by nickel column chromatography (Fig. 3A). Addition of increasing amounts of rTRP-185 to the junction formation reaction mixture resulted in clear inhibition of junction formation in a dose-dependent manner (Fig. 3B, lanes 3 to 5), whereas heat-denatured rTRP-185 or BSA had no appreciable effect (Fig. 3B, lanes 6 and 7).
To further confirm the involvement of TRP-185 in junction formation, TRP-185 was immunodepleted from HeLa NE using anti-TRP-185 antibody. HeLa NEs were passed three times through an anti-TRP-185 antibody-immobilized column or a control column, and the presence of TRP-185 was monitored by immunoblotting (Fig. 3C). After three rounds of immunodepletion, more than 90% of TRP-185 was removed from the NE, whereas little change in the level of a control protein, HMG-1, was observed. Addition of control NE to the junction formation assay strongly inhibited junction formation (Fig. 3D, lanes 2 and 3). In contrast, the addition of TRP-185-immunodepleted NE had only a weak inhibitory effect (Fig. 3D, lane 4), and simultaneous addition of rTRP-185 restored strong inhibition of junction formation (Fig. 3D, lanes 5 and 6). These results demonstrated that TRP-185 inhibits Rep68-dependent AAV-AAVS1 junction formation at the RBS in vitro.
TRP-185 alters AAV integration sites from the RBS to a downstream region in vivo.
A previous report showed that in vivo AAV integration sites on AAVS1 are scattered within ca. 1 kb downstream of the RBS (21). This prompted us to examine whether TRP-185 affected determination of the AAV integration site. To this end, we generated TRP-185-knockdown HeLa cells using a lentiviral expression vector encoding an shRNA targeting TRP-185. At 7 days post-lentivirus infection, the protein level of TRP-185 in knockdown cells was reduced to approximately 20% of the control level (Fig. 4A). The level of TRP-185 mRNA was also reduced, to approximately 7% of the control level, as determined by real-time RT-PCR analysis (Fig. 4B). Downregulation of TRP-185 to this level was maintained for at least 4 weeks postinfection without any significant effect on cell proliferation (data not shown).
FIG. 4.
TRP-185 alters AAV integration sites from the RBS to a downstream region in vivo. (A, B) HeLa cells were infected with a lentiviral expression vector encoding an shRNA that targeted TRP-185 (deltaTRP) or a control vector (ctrl). Seven days postinfection, whole cell extracts were prepared and immunoblotted with anti-TRP-185 and anti-β-tubulin antibodies (A). Alternatively, total RNA was prepared, and the mRNA level (arbitrary units) of TRP-185 was quantified by real-time RT-PCR (B). (C) In vivo integration assays were performed in control and deltaTRP HeLa cells. Following AAV or mock infection, AAV integration into the RBS, a region downstream of the RBS, and Alu repeat regions was analyzed by PCR using specific primer sets for each region, followed by Southern blot analysis. AAV-infected samples were analyzed in duplicate. The positions of DNA size markers are shown on the right. (D) Sequence analysis of AAV-AAVS1 junctions at the RBS (in deltaTRP cells) and downstream of the RBS (in control cells). In the top diagram, the positions of the PCR primers and Southern probe used are indicated, and integration events are denoted by arrows. nt, nucleotide. In the RBS junction sequences, the RBS sequences are indicated by letters, and overlapping sequences between AAV and AAVS1 are indicated by open boxes. In the downstream junction sequences, overlapping sequences between AAV and AAVS1 are indicated by letters.
Control and knockdown cells were infected with AAV at a multiplicity of infection of 500. Since it was reported that AAV site-specific integration becomes detectable between 8 and 16 h postinfection (11), total DNA was prepared 16 h postinfection and subjected to PCR and Southern blot analysis. For PCR, three different primer sets were used to analyze integration at the AAVS1 RBS, at a region downstream of the RBS, and near Alu repeat regions. Since Alu repeats are distributed throughout the human genome (1), we considered that AAV-Alu signals would represent nonspecific AAV integration into the genome outside AAVS1, which is known to occur occasionally in AAV-infected cells (22). Using these primer sets, PCR products of various lengths should be generated due to the heterogeneity of junction points, especially in the case of the Alu primer set. In control cells, junction formation occurred efficiently at a region approximately 1 kb downstream of the RBS (Fig. 4C), consistent with previous studies (11, 21). Strikingly, however, in TRP-185-depleted cells, there was a reduction in junction formation at the RBS downstream region and a concomitant increase in junction formation at the RBS. By contrast, nonspecific integration around Alu repeats was unaffected in the TRP-185 knockdown cells. These results were consistent with the results of our in vitro junction formation assays and suggested that TRP-185 positively or negatively affects AAV integration around the RBS in cells.
Sequence analysis of cloned products revealed that in TRP185-depleted cells, junctions were formed precisely at the RBS of AAV and AAVS1, as observed in our in vitro assay. In control cells, however, junction points were distributed over a 50-bp region spanning the AAV RBS and a 600-bp region downstream of the AAVS1 RBS, and there were overlapping sequences of 2 to 11 bp between AAV and AAVS1.
TRP-185 promotes Rep68 helicase activity in an RBS-dependent manner.
According to a model presented by Linden et al. (21), the 3′→5′-helicase activity of Rep is involved in junction formation at the RBS downstream region. To determine whether TRP-185 affects Rep68 helicase activity, we carried out helicase assays using a partially double-stranded linear DNA substrate containing the AAVS1 RBS. As previously described (37), rRep68 unwound the AAVS1 substrate in a dose-dependent manner (Fig. 5B), while the K340H mutant (Fig. 5A), which lacks ATPase/helicase activity (18), had no appreciable effect (Fig. 5B), demonstrating that the observed helicase activity is due to Rep68. In the presence of 45 fmol of rRep68, which had modest helicase activity, further addition of rTRP-185 increased DNA unwinding in a dose-dependent manner (Fig. 5C). No such effect was observed in the absence of rRep68 (Fig. 5C) or when the RBS was mutated (Fig. 5D), suggesting that TRP-185 stimulates Rep68 helicase activity.
FIG. 5.
TRP-185 promotes Rep68 helicase activity in an RBS-dependent manner. (A) Wild-type rRep68 and its K340H mutant were visualized by Coomassie staining. The positions of molecular mass markers are shown on the left. (B to F) Helicase assays were performed using 12.5 fmol of 32P-labeled linear AAVS1 substrate containing wild-type (B, C) or mutant (D) RBS or 10 fmol of M13 circular substrate (E, F) and the indicated amounts of rRep68 and rTRP-185. The products were then electrophoresed on a nondenaturing polyacrylamide gel. “Boil” samples were heated to 98°C for 5 min immediately before electrophoresis. Quantitation of the unwound products is shown below each panel. Schematic structures of the substrates and the unwound products are shown between or to the right of the gels.
It remained to be determined whether the stimulatory function of TRP-185 requires intact RBS, because with the mutant RBS substrate, Rep68 helicase activity was too low to be quantified (Fig. 5D). We therefore used circular M13 DNA substrate, which was efficiently unwound by rRep68 in an RBS-independent manner (Fig. 5E). With this substrate, rTRP-185 did not appreciably promote Rep68-mediated DNA unwinding (Fig. 5F). These results indicated that TRP-185 enhances Rep68 helicase activity in an RBS-dependent manner.
Rep68 and TRP-185 do not bind to AAVS1 DNA simultaneously.
Since both TRP-185 and Rep68 bind to AAVS1 in a sequence-specific manner, we examined whether these proteins bind to AAVS1 simultaneously by performing gel-shift assays (Fig. 6A). rRep68 alone formed protein-DNA complexes, whose sizes increased in a concentration-dependent manner (Fig. 6A, lanes 2 to 6). rTRP-185 alone formed a slower-migrating protein-DNA complex (Fig. 6A, lane 7), and simultaneous addition of rRep68 and rTRP-185 resulted in the generation of a novel mobility species (Fig. 6A, lanes 8 to 12).
FIG. 6.
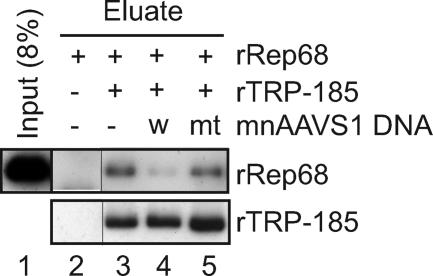
Rep68 and TRP-185 do not bind to AAVS1 DNA simultaneously. (A, B) Gel-shift assays were performed with 10 fmol of 32P-labeled mnAAVS1 wild-type probe and the indicated amounts of rRep68 and rTRP-185. In (B), anti-His (H) and anti-Flag (F) antibodies were included in the binding reaction, as indicated (−, absent; +, present). The reaction mixtures were electrophoresed on a 4% nondenaturing polyacrylamide gel. The asterisks indicate the positions of the wells. (C) rTRP-185, rRep68, and mnAAVS1 DNA were incubated either individually or in combination and then subjected to gel filtration analysis. Fractionated samples and input materials were analyzed by immunoblotting using anti-Flag and anti-His antibodies for rRep68 and rTRP-185, respectively, or by Southern blotting using 32P-labeled mnAAVS1-specific probe. The positions of molecular mass markers are shown below. (D) The indicated amounts of rTRP-185 and rRep68 were examined, either individually or in combination, for AAVS1 binding using latex beads onto which wild-type mnAAVS1 DNA was immobilized. Eluted proteins were visualized by silver staining. The positions of molecular mass markers are shown on the left.
We next examined the components of the protein-DNA complexes using anti-His and anti-Flag antibodies which were specific for rTRP-185 and rRep68, respectively. Anti-Flag antibody caused a supershift of the Rep68-DNA complex and the newly appeared complex (Fig. 6B, lanes 5, 7, 8, and 10). On the other hand, anti-His antibody seemed to dissociate the slow-migrating TRP-185-DNA complex but had no appreciable effect on the newly appeared complex (Fig. 6B, lanes 2, 3, 8, and 9). These results indicated that at least Rep68 is contained in the newly appeared complex. TRP-185 may also be incorporated into the complex, in such a way that the His tag is not recognized by the antibody. Alternatively, TRP-185 may not be incorporated into the complex but instead may affect the Rep68-DNA complex structure to cause its mobility shift, by transiently interacting with the Rep68-DNA complex.
To discriminate between these possibilities, we performed gel filtration analysis (Fig. 6C). In the presence of mnAAVS1 DNA, the rRep68 peak was shifted from fractions 11 and 12 to fractions 9 and 10, which most likely reflects DNA binding of rRep68 (Fig. 6C, panels 1 to 3). As for rTRP-185, the protein was fractionated broadly in high-molecular-weight fractions in the absence of DNA but with a peak at fractions 8 and 9 in the presence of mnAAVS1 DNA, probably reflecting DNA binding of rTRP-185 (Fig. 6C, panels 4 to 6). Remarkably, in the presence of rRep68, rTRP-185, and mnAAVS1 DNA, formation of the rTRP-185-DNA complex seemed to be abrogated, while the rRep68-DNA complex remained intact (Fig. 6C, panels 7 to 9). These results are consistent with the idea that Rep68 binds to AAVS1 with higher affinity than TRP-185 and that Rep68 and TRP-185 do not bind to AAVS1 DNA simultaneously.
The same issue was examined in a different way, i.e., by DNA pull-down assays using AAVS1-immobilized latex beads. As described above, rTRP-185 bound to AAVS1 DNA in the absence of rRep68 (Fig. 6D, lane 5). When increasing amounts of rRep68 were added together with rTRP-185 (Fig. 6D, lanes 6 to 8), rRep68 bound to the AAVS1-immobilized beads, and concomitantly, rTRP-185 was released from the beads in an rRep68 dose-dependent manner. These results supported the above conclusion that Rep68 and TRP-185 do not bind to AAVS1 DNA simultaneously and, together with the results of the gel-shift assays, suggested that TRP-185 affects the Rep68-DNA complex structure, possibly by transiently interacting with the Rep68-DNA complex.
TRP-185 directly interacts with Rep68.
The results obtained thus far indicated that TRP-185 and Rep68 physically interact with each other. To test this idea, we carried out an in vitro binding assay using His-tagged rTRP-185. Ni-NTA beads loaded with rTRP-185 were incubated with purified rRep68 and washed extensively, and bound proteins were subjected to immunoblot analysis. As expected, rRep68 bound to the Ni-NTA beads in an rTRP-185-dependent manner (Fig. 7, lanes 2 and 3), indicating that TRP-185 directly associates with Rep68.
FIG. 7.
TRP-185 directly interacts with Rep68. rTRP-185 (1.2 pmol) was coupled to Ni-NTA beads and incubated with 3.75 pmol of rRep68 in the absence (−) or presence (+) of 2.5 pmol of mnAAVS1 wild-type (w) or mutant (mt) DNA. Eluted samples and input material were subjected to immunoblot analysis using anti-Flag and anti-His antibodies for rRep68 and rTRP-185, respectively.
We then examined whether AAVS1 DNA has any effect on the interaction between TRP-185 and Rep68. As expected, the TRP-185-Rep68 interaction was significantly reduced by the presence of wild-type mnAAVS1 DNA, but not by the mutant mnAAVS1 DNA (Fig. 7, lanes 4 and 5). These results were fully consistent with the findings shown in Fig. 6 and suggested that the direct Rep68-TRP185 interaction is abrogated by the presence of AAVS1 DNA.
DISCUSSION
The mechanism of AAV site-specific integration, particularly, the mechanism of integration site selection and the proteins that regulate this complicated process, has not yet been fully elucidated. In the present study, we identified TRP-185 as an AAVS1-binding protein in HeLa NEs. Immunodepletion experiments in vitro and RNA interference-mediated knockdown analysis in vivo suggested that TRP-185 alters selection of the AAV integration site from the RBS to a downstream region. We also provided evidence that TRP-185 binds to Rep68 directly, changes the Rep68 DNA binding property, and stimulates Rep68 helicase activity on AAVS1 DNA.
Identification of TRP-185 as an AAVS1-binding protein.
Previous studies (32, 33) have shown that TRP-185 stimulates human immunodeficiency virus type 1 gene expression from the long terminal repeat by binding to TAR RNA. Although TRP-185 is expressed in a variety of human tissues, its function in other processes remains largely unknown. In this study, we showed that TRP-185 binds to AAVS1 in vitro and in vivo (Fig. 1). Whereas TRP-185 binding to TAR RNA is critically dependent on a group of cellular cofactors (33, 34), we showed by using a highly purified recombinant protein that TRP-185 directly binds to AAVS1 DNA (Fig. 6). TRP-185 does not contain a classical nucleic acid-binding domain. However, it contains multiple basic amino acid residues and a leucine zipper motif in its N terminus, which may be involved in mediating binding to AAVS1 DNA.
Rep68 alone is sufficient for mediating AAV-AAVS1 junction formation at the RBS.
We showed that Rep68 alone is sufficient for the AAV-AAVS1 junction formation at the RBS (Fig. 2). Although previous studies have shown that crude cell extract or DNA replication promotes low-level junction formation by Rep68 (6, 28), in our in vitro integration assay, Rep68 alone was sufficient to support junction formation at the RBS, and cellular proteins other than TRP-185 in HeLa cell NE had no appreciable effect on junction formation (Fig. 3). The apparent discrepancy between the previous and our study results may arise from differences in the methods used to prepare cell extracts. It may be that cellular factors promoting AAV integration were not enriched in our extract. Junction formation at the RBS by Rep68 is possible in the absence of any additional protein factors because Rep68 has all the activities expected to be required for the process; i.e., Rep68 is capable of sequence-specific DNA binding, mediating interactions between AAV and AAVS1, creating a nick, and unwinding and ligating DNA (13, 23, 27, 31).
TRP-185 may regulate AAV integration site determination through its chaperone-like activity for Rep68.
We propose that TRP-185 acts as a molecular chaperone that facilitates the formation of helicase-competent, oligomerized Rep68-AAVS1 complexes through its transient interactions with Rep68 and AAVS1 DNA. Our data showed that whereas a high concentration of Rep is necessary for its helicase activity in the absence of TRP-185, a low concentration of Rep is sufficient for its helicase activity in the presence of TRP-185 (Fig. 5). We also showed that the presence of TRP-185 and a high concentration of Rep68 both contribute to the formation of slow-migrating Rep68-DNA complexes (Fig. 6). Previous biochemical studies have shown that Rep78/68 can form oligomerized complexes on DNA and that this oligomerization enhances its nicking and helicase activities (15, 20). It is therefore likely that TRP-185 enhances Rep68 helicase activity by promoting its oligomerization.
We provided evidence that TRP-185 regulates AAV integration site determination through regulation of Rep's DNA binding property and sequence-specific helicase activity. We showed that TRP-185, as well as a high concentration of Rep68, inhibits Rep-dependent integration at the RBS in vitro (Fig. 2 and 3). This inhibition probably reflects a shift in the integration site from the RBS to a downstream region, caused by Rep68 oligomerization and activation of its helicase activity. Since the expression of Rep78/68 is strictly regulated to a low level during latent infection by AAV (36), we assume that this TRP-185-dependent mechanism operates during the viral life cycle. This assumption is supported by the results of our in vivo assays (Fig. 1C and 4).
Here we present a model as to how TRP-185 affects AAV integration site determination. Following AAV infection, Rep78/68 proteins are synthesized and associate with the AAVS1 RBS, which is already occupied by TRP-185. Through protein-protein and protein-DNA interactions, TRP-185 affects the DNA binding property of Rep78/68, activating Rep's helicase activity, and then dissociates from the DNA. Subsequently, Rep introduces a nick at the trs; proceeds downstream, unwinding the DNA through its helicase activity; and induces AAV integration at the downstream region. In the absence of TRP-185, Rep's helicase activity is not activated, and therefore, after introducing a nick at the trs, Rep remains bound to the AAVS1 RBS and facilitates AAV integration at the RBS.
Possible interplay of TRP-185 and other cellular proteins during AAV integration.
It has been shown that the high-mobility-group protein HMG-1 promotes the formation of Rep-DNA complexes (3). Unlike TRP-185, HMG-1 enhances AAV-AAVS1 junction formation at the RBS (6). HMG-1 and TRP-185 seem to have different effects on Rep68, as HMG-1, but not TRP-185, promotes its endonuclease activity (3).
In a previous study, the zinc finger protein ZF5 was identified as a cellular protein that binds to the RBS of the AAV ITR (2). A transient overexpression study indicated that ZF5 inhibits the transcription, replication, and production of AAV, possibly by competing with Rep for binding to the ITR (2). It seems possible that ZF5 also regulates, either positively or negatively, site-specific integration through its binding to the RBS on the AAV genome and/or AAVS1, although this issue has not been examined. Thus, it will be interesting to investigate the possible functional interplay of TRP-185, HMG-1, and ZF5 in the AAV integration process in future studies.
The biological significance of TRP-185 in the AAV life cycle.
What is the physiological significance of the role of TRP-185 in AAV integration site determination? From an evolutionary perspective, there is no evidence for AAV site-specific integration in mice. Interestingly, whereas an ortholog of human AAVS1 was recently discovered in the mouse genome (5), the homologue of the gene encoding TRP-185 does not exist in the mouse genome (33). These points suggest that TRP-185 plays a role in AAV site-specific integration specifically in humans. Second, abnormal integration at the AAVS1 RBS may be harmful to host cells and may be prevented by TRP-185. In fact, however, infection and abnormal integration of AAV had no appreciable effect on the proliferation of TRP-185-depleted cells (data not shown). This may be due to the partial downregulation of TRP-185, and a more significant effect of abnormal integration would be evident upon complete downregulation. Unfortunately, this hypothesis could not be tested, because greater than 90% downregulation of TRP-185 using a different shRNA construct caused cell death (data not shown), indicating that TRP-185 is essential for viability in humans. Third, AAV integration at the RBS downstream region may be important for the rescue of AAV from host cells. It remains our future challenge to address the significance of TRP-185 function for both the host cell and AAV.
Finally, it is important to note that AAV is a prospective vector for gene therapy because of its property of integrating into a specific locus. Further analyses of AAV site-specific integration will make it possible to control the site-specific integration of a desired gene and will lead to significant developments in gene therapy.
Acknowledgments
We thank Richard B. Gaynor for providing anti-TRP-185 monoclonal antibodies, Tadahito Kanda for providing pUC18 (HindIII) AAVS1 plasmid DNA, Yasunori Tsuboi for technical support, and Masasuke Yoshida and Akira Nakanishi for discussions and comments on the manuscript.
This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas and by the Grant of the 21st Century COE program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 2.Cathomen, T., T. H. Stracker, L. B. Gilbert, and M. D. Weitzman. 2001. A genetic screen identifies a cellular regulator of adeno-associated virus. Proc. Natl. Acad. Sci. USA 98:14991-14996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costello, E., P. Saudan, E. Winocour, L. Pizer, and P. Beard. 1997. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 16:5943-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutheil, N., M. Yoon-Robarts, P. Ward, E. Henckaerts, L. Skrabanek, K. I. Berns, F. Campagne, and R. M. Linden. 2004. Characterization of the mouse adeno-associated virus AAVS1 ortholog. J. Virol. 78:8917-8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyall, J., P. Szabo, and K. I. Berns. 1999. Adeno-associated virus (AAV) site-specific integration: formation of AAV-AAVS1 junctions in an in vitro system. Proc. Natl. Acad. Sci. USA 96:12849-12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endoh, M., W. Zhu, J. Hasegawa, H. Watanabe, D. K. Kim, M. Aida, N. Inukai, T. Narita, T. Yamada, A. Furuya, H. Sato, Y. Yamaguchi, S. S. Mandal, D. Reinberg, T. Wada, and H. Handa. 2004. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 24:3324-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilton, H., J. Gomos, K. I. Berns, and E. Falck-Pedersen. 2004. Adeno-associated virus site-specific integration and AAVS1 disruption. J. Virol. 78:7874-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han, S. I., M. A. Kawano, K. Ishizu, H. Watanabe, M. Hasegawa, S. N. Kanesashi, Y. S. Kim, A. Nakanishi, K. Kataoka, and H. Handa. 2004. Rep68 protein of adeno-associated virus type 2 interacts with 14-3-3 proteins depending on phosphorylation at serine 535. Virology 320:144-155. [DOI] [PubMed] [Google Scholar]
- 10.Huser, D., and R. Heilbronn. 2003. Adeno-associated virus integrates site-specifically into human chromosome 19 in either orientation and with equal kinetics and frequency. J. Gen. Virol. 84:133-137. [DOI] [PubMed] [Google Scholar]
- 11.Huser, D., S. Weger, and R. Heilbronn. 2002. Kinetics and frequency of adeno-associated virus site-specific integration into human chromosome 19 monitored by quantitative real-time PCR. J. Virol. 76:7554-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huser, D., S. Weger, and R. Heilbronn. 2003. Packaging of human chromosome 19-specific adeno-associated virus (AAV) integration sites in AAV virions during AAV wild-type and recombinant AAV vector production. J. Virol. 77:4881-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Im, D. S., and N. Muzyczka. 1990. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61:447-457. [DOI] [PubMed] [Google Scholar]
- 14.Inomata, Y., T. Wada, H. Handa, K. Fujimoto, and H. Kawaguchi. 1994. Preparation of DNA-carrying affinity latex and purification of transcription factors with the latex. J. Biomater. Sci. Polym. Ed. 5:293-302. [DOI] [PubMed] [Google Scholar]
- 15.James, J. A., C. R. Escalante, M. Yoon-Robarts, T. A. Edwards, R. M. Linden, and A. K. Aggarwal. 2003. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure 11:1025-1035. [DOI] [PubMed] [Google Scholar]
- 16.Kotin, R. M., R. M. Linden, and K. I. Berns. 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 11:5071-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotin, R. M., M. Siniscalco, R. J. Samulski, X. D. Zhu, L. Hunter, C. A. Laughlin, S. McLaughlin, N. Muzyczka, M. Rocchi, and K. I. Berns. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 87:2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyostio, S. R., and R. A. Owens. 1996. Identification of mutant adeno-associated virus Rep proteins which are dominant-negative for DNA helicase activity. Biochem. Biophys. Res. Commun. 220:294-299. [DOI] [PubMed] [Google Scholar]
- 19.Laughlin, C. A., J. D. Tratschin, H. Coon, and B. J. Carter. 1983. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene 23:65-73. [DOI] [PubMed] [Google Scholar]
- 20.Li, Z., J. R. Brister, D. S. Im, and N. Muzyczka. 2003. Characterization of the adenoassociated virus Rep protein complex formed on the viral origin of DNA replication. Virology 313:364-376. [DOI] [PubMed] [Google Scholar]
- 21.Linden, R. M., P. Ward, C. Giraud, E. Winocour, and K. I. Berns. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 93:11288-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muzyczka, N. 1992. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 158:97-129. [DOI] [PubMed] [Google Scholar]
- 23.Owens, R. A., M. D. Weitzman, S. R. Kyostio, and B. J. Carter. 1993. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J. Virol. 67:997-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Philpott, N. J., J. Gomos, K. I. Berns, and E. Falck-Pedersen. 2002. A p5 integration efficiency element mediates Rep-dependent integration into AAVS1 at chromosome 19. Proc. Natl. Acad. Sci. USA 99:12381-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samulski, R. J., X. Zhu, X. Xiao, J. D. Brook, D. E. Housman, N. Epstein, and L. A. Hunter. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu, N., K. Sugimoto, J. Tang, T. Nishi, I. Sato, M. Hiramoto, S. Aizawa, M. Hatakeyama, R. Ohba, H. Hatori, T. Yoshikawa, F. Suzuki, A. Oomori, H. Tanaka, H. Kawaguchi, H. Watanabe, and H. Handa. 2000. High-performance affinity beads for identifying drug receptors. Nat. Biotechnol. 18:877-881. [DOI] [PubMed] [Google Scholar]
- 27.Smith, R. H., and R. M. Kotin. 2000. An adeno-associated virus (AAV) initiator protein, Rep78, catalyzes the cleavage and ligation of single-stranded AAV ori DNA. J. Virol. 74:3122-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song, S., Y. Lu, Y. K. Choi, Y. Han, Q. Tang, G. Zhao, K. I. Berns, and T. R. Flotte. 2004. DNA-dependent PK inhibits adeno-associated virus DNA integration. Proc. Natl. Acad. Sci. USA 101:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urcelay, E., P. Ward, S. M. Wiener, B. Safer, and R. M. Kotin. 1995. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J. Virol. 69:2038-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada, T., T. Takagi, Y. Yamaguchi, H. Kawase, M. Hiramoto, A. Ferdous, M. Takayama, K. A. Lee, H. C. Hurst, and H. Handa. 1996. Copurification of casein kinase II with transcription factor ATF/E4TF3. Nucleic Acids Res. 24:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weitzman, M. D., S. R. Kyostio, R. M. Kotin, and R. A. Owens. 1994. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc. Natl. Acad. Sci. USA 91:5808-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu, F., J. Garcia, D. Sigman, and R. Gaynor. 1991. tat regulates binding of the human immunodeficiency virus trans-activating region RNA loop-binding protein TRP-185. Genes Dev. 5:2128-2140. [DOI] [PubMed] [Google Scholar]
- 33.Wu-Baer, F., W. S. Lane, and R. B. Gaynor. 1995. The cellular factor TRP-185 regulates RNA polymerase II binding to HIV-1 TAR RNA. EMBO J. 14:5995-6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu-Baer, F., W. S. Lane, and R. B. Gaynor. 1996. Identification of a group of cellular cofactors that stimulate the binding of RNA polymerase II and TRP-185 to human immunodeficiency virus 1 TAR RNA. J. Biol. Chem. 271:4201-4208. [DOI] [PubMed] [Google Scholar]
- 35.Yamada, T., Y. Yamaguchi, N. Inukai, S. Okamoto, T. Mura, and H. Handa. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21:227-237. [DOI] [PubMed] [Google Scholar]
- 36.Young, S. M., Jr., D. M. McCarty, N. Degtyareva, and R. J. Samulski. 2000. Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J. Virol. 74:3953-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, X., I. Zolotukhin, D. S. Im, and N. Muzyczka. 1999. Biochemical characterization of adeno-associated virus Rep68 DNA helicase and ATPase activities. J. Virol. 73:1580-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]