Abstract
In order to understand the evolutionary mechanisms of persistence and diversification within the Caliciviridae, we have been exploiting endemic infection of feline calicivirus within five geographically distinct household groups of cats. By sequencing immunodominant and variable regions of the capsid gene, we identified the relative contribution of the different evolutionary processes employed by the virus to ensure its long-term survival in the host population. Such strategies included progressive evolution of a given variant of a strain through mutation accumulation within an individual, sequential reinfection with either a variant of the same strain or with a different strain, and mixed infection. Recombination between different strains in this study has been reported in detail elsewhere (K. P. Coyne et al., J. Gen. Virol. 87:921-926, 2006). Here, we provide evidence to suggest that true long-term persistent infection in individuals is relatively rare, with the majority of apparent viral carriers undergoing a combination of progressive evolution and cyclical reinfection. Progressive evolution at the individual level and variant reinfection at both the individual and population levels were associated with positive selection. Two measures of evolution rate were determined; for a virus progressively evolving within an individual (1.32 × 10−2 to 2.64 × 10−2 substitutions per nucleotide per year, i.e., no transmission) and for a strain circulating within a population (3.84 × 10−2 to 4.56 × 10−2 substitutions per nucleotide per year, i.e., including transmission). Reiteration of both progressive evolution and variant reinfection appeared to lead to a gradual increase in the diversity of a given strain of virus, both in the individual and in the population, until eventually new strains emerged.
The Caliciviridae are an important group of human and animal RNA pathogens, causing a wide range of diseases, including acute outbreaks of gastroenteritis in humans (noroviruses and sapoviruses) and vesicular and other diseases in animals (vesiviruses and lagoviruses) (19). The Caliciviridae are a highly variable family of small positive-strand RNA viruses, demonstrating considerable antigenic and genetic diversity both between and within genera. Such variability has been associated with the emergence of highly transmissible globally dominant strains of human caliciviruses (36) and hypervirulent animal caliciviruses that are frequently lethal (9, 33, 39). Despite the obvious significance of such variation, the origins of this diversity and the evolutionary mechanisms by which new, and possibly more virulent strains evolve remain unclear.
Feline calicivirus (FCV) belongs to the genus Vesivirus and is a highly infectious oral and respiratory pathogen of domestic cats (18). Following acute infection, some cats remain persistently infected with the virus (11, 40, 48, 59). Such carriers appear to be relatively common in the general population, with prevalences ranging from 15 to 91%, and they play an important role in the epidemiology of the disease (1, 3, 7, 8, 21, 22, 42). Persistent infections are also recognized in other caliciviruses, although in general these are not as well characterized as in feline calicivirus, and their role in the epidemiology of the disease is not as clear (15, 17, 35, 63).
The mechanisms of viral persistence in the Caliciviridae, both within an individual and at the population level, are not fully elucidated. Persistence within an individual is thought to be largely via immune-mediated positive selection (29, 35, 48), although other mechanisms may also play a role. Key questions that need to be resolved are the following: what contribution do such carriers make to the long-term survival of the virus at the population level, and to what extent do they drive virus evolution and diversification.
In order to address these issues, we have been exploiting endemic infection of feline calicivirus within stable household groups of cats. We have previously reported that such households tend to be infected with a small number of distinct strains of feline calicivirus (8) and provide a useful model system to monitor calicivirus evolution and persistence over long periods of time, in a defined geographic space within a natural population. In contrast, in other caliciviruses, most sampling is limited to investigation of disease outbreaks over relatively short periods, where opportunities for virus evolution are likely to be limited (15, 16, 32, 57, 64). The shape of resulting phylogenies therefore tends to be a reflection of the sampling strategy employed and, thus, intermediate evolutionary steps can only be conjectured (20). Here we use structured long-term sampling of endemically infected groups of cats to identify the relative contribution of the different evolutionary processes by which caliciviruses generate viral diversity and ensure their long-term survival in the host population.
MATERIALS AND METHODS
Virus origins and isolation.
The origins of the 144 FCV isolates used in this study have been described elsewhere (8). Briefly, virus isolates were sequentially collected from cats within five different geographically located communities (designated A to E) over a 15- to 46-month study period. Oropharyngeal swabs were collected into 2 ml of virus transport medium. Viruses were isolated as described previously on feline embryo A cells and Crandall Reese feline kidney cells (27, 61) and stored at −80°C for further analysis.
RNA extraction, reverse transcription-PCR, and consensus sequencing.
RNA was extracted from positive samples (second passage or less; QIAmp viral RNA mini kit; QIAGEN) and transcribed into cDNA (Superscript III; Invitrogen) using 500 ng of specific reverse primer (Table 1) as the first-strand primer according to the manufacturers' instructions. A 529-nucleotide region of the capsid gene, equivalent to residues 6406 to 6934 of the FCV strain F9 (5) and incorporating immunodominant region E (34, 49, 53), was amplified using Pfu DNA polymerase (Stratagene) according to the manufacturer's instructions. Each 50-μl reaction mixture contained 2 μl of cDNA and 100 ng of each PCR primer (Table 1). Thermal cycling conditions consisted of DNA denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 30 seconds, primer annealing at 50°C for 30 seconds, and primer extension at 72°C for 90 seconds. A final extension was performed at 72°C for 5 min.
TABLE 1.
Primers used for amplification and sequencing of variable regions C and E of the FCV capsid gene
| Household | RT primer | PCR primer | Sequence (5′-3′) | Binding sitea |
|---|---|---|---|---|
| A | A2 (reverse) | A1 (forward) | CCCTTCGTCTTTCAGGCCAACCG | 6406-6428 |
| C | A2 (reverse) | A2 (reverse) | CCTCGCCAATCCCAGTGTAGCC | 6934-6913 |
| D | P2 (reverse) | P1 (forward) | CCGTTTGTGTTTCAAGCAAACCG | 6406-6428 |
| E | P2 (reverse) | P2 (reverse) | CCTCACCTATACCAGTGTAACC | 6934-6913 |
| B | A2/P2 | A1/A2 or P1/P2 | As above |
Binding sites relate to FCV strain F9.
Amplicons were purified (QIAquick PCR purification kit; QIAGEN Ltd.) and sequenced bidirectionally using the corresponding PCR primers (Table 1) according to standard protocols (ABI Prism BigDye terminators version 3.0 cycle sequencing kits; Applied Biosystems). For each amplicon, a consensus sequence was produced using ChromasPro version 1.32 (Technelysium Ptl. Ltd.). Ambiguities identified consistently in both strands of sequence were included in the consensus sequence. All primer sites were removed prior to sequence analysis, resulting in a final useable sequence of 420 nucleotides corresponding to codons 383 to 522 of the FCV capsid (53) and including variable regions C and E. In order to determine the impact of cell culture, virus isolation, passage, and reverse transcription-PCR on consensus sequences, we compared the cell culture-passaged sequence to the sequence determined directly from the oropharyngeal swab for six viruses. These repeat sequences were 99.3 to 100% identical (data not presented).
Cloning of PCR products.
In order to characterize the diversity of FCV within individual cats and to attempt to identify transmission events, multiple clones of 20 FCV isolates were obtained from three representative cats for which a number of serial samples were available. Cloning of PCR products was performed using the Zero Blunt TOPO PCR cloning kit (Invitrogen life technologies) essentially according to the manufacturer's instructions. Plasmids were purified using a Hamilton Microlab Star robot running the Millipore plasmid extraction kit (Montage Plasmid MiniprepB96B kit) as per the manufacturers' recommendations. Plasmids containing an insert of the appropriate size were sequenced using standard protocols.
Nucleotide sequence and phylogenetic analysis.
Nucleotide distance calculations and sequence alignments were performed using the programs Distance and Pileup from version 8 of the Wisconsin package, Genetics Computer Group (13), using the Jukes-Cantor model. This model accounts for multiple substitutions and ambiguities in the sequence data, which for FCV are attributable to the quasispecies nature of the virus (48). All distance values are corrected unless otherwise specified. Where appropriate, uncorrected analyses were also carried out to allow comparison with previous epidemiological studies (44, 45). Phylogenetic analysis was performed using the program GROWTREE (Jukes-Cantor distance analysis and neighbor joining) also available in the Genetics Computer Group package and viewed using TREEVIEW (38). Support for individual nodes was sought by bootstrap analysis using 1,000 repetitions (MEGA version 3.0) (30).
Clonal sequence analysis.
For those isolates for which clonal sequences were available, phylogenetic analyses and average pair-wise genetic diversity were performed using the Jukes-Cantor and neighbor-joining models (MEGA version 3.0) (30). No ambiguous sites were present in the cloned sequence data. Support for individual nodes was sought by bootstrap analysis using 1,000 repetitions (MEGA version 3.0) (30).
Simpson's equitability index.
Simpson's equitability index (E) was used in addition to the average pair-wise genetic diversity to give a further measure of intraisolate quasispecies diversity (2, 54).
 |
where Pi is the proportion of identical clones making up the population and S is the total number of clones within the population. This returns a value of 1 for maximum diversity (all clones different) and tends toward zero as the diversity decreases and the clonal population size increases.
Statistical analysis.
Statistical analysis was carried out using Minitab for Windows 14.1 (Minitab Inc.). Comparisons between nucleotide distance values were analyzed using the Mann-Whitney test. The critical probability was taken as a P value of ≤0.05 for a two-sided alternative hypothesis.
Estimating evolution rates.
Where appropriately sized subdata sets of sequences were available, evolution rates were calculated using Tipdate v. 1.01 (50). These data sets were based on consensus sequences from those cats that were persistently infected with their own strain of virus (progressive evolvers; group D, cat 4 and cat 2) (8) and from those virus strains that arrived in the group after the start of the study and for which the group ancestor sequence was available (strains D2 and E2).
Evidence for positive selection.
In order to identify the mechanisms underlying the diversification of strains within individual groups, evidence of positive selection was sought using a codon-based approach as implemented in Datamonkey (41). These offer several advantages over previously described methods, including not needing to assume equal synonymous substitution rates throughout the sequence and allowing the user to choose the most appropriate model for nucleotide substitution from the original data set. Because of the different sizes of the data sets, a combination of three approaches was used. For larger data sets, a single likelihood ancestor counting (SLAC) analysis was performed suing a P value of 0.1. For smaller data sets, where type I errors may be more common, an integrative approach was taken using SLAC, fixed effects likelihood (FEL), and random effects likelihood (REL) (P = 0.25 for SLAC and FEL; Bayes Factor of 10 for REL). In all cases, ambiguities in the consensus sequence were averaged in the analysis.
Predicted structure studies.
The regions of the FCV capsid predicted to be evolving under positive selection were mapped onto a predicted structure for the FCV F9 capsid protein (GenBank accession number M86379), which was modeled against the known structures for San Miguel sea lion virus (SMSV), which also belongs to the Vesivirus genus (PDB entry 2gh8.pdb) (6) and human norovirus (PDB entry1ihm.pdb) (43) using SWISS-MODEL via the ExPASy web server (52). The predicted FCV capsid was viewed and manipulated using PyMol 0.99 (12).
Nucleotide sequence accession numbers.
All consensus sequences have been submitted to GenBank (accession numbers DQ397674 to DQ397814). Each sequence is identified by the following strain identification key: group and strain number, cat number, and time in months, e.g., b1c7t09 indicates group B, strain 1, from cat 7, 9 months after the start of the study.
RESULTS
Viral prevalence.
The mean prevalence of FCV observed from the five households sampled ranged from 6 to 75% (overall mean FCV prevalence, 35%): further details of prevalence dynamics are reported elsewhere (8). Within each household, cats were broadly divided into three categories: those that shed virus consistently for prolonged periods (“consistent shedders”), those that shed intermittently (“intermittent shedders”), and those that appeared to be resistant to infection over the study period (“nonshedders”) (8).
Strain identification and variation.
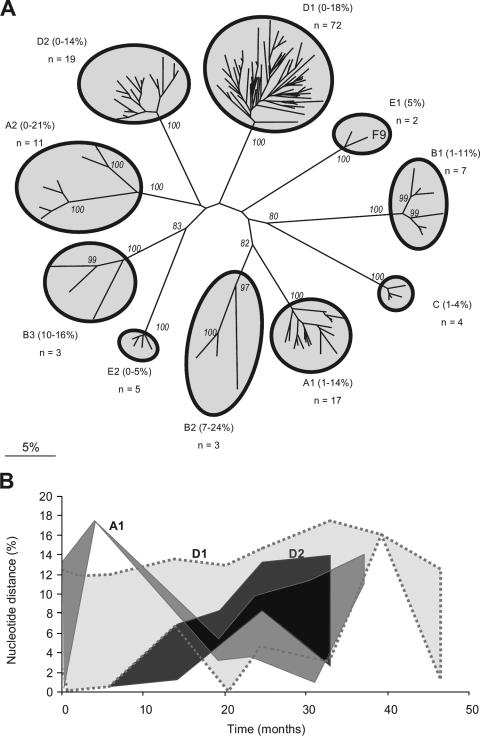
Phylogenetic analysis based on consensus sequences of variable immunodominant regions of the FCV capsid gene identified a total of 10 distinct viral strains among the five households. Each of these strains was separated by distance values of 26 to 51% (Fig. 1A). Their uncorrected distance values (which allow direct comparison with previous studies) ranged from 21 to 39% (data not shown), as previously reported for distinct epidemiologically unrelated strains of FCV (44). Within households, one to three distinct strains of FCV were identified. The maximum variability within each strain ranged from 5 to 24% (Fig. 1A) (with uncorrected distances of 4 to 20% [data not shown]). This is in contrast to previous studies where variability within a strain, during either disease outbreaks or persistent infection, was generally relatively low (up to 6% uncorrected distance) (44, 46). Viral strains present within households generally sustained considerable levels of diversity over time (Fig. 1B), though an occasional reduction in diversity (i.e., a putative transmission bottleneck), followed by an increase, also occurred (e.g., strain A1 at 19 months [Fig. 1B]). In each of two households (D and E), new strains were identified during the course of the study. Virus variability in these two strains increased from 0 to 5% (strain E2) (Fig. 1A) and from 0 to 14% (strain D2) (Fig. 1A and B) over 6 and 27 months, respectively, showing that in this situation viral diversity tends to increase over time.
FIG. 1.
(A) Unrooted neighbor-joining tree of 143 FCV consensus sequences (including the published sequence FCV F9, GenBank accession number M86379), for a 420-nucleotide region of the FCV capsid gene. One consensus sequence was excluded, as it contained two strains. Shaded areas represent the 10 distinct virus strains identified in this study. Strain names, percent diversity, and the number of variants within each strain are indicated next to the corresponding cluster. Evolutionary distances were calculated using the Jukes-Cantor model, with the scale bar indicating percent divergence. Numbers at major nodes are the bootstrap values of ≥80% out of 1,000 replicates (MEGA 3.0). (B) Changes in viral diversity over time for the three strains (A1, D1, and D2) for which the most data were available. Shaded areas represent the ranges of viral strain diversity (Jukes-Cantor distance analysis) present in the cat groups at the time of sampling, with putative bottlenecks at 19 months (strain A) and 6 months (strain D2).
Mechanisms of persistence.
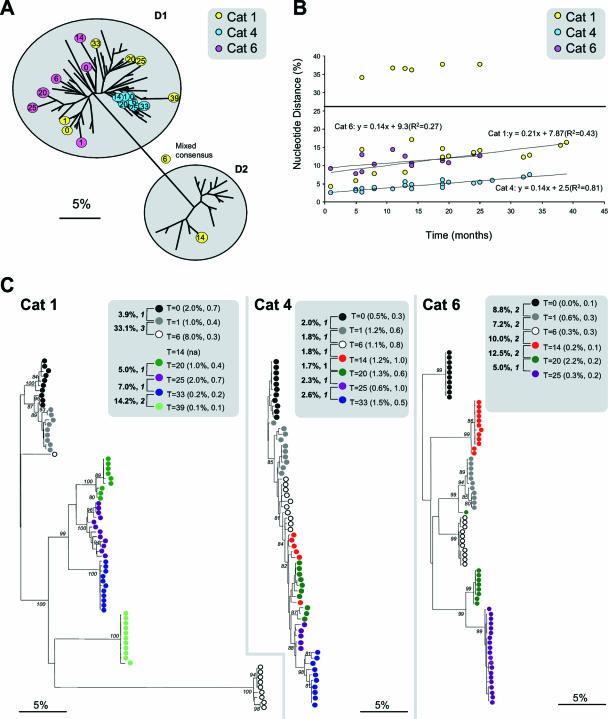
Having established the genetic identities of the strains within each household, patterns of evolution, both within individuals and at the population level, were characterized. A total of 130 partial FCV capsid consensus sequences were compared from 31 individual cats which shed virus over prolonged periods (8). Within these individuals, two mechanisms for this persistent virus shedding were identified. The first mechanism, characterized by mutation accumulation in a given variant of a strain within an individual (“progressive evolution”), was only evident in four cats. Here, all consecutive consensus sequences isolated from an individual cat clustered together phylogenetically (e.g., cat 4) (Fig. 2A), consistently demonstrating low interisolate variability between consecutive samples and clear evidence of temporal evolution (Fig. 2A and B, cat 4). Clonal samples from this individual cat showed that low interisolate variability was coupled with relatively high intraisolate variability along with low bootstrap support for the clustering of clones from individual isolates at each time point (Fig. 2C, cat 4).
FIG. 2.
(A) Unrooted neighbor-joining tree of 91 FCV consensus sequences from samples isolated from colony D. Phylogenetic relationships are highlighted for three representative cats, numbers 1, 4, and 6. Colored circles represent viruses isolated from each cat, with the time at which each isolate was collected shown in months. Evolutionary distances were calculated using the Jukes-Cantor model, with the scale bar indicating percent divergence. (B) All possible pair-wise comparisons of genetic distances against time for all consensus sequences for cats 1, 4, and 6. Symbols below the line represent comparisons made within a strain and were used to calculate lines of best fit. Symbols above the line for cat 1 represent comparisons made between strains. (C) Unrooted neighbor-joining trees of cloned sequences derived for viruses isolated from cats 1, 4, and 6. Colored circles represent times at which each FCV sample was obtained. The mean interisolate variability is shown in bold, and the pair-wise ranking score is in italics. Mean intraisolate variability and E values are shown in brackets. Numbers at major nodes are the bootstrap values of ≥80% out of 1,000 replicates (MEGA 3.0). The scale bar indicates percent divergence.
In contrast, for the remaining 27 cats, progressive evolution was interrupted by relatively high interisolate variability over short periods of time, where consecutive isolates from individual cats largely mixed together phylogenetically with viruses from other cats (e.g., cat 1 at 33 and 39 months and cat 6 at all time points except 20 and 25 months) (Fig. 2A and B). Clonal samples from these two individual cats demonstrated high interisolate variability, coupled with low intraisolate variability, with high bootstrap support for the clustering of clones from individual isolates at these time points (Fig. 2C, cats 1 and 6). This suggested a second mechanism of persistent infection within individuals, in which cats were being periodically reinfected at some time points from within the household (“sequential reinfection”). These transmission events were either with a diverse variant of the same strain (cats 1 and 6 [Fig. 2]) or with a distinct strain (cat 1 at 14 months [Fig. 2]).
Interestingly, consensus and cloned sequence analyses showed that at least one cat was infected with two distinct strains of virus simultaneously (e.g., cat 1 at 6 months [Fig. 2C]).
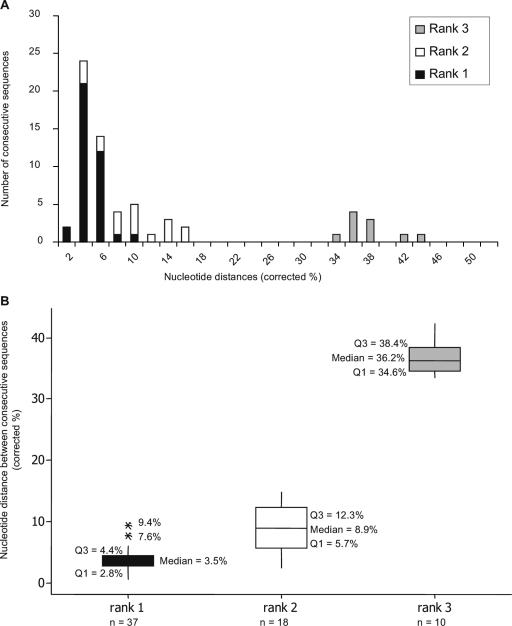
Further evidence was sought for these two mechanisms of persistence by ranking interisolate distances between consecutive pairs of isolates from individual cats. Pairs were ranked as 1 if they were more closely related to each other than to any other viruses cocirculating in the household, suggestive of progressive evolution of a given variant within an individual. Pairs were ranked as 2 if they were more closely related to other viruses of the same strain cocirculating in the household than to each other, suggestive of transmission of variants within a strain. Pairs were ranked as 3 if they were distinct strains, suggesting between-strain transmission (Fig. 3). Significant differences were found between all three categories (P < 0.001). For the four cats that we hypothesized to be persistently infected by progressive evolution, interisolate distances were consistently ranked one (e.g., cat 4 [Fig. 2c and 3]). In contrast, for the remaining 27 cats which we hypothesized to be persistently infected by the processes of progressive evolution as well as sequential reinfection, their interisolate ranks were a mixture of 1, 2, and 3 (e.g., cats 1 and 6 [Fig. 2c and 3]).
FIG. 3.
(A) Bar chart showing nucleotide distance comparisons for 65 pairs of FCV isolates that were isolated on consecutive samplings from 24 cats (refer to the text for an explanation of the ranking system). (B) Box and whisker plot comparing the distributions of nucleotide distances for sequential sequences ranked 1 (progressive evolution), 2 (within-strain transmissions), and 3 (between-strain transmissions). The number of paired sequences that were placed within each ranking system is indicated. Median upper quartile (Q3, where 75% of data are less than or equal to this value) and lower quartile (Q1, where 25% of data are less than or equal to this value) distance values are shown for each rank. *, distance values outside the whiskers are classified as outliers.
Evolution rates.
From these data it was possible to determine two measures of FCV strain evolution rate: progressive evolution firstly within individual cats (no transmission) and secondly within the population (including transmission). For two of the four cats putatively undergoing progressive evolution of the same strain of virus (cat 4 [Fig. 2] and cat 2 [reference 8]), evolution rates were 1.32 × 10−2 and 2.64 × 10−2 substitutions per nucleotide per year, respectively. For two strains (D2 and E2) that were putatively introduced into their households during the course of the study, thereby allowing analyses of the founding sequence, evolution rates within the population were 3.84 × 10−2 and 4.56 × 10−2 substitutions per nucleotide per year, respectively.
Analysis of positive selection.
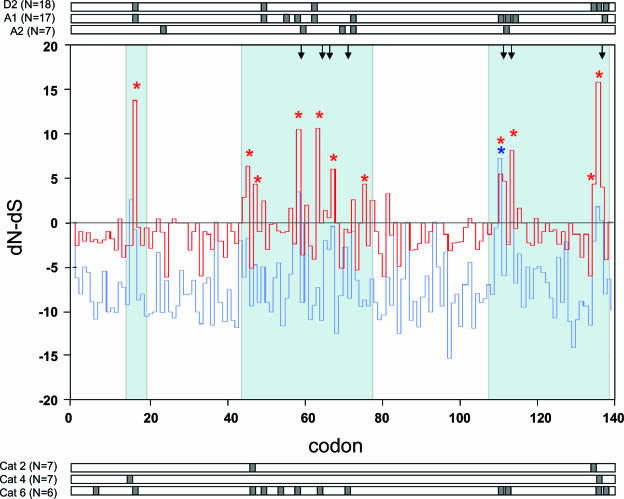
Patterns of evolution were clearly associated with positive selection at known antigenic sites, suggesting an immune-mediated mechanism for viral evolution and persistence (Fig. 4). This was clear not only in the small number of individuals progressively evolving virus (e.g., cats 2 and 4), but also in those cats undergoing variant reinfection (e.g., cat 6) and for all variants of a strain at the population level (strains D2, A1, and A2) (Fig. 4). In contrast, when published sequences from epidemiologically unrelated FCV strains were compared, there was little evidence for positive selection (Fig. 4). Codons predicted to be under positive selection mapped to the 5′ and 3′ hypervariable regions (HVRs) of region E of the FCV capsid (Fig. 4), which have been shown to contain linear and conformational B-cell epitopes, respectively (49, 55). Positive selection was also evident in variable region C, providing for the first time evidence that this region of the capsid may also be antigenic.
FIG. 4.
Codon-based analysis of selection pressures on regions C and E of the FCV capsid gene (Datamonkey). The main figure shows a plot of dN-dS values for each predicted codon of the consensus sequences. The red line represents all variants of strain D1. The blue line represents previously published sequences from epidemiologically unrelated strains of FCV. For each data set, the codons predicted to be evolving under positive selection by SLAC (P = 0.1) are indicated by red and blue asterisks above the plot. Codon mutations known to disrupt neutralizing B-cell epitopes are indicated by downward-pointing arrows above the plot. The blue shaded areas correspond to FCV capsid variable regions (C, 5′ and 3′ HVRs). Codon 1 is equivalent to codon 383 of the FCV capsid protein (53). Bars above and below this main plot represent the results of integrated analyses (SLAC, FEL, and REL) for three other strains (D2, A1, and A2) and for three individual cats (progressive evolvers, cat 4 and cat 2 [8], and sequential reinfection with a variant of a strain [cat 6]), respectively. Gray areas correspond to those regions predicted to be evolving under positive selection in each data set (P = 0.25 for SLAC and FEL; Bayes factor of 10 for REL).
Predicted structure.
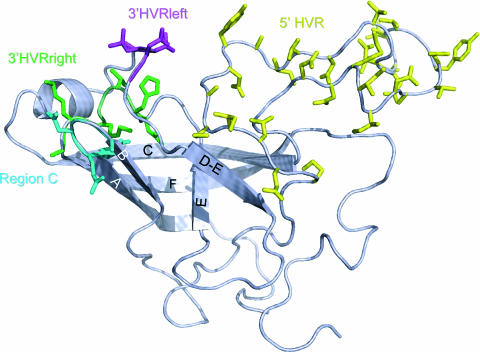
Regions showing the most consistent evidence of positive selection are mapped onto the predicted structure of the FCV P2 surface domain (6) (Fig. 5). The regions predicted to be under positive selection in the 5′ HVR of the capsid gene mapped to a long and largely flexible loop, a region known to contain linear epitopes (49, 55). In contrast, two small discrete regions under positive selection at the extremes of the 3′ HVR of the capsid gene mapped close to one another in the structure. The close proximity of these two regions may provide a structural explanation for the conformational nature of epitopes previously identified in this region (55).
FIG. 5.
Predicted structure of the FCV F9 capsid P2 surface domain (6), highlighting regions identified in this study that are predicted to be under positive selection. The yellow region corresponds to the 5′ HVR, known to contain linear epitopes. The magenta and green regions correspond to the left and right boundaries of the 3′ HVR, known to contain conformational epitopes. The cyan region contains variable region C and maps close to the 3′ HVR. The FCV P2 domain as modeled contains six β-strands, labeled A, B, C, D-E, E, and F.
Those amino acids predicted to be under positive selection in region C mapped structurally close to previously identified epitopes in the 3′ HVR (55). The FCV P2 domain as modeled contains six β-strands labeled A, B, C, D-E, E, and F (Fig. 5). Strands A, B, C, D, and F map closely with similar structures in SMSV and Norwalk virus (data not shown). A corresponding structure to SMSV and Norwalk virus strand D was not predicted in FCV. Instead, a strand D-E corresponds closely with a similar strand predicted between D and E in SMSV (6).
DISCUSSION
For viruses belonging to the family Caliciviridae, there are limited opportunities to study their evolution, diversification, and mechanisms of persistence in their natural host populations. In this study, we have shown how feline calicivirus infection within stable household groups of cats provides a useful model system to characterize evolutionary strategies employed by caliciviruses to ensure long-term survival in their host population. Structured sampling of these groups over long periods of time enabled us to identify the relative contribution of the different evolutionary mechanisms employed by feline calicivirus to ensure its persistence both in the individual and at the population level.
The first evolutionary mechanism identified was progressive evolution of a given variant of a strain through mutation accumulation within an individual. This was consistently present in only a small number of cats, suggesting that although a large proportion of the cat population appear to be viral carriers, only a minority of animals appear to be truly persistently infected, progressively evolving their own variant of a strain over prolonged periods of time. This helps explain the anomaly that has been observed in experimental studies, whereby in small groups of cats, persistent infections cannot always be induced (11, 28, 40, 60, 61).
The second evolutionary mechanism, sequential reinfection, which in a number of cases was interspersed with periods of progressive evolution, accounted for the majority of animals that appeared to be persistently infected. In this situation individual animals were undergoing reinfection from within the household either with a diverse variant of the same strain or with a distinct strain of virus. Reinfection with distinct strains of FCV is consistent with previous experimental studies which have shown that cats do appear to eliminate virus and that they remain susceptible to reinfection with a different strain (28, 60). However, this is the first report where sequential reinfection has been shown to occur with closely related variants of the same strain. This has important implications for both viral evolution and the epidemiology of the disease, particularly within groups of cats. Clearly, however, we cannot exclude the possibility that the original virus may still be present in such cats and is being out-competed at intervals by a new incoming virus.
Mixed infection with two distinct strains within an individual cat was also identified in one household, and although putative mixed infections have previously been reported (47), this is the first time this has been confirmed for caliciviruses by analysis of a cloned sequence. Such mixed infections are a prerequisite for the generation of recombinant viral strains, which together with the putative parent viruses have also been identified in this household and have been reported in detail elsewhere (10). Recombination in other calicivirus genera has also been described, but parental strains have not been identified (4, 25, 26, 37, 58). Recombination between variants of the same strain may also occur, but because of the close relationship between variants within a strain, such events would be more difficult to detect.
This study also demonstrated the high levels of viral diversity that can occur within a strain circulating within a population over extended periods of time. Using a corrected substitution model, levels of up to 24% (20% uncorrected) were observed within a strain within a household and up to 21% (19% uncorrected) was observed within individual animals that were persistently infected over extended periods of time. This suggests that within the overlapping population of viral variants present, new strains of FCV appear to be emerging, presumably through a combination of the evolutionary mechanisms described above. In cross-sectional studies, levels of diversity of up to 16% (uncorrected distance values) have been reported previously in some of these households (45). It is clear that such closed or semiclosed communities of endemically infected animals provide ideal conditions for the generation of viral biodiversity, and possible increased virulence, and may have parallels in other caliciviruses.
How long these strains had been circulating in each household and how this contributed to viral diversity are not known. However, in two colonies it appeared that a new strain was introduced during the course of the study, thus providing a natural opportunity to monitor viral evolution and diversification from a presumed single ancestor. In these two cases, virus variability increased from 0 to 5% and from 0 to 14% over 6 and 27 months, respectively. Thus, it appears that transmission of a new strain of virus into an individual within a household may represent a bottleneck to virus variability. Such reductions in variability may be caused either by low-dose virus transmission or by preferential selection of the fittest variant from the viral population at the time of transmission. Reductions in viral diversity by transmission bottlenecks have not previously been reported for caliciviruses but have been shown during natural transmission events for other RNA viruses (62, 65).
This study also allowed the calculation of viral evolution rates for the partial capsid region analyzed; the rate appeared to be higher (3.84 × 10−2 to 4.56 × 10−2 substitutions per nucleotide per year) for a strain circulating within a household (i.e., including transmission) compared to that for an individual cat (1.32 × 10−2 to 2.64 × 10−2 substitutions per nucleotide per year, i.e., progressive evolver, no transmission). The rate at which feline calicivirus evolved within an individual cat was lower than has been previously reported (48), although this latter study was based on the 5′ hypervariable region alone. There are no other estimates available for the rate at which caliciviruses evolve within populations, although estimates have been made for other RNA viruses (24). Evolution rates depend on a number of factors, including the region of the genome selected and the various selection pressures that may be operating. In this study, the evolution rates for feline calicivirus appear to be relatively high compared to other RNA viruses, which may be in part a reflection of the impact of positive selection acting on these regions of the genome.
Positive selection in the immunodominant 5′ and 3′ hypervariable regions of the capsid gene was found within strains at both the individual and population level. Positive selection in the 5′ HVR has been shown previously in an individual infected cat, where it was also demonstrated that such changes alter the neutralization profile (48). In other caliciviruses, such as human norovirus, immune-mediated positive selection in the P2 domain (analogous to the hypervariable regions of the FCV capsid gene) has also been suggested as a mechanism for facilitating persistent infection in an individual (35).
Positive selection within a strain at the population level has not previously been reported but can probably occur because of the closely matched immune response within the endemically infected host population. In contrast, there was little evidence of positive selection between distinct FCV strains at the population level, which is similar to the situation in other RNA viruses (31). It has been suggested that viral evolution at the population level may be a reflection of the demographic and spatial history of transmission within such populations rather than positive immune selection (23, 51). However, such conclusions may to some extent be a consequence of the sampling strategy used (20). Where in-depth sampling of host populations is carried out, as in this study, the intermediate evolutionary steps associated with positive immune selection can be clearly identified.
Interestingly, evidence of positive selection was also found in region C of the capsid, which has not previously been reported, and may suggest a possible antigenic role for this region. Previous studies have failed to identify any linear epitopes in region C (49), suggesting that if this region is antigenic it may contain either conformational B-cell or T-cell epitopes. Further support for the conformational nature of such epitopes was found in the predicted structural studies, where the codons predicted to be under positive selection within this region mapped structurally close to other known conformational epitopes in the 3′ HVR (49, 55).
This study has generated insights into the evolutionary mechanisms of FCV persistence and diversification, which are likely to be generalizable to other caliciviruses. It appears that true long-term persistent infection in an individual, characterized by continuous progressive evolution, is relatively rare and that the majority of apparent carriers in the population are undergoing a combination of progressive evolution and cycles of reinfection. In human norovirus infections, persistently infected people also appear to be relatively rare, and population persistence is largely thought to be through environmental contamination and epidemic spread (14, 56). It may be, however, that in some situations the two viruses employ a similar range of evolutionary strategies and that reinfection with variants of the same strain or with a different strain of norovirus might be more common than was previously thought. Nevertheless, the minority of individuals that are true carriers may provide an important fail-safe mechanism to ensure the virus's long-term survival within a population. It will be critical, therefore, to identify these individuals to control disease and further understand the evolution of the Caliciviridae.
Acknowledgments
This work was funded by the PetPlan Charitable Trust.
We thank the cat owners for allowing us to sample their cats, Chris McCracken and Ruth Ryvar for technical assistance, and Sergei Kosakovsky Pond (University of California), Dan Haydon (University of Glasgow), and Bryan Grenfell (Pennsylvania State University) for helpful discussions.
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Bannasch, M. J., and J. E. Foley. 2005. Epidemiologic evaluation of multiple respiratory pathogens in cats in animal shelters. J. Feline Med. Surg. 7:109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begon, M., J. L. Harper, and C. R. Townsend. 1990. Ecology: individuals, populations and communities, 2nd ed. Blackwell Scientific Publications, Cambridge, Mass.
- 3.Binns, S. H., S. Dawson, A. J. Speakman, L. E. Cuevas, C. A. Hart, C. J. Gaskell, K. L. Morgan, and R. M. Gaskell. 2000. A study of feline upper respiratory tract disease with reference to prevalence and risk factors for infection with feline calicivirus and feline herpesvirus. J. Feline Med. Surg. 2:123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bull, R. A., G. S. Hansman, L. E. Clancy, M. M. Tanaka, W. D. Rawlinson, and P. A. White. 2005. Norovirus recombination in ORF1/ORF2 overlap. Emerg. Infect. Dis. 11:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter, M. J., I. D. Milton, J. Meanger, M. Bennett, R. M. Gaskell, and P. C. Turner. 1992. The complete nucleotide sequence of a feline calicivirus. Virology 190:443-448. [DOI] [PubMed] [Google Scholar]
- 6.Chen, R., J. D. Neill, M. K. Estes, and B. V. Prasad. 2006. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. USA 103:8048-8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coutts, A. J., S. Dawson, K. Willoughby, and R. M. Gaskell. 1994. Isolation of feline respiratory viruses from clinically healthy cats at UK cat shows. Vet. Rec. 135:555-556. [PubMed] [Google Scholar]
- 8.Coyne, K. P., S. Dawson, A. D. Radford, P. J. Cripps, C. J. Porter, C. M. McCracken, and R. M. Gaskell. 2006. Long term analysis of feline calicivirus prevalence and viral shedding patterns in naturally infected colonies of domestic cats. Vet. Microbiol. 118:12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coyne, K. P., B. R. D. Jones, A. Kipar, J. Chantrey, C. J. Porter, P. Barber, S. Dawson, R. M. Gaskell, and A. D. Radford. 2006. Lethal outbreak of disease associated with feline calicivirus infection in cats. Vet. Rec. 158:544-550. [DOI] [PubMed] [Google Scholar]
- 10.Coyne, K. P., F. C. Reed, C. J. Porter, S. Dawson, R. M. Gaskell, and A. D. Radford. 2006. Recombination of feline calicivirus within an endemically-infected cat colony. J. Gen. Virol. 87:921-926. [DOI] [PubMed] [Google Scholar]
- 11.Dawson, S., N. R. Smyth, M. Bennett, R. M. Gaskell, C. M. McCracken, A. Brown, and C. J. Gaskell. 1991. Effect of primary-stage feline immunodeficiency virus infection on subsequent feline calicivirus vaccination and challenge in cats. AIDS 5:747-750. [DOI] [PubMed] [Google Scholar]
- 12.DeLano, W. L. 2002. The PyMOL molecular graphics system. Scientific, San Carlos, Calif.
- 13.Deveraux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Wit, M. A., M. P. Koopmans, L. M. Kortbeek, N. J. van Leeuwen, J. Vinje, and Y. T. van Duynhoven. 2001. Etiology of gastroenteritis in sentinel general practices in The Netherlands. Clin. Infect. Dis. 33:280-288. [DOI] [PubMed] [Google Scholar]
- 15.Dingle, K. E. 2004. Mutation in a Lordsdale norovirus epidemic strain as a potential indicator of transmission routes. J. Clin. Microbiol. 42:3950-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 17.Forrester, N. L., B. Boag, S. R. Moss, S. L. Turner, R. C. Trout, P. J. White, P. J. Hudson, and E. A. Gould. 2003. Long-term survival of New Zealand rabbit haemorrhagic disease virus RNA in wild rabbits, revealed by RT-PCR and phylogenetic analysis. J. Gen. Virol. 84:3079-3086. [DOI] [PubMed] [Google Scholar]
- 18.Gaskell, R. M., A. D. Radford, and S. Dawson. 2006. Feline infectious respiratory disease, p. 145-154. In E. A. Chandler, C. J. Gaskell, and R. M. Gaskell (ed.), Feline medicine and therapeutics, 3rd ed. Blackwell Publishing, Ames, Iowa.
- 19.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181(Suppl. 2):S322-S330. [DOI] [PubMed] [Google Scholar]
- 20.Grenfell, B. T., O. G. Pybus, J. R. Gog, J. L. Wood, J. M. Daly, J. A. Mumford, and E. C. Holmes. 2004. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303:327-332. [DOI] [PubMed] [Google Scholar]
- 21.Harbour, D. A., P. E. Howard, and R. M. Gaskell. 1991. Isolation of feline calicivirus and feline herpesvirus from domestic cats 1980 to 1989. Vet. Rec. 128:77-80. [DOI] [PubMed] [Google Scholar]
- 22.Helps, C. R., P. Lait, A. Damhuis, U. Bjornehammar, D. Bolta, C. Brovida, L. Chabanne, H. Egberink, G. Ferrand, A. Fontbonne, M. G. Pennisi, T. Gruffydd-Jones, D. Gunn-Moore, K. Hartmann, H. Lutz, E. Malandain, K. Mostl, C. Stengel, D. A. Harbour, and E. A. Graat. 2005. Factors associated with upper respiratory tract disease caused by feline herpesvirus, feline calicivirus, Chlamydophila felis and Bordetella bronchiseptica in cats: experience from 218 European catteries. Vet. Rec. 156:669-673. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, E. C. 2004. The phylogeography of human viruses. Mol. Ecol. 13:745-756. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:156-165. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, X., C. Espul, W. M. Zhong, H. Cuello, and D. O. Matson. 1999. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch. Virol. 144:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katayama, K., T. Miyoshi, K. Uchino, T. Oka, T. Tanaka, N. Takeda, and G. S. Hansman. 2004. Novel recombinant sapovirus. Emerg. Infect. Dis. 10:1874-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knowles, J. O., S. Dawson, R. M. Gaskell, C. J. Gaskell, and C. E. Harvey. 1990. Neutralisation patterns among recent British and North American feline calicivirus isolates from different clinical origins. Vet. Rec. 127:125-127. [PubMed] [Google Scholar]
- 28.Knowles, J. O., F. McArdle, S. Dawson, S. D. Carter, C. J. Gaskell, and R. M. Gaskell. 1991. Studies on the role of feline calicivirus in chronic stomatitis in cats. Vet. Microbiol. 27:205-219. [DOI] [PubMed] [Google Scholar]
- 29.Kreutz, L. C., R. P. Johnson, and B. S. Seal. 1998. Phenotypic and genotypic variation of feline calicivirus during persistent infection of cats. Vet. Microbiol. 59:229-236. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 31.Lemey, P., I. Derdelinckx, A. Rambaut, K. Van Laethem, S. Dumont, S. Vermeulen, E. Van Wijngaerden, and A. M. Vandamme. 2005. Molecular footprint of drug-selective pressure in a human immunodeficiency virus transmission chain. J. Virol. 79:11981-11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy, M., M. K. Estes, and K. C. Hyams. 2000. Norwalk-like virus infection in military forces: epidemic potential, sporadic disease, and the future direction of prevention and control efforts. J. Infect. Dis. 181(Suppl. 2):S387-S391. [DOI] [PubMed] [Google Scholar]
- 33.Moss, S. R., S. L. Turner, R. C. Trout, P. J. White, P. J. Hudson, A. Desai, M. Armesto, N. L. Forrester, and E. A. Gould. 2002. Molecular epidemiology of Rabbit haemorrhagic disease virus. J. Gen. Virol. 83:2461-2467. [DOI] [PubMed] [Google Scholar]
- 34.Neill, J. D. 1992. Nucleotide sequence of the capsid protein gene of two serotypes of San Miguel sea lion virus: identification of conserved and non-conserved amino acid sequences among calicivirus capsid proteins. Virus Res. 24:211-222. [DOI] [PubMed] [Google Scholar]
- 35.Nilsson, M., K. O. Hedlund, M. Thorhagen, G. Larson, K. Johansen, A. Ekspong, and L. Svensson. 2003. Evolution of human calicivirus RNA in vivo: accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J. Virol. 77:13117-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334-1344. [DOI] [PubMed] [Google Scholar]
- 37.Oliver, S. L., D. W. Brown, J. Green, and J. C. Bridger. 2004. A chimeric bovine enteric calicivirus: evidence for genomic recombination in genogroup III of the Norovirus genus of the Caliciviridae. Virology 326:231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen, N. C., J. B. Elliott, A. Glasgow, A. Poland, and K. Keel. 2000. An isolated epizootic of hemorrhagic-like fever in cats caused by a novel and highly virulent strain of feline calicivirus. Vet. Microbiol. 73:281-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedersen, N. C., and K. F. Hawkins. 1995. Mechanisms for persistence of acute and chronic feline calicivirus infections in the face of vaccination. Vet. Microbiol. 47:141-156. [DOI] [PubMed] [Google Scholar]
- 41.Pond, S. L., and S. D. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 42.Povey, R. C., R. C. Wardley, and H. Jessen. 1973. Feline picornavirus infection: the in vivo carrier state. Vet. Rec. 92:224-229. [DOI] [PubMed] [Google Scholar]
- 43.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 44.Radford, A. D., M. Bennett, F. McArdle, S. Dawson, P. C. Turner, M. A. Glenn, and R. M. Gaskell. 1997. The use of sequence analysis of a feline calicivirus (FCV) hypervariable region in the epidemiological investigation of FCV related disease and vaccine failures. Vaccine 15:1451-1458. [DOI] [PubMed] [Google Scholar]
- 45.Radford, A. D., S. Dawson, R. Ryvar, K. Coyne, D. R. Johnson, M. B. Cox, E. F. Acke, D. D. Addie, and R. M. Gaskell. 2003. High genetic diversity of the immunodominant region of the feline calicivirus capsid gene in endemically infected cat colonies. Virus Genes 27:145-155. [DOI] [PubMed] [Google Scholar]
- 46.Radford, A. D., L. Sommerville, R. Ryvar, M. B. Cox, D. R. Johnson, S. Dawson, and R. M. Gaskell. 2001. Endemic infection of a cat colony with a feline calicivirus closely related to an isolate used in live attenuated vaccines. Vaccine 19:4358-4362. [DOI] [PubMed] [Google Scholar]
- 47.Radford, A. D., L. M. Sommerville, S. Dawson, A. M. Kerins, R. Ryvar, and R. M. Gaskell. 2001. Molecular analysis of isolates of feline calicivirus from a population of cats in a rescue shelter. Vet. Rec. 149:477-481. [DOI] [PubMed] [Google Scholar]
- 48.Radford, A. D., P. C. Turner, M. Bennett, F. McArdle, S. Dawson, M. A. Glenn, R. A. Williams, and R. M. Gaskell. 1998. Quasispecies evolution of a hypervariable region of the feline calicivirus capsid gene in cell culture and in persistently infected cats. J. Gen. Virol. 79:1-10. [DOI] [PubMed] [Google Scholar]
- 49.Radford, A. D., K. Willoughby, S. Dawson, C. McCracken, and R. M. Gaskell. 1999. The capsid gene of feline calicivirus contains linear B-cell epitopes in both variable and conserved regions. J. Virol. 73:8496-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rambaut, A. 2000. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 16:395-399. [DOI] [PubMed] [Google Scholar]
- 51.Rambaut, A., D. Posada, K. A. Crandall, and E. C. Holmes. 2004. The causes and consequences of HIV evolution. Nat. Rev. Genet. 5:52-61. [DOI] [PubMed] [Google Scholar]
- 52.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seal, B. S., J. F. Ridpath, and W. L. Mengeling. 1993. Analysis of feline calicivirus capsid protein genes: identification of variable antigenic determinant regions of the protein. J. Gen. Virol. 74:2519-2524. [DOI] [PubMed] [Google Scholar]
- 54.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 55.Tohya, Y., N. Yokoyama, K. Maeda, Y. Kawaguchi, and T. Mikami. 1997. Mapping of antigenic sites involved in neutralization on the capsid protein of feline calicivirus. J. Gen. Virol. 78:303-305. [DOI] [PubMed] [Google Scholar]
- 56.Tompkins, D. S., M. J. Hudson, H. R. Smith, R. P. Eglin, J. G. Wheeler, M. M. Brett, R. J. Owen, J. S. Brazier, P. Cumberland, V. King, and P. E. Cook. 1999. A study of infectious intestinal disease in England: microbiological findings in cases and controls. Commun. Dis. Public Health 2:108-113. [PubMed] [Google Scholar]
- 57.van Duynhoven, Y. T., C. M. de Jager, L. M. Kortbeek, H. Vennema, M. P. Koopmans, F. van Leusden, W. H. van der Poel, and M. J. van den Broek. 2005. A one-year intensified study of outbreaks of gastroenteritis in The Netherlands. Epidemiol. Infect. 133:9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vidal, R., P. Roessler, V. Solari, J. Vollaire, X. Jiang, D. O. Matson, N. Mamani, V. Prado, and M. L. O'Ryan. 2006. Novel recombinant norovirus causing outbreaks of gastroenteritis in Santiago, Chile. J. Clin. Microbiol. 44:2271-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wardley, R. C. 1976. Feline calicivirus carrier state. A study of the host/virus relationship. Arch. Virol. 52:243-249. [DOI] [PubMed] [Google Scholar]
- 60.Wardley, R. C., and R. C. Povey. 1977. The clinical disease and patterns of excretion associated with three different strains of feline caliciviruses. Res. Vet. Sci. 23:7-14. [PubMed] [Google Scholar]
- 61.Wardley, R. C., and R. C. Povey. 1977. The pathology and sites of persistence associated with three different strains of feline calicivirus. Res. Vet. Sci. 23:15-19. [PubMed] [Google Scholar]
- 62.Weiner, A. J., M. M. Thaler, K. Crawford, K. Ching, J. Kansopon, D. Y. Chien, J. E. Hall, F. Hu, and M. Houghton. 1993. A unique, predominant hepatitis C virus variant found in an infant born to a mother with multiple variants. J. Virol. 67:4365-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White, P. J., R. C. Trout, S. R. Moss, A. Desai, M. Armesto, N. L. Forrester, E. A. Gould, and P. J. Hudson. 2004. Epidemiology of Rabbit haemorrhagic disease virus in the United Kingdom: evidence for seasonal transmission by both virulent and avirulent modes of infection. Epidemiol. Infect. 132:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Widdowson, M. A., E. H. Cramer, L. Hadley, J. S. Bresee, R. S. Beard, S. N. Bulens, M. Charles, W. Chege, E. Isakbaeva, J. G. Wright, E. Mintz, D. Forney, J. Massey, R. I. Glass, and S. S. Monroe. 2004. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus—United States, 2002. J. Infect. Dis. 190:27-36. [DOI] [PubMed] [Google Scholar]
- 65.Wolinsky, S. M., C. M. Wike, B. T. Korber, C. Hutto, W. P. Parks, L. L. Rosenblum, K. J. Kunstman, M. R. Furtado, and J. L. Munoz. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134-1137. [DOI] [PubMed] [Google Scholar]