Abstract
We had previously identified active autonomous copies of the MusD long terminal repeat-retrotransposon family, which have retained transpositional activity. These elements are closely related to betaretroviruses but lack an envelope (env) gene. Here we show that these elements encode strictly intracellular virus-like particles that can unambiguously be identified by electron microscopy. We demonstrate intracellular maturation of the particles, with a significant proportion of densely packed cores for wild-type MusD but not for a protease mutant. We show that the molecular origin of this unexpected intracellular localization is solely dependent on the N-terminal part of the Gag protein, which lacks a functional sequence for myristoylation and plasma membrane targeting: replacement of the N-terminal domain of the MusD matrix protein by that of its closest relative—the Mason-Pfizer monkey virus—led to targeting of the MusD Gag to the plasma membrane, with viral particles budding and being released into the cell supernatant. These particles can further be pseudotyped with a heterologous envelope protein and become infectious, thus “reconstituting” a functional retrovirus prone to proviral insertions. Consistent with its retroviral origin, a sequence with a constitutive transport element-like activity can further be identified at the MusD 3′ untranslated region. A molecular scenario is proposed that accounts for the transition, during evolution, from an ancestral infectious betaretrovirus to the strictly intracellular MusD retrotransposon, involving not only the loss of the env gene but also an inability to escape the cell—via altered targeting of the Gag protein—resulting de facto in the generation of a very successful “intracellularized” insertional mutagen.
Mammalian genomes contain a high load of so-called endogenous retroviruses (ERVs). These elements are suspected to be the genomic traces of infections of the germ line of remote ancestors by bona fide retroviruses which have been transmitted in a Mendelian manner and, for some of these founder elements, amplified by reinfection and/or retrotransposition. This scenario has most probably generated the large set of elements that can be found in the present-day human or murine genome (23, 40) and which actually discloses similarly diverse structures and, when still active, life cycles as observed for the present-day infectious retrovirus population. However, it should be pointed out that most ERVs are now defective due to the accumulation of mutations, and only a few have been identified as functional elements. Although there remains a possibility that some ERVs from humans are still active, with evidence for positional polymorphism of the HERV-K(HML2) elements within the human population (3, 36, 37) and of related virus-like particles (VLPs) released from some tumor cells (6, 8, 24, 26), the most active ERV families have been characterized in mice. These include the IAP and MusD families, which are responsible for a large set of mutations by proviral insertions, both at the germ line and at the somatic cell levels (4, 22, 27). Interestingly, we have previously succeeded in identifying, within these two families of elements, “master” copies that are competent and autonomous for retrotransposition (14, 30). IAP elements, present at a thousand copies in the mouse genome, proved to be very active retrotransposons in an ex vivo assay initially devised in the laboratory (14, 19, 20), with at least a hundred elements still fully active. The MusD family comprises a hundred copies in the C57BL/6 mouse genome, among which only three are retrotransposition competent (30). These elements are also very active retrotransposons and can further mediate in trans the retrotransposition of the ETn elements, which are closely related retrotransposons without any coding sequence (25, 35) but that very efficiently use the MusD machinery for their mobilization (30). It is noteworthy that the IAP, MusD, and ETn elements should have already quite diverged from their putative retroviral progenitors, since they lack, for instance, an env gene. However, other recently amplified elements, such as some human-specific HERV-K(HML2) family members, still possess an intact and functional env gene, which can further confer infectivity on pseudotypes (13, 15). In addition to this genomic diversity, other differences have been found, with evidence for diverse localizations of the VLPs associated with some of these elements: the murine IAPs, for instance, assemble at the endoplasmic reticulum (ER) membrane, where they bud to release particles of immature structure into the lumen of the ER (14, 16, 22), whereas human HERV-K particles assemble at the cell membrane, where they are further released into the cell supernatant, as observed, for instance, for classical type C retroviruses (6, 15). In fact, retroviruses, once they have succeeded in inserting into a host genome, should evolve in such a way as to preserve their replicative efficacy; otherwise, they would simply “fade out” due to genetic drift and the absence of selective pressure for their maintenance. Analysis of the endogenization process of one of the still active ERVs for which autonomous proviral copies have been identified, namely, MusD, has therefore been performed, resulting in the characterization of its life cycle and the unraveling of the molecular steps that have led from an infectious retrovirus to an intracellular retrotransposon.
MATERIALS AND METHODS
Plasmids.
pCMV-MusDwt is an expression vector for the MusD-6 copy (AC124426, nucleotide [nt] positions 9078 to 16569) under the control of cytomegalovirus (CMV) promoter (30). pCMV-MusDmyr was obtained by adding the first 21 nt of the Mason-Pfizer monkey virus (MPMV) gag gene to the MusD gag gene of pCMV-MusDwt (the resulting MusD Gag product thus contains the sequence MGQELSQ added at the N terminus). pCMV-MusDmyr+basic was obtained by replacing the first 105 nt of the MusD gag gene of pCMV-MusDwt by the first 123 nt of the MPMV gag gene (the resulting MusD Gag product will contain the first 41 N-terminal residues of the MPMV Gag protein). The pDM128/PL plasmid contains the cat gene and a polylinker flanked by a donor and an acceptor splice site (17, 21). pDM128/MPMV-CTE contains the full-length CTE of MPMV inserted into the polylinker BglII site present in pDM128/PL (7). MusD 3′ end noncoding sequences and deletion derivatives were PCR amplified and inserted into pDM128/PL between the polylinker BglII and KpnI sites.
Cell culture, transposition, and infectivity assay.
Human HeLa and 293T cells and feline fetal astrocyte G355.5 cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (VWR), 100 μg of streptomycin/ml, and 100 U of penicillin/ml. The day prior to transfection, cells were seeded at 5.5 × 105, 7.5 × 105, or 6 × 105 cells per 60-mm dish for HeLa, 293T, or G355.5 cells, respectively. Transfection of G355.5 or HeLa cells was performed with 1 or 3 μg of DNA, respectively, using Lipofectamine (Invitrogen). Transfection of 293T cells was performed with 4.5 μg of DNA using the MBS transfection kit (Stratagene).
To assay for retrotransposition, transfected HeLa cells were expanded for 6 days, seeded at 5 × 105 cells per 100-mm dish and allowed to settle for 24 h before G418 (560 μg/ml; Gibco-BRL) was added. After a 15-day selection, G418-resistant (G418r) foci were fixed, stained, and counted.
To assay for infection, 293T cells were transfected with pCMV-MusDwt, pCMV-MusDmyr, or pCMV-MusDmyr+basic plus a neoTNF-marked defective MusD vector (30) and an expression vector for the pseudotyping vesicular stomatitis virus G (VSV-G) envelope protein (GenBank accession no. AJ318514, a gift from F.-L. Cosset, Ecole Normale Supérieure, Lyon, France). At 24 h posttransfection, target HeLa cells were seeded in six-well plates at 1.2 × 105 cells per well. Supernatants from 293T cells were harvested 48 h posttransfection, filtered through 0.45-μm-pore-size membranes, supplemented with Polybrene (4 μg/ml), and transferred to target HeLa cells. To increase sensitivity, target cells were subjected to spinoculation at 1,200 × g for 2.5 h at 25°C. After removal of the supernatants, cells were incubated in regular medium for 72 h at 37°C. HeLa cells were then split into 100-mm dishes (5 × 105 cells per dish), allowed to settle for 24 h, and subjected to G418 selection. G418r foci were fixed, stained, and counted or individually picked and expanded.
Immunoblotting.
Transfected HeLa or 293T cells were lysed 48 h posttransfection in Laemmli buffer and whole-cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using gels containing 12% polyacrylamide (37.5:1 ratio of acrylamide to N,N-methylenebisacrylamide). Proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) and incubated with a polyclonal rabbit antiserum anti-Gag MusD (30). A goat antibody to rabbit immunoglobulin G (IgG) conjugated to horseradish peroxidase (Amersham) was used as the secondary antiserum.
For analysis of cell supernatants, the culture medium of transfected 293T cells was harvested 48 h postinfection, and the particulate material was concentrated by ultracentrifugation (38,000 rpm for 1.5 h in a Beckman SW41 rotor) through a 20% sucrose cushion. Virions were lysed in Laemmli buffer before analysis by SDS-PAGE and immunoblotting.
Metabolic radiolabeling.
For fatty acid labeling, [9,10(n)-3H]myristic acid (50 Ci/mmol; Amersham) was added to complete growth medium (0.1 mCi/ml). 293T cells were labeled 48 h posttransfection for 6 h at 37°C. Cells were then washed twice in phosphate-buffered saline (PBS) and lysed in Laemmli buffer. Whole-cell extracts were separated by SDS-PAGE, and labeled proteins were detected by fluorography using the amplify fluorographic reagent (Amersham).
CAT activity assay.
293T cells transfected with pDM128/PL or its derivatives containing MusD sequences were lysed 48 h posttransfection in 0.25 M Tris-HCl (pH 7.0) by three freeze-thaw cycles. Cell extracts were incubated for 1 h at 37°C in the presence of 0.625 μCi of d-threo-[dichloroacetyl-1,2-14C]chloramphenicol (60 mCi/mmol; Perkin-Elmer Life Sciences)/ml and 1.25 mM N-acetyl-coenzyme A (Roche Diagnostics). Chloramphenicol and its derivatives were extracted with ethyl acetate. Acetylated and unacetylated forms of chloramphenicol were separated by thin-layer chromatography on TLC silica plates (Macherey-Nagel) using chloroform-methanol (19:1). The chloramphenicol acetyltransferase (CAT) activity was measured by PhosphorImager quantitation using a FLA-3000 scanner.
Immunofluorescence microscopy.
Transfected HeLa cells were grown on 14-mm glass coverslips, fixed in 4% paraformaldehyde in PBS at room temperature, and permeabilized in PBS-0.1% SDS. After a blocking step in PBS-3% bovine serum albumin, the cells were immunostained with the polyclonal rabbit antiserum anti-Gag MusD. After subsequent washes in PBS, the cells were incubated with Alexa Fluor 488 goat anti-rabbit IgG (Molecular Probes) as a secondary antibody. Cell nuclei were stained with TO-PRO-3 iodide (Molecular Probes). Fluorescence patterns were visualized with a Zeiss LSM 510 laser-scanning confocal microscope.
Electron microscopy.
For ultrastructural studies, transfected G355.5, HeLa, and 293T cells were fixed in phosphate buffer (pH 7.2)-1.6% glutaraldehyde for 1 h and postfixed in 0.1 M cacodylate buffer-1% osmium tetroxide for 2 h. After being rinsed for 5 min in water and 15 min in 0.1 M cacodylate buffer, the cells were transferred to 0.2 M cacodylate buffer for 30 min. The cells were washed in 30% methanol for 10 min, stained in 2% uranyl acetate in 0.1 M cacodylate buffer-30% methanol for 1 h, and washed in 30% methanol. The cells were then dehydrated through a graded ethanol series and embedded in Epon 812. Ultrathin sections were stained with uranyl acetate and lead citrate and examined with a Zeiss 902 microscope at 80 kV.
For immunoelectron microscopy experiments, transfected cells were fixed for 1 h at 4°C with either 4% formaldehyde or 1.6% glutaraldehyde in 0.1 M phosphate buffer (pH 7.3). The cell pellets were dehydrated in increasing concentrations of methanol and embedded in Lowicryl K4M at a low temperature. Polymerization was carried out for 5 days at −30°C under long-wavelength UV light. Ultrathin sections of Lowicryl-embedded material were used for labeling with the rabbit antiserum specific for MusD Gag, followed by incubation with a goat anti-rabbit IgG conjugated to gold particles, 10 nm in diameter. Sections were contrasted with uranyl acetate.
RESULTS
MusD generates intracellular VLPs.
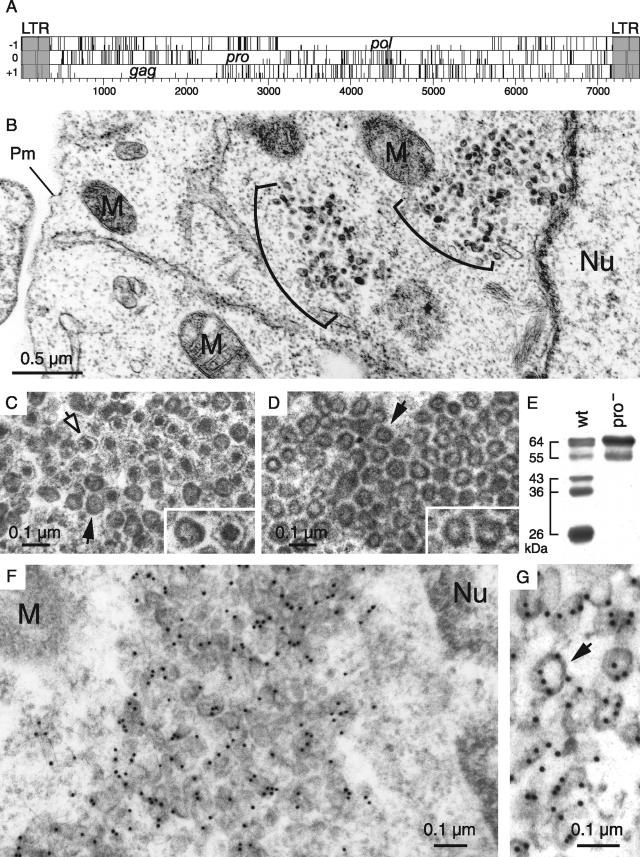
Among the hundred copies of MusD elements that can be found in the murine genome, we have previously identified three autonomous MusD proviruses that are still functional for retrotransposition. As illustrated in Fig. 1A, these elements contain only three open reading frames (ORFs), namely, gag, pro, and pol, all of which were demonstrated to be absolutely required for MusD retrotransposition in an assay using an element marked with the neoTNF indicator gene for retrotransposition (30). To identify the VLPs that are expected to be associated with the expression and retrotransposition of MusD, we transfected heterologous feline or human cells (devoid of endogenous MusD elements) with expression vectors for the retrotransposition-competent MusD elements. Cells were fixed 2 days posttransfection after partial removal of the supernatant, and cross-sections of the whole pellet were analyzed by electron microscopy. Identical results were obtained with human HeLa and 293T cells and feline G355.5 cells. As illustrated in Fig. 1B for HeLa cells, VLPs can be easily detected in cells transfected with the MusD-6 copy (the same results were obtained with another functional MusD copy [data not shown]) and were not observed in cells transfected with a control plasmid (pCMV-β [data not shown]). In no instance could particles be detected outside the cells (see also a demonstration of the strictly intracellular nature of the MusD retrotransposition cycle in Supplementary Figure 1 at http://www.igr.fr/jvi_ribet_suppl/). Particles are only found within the cytosol and are not associated with either the cell or the ER membrane. They appear in clusters, and two morphologies can be distinguished: relatively homogeneous dark particles with no discernible internal structures and particles containing a contrasted central core separated by an electron-lucent zone from a thin peripheral dark layer. The latter particles represent ca. 15% of all VLPs (Fig. 1C). These two structures, which are mixed within the clusters, have a comparable size, with a mean diameter of 75 to 80 nm. To confirm that the observed particles are actually composed of MusD proteins, we used a specific rabbit antiserum that we had raised against the MusD full-length Gag polyprotein, and we analyzed the cells by electron microscopy after immunogold labeling. As illustrated in Fig. 1F and G, the particles were specifically labeled with the antiserum, as expected.
FIG. 1.
Structure of the MusD genome and associated VLPs. (A) Genomic organization of autonomous MusD elements, with the LTRs (gray) bordering the three ORFs homologous to the retroviral gag, pro, and pol genes. No ORF for an envelope protein can be identified. (B to D) Electron microscopy of the VLPs encoded by the retrotransposition competent MusD-6 copy (B and C) and a MusD-6 protease mutant (D), observed in human cells 48 h after transfection. (B) Representative low-magnification image of a transfected HeLa cell, with numerous particles organized in intracellular clusters. The nucleus (Nu), mitochondria (M), and plasma membrane (Pm) are indicated. No particle can be observed at the level of the cell membrane or in the extracellular space. (C) High-magnification view of a cluster of particles with two different morphologies: immature (solid arrow) with an electron-lucent core and mature (open arrow) with a condensed central core. (D) Same magnification as in panel C for a cluster of particles observed in cells transfected with a protease-deficient MusD-6 mutant. Only immature particles can be observed (arrow). (E) Western blot analysis of whole-cell lysates of HeLa cells transfected with the MusD-6 copy (wt, lane 1) or the protease-deficient mutant (pro−, lane 2) used in panels B to D. The proteins were separated by SDS-PAGE, and Gag products were revealed using a rabbit antiserum directed against a MusD Gag recombinant protein (30). The molecular masses of the Gag precursor and protein cleavage products are indicated in kilodaltons. (F and G) Immunogold labeling of MusD particles in transfected cells using the anti-Gag MusD antiserum and a secondary antibody linked to gold beads, observed by electron microscopy; labeling is seen within the clusters (F), at the level of the MusD particles (a higher-magnification view is seen in panel G [see arrow]).
The two observed VLP morphologies are reminiscent of the “mature” and “immature” particles that are commonly found among infectious retroviruses (reviewed in reference 38), except that, in the latter case, matured particles are found only outside of the cells. Since maturation is the consequence of viral polyprotein cleavage by the retroviral protease, cells were transfected with an expression vector for a MusD protease-deficient mutant, derived from the MusD-6 copy (see reference 30 and the Western blot in Fig. 1E). VLPs were also observed with this mutant, organized in dense clusters containing 10 to 100 units (Fig. 1D) and closely resembling the “immature” particles generated by wild-type MusD. The VLPs appear to be rather homogeneous in size, with a mean diameter of 75 nm (most particles having diameters ranging between 60 and 90 nm). Particles with a condensed central core were never observed with the protease mutant, thus strongly suggesting that they indeed correspond to the mature MusD VLPs.
MusD VLPs have lost a cell membrane-targeting signal.
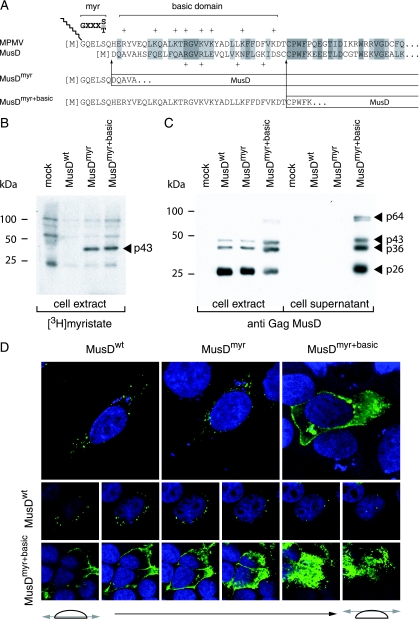
The localization of the observed VLPs is rather unexpected for an element of retroviral origin, which should either directly assemble at the cell membrane (as for type C retroviruses) or assemble in the cytoplasm, but then be targeted to the cell membrane (as for type B/D retroviruses). As in the latter case—and consistent with its close phylogenetic relationship to type B/D retroviruses—MusD particles assemble in the cytoplasm but fail to be transported to the cell membrane. An overall sequence comparison between MusD and the prototypic type-D retrovirus MPMV actually discloses significant similarities, which extend to the entire pro and pol genes and a large fraction of the gag gene, including the capsid and nucleocapsid Gag moieties (25). Interestingly, amino acid sequences at the Gag N terminus show significant divergences, which include the absence of a canonical plasma membrane targeting signal. This signal, which has been clearly identified for MPMV (see reference 11 and Fig. 2A), is a classical membrane targeting signal of retroviruses, with a bipartite structure. Its N-terminal domain has a [M]GXXXS/T consensus sequence (the first M corresponding to the Gag initiation codon), allowing covalent binding of a myristic acid to the glycine residue by fatty acylation (for a review, see reference 34). The second half contains positively charged basic residues (indicated by a “+” symbol in Fig. 2A), which interact with the negatively charged phospholipids at the plasma membrane (32, 41). The complete bipartite signal, with both the myristate anchor and the basic residues, is required for efficient membrane targeting and subsequent budding of the retroviral particles (10, 28, 29, 42). Although the MusD N-terminal sequence discloses a few positive charges, it does not display the canonical [M]GXXXS/T myristoylation signal. We therefore hypothesized that this signal had simply been lost during MusD endogenization, thus preventing myristoylation and targeting of the MusD particles to the cell membrane, and we tried to restore it in order to re-address the MusD VLPs toward the plasma membrane and tentatively “resuscitate” the retroviral MusD ancestor. To do so, two chimeras were constructed (see Fig. 2A): one with only the first six N-terminal residues of MPMV Gag added to the MusD sequence (MusDmyr) and one with the entire MPMV plasma membrane targeting signal, including the basic residues (MusDmyr+basic), inserted tentatively at an homologous position into the MusD sequence (Fig. 2A). In a first series of experiments, we assayed the wild-type MusD and both mutants for Gag protein myristoylation by adding [3H]myristate to cells transfected with the respective MusD expression vectors. As illustrated in Fig. 2B, Gag myristoylation was not observed, as expected, with wild-type MusD (MusDwt) but was unambiguously detected with the two chimeras. In a second series of experiments, we analyzed MusD targeting by immunofluorescence, using Gag-specific antibodies (Fig. 2D). As clearly illustrated in the figure and as expected from the electron microscopy analysis, cells transfected with MusDwt display a cytoplasmic staining with a punctuated distribution pattern, which was also observed with the MusDmyr construct, whereas a complete redistribution is observed in cells transfected with MusDmyr+basic. In the latter case, part of the staining is localized at the cell membrane, as further confirmed by serial confocal images. Conclusively, the mere introduction of a myristoylation signal is not sufficient for redirecting MusD Gag to the cell membrane, which actually requires a complete plasma membrane targeting signal, including the basic residue domain.
FIG. 2.
Consequences of restoring myristoylation and membrane targeting signals on MusD Gag protein addressing. (A) Sequence of the N-terminal domain of the MusD Gag protein and comparison with the corresponding domain from its closest relative, the type-D MPMV retrovirus. The consensus sequence required for myristoylation (myr, [M]GXXXS/T) and the MPMV domain rich in basic residues (basic domain, +) are indicated. Arrows indicate the limits of the replacements between the N-terminal residues of MPMV and MusD Gag proteins to generate the MusDmyr and MusDmyr+basic proviruses, respectively. (B) Restoration of MusD Gag myristoylation. Whole-cell lysates were prepared from human 293T cells transfected with the indicated MusD constructs and labeled with [3H]myristate. Myristoylated proteins were revealed by fluorography after separation by SDS-PAGE. (C) Western blot analysis of whole-cell lysates or cell supernatants from 293T cells transfected as in panel B with the indicated MusD constructs, using a rabbit antiserum directed against the MusD Gag polyprotein. The molecular masses of the Gag-specific bands are indicated in kilodaltons, as in Fig. 1E. (D) Immunofluorescence confocal analysis of human HeLa cells transfected with the indicated MusD constructs. Cells were grown on glass coverslips approximately 48 h posttransfection, fixed, and permeabilized, and MusD Gag proteins were detected by using the same rabbit anti-Gag MusD antiserum as in panel C and an Alexa Fluor 488-conjugated anti-rabbit secondary antibody. Nuclei were stained with TO-PRO-3 iodide. The two rows at the bottom show the successive confocal images of stained cells.
Bona fide retrovirus as a MusD progenitor.
To further determine whether the MusD element with a restored plasma membrane targeting signal indeed behaves as a bona fide retrovirus, a series of experiments was performed to assay (i) the viral particle release from the cells and (ii) the infectivity of the released particles when pseudotyped with a heterologous envelope protein.
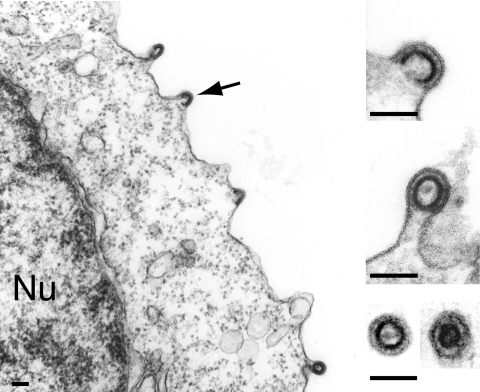
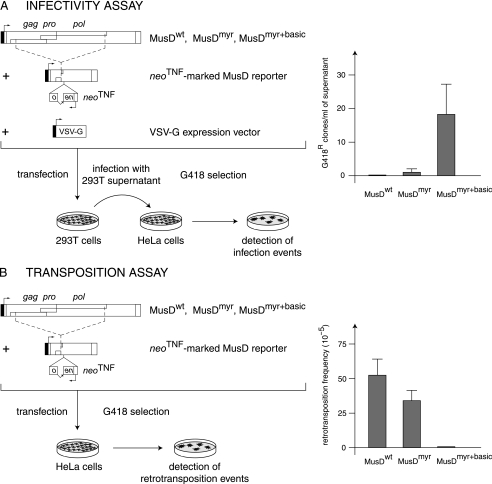
Western blot analyses were first performed on both whole-cell lysates and cell supernatants, upon transfection of 293T cells with expression vectors for the MusDwt, MusDmyr, and MusDmyr+basic constructs. As a control, and as illustrated in Fig. 2C, closely related levels of Gag proteins, with the expected protein cleavage products, were produced by all three constructs in whole-cell extracts. However, only the MusDmyr+basic construct resulted in the release of Gag proteins into the cell supernatant, strongly suggesting the release of viral particles. This latter point was assessed by electron microscopy analysis of the corresponding cells (Fig. 3), which disclosed viral particles budding at the cell membrane, as well as images of both immature and mature particles released from the cell surface, that were not observed with the control MusDwt construct. As a next step, we tested whether the released particles could be rendered infectious upon the addition of a heterologous envelope protein. To do so, as illustrated in the scheme in Fig. 4A, we used an expression vector for the VSV-G envelope protein cotransfected with the various MusD constructs and an internally deleted neoTNF-marked MusD reporter (30). At 2 days posttransfection, 293T cell supernatants were collected and used to tentatively infect naive target cells. Infectivity was quantitated by subjecting the target cells to selection for resistance to the neomycin derivative G418 and counting the number of G418r cell foci. Under these conditions (Fig. 4A), it can be clearly shown that the MusDmyr+basic is infectious when pseudotyped with the VSV-G envelope protein, whereas only very low infectivity is displayed by the MusDmyr construct and none is shown by the wild-type MusDwt construct. Some of the G418r clones resulting from HeLa cell infection with MusDmyr+basic were recovered and propagated to analyze the structure of the newly integrated MusD copies. PCR amplifications performed on the genomic DNA from two independent clones, followed by sequencing, revealed the canonical features of a retroviral integration (see Supplementary Fig. 2 at http://www.igr.fr/jvi_ribet_suppl/) with regeneration of complete long terminal repeats (LTRs) and target site duplications of 6 bp, as observed for the genomic MusD copies. Conclusively, restoring a functional plasma membrane targeting signal to MusD appears to be sufficient to generate functional viral particles that can be released from the cells, whereas further addition of an envelope protein gene ultimately “resuscitates” a functional retrovirus prone to proviral insertions.
FIG. 3.
Plasma membrane budding and release of MusD particles upon restoration of the Gag myristoylation signal and basic domain. Electron microscopy analysis of human 293T cells transfected with the MusDmyr+basic construct. Same experimental conditions as in Fig. 1B and C. A low-magnification image is shown on the left with budding particles (arrow), and enlarged views are shown on the right, with two free particles (immature and mature) in the cell supernatant (bottom image). Bars, 0.1 μm.
FIG. 4.
Functional characterization of MusD and its variants with modified myristoylation and membrane targeting signals. (A) Infectivity assay. 293T cells were transiently transfected with an expression vector for the indicated MusD constructs and a neoTNF-marked defective MusD reporter (in which a “backward” neomycin resistance gene [with the neo ORF interrupted by a “forward” TNF intron] becomes functional only after a complete replicative cycle) (30), plus an expression vector for the VSV-G envelope protein. Supernatants from the transfected cells were collected 48 h posttransfection and used to infect naive target HeLa cells. After a 3-day growth period, infection events were detected upon G418 selection of the cells and quantitated by counting the number of G418r clones. No clone was detected in the absence of the VSV-G expression vector. (B) Transposition assay. HeLa cells were transfected with the same MusD plasmids as in panel A but without the VSV-G envelope protein expression vector; after a 6-day growth period, the cells were seeded (5 × 105 cells per plate) and subjected to G418 selection to detect retrotransposition events (number of G418r clones per cells in selection [see reference 30]).
Extracellular retrovirus versus intracellular MusD: consequences for retrotransposition efficiency.
To determine the impact of the “intracellularization” of the MusD progenitor on the mode of proviral amplification and on retrotransposition efficiency, we devised experiments aimed at measuring the retrotransposition rate of the three above-mentioned constructs, namely, MusDwt, MusDmyr, and MusDmyr+basic.
Each of the three constructs was introduced by transfection into HeLa cells together with the internally deleted neoTNF-marked MusD reporter. Retrotransposition—in the absence of any added envelope protein expression vector—was then measured by counting the number of G418r colonies that were recovered, under conditions identical to those described above, after G418 selection of the cells (see the scheme in Fig. 4B). As illustrated in the figure, restoring a functional plasma membrane targeting signal to MusD (MusDmyr+basic) resulted in a dramatic decrease (∼100-fold) in the retrotransposition efficiency of the corresponding construct, whereas only a limited decrease was observed with the MusDmyr construct. It therefore clearly appears that targeting the Gag protein to the plasma membrane and release of the MusD VLPs outside the cells is detrimental to retrotransposition, which is to say, in terms of evolution, that “intracellularization” of MusD has most probably been beneficial for MusD provirus amplification.
Identification of a sequence with a CTE-like activity within the MusD provirus 3′end.
As already noted, the MusD sequence lacks an env gene. However, a 1.4-kb noncoding sequence can be found at the provirus 3′ end, at the expected location for env (Fig. 1A). Furthermore, this sequence is conserved at the same relative position in retrotransposons of the MusD-derived ETn family, which is a family of defective elements with the gag, pro, and pol genes deleted but that can still be mobilized in trans by the MusD machinery (25, 30). Consistent with the “conservation” of this noncoding sequence, we have demonstrated that it is required for efficient retrotransposition, since its deletion from the MusD sequence resulted in a strong inhibition of MusD mobility (data not shown). In many retroviruses, a sequence in the 3′ untranslated region of the viral genome—the so-called CTE (for constitutive transport element)—has been found to play an important role in the viral life cycle by allowing nuclear export of the unspliced viral RNAs to be packaged, which would otherwise be subjected to nuclear retention by cellular splicing factors (reviewed in reference 18). By analogy, again, with what was found for bona fide retroviruses, we determined whether such functional elements are present in the MusD genome and, more precisely, within the noncoding sequence identified above as stringently required for MusD retrotransposition.
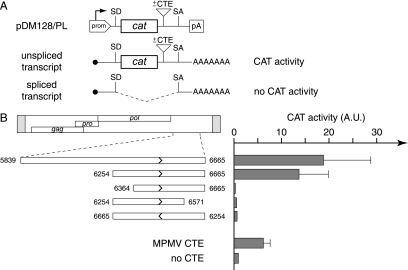
To do so, we cloned this sequence and its deletion derivatives (Fig. 5A) within a previously characterized indicator construct (pDM128/PL [17, 21]). As illustrated in the figure, this indicator contains the cat gene flanked by a donor and an acceptor splice site, with a polylinker for inserting the sequences to be tested placed in-between the cat gene and the acceptor splice site. In this construct, CAT enzyme expression is indeed highly dependent on efficient nuclear export of the unspliced RNA, with the insertion of a CTE sequence resulting in enhanced CAT activity. As illustrated in Fig. 5B, the transfection of 293T cells with the pDM128/PL vector containing the previously identified MPMV CTE sequence clearly demonstrates a strong enhancement of CAT activity. Similarly, it can be seen that the MusD 3′-end noncoding sequence also has a strong stimulatory effect. As illustrated in the figure, assays for various deletion derivatives finally allowed the delineation of an ∼400-bp fragment that discloses strong CTE activity, which is abrogated when that same sequence is inserted in reverse orientation within the indicator vector, as expected for a sequence acting at the RNA (i.e., plus strand) level.
FIG. 5.
Identification of a sequence with a CTE-like activity in the MusD 3′ untranslated region. (A) Rationale of the assay and structure of the reporter pDM128/PL vector. pDM128/PL contains a cat gene flanked by donor and acceptor splice sites (SD and SA) and placed under the control of a simian virus 40 promoter and polyadenylation signal (prom, pA). Sequences to be tested for CTE activity are inserted as indicated. The presence of a CTE should promote the export of unspliced RNA from the nucleus of cells transfected with the reporter plasmid, leading to a detectable CAT activity. The absence of CTE-like activity should lead to the export of spliced RNA and a lack of CAT activity. (B) 293T cells were transiently transfected with pDM128/PL vectors containing the indicated MusD fragments, placed in the forward or reverse orientation (termini are indicated by nucleotide positions in the MusD sequence, and orientation is indicated by arrows). At 48 h posttransfection, cells were lysed and CAT activity was determined by using [14C]chloramphenicol as described in Materials and Methods. The mean ratio of CAT activity between pDM128/PL containing a MusD (or control MPMV CTE) sequence versus the empty vector (no CTE) was calculated from two to four independent experiments.
DISCUSSION
Based on our previous identification of a proviral MusD copy functional for retrotransposition (30), we have now characterized the structure, life cycle, and evolutionary history of one of the most productive family of ERVs. The data strongly suggest a molecular scenario for the endogenization of this element, in which a very significant gain in retrotransposition efficiency is achieved via the “intracellularization” of the encoded viral particles, with the loss of the plasma membrane targeting signal within the matrix Gag protein resulting in retention of the particles within the cells and therefore enhancing insertional potency.
A first issue of the present study is the characterization of VLPs unambiguously associated with MusD, which are found exclusively within the cell and which disclose a change in morphology upon MusD protease-dependent maturation. Electron microscopy uncovers particles with a dense core corresponding to the mature particles, which originate from immature particles that are the only ones observed with a protease mutant. The occurrence of this maturation process is consistent with a Western blot analysis of cell lysates, disclosing cleavage of the Gag precursor (again not observed with a protease mutant), as well as with the functionality of the MusD retroelement, as previously demonstrated by monitoring its transpositional activity (30). The observed intracellular localization of the MusD particles is also consistent with coculture experiments indicating that transposition activity does not involve any extracellular intermediate. Finally, it is noteworthy that the currently unraveled structure of the MusD particles is not compatible with that suggested by a previous publication by Ristevski et al. (31), in which particles with a type D morphology, budding at the cell membrane, were observed by using a cell line derived from a thymic lymphoma. Although these cells indeed expressed MusD transcripts, it is likely that the observed particles were not related to MusD and possibly derived from contaminants—or recombinants—as often observed within cell lines.
A second important issue concerns the origin and molecular evolution of the MusD retroelement. Clearly, as illustrated by phylogenetic analyses based on the reverse transcriptase domain (1), the MusD element belongs to the Betaretrovirus genus, distinct from the retrotransposon branch. In this respect, and despite the absence of an env gene, it can be ruled out that MusD is a Ty1- or copia-like primitive retrotransposon (both of which never possessed an env gene). Most probably, MusD is the genomic trace of infection of a rodent ancestor by a bona fide retrovirus. This scheme is also consistent with the known entry date of MusD in the germ line of a Muridae, which actually is late in evolution, with elements found only in the Mus genus (i.e., <5 Myr [25, 35]). We were then able to unravel the likely steps that have led from a primitive infectious retrovirus to the present-day MusD elements. One of the major events in the endogenization process is the loss of a Gag-associated plasma membrane targeting signal. Indeed, although we have shown that MusD can assemble VLPs in the cytoplasm, these particles are not targeted to the cell membrane, nor do they bud out of the cells. Along this line, we have unambiguously demonstrated that this departure from the behavior of an infectious retrovirus is due to an alteration of the myristoylation signal and basic domain that are normally present at the N-terminal end of retroviral matrix Gag proteins. Actually, the MusD matrix sequence, compared to that of its closest relative, MPMV, only disclosed a few positive charges but no G residue after the initiation codon, therefore precluding Gag myristoylation. Indeed, Gag targeting requires a functional bipartite signal, with the myristate bound by acylation of the glycine residue conferring a hydrophobic pole to the Gag polyprotein and the positively charged residues interacting with the negatively charged phospholipids at the plasma membrane. The series of data presented in Fig. 2 clearly demonstrate that the alteration of the myristoylation and basic domain in MusD is entirely and solely responsible for the loss of cell membrane targeting of the viral particles. Indeed, replacement of a few N-terminal residues of the MusD matrix protein by those of the MPMV retrovirus led to the complete redirection of the particles toward the cell membrane, as illustrated by both immunofluorescence and electron microscopy analyses. We also show that the mere addition of a myristoylation signal at the appropriate position in MusD is not sufficient for the targeting effect, albeit our experiments using [3H]myristate demonstrate Gag myristoylation. Clearly, the basic residues of the bipartite signal are also stringently required. As expected, we also show that the “resuscitation” of an active plasma membrane targeting signal results in viral particles budding at the cell surface and released in the supernatant. Moreover, we demonstrate that these extracellular particles are functional since, when pseudotyped with a heterologous envelope protein, they become infectious. This strongly suggests that the MusD progenitor was most probably an infectious retrovirus, which then lost its membrane addressing signal, which de facto resulted in the complete “intracellularization” of the viral particles. This should have been a founding event in the generation of MusD, for reasons that most probably include (see below) an improved retrotransposition efficiency of the captured retrovirus. In addition to this founding molecular event, other alterations in the “progenitor” retrovirus have taken place. The most visible one is the loss of the env gene, with only a 1.4-kb noncoding sequence being left at the 3′ end of the MusD genome. Lack of an env gene is indeed fully consistent with the strictly intracellular life cycle of the MusD retroelement and most probably results from the absence of any selective pressure for its maintenance. However, a puzzling feature of the 3′ end of the MusD genome is that this domain has been conserved within the ETn retroelements—the noncoding transpositionally competent derivatives of MusD—and that deletion of this domain severely reduces MusD transposition activity. Actually, the data in the present study gave us a clue by showing that this domain contains a CTE-like activity like that observed in the 3′ region of many retroviral genomes. For the latter, this sequence promotes the nuclear export of unspliced, full-length viral genomic RNAs, allowing them to escape the splicing machinery and to be packaged within the newly assembled viral particles (18), whereas the spliced transcripts are essentially devoted to translation of the env gene. Although the presence of such a sequence within MusD is therefore fully consistent with its suggested viral origin, the situation remains paradoxical since there is no clear evidence for spliced transcripts encoded by MusD (actually needless after the loss of the env gene). A plausible interpretation is therefore that the identified CTE-like sequence has been conserved to promote high-levels of MusD transcripts and proteins for retrotransposition, which is also consistent with reports on noncanonical roles for CTE sequences not directly related to the preservation of full-length unspliced RNA but associated with translation enhancement (9, 12).
A final important issue of the present study concerns the outcome of the identified molecular events targeted to the MusD progenitor, in terms of “function.” In other words, what could have been the benefit for the MusD progenitor to become an “intracellularized” retroelement and not simply persist as an ERV, such as occurred, for instance, in the case of the human HERV-K elements, which still possess a functional env gene and can generate extracellular viral particles (13; reviewed in reference 2). Basically, an answer is suggested in Fig. 4, which unambiguously demonstrates that “intracellularization” of the MusD progenitor (via the loss of a functional membrane targeting signal [see above]) is associated with a dramatic increase (∼100-fold) in retrotransposition efficiency. This result can be simply accounted for by the retention of the MusD viral particles within the cells that makes them prone to direct reintegration—after reverse transcription—of their genome into the proper cell, precisely that from which they are issued, without any dispersion in the cell supernatant. This “physical” constraint, as similarly observed for classical bona fide retrotransposons such as the yeast Ty1 and Ty3 retrotransposons (reviewed in references 5, 33, and 39), is most probably the key feature that allowed this ancestral retrovirus to very efficiently transpose and consequently to maintain within the host genome, despite the genetic drift, the few copies still active that are found today. A similar strategy most probably also holds for the members of another very successful murine ERV family, the IAP elements, which also make particles that do not escape the cell and transpose at a very high rate (14, 22).
In summary, based on our experimental data, a model can be proposed (see the scheme in Fig. 6 and the accompanying legend) that accounts for the generation of the MusD retrotransposon from an infectious retroviral progenitor. The model includes “intracellularization” of the endogenized retrovirus, via alteration of its membrane-targeting signal and loss of its env gene rendered needless by the intracellular life cycle. It results in the generation of elements with high transpositional efficacy and their conservation in evolution.
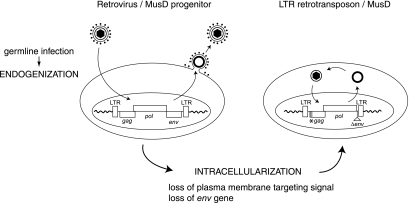
FIG. 6.
Model for the “intracellularization” of infectious retroviruses. A bona fide infectious retrovirus, e.g., the MusD progenitor, is endogenized upon infection of the germ line of a remote ancestor and Mendelian transmission to the following generations, thus resulting in a so-called ERV. This ERV may retain the characteristic features of retroviruses, i.e., produce infectious extracellular particles with a functional envelope protein, and be prone to horizontal transmission. Intracellularization is expected to correspond to an additional adaptation of the progenitor retrovirus, in which the produced VLPs are no longer able to exit or reenter the cell. For MusD, this has been achieved via the loss of the plasma membrane targeting function (degeneration of the myristoylation signal and basic domain at the N-terminal end of the matrix Gag protein) and the loss of the env gene. The resulting strictly intracellular life cycle resembles that of primitive Ty-like LTR-retrotransposons (although both classes of elements should be evolutionarily distinct) and is associated—at least in the case of the MusD element—with high-efficiency retrotransposition.
Acknowledgments
We thank Bryan R. Cullen for the pDM128 plasmids, A. Jalil for assistance with the confocal microscopy, E. Pichard for technical assistance, and C. Lavialle for critical reading of the manuscript.
This study was supported by the CNRS and by grants from the Ligue Nationale Contre le Cancer (Equipe Labellisée).
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Baillie, G. J., L. N. van de Lagemaat, C. Baust, and D. L. Mager. 2004. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J. Virol. 78:5784-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannert, N., and R. Kurth. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. USA 13:14572-14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. [DOI] [PubMed] [Google Scholar]
- 4.Baust, C., G. J. Baillie, and D. L. Mager. 2002. Insertional polymorphisms of ETn retrotransposons include a disruption of the wiz gene in C57BL/6 mice. Mamm. Genome 13:423-428. [DOI] [PubMed] [Google Scholar]
- 5.Beliakova-Bethell, N., C. Beckham, T. H. Giddings, Jr., M. Winey, R. Parker, and S. Sandmeyer. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieda, K., A. Hoffmann, and K. Boller. 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J. Gen. Virol. 82:591-596. [DOI] [PubMed] [Google Scholar]
- 7.Bogerd, H. P., A. Echarri, T. M. Ross, and B. R. Cullen. 1998. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J. Virol. 72:8627-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boller, K., H. König, M. Sauter, N. Mueller-Lantzsch, R. Löwer, J. Löwer, and R. Kurth. 1993. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 196:349-353. [DOI] [PubMed] [Google Scholar]
- 9.Bor, Y. C., J. Swartz, A. Morrison, D. Rekosh, M. Ladomery, and M. L. Hammarskjold. 2006. The Wilms' tumor 1 (WT1) gene (+KTS isoform) functions with a CTE to enhance translation from an unspliced RNA with a retained intron. Genes Dev. 20:1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conte, M. R., M. Klikova, E. Hunter, T. Ruml, and S. Matthews. 1997. The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus, and implications for the morphology of retroviral assembly. EMBO J. 16:5819-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coyle, J. H., B. W. Guzik, Y. C. Bor, L. Jin, L. Eisner-Smerage, S. J. Taylor, D. Rekosh, and M. L. Hammarskjold. 2003. Sam68 enhances the cytoplasmic utilization of intron-containing RNA and is functionally regulated by the nuclear kinase Sik/BRK. Mol. Cell. Biol. 23:92-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewannieux, M., S. Blaise, and T. Heidmann. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J. Virol. 79:15573-15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewannieux, M., A. Dupressoir, F. Harper, G. Pierron, and T. Heidmann. 2004. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat. Genet. 36:534-539. [DOI] [PubMed] [Google Scholar]
- 15.Dewannieux, M., F. Harper, A. Richaud, C. Letzelter, D. Ribet, G. Pierron, and T. Heidmann. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16:1548-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehrmann, F., M. Jung, R. Zimmermann, and H. G. Krausslich. 2003. Transport of the intracisternal A-type particle Gag polyprotein to the endoplasmic reticulum is mediated by the signal recognition particle. J. Virol. 77:6293-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fridell, R. A., K. M. Partin, S. Carpenter, and B. R. Cullen. 1993. Identification of the activation domain of equine infectious anemia virus rev. J. Virol. 67:7317-7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammarskjold, M. L. 2001. Constitutive transport element-mediated nuclear export. Curr. Top. Microbiol. Immunol. 259:77-93. [DOI] [PubMed] [Google Scholar]
- 19.Heidmann, O., and T. Heidmann. 1991. Retrotransposition of a mouse IAP sequence tagged with an indicator gene. Cell 64:159-170. [DOI] [PubMed] [Google Scholar]
- 20.Heidmann, T., O. Heidmann, and J. F. Nicolas. 1988. An indicator gene to demonstrate intracellular transposition of defective retroviruses. Proc. Natl. Acad. Sci. USA 85:2219-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hope, T. J., X. J. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuff, E. L., and K. K. Lueders. 1988. The intracisternal A-particle gene family: structure and functional aspects. Adv. Cancer Res. 51:183-276. [DOI] [PubMed] [Google Scholar]
- 23.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 24.Löwer, R., K. Boller, B. Hasenmaier, C. Korbmacher, N. Mueller-Lantzsch, J. Löwer, and R. Kurth. 1993. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc. Natl. Acad. Sci. USA 90:4480-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mager, D. L., and J. D. Freeman. 2000. Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences. J. Virol. 74:7221-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muster, T., A. Waltenberger, A. Grassauer, S. Hirschl, P. Caucig, I. Romirer, D. Fodinger, H. Seppele, O. Schanab, C. Magin-Lachmann, R. Lower, B. Jansen, H. Pehamberger, and K. Wolff. 2003. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 63:8735-8741. [PubMed] [Google Scholar]
- 27.Ostertag, E. M., and H. H. J. Kazazian. 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35:501-538. [DOI] [PubMed] [Google Scholar]
- 28.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhee, S. S., and E. Hunter. 1987. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J. Virol. 61:1045-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribet, D., M. Dewannieux, and T. Heidmann. 2004. An active murine transposon family pair: retrotransposition of “master” MusD copies and ETn trans-mobilization. Genome Res. 14:2261-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ristevski, S., D. F. Purcell, J. Marshall, D. Campagna, S. Nouri, S. P. Fenton, D. A. McPhee, and G. Kannourakis. 1999. Novel endogenous type D retroviral particles expressed at high levels in a SCID mouse thymic lymphoma. J. Virol. 73:4662-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saad, J. S., J. Miller, J. Tai, A. Kim, R. H. Ghanam, and M. F. Summers. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 103:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandmeyer, S. B., M. Aye, and T. Menees. 2002. Ty3, a position-specific, gypsy-like element in Saccharomyces cerevisiae, p. 663-683. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 34.Schultz, A. M., L. E. Henderson, and S. Oroszlan. 1988. Fatty acylation of proteins. Annu. Rev. Cell Biol. 4:611-647. [DOI] [PubMed] [Google Scholar]
- 35.Sonigo, P., S. Wain-Hobson, L. Bougueleret, P. Tiollais, F. Jacob, and P. Brûlet. 1987. Nucleotide sequence and evolution of ETn elements. Proc. Natl. Acad. Sci. USA 84:3768-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinhuber, S., M. Brack, G. Hunsmann, H. Schwelberger, M. P. Dierich, and W. Vogetseder. 1995. Distribution of human endogenous retrovirus HERV-K genomes in humans and different primates. Hum. Genet. 96:188-192. [DOI] [PubMed] [Google Scholar]
- 37.Turner, G., M. Barbulescu, M. Su, M. I. Jensen-Seaman, K. K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531-1535. [DOI] [PubMed] [Google Scholar]
- 38.Vogt, V. M. 1997. Retroviral virions and genome, p. 27-69. In J. M. Coffin, C. L. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, NY. [PubMed]
- 39.Voytas, D. F., and J. D. Boeke. 2002. Ty1 and Ty5 of Saccharomyces cerevisiae, p. 631-662. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 40.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, S. E. Antonarakis, J. Attwood, R. Baertsch, J. Bailey, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 41.Yuan, X., X. Yu, T. H. Lee, and M. Essex. 1993. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J. Virol. 67:6387-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]