Abstract
Many viruses (herpes simplex virus type 1, polyomavirus, and human immunodeficiency virus type 1) require the activation of ataxia telangiectasia mutated protein (ATM) and/or Mre11 for a fully permissive infection. However, the longer life cycle of human cytomegalovirus (HCMV) may require more specific interactions with the DNA repair machinery to maximize viral replication. A prototypical damage response to the double-stranded ends of the incoming linear viral DNA was not observed in fibroblasts at early times postinfection (p.i.). Apparently, a constant low level of phosphorylated ATM was enough to phosphorylate its downstream targets, p53 and Nbs1. p53 was the only cellular protein observed to relocate at early times, forming foci in infected cell nuclei between 3.5 and 5.5 h p.i. Approximately half of these foci localized with input viral DNA, and all localized with viral UL112/113 prereplication site foci. No other DNA repair proteins localized with the virus or prereplication foci in the first 24 h p.i. When viral replication began in earnest, between 24 and 48 h p.i., there were large increases in steady-state levels and phosphorylation of many proteins involved in the damage response, presumably triggered by ATM-Rad3-related kinase activation. However, a sieving process occurred in which only certain proteins were specifically sequestered into viral replication centers and others were particularly excluded. In contrast to other viruses, activation of a damage response is neither necessary nor detrimental to infection, as neither ATM nor Mre11 was required for full virus replication and production. Thus, by preventing simultaneous relocalization of all the necessary repair components to the replication centers, HCMV subverts full activation and completion of both double-stranded break and S-phase checkpoints that should arrest all replication within the cell and likely lead to apoptosis.
Human cytomegalovirus (HCMV) is the major viral cause of birth defects. Each year approximately 1 to 2% of all newborns are congenitally infected, and of these infants, 5 to 10% manifest signs of serious neurological defects (6, 8, 16, 31). HCMV infection is also a major medical problem in immunocompromised individuals (8).
Recent literature points to HCMV as a potential causative agent for the development of certain types of cancers including malignant gliomas, prostate carcinomas, and colorectal cancers (17, 35, 74). Studies of HCMV infection in nonpermissive rodent cells indicate that the virus can act as a mutagen (1, 5, 78). In fact, in permissive human cells expression of the immediate-early (IE) gene products by themselves can drive cells into S phase (57). This has led to the suggestion that the IE proteins of HCMV may cause “hit-and-run” mutagenesis. In addition, our earlier findings showed that 15% of fibroblasts infected in S phase were specifically broken in the long arm of chromosome 1 when analyzed during the subsequent mitosis (28). Due to the clinical ramifications of HCMV infection, the study of its interactions with the host cell, and particularly with the DNA damage machinery of the cell, is well warranted.
After penetration of the plasma membrane, components of the virion, including its 240-kb linear double-stranded DNA (dsDNA) genome (which consists of two unique coding sequences [UL and US] flanked by a series of inverted repeats [54]), are rapidly transported to the nucleus, where viral transcription and replication take place (54). A fraction of the input viral genomes are deposited at nuclear domain 10 (ND10) sites, and only these genomes express IE transcripts (37), illustrating the importance of this localization in the establishment of a lytic infection.
It has been shown that circularization of the input linear viral genome occurs at approximately 4 h postinfection (p.i.), indicating a requirement for the assistance of some viral and/or cellular proteins to accomplish this task (52). After circularization, herpesviral DNA replication is thought to proceed in a biphasic manner within virus-established replication centers in the cell nucleus. These centers progress from small foci (termed prereplication centers, visible between 12 and 24 h p.i.), to two larger foci located at the poles of the nucleus (48 h p.i.), and finally (during late stages of infection, after 72 h p.i.) to a single large focus, which covers the majority of the infected cell nucleus (66).
The initial phase of infection is characterized by origin-specific replication from the input circularized genome, which leads to single copies of the virus. Later during infection, a switch is made to a rolling-circle mechanism of replication, during which large concatemers of single-length units are formed. Homologous recombination occurs between concatemeric DNA, as evidenced by the observation of inversions of UL and US genome segments in successive monomeric units. Recombination also appears to occur between free cleaved monomers and the concatemeric structure, producing the branched intermediate structures observed at later stages of infection (see references in references 19, 76, and 85). It has been suggested that these recombinogenic structures may trigger a host cell DNA damage response during herpesviral replication (33, 43, 47, 80, 87, 92).
The eukaryotic cell has two double-strand break (DSB) repair systems. The first pathway is homologous recombination (HR), and the second is nonhomologous end joining (NHEJ) (as reviewed in reference 48). In all eukaryotic cells DSB sensing and cell cycle arrest are coupled to promote damage repair. The current understanding of this process in eukaryotes (as reviewed in reference 79) is that the Mre11-Rad50-Nbs1 (MRN) complex binds to the linear ends of the break, bringing them adjacent to each other, with each protein in the complex serving a distinct function (22, 36, 63, 88). The Nbs1 protein appears to be the glue in this complex. Once Nbs1 is bound to the break site, it attracts a dimeric form of the ataxia telangiectasia mutated protein (ATM), the key transducing molecule for the DSB response. ATM is subsequently autophosphorylated and becomes disassociated to an active monomeric form (3), which can then phosphorylate the histone variant H2AX (known as γH2AX) (64, 70), marking the location of damage. γH2AX is thought to act as an anchor point for the MRN complex as well as for several other protein sensors involved in damage signaling, namely, 53BP1 and Mdc1. The focus expands as additional MRN complex, 53BP1, and other signaling proteins are then recruited in a sequential fashion to the site. The sensor proteins Nbs1 and 53BP1 are also substrates for the activated phospho-ATM (pATM) kinase, as are the downstream targets p53 and Chk2. Through the activation of p53, cells in G1 can be arrested via p21 upregulation to allow repair, generally via NHEJ. In this case another trimolecular complex, known as the DNA-dependent protein kinase (DNA-PK) (82), must be attracted to the site of damage. If cells are arrested via a DSB response in S or G2/M, the HR machinery, including Rad51 and BRCA1 and -2, is more likely recruited to the site.
The second type of damage response that viral replication can trigger is related to stalled replication forks, the main transducer of which is the ATM-Rad3-related kinase (ATR) (60, 77, 95). Sensing the presence of single-stranded DNA coated with replication protein A (RPA), the small binding partner of ATR, ATRIP, helps to localize monomeric ATR to the site of damage. Further interactions with Rad9, Rad1, and Hus1 (the 9-1-1 complex) serve to fully activate ATR, which can arrest the cell by activation of its downstream effectors, p53 and Chk1.
Elevated steady-state levels of p53 are observed in many HCMV-infected cell types by 24 h p.i. (39, 42, 49, 56, 83). However, p53 cellular targets are not activated after infection (7, 39). We have proposed that sequestration of p53 into viral replication centers, which occurs relatively early (29), provides a mechanism for silencing p53 activity during infection. We have also shown that the transactivation functions of p53 may be coopted by the virus (73) and that infection in the absence of p53 delays and reduces viral yields (11).
It is clear from the literature that viral interaction with the host cell, whether upon initial entry or through successive rounds of replication, often triggers a DNA damage response. In the case of adenoviral infection, this response can be detrimental to virus replication and is dampened by virus-mediated degradation of the MRN components (84). In the case of herpes simplex virus (HSV) type 1, an ATM-mediated damage response is evoked as soon as prereplication centers are formed (47, 80). This response appears to be essential for productive replication of the virus, as viral titers are decreased in both ATM− and Mre11− cells (47). Polyomavirus also appears to require an ATM-generated S-phase arrest in order to replicate at its full capacity (20), and there are reports that human immunodeficiency virus integration may depend upon the activation of ATM and/or ATR within the infected cell (21, 45).
We have undertaken a thorough investigation of the response of the permissively infected fibroblast to HCMV infection. We have analyzed the interaction of the virus from the time of entry and viral deposition, through establishment of replication centers, and into late stages of infection. The novel technique of labeling viral DNA (72) to track input virus was used to visualize input viral DNA and colocalized cellular proteins, which may be aiding in circularization and/or involved in the DSB damage response. Our results indicate that after HCMV genome deposition and again after commencement of replication in permissive human foreskin fibroblasts (HFFs), damage responses are initiated. However, the virus is able to subvert these responses by thwarting relocalization of the necessary components sufficient to cause cell cycle arrest. In this way, optimal viral replication conditions are obtained.
MATERIALS AND METHODS
Cells and cell culture.
Primary HFFs and T98G (human glioblastoma cell line, ATCC CRL-1690) cells were grown in Eagle's minimal essential medium (Gibco BRL) supplemented with 10% fetal bovine serum, penicillin (200 U/ml), streptomycin (200 μg/ml), l-glutamine (2 mM), and amphotericin B (Fungizone; 1.5 μg/ml). The ATM− cell line GM02530 (Coriell Institute for Medical Research) was grown in minimal essential medium supplemented with 15% fetal bovine serum and l-glutamine (2 mM). A-TLD1 (Mre11−) and A-TLD1/Mre11 (reconstituted with Mre11) cells (generous gifts from Caroline Lilley, Salk Institute of Biological Research) were cultured as previously described (47), as were the BT leukemia cells (a generous gift from Susan Lees-Miller, University of Calgary) (44). All of the cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Drug treatments.
BT cells, used as a positive control for immunoblotting, were treated with doxorubicin (1 μM) for 2 h to induce a DSB response and then harvested for cell lysate preparation (44). For immunofluorescence positive controls, doxorubicin (10 μM) was added for 1 h to HFF cells growing asynchronously. The cells were then washed with phosphate-buffered saline (PBS), and fresh medium was added for 4 h, after which cells were harvested, washed, extracted, and fixed as described below.
Virus infection.
Cells were synchronized in G0 (by either confluence arrest or serum starvation) so as to provide for synchronous infection as has been described previously (11, 29) and as judged by nuclear staining of pp65 and IE1 in >90% of cells by 12 h p.i. Both methods of synchronization gave essentially identical results. Cells were trypsinized and replated to 100-mm dishes at a density of 5 × 105 cells/dish. After attachment (for 1 to 2 h), the cells were infected with HCMV (Towne strain ATCC VR977) at a multiplicity of infection (MOI) of 5 unless otherwise noted. The cells were then incubated with virus for 1 h to allow for adsorption, after which the inoculum was removed and cells were refed with growth medium. Cells were harvested at the indicated h postinfection. Bromodeoxyuridine-labeled HCMV (BrdU-HCMV) was prepared and detected as described previously (72).
Immunoblotting.
For immunoblot analysis, infected cells were harvested at 4, 8, 12, 24, 48, 72, and 96 h p.i. as previously described (73), with the following differences: cell pellets were resuspended in RIPA buffer (50 mM Tris, pH 7.5, 1 mM EDTA, 0.5% NP-40, 10% glycerol, 150 mM NaCl, 2 mM dithiothreitol) containing a protease and phosphatase inhibitor cocktail (20 mM NaF; 1 mM NaVO3; and 8 mM β-glycerophosphate, aprotinin, and leupeptin [2 μg/ml each]), sonicated, and spun to clear debris, and then an equal volume of 2× sample buffer (4% sodium dodecyl sulfate, 20% glycerol, 100 mM Tris-HCl, pH 6.8, 10% β-mercaptoethanol) was added. Equivalent amounts of cell lysate (2 × 105 cells) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently transferred to PROTRAN membranes (Schleicher & Schuell). The membranes were then incubated with antibody (Ab) for protein detection as previously described (11).
Immunofluorescence analysis (IFA).
T98G, ATM−, or HFF cells arrested in G0 were trypsinized and replated to 100-mm dishes (5 × 105 cells/dish) containing glass coverslips (poly-l-lysine coated for T98G). After infection the coverslips were harvested at 4.5 to 6 and 48 h p.i. and rinsed twice in PBS. Coverslips were fixed in methanol for use with the γH2AX, Chk2, Ku70/80 (at 48 h p.i.), and DNA-PKcs (at 48 h p.i.) Abs. All other coverslips were extracted first in a CSK buffer solution {10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 100 mM NaCl, 300 mM sucrose, and 3 mM MgCl2} containing 0.5% Triton X-100 (50). Cells were then rinsed in CSK twice and fixed with 3% formaldehyde in PBS (with 0.5 mM MgCl2 and 0.5 mM CaCl2) for 10 min. Incubation of coverslips with Abs was carried out as described previously (73). The images were obtained by using a Nikon Eclipse E800 fluorescence microscope equipped with a Nikon DXM camera and Metavue software. Early colocalization studies were confirmed using a Zeiss LSM5 confocal microscope equipped with one argon and two HeNe lasers.
Antibodies.
Primary Abs against DNA damage response proteins were as follows: mouse monoclonal anti-Rad50 (clone 13B3) and anti-ATR (clone 2B5) Abs and rabbit polyclonal anti-Nbs1, anti-pNbs1 (Ser343), and anti-Mre11 Abs (Novus Biologicals); anti-ATM rabbit polyclonal, anti-pATM (Ser1981) mouse monoclonal (clone 10H11.E12), and anti-53BP1 mouse monoclonal (clone 19) Abs (BD Bioscience, Pharmingen); anti-pp53 (Ser15), anti-pChk2 (Tyr68), anti-p53BP1 (Ser25/29), and anti-pChk1 (Ser317) rabbit polyclonal Abs and anti-pp53 mouse monoclonal Ab (clone 16G8) (Cell Signaling Technology); anti-γH2AX (Ser139) polyclonal Ab (Upstate Biologicals); anti-Chk2 (clone 7), anti-p53 (clone DO-1), anti-DNA-PKcs (clone 25-4), anti-Ku80 (clone 111), and anti-Ku70 (clone N3H10) mouse monoclonal Abs (NeoMarkers); anti-ATRIP and anti-Chk1 rabbit Abs (Abcam). Primary Abs against viral proteins were as follows: anti-IE1 and anti-IE2 monoclonal Abs (kind gifts from Bill Britt, University of Alabama, Birmingham), anti-UL44 monoclonal Ab (Rumbaugh-Goodwin Institute for Cancer Research), and UL112/113 monoclonal Ab (a kind gift from Masaki Shirakata, Japan). Rat anti-BrdU monoclonal Ab (Harlan Sera-Lab) was used to detect labeled viral particles as described previously (72). Secondary Abs used were as follows: for immunoblot detection, horseradish peroxidase-linked sheep anti-mouse and donkey anti-rabbit secondary Abs (Amersham Bioscience); for IFA detection, tetramethyl rhodamine isothiocyanate-conjugated anti-rat and anti-mouse immunoglobulin G1 (IgG1), IgG2a, and IgG2b (Jackson ImmunoResearch Laboratories) and Alexa Fluor 488-conjugated goat anti-mouse IgG1, IgG2a, and IgG3 (Molecular Probes).
Virus titration.
Cells were seeded and infected at an MOI of 5 as described above. During each time course, duplicate infections were performed for each cell type and each cell type was tested at least twice. At the indicated times p.i., a small aliquot (150 μl) of supernatant was harvested from each dish and stored at −80°C (with 1% dimethyl sulfoxide). Virus titers were then determined on HFFs using standard techniques. Plaques were counted at days 7 and 9 postplating, with multiple wells seeded for each dilution in the series, so that an average could be obtained for that dilution. The numbers for the duplicate infections within the experiment were then compared, and an average titer was derived. In ATM− cells low-MOI infections (0.25) were also performed, with aliquots removed between days 1 and 11 p.i. and titers determined as described above.
RESULTS
No relocalization to the site of viral deposition was observed in HFF cells for the double-strand break-sensing complex MRN at early times p.i.
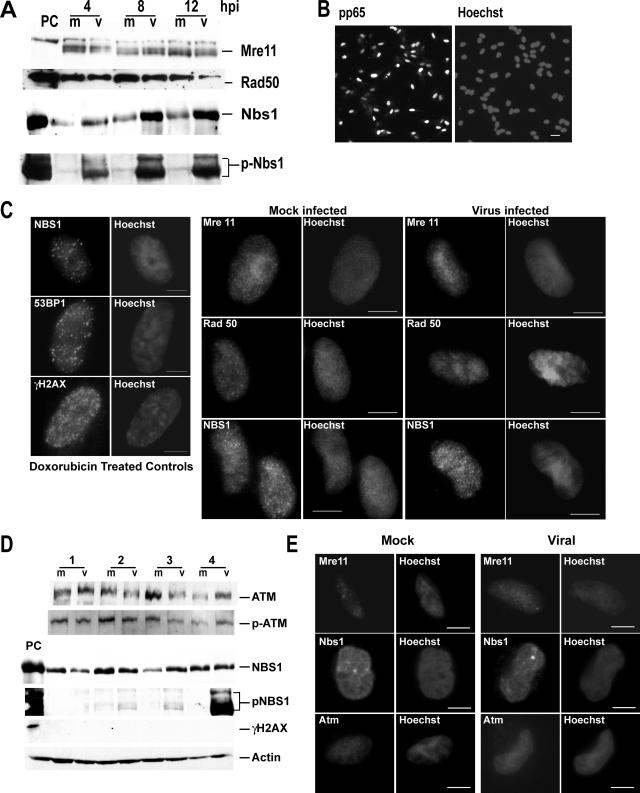
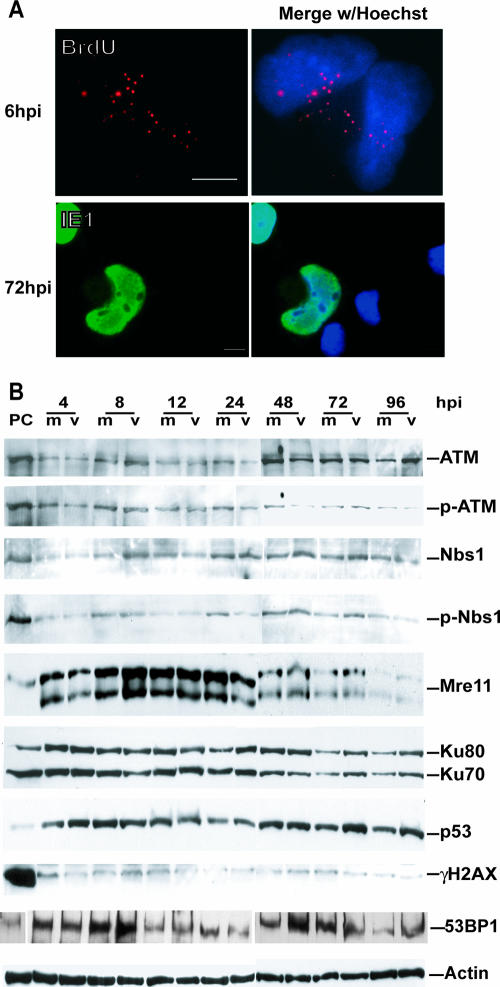
HCMV has a large linear dsDNA genome. In theory, once this genome enters a cell's nucleus its ends should be recognized as dsDNA breaks by the cell's damage machinery and trigger a DSB response. The MRN complex is essential in the DSB response as a primary sensor. To assess the activation of the DSB response pathway, G0-synchronized HFF cells were mock or virus infected with HCMV at an MOI of 5, and cells were harvested at the times indicated (between 0 and 12 h p.i.). We first assessed the steady-state levels of the MRN complex by immunoblotting (Fig. 1A). Mre11 was detected as a doublet, while Rad50 and Nbs1 were distinct single bands. There were no appreciable differences in protein levels between the virus- and mock-infected groups during early times p.i. (4 to 12 h p.i.) for either Mre11 or Rad50. However in virus-infected cells both total Nbs1 and pNbs1 (Ser343) levels increased during early times p.i. (Fig. 1A). This Nbs1 phosphorylation is typical of a DSB response, as can be viewed in the BT positive controls that have been treated with doxorubicin for 2 h to induce such a response (Fig. 1A) (44). This phosphorylation is generally carried out by an activated form of the transducing kinase, ATM (see below).
FIG. 1.
Activation of the MRN DSB-sensing complex is incomplete after HCMV infection. (A) Immunoblot analysis of the MRN complex of proteins at 4 to 12 h p.i. For all figures, PC indicates positive-control BT cells treated with doxorubicin to induce a DSB response, m indicates mock infected, and v indicates virus infected. (B) IFA localization of viral protein pp65 in the nucleus of infected cells to show synchrony of infection. Variation in pp65 levels is due to the harsh CSK extraction procedures used for all experiments (see Materials and Methods). (C) Left panels: IFA localization of DSB-sensing proteins after doxorubicin treatment (10 μM for 1 h and then 4 h of recovery) to provide a positive control for focus formation. Protein localization is on the left; nuclei stained with Hoechst stain are on the right. Right panels: IFA localization of the MRN proteins 4 to 6 h p.i. in either mock (left two columns)- or virus (right two columns)-infected cells. (D) Immunoblot analysis of cells between 1 and 4 h p.i. shows that the only change in these proteins is the phosphorylation of Nbs1 at 4 h p.i. An actin loading control is included. (E) IFA localization of the MRN complex proteins Mre11 and Nbs1 and the ATM kinase at 1 h p.i. No change in localization was observed for any of these proteins between 1 and 4 h p.i. Abs and fixation for all figures are as described in Materials and Methods. Bars for all panels, 5 μm.
Phosphorylation of Nbs1 indicated a possible initiation of a DSB response, so we looked for relocalization of the MRN complex to the site of damage within the nucleus, as is required for a functional response (59). IFA was used to detect localization of the MRN complex after 4 to 6 h p.i. All infections were performed at a high MOI to ensure that >95% of cells would be positive for the tegument protein pp65 within the nucleus after 4 to 6 h p.i. A low-magnification example of this staining is provided in Fig. 1B. In order to better visualize protein focus formation, cells were extracted with CSK buffer containing Triton X-100 prior to fixation, as has been previously described (50). This extraction removes proteins that are not attached to the chromatin or scaffolding substructure of the nucleus and thus provides a clearer view of protein relocalization within this compartment.
We treated HFFs with doxorubicin to induce DSBs and provide a model of DSB damage protein relocalization (see Materials and Methods). Figure 1C (left panels) illustrates relocalization of Nbs1, as well as the additional sensing proteins 53BP1 and γH2AX, into the prototypical foci induced by doxorubicin treatment. No such relocalization was observed when dimethyl sulfoxide-alone control cells were examined (data not shown). As can be observed in Fig. 1C (right panels), all components of the MRN complex remained in the nucleus of infected cells and showed no change in localization from mock-infected cells at these early times p.i. Occasionally, small specks were observed in the nuclei of both mock- and virus-infected cells, particularly when Nbs1 localization was examined. However, these patterns were essentially identical for both mock and viral samples and were in agreement with what has been reported for unirradiated/uninfected cells previously (50, 53).
By 4 h p.i. ample time had elapsed for relocalization to occur; in fact, by 1 h p.i. the viral genome had been deposited into the nucleus of infected cells (72). We therefore looked for foci at earlier times p.i. (1 to 4 h p.i.) and saw no relocalization occurring, nor did we see changes in protein levels in the sensing molecules or several other proteins (Fig. 1D and E show examples). No relocalization was seen using an even higher virus input (MOI of 15) and harsher extraction conditions (53) (not shown). It appears that the main DSB damage-sensing complex (MRN) does not detect input viral genomes at early times p.i. as measured by relocalization to the site of deposition.
Although other sensing molecules formed foci, they did not appear to colocalize with input viral genomes at early times p.i.
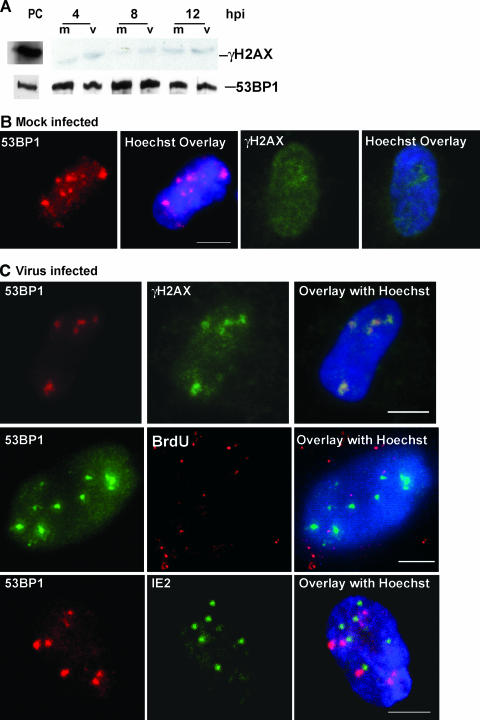
As there was evidence for active ATM within the virus-infected cells, we also investigated two other proteins involved in DSB sensing, 53BP1 and the histone variant H2AX, to look for changes in steady-state protein levels and localization. The histone variant H2AX comprises 2 to 20% of the H2A pool. It is a target of the ATM signaling kinase, and its phosphorylated form (γH2AX) marks the chromatin on either side of a DSB in a region that can span up to 2 megabases (26). γH2AX interacts with Nbs1 in the MRN complex forming nuclear foci at DNA damage sites (41) and is believed to act as a nucleator/scaffolding protein for several other DSB-sensing molecules, particularly at low levels of DSB development (12, 25). It has been shown that even a single DSB can cause the induction of γH2AX foci (as reviewed in reference 26). Generally, high levels of γH2AX can be detected via immunoblotting upon its activation (71). Figure 2A shows that although γH2AX was present at low levels, the steady-state level of γH2AX did not increase above that in mock-infected cells in the first 12 h p.i. (also Fig. 1D).
FIG. 2.
Localization of γH2AX and 53BP1 into foci juxtaposed to input viral DNA at early times p.i. (A) Immunoblot analysis of γH2AX and 53BP1 at early times p.i. PCs (see Fig. 1 legend for definition) for these two blots were run on parallel gels. (B) IFA localization of γH2AX and 53BP1 in mock-infected cells between 4 and 6 h p.i. (C) IFA localization of γH2AX and 53BP1 in virus-infected cells at 4 to 6 h p.i. The first row shows γH2AX and 53BP1 colocalization during this time period. The second and third rows show the lack of colocalization of 53BP1 with either BrdU-labeled viral DNA (second row) or the viral IE2 antigen (third row).
53BP1 is another sensor of DSBs thought to relocalize quickly to the site of damage and be anchored there via binding to γH2AX (25, 91). This protein can bind single-strand breaks, DSBs, and chromatin in vitro (38) and is a substrate for ATM. In the absence of 53BP1, some downstream targets of ATM are not phosphorylated, indicating the importance of this sensor for proper repair (4). The relatively high basal levels of 53BP1 did not deviate from those observed in mock-infected cells at early times p.i. (Fig. 2A). We also checked for phosphorylation of 53BP1 on Ser25/29 and saw no changes in these levels in virus-infected cells compared to mock-infected cells (data not shown).
We then used IFA to examine the localization of these two proteins early in infection (Fig. 2C). Even though only low levels of γH2AX were present in infected cells, this protein became transiently relocalized into diffuse foci between 4 and 6 h p.i. The quantity and size of γH2AX foci increased during this period until over 50% of the infected cells had detectable foci. Although γH2AX did form foci in mock cells, they were usually only single or double foci and very faint (Fig. 2B), as opposed to the infected cells' γH2AX foci, which were larger, more numerous, and brighter (Fig. 2C).
IFA of 53BP1 at early times showed no obvious differences between the mock-infected and virus-infected cells, with both groups displaying a wide range of staining patterns, from diffuse nuclear to multiple large foci in the nucleus (compare Fig. 2B and C) (2, 68). Foci of 53BP1 have been reported previously in the literature in unirradiated cells, with the ratio of large to small foci changing as cells cycle from G1 to S phase (2, 68). Of particular note is that when virus-infected cells displayed large, diffuse γH2AX foci, they were always colocalized with 53BP1 foci.
The IFA staining suggested that at least a subset of γH2AX might be localized near viral genome deposition sites. Unfortunately, we have not been successful in staining γH2AX with the viral genome. We surmise that the acid treatment required for the exposure of BrdU residues may displace γH2AX from the DNA or alter the epitope required for Ab binding. However, costaining of 53BP1 with the input genomes (labeled with BrdU and detected using anti-BrdU Ab [72]) indicated that this protein did not colocalize with viral genomes but was rather juxtaposed to a subset (Fig. 2C). In addition, costaining of 53BP1 with other viral proteins involved in establishing the prereplication centers (IE2 and UL112/113) showed no colocalization with these proteins either (Fig. 2C and data not shown). 53BP1 was juxtaposed to these viral proteins (and the input viral genome) when they were present. Similar staining for juxtaposition of γH2AX and IE2 was observed (not shown). In sum, no direct relocalization to the sites of viral deposition was observed for any of the DSB-sensing machinery within the first 6 h p.i.
No changes in phosphorylation status or localization of the main transducer of the DSB response, the ATM signaling kinase, were observed in the early times of HCMV infection in HFFs.
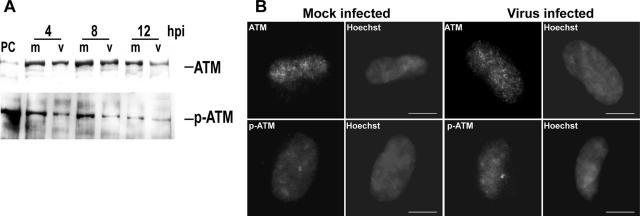
Although the Nbs1 protein of the MRN complex was phosphorylated quickly after infection, we observed no distinct phosphorylation of the ATM signaling kinase at early times p.i. (Fig. 3A). During the prototypical DSB response, ATM is recruited to the sites of DSB by binding to the MRN complex and is then activated through autophosphorylation on serine 1981 (3, 14, 24, 93). Figure 3A shows that a clear basal level of pATM could be detected, but there was no increase in this level, or of total ATM levels, in the virus-infected cells at early times p.i. (also Fig. 1D). This is in contrast to what is observed during HSV infection, where large increases in pATM (Ser1981) levels are observed very early p.i. (47). Also unlike HSV, during HCMV infection, no relocalization into foci was observed for either total ATM or pATM compared to mock-infected cells at these early times (Fig. 3B and Fig. 1E).
FIG. 3.
ATM is not phosphorylated in response to viral genome deposition. Shown are immunoblot analyses (A) and IFAs (B) of both total and phosphorylated levels of the transducing kinase, ATM, at early times p.i. Abbreviations are defined in the Fig. 1 legend.
One downstream effector of ATM was activated by viral infection at early times p.i. in HFFs.
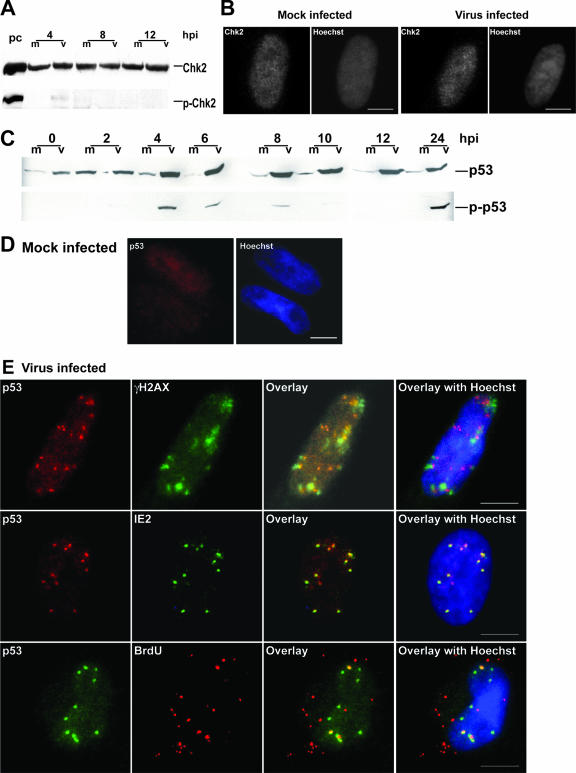
Since the immunoblotting results revealed a basal level of pATM that could be active in these cells, we investigated the activation of downstream effector molecules in the DSB response pathway in virus-infected cells. The checkpoint kinase Chk2 is a critical component of the signaling pathway following ATM activation. Steady-state protein levels of Chk2 and pChk2 showed no changes from those of mock-infected cells at early times p.i. (Fig. 4A), and no foci were observed when a total Chk2 Ab was used to localize the protein (Fig. 4B).
FIG. 4.
p53, the downstream target of ATM, is activated and does relocalize to the sites of viral deposition. (A) Immunoblot analysis of both total and phosphorylated levels of the checkpoint kinase Chk2 at early times p.i. (B) IFA localization of total Chk2 in mock- and virus-infected cells between 4 and 6 h p.i. (C) Immunoblot analysis of total p53 and pp53 for the first 24 h p.i., showing dramatic increases in steady-state levels of both forms of the protein. (D) IFA of p53 in mock-infected cells. (E) IFA localization of p53 with γH2AX (first row, no colocalization), viral protein IE2 (second row, complete colocalization), and BrdU-labeled virus (third row, partial colocalization) between 4 and 6 h p.i. For definitions of abbreviations, see the Fig. 1 legend.
In dramatic contrast, total steady-state levels of the cellular transcription factor p53 were rapidly increased by 4 h p.i. and remained high throughout the entire infection (Fig. 4C; see also Fig. 6D). Immunoblot analysis also revealed two distinct Ser15 phosphorylation events of the p53 protein, the first wave occurring between 4 and 8 h p.i. (Fig. 4C) and the second starting between 18 and 24 h p.i. (data not shown).
FIG. 6.
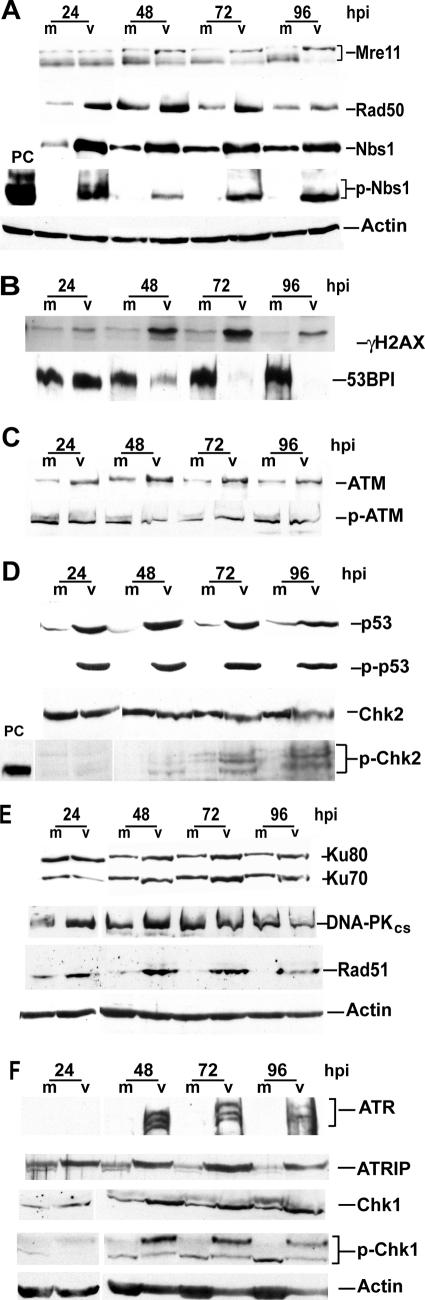
Changes to steady-state levels and phosphorylation states of several DNA repair proteins are observed at late times p.i. Shown are results of immunoblot analyses for the MRN complex (A), the additional DSB-sensing proteins γH2AX and 53BP1 (B), the transducing kinase ATM (C), ATM downstream targets p53 and Chk2 (D), NHEJ and HR machinery proteins (E), and the stalled replication fork response proteins ATR, ATRIP, and Chk1 (F). Actin loading controls are included with several panels. For definitions of abbreviations, see the Fig. 1 legend.
IFA localization of p53 also displayed interesting results. p53 formed foci similar to those of γH2AX very early p.i., between 3.5 and 5.5 h p.i. (Fig. 4E). These p53 foci were observed in approximately 30% of the infected cells at all times between 3.5 and 5.5 h p.i. and were not observed in mock-infected cells (Fig. 4D). Staining for pp53 revealed that this form was also localized within these foci (data not shown). It is important that γH2AX and p53 foci did not colocalize but instead were juxtaposed in these cells (Fig. 4E). Further examination of the localization of p53 revealed complete colocalization with two viral proteins, the IE2 transactivator (Fig. 4E) and UL112/113 (not shown). IE2 and UL112/113 are believed to mark the sites of prereplication centers during these early times p.i., as ICP8 does during an HSV infection (66, 87, 92). Infection with BrdU-labeled virus (72) allowed us to visualize p53 and the labeled genome in relation to each other. In cells exhibiting the p53 punctate staining pattern, the protein colocalized with approximately half the input viral genomes (Fig. 4E). IE2 also colocalized with approximately half the genomes (data not shown). Thus, of the DSB pathway proteins, only p53 appeared to be specifically relocalized to the site of viral deposition at these early times p.i.
An assessment of the DSB repair machinery at early times p.i. revealed no changes.
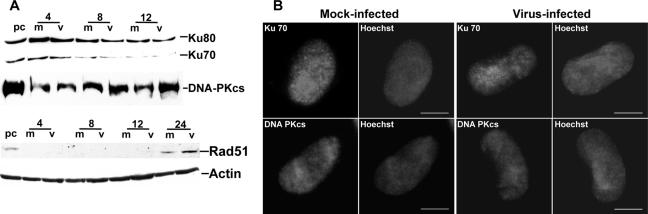
There are two evolutionarily conserved pathways, NHEJ and HR, for DSB repair. The NHEJ machinery was not activated in the early times of HCMV infection, with no differences in steady-state levels of any of the three components, Ku70, Ku80, and DNA-PKcs, compared to the mock-infected cells (Fig. 5A). IFA of the three proteins (Ku70 and DNA-PKcs shown) at early times revealed no change in the diffuse nuclear staining patterns observed in both infected and mock-infected cells (Fig. 5B). The HR machinery also does not appear to change in the early times of HCMV infection (4 to 12 h p.i.) as judged by the absence of an increase from the below-detectable steady-state levels of the crucial protein, Rad51, in a comparison of mock-infected to virus-infected samples (Fig. 5A). This is in marked contrast to what is observed at late times p.i. (as seen at 24 h p.i. in Fig. 5A and in Fig. 6E). IFAs were uninformative for Rad51 at these early times p.i. as no or very low protein levels were present. This very low level of protein is consistent with earlier reports of undetectable levels of protein in G1 phase of the cell cycle, where our cells reside at this point p.i. (27, 53). Taken together, our results analyzing early times p.i. indicate that although initiated, a prototypical and functional DSB damage response is not completed in response to the input viral genomes within the nucleus.
FIG. 5.
No changes to the NHEJ or HR proteins are observed at early times p.i. (A) Immunoblot analysis of the components of the DNA-PK complex and the HR protein Rad51 at early times p.i. An actin loading control is included. The 24-h-p.i. time point is included with the Rad51 blot to show increases that occur later in infection. For definitions of abbreviations, see the Fig. 1 legend. (B) IFA localization of the DNA-PK complex proteins Ku70 and DNA-PKcs between 4 and 6 h p.i. in either mock (left two columns)- or virus (right two columns)-infected cells.
Steady-state-level changes occurred in many of the DSB DNA damage repair proteins in the latter stages of HCMV infection of HFFs.
To investigate if the DSB DNA damage repair proteins were stabilized, activated, and involved in the later stages of HCMV infection, we performed immunoblot analysis of HFFs between 24 and 96 h p.i. (Fig. 6). Analysis of the MRN sensor complex revealed substantial changes to all three proteins. Total Nbs1 and pNbs1 levels remained elevated, although there was a small decrease in the second beginning at 48 h p.i. (Fig. 6A). In addition, Rad50 levels increased between 24 and 72 h p.i. and a shift to the slower-migrating form of Mre11 was observed in virus-infected cells beginning at approximately 48 h p.i. This may be a phosphorylation event of Mre11, as has been reported elsewhere (13, 69).
When the additional sensor proteins γH2AX and 53BP1 were examined, significant effects were observed (Fig. 6B). The level of γH2AX showed increases beginning at 24 h p.i. In contrast, the high basal level of 53BP1 in the virus-infected cells began to decline at 48 h p.i. and by 72 h p.i. was completely absent. Levels of p53BP1 declined as well, and a distinct band was no longer present after 48 h p.i. (data not shown).
Steady-state levels of the ATM transducing protein rose slightly above the basal levels seen in mock-infected cells beginning at 24 h p.i. (Fig. 6C), but this change was not accompanied by any distinguishable increase above the basal level of pATM within infected cells. The downstream effector of ATM, Chk2, showed no distinct changes in total protein levels but did show an increase in the pChk2 (Thr68) level beginning between 48 and 72 h p.i. (Fig. 6D). One of these phosphorylated forms of the protein migrated more slowly than pChk2 in our control doxorubicin-treated cells (which may indicate additional phosphorylated residues within the protein). As was illustrated earlier, total p53 had rapidly increased at 4 h p.i. and remained high throughout the rest of the infection, as did the Ser15-phosphorylated form of the protein (Fig. 6D).
In examining the DSB machinery (Fig. 6E), minor increases in both NHEJ proteins, Ku70 and Ku80, were seen beginning at about 48 h p.i. DNA-PKcs protein levels did not change until 96 h p.i., when a small decrease was observed. This last change in DNA-PKcs was in contrast to HSV infection, where the protein is rapidly degraded in some cell types (46, 62). Rad51, a component of the HR machinery, had substantially increased steady-state levels at all late time points while the levels in the mock-infected cells remained essentially undetectable (except at 24 h p.i., where levels showed the indicative increase of wild-type [wt] cells moving into S phase [27]).
During late time points replication of the virus converts to a rolling-circle mechanism, potentially leading to branched intermediates that could activate a stalled replication fork response within the cell. We therefore examined some of the components of this response at these later times p.i. (Fig. 6F). The main transducing kinase for this type of damage response is ATR, which, in conjunction with its binding partner, ATRIP, relocalizes to the site of damage. ATRIP is attracted to the site by an excess of RPA bound to single-stranded DNA at the fork (60, 77, 95). Activated ATR then acts through its effector molecules, p53 and Chk1, to arrest the cell so that repair can occur. The steady-state level of ATR increased at 48 h p.i., accompanied by a shift in mobility (Fig. 6F). This shift may be due to phosphorylation, although levels of pATR (Ser428) did not appear to increase during infection (data not shown). Levels of ATR's binding partner, ATRIP, also increased in a coincident manner. The small shift in mobility of this protein indicated that an activating phosphorylation event had taken place (18). Total levels of the downstream kinase Chk1 also showed variation, and the protein showed an additional slower-migrating form phosphorylated on Ser317 beginning at 48 h p.i., indicating an initial activation of this pathway.
Association of the DNA damage proteins with HCMV replication centers.
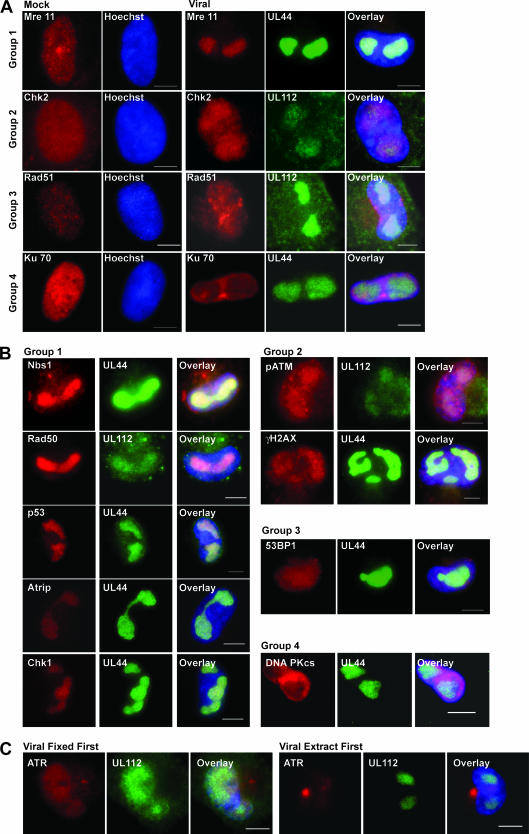
As there were changes in levels and phosphorylation patterns of many of the proteins involved in both the DSB and stalled replication fork responses to damage at later times p.i., we next examined where these proteins were located. We examined the infected cell nucleus to ascertain these proteins' proximity to the replicating viral genomes. Arrest of viral replication should occur only if these cellular proteins were localized with the virus. Therefore, association of DNA damage proteins with HCMV viral replication centers was investigated at 48 h p.i., when replication centers were clearly formed and visible. Except where noted in Materials and Methods, extraction in CSK buffer procedures was utilized to clearly visualize the replication centers and proteins localized within them. Viral replication centers were visualized in all cells by costaining with either UL44 (the viral processivity factor) or UL112/113, which is essential for viral replication and localized within the replication centers (29, 61, 66). In general, we categorized the localization of DSB and stalled replication fork repair proteins within the replication centers into four groups. Mock and viral staining patterns of one member from each group are shown in Fig. 7A. Localization of all other proteins tested can be seen in Fig. 7B (virus-infected cells only). The groups were as follows. Group 1 consisted of strongly sequestered proteins (p53, Nbs1, Mre11, Rad50, ATRIP, and Chk1); this category was defined by clear, strong staining within the replication centers and complete colocalization with the viral protein control. All components of the MRN complex were sequestered into the viral replication centers as were ATRIP and Chk1. Group 2 consisted of weakly sequestered proteins (ATM, pATM, Chk2, and γH2AX) that were all localized within the nucleus, with only partial (weak) sequestration into the viral replication centers. Group 3 showed diffused localization (Rad51 and 53BP1); these proteins showed no specific relocalization into the replication centers but were diffused throughout the nucleus. In group 4, proteins (Ku70 and Ku80, DNA-PKcs, and ATR) were specifically excluded from the viral replication centers although some did stain the rest of the nucleoplasm at late times (as illustrated by Ku70 colocalized with UL44 [Fig. 7A]). Although ATR exhibited a diffuse staining throughout the nucleus if fixation was performed prior to extraction (as seen for Rad51 in Fig. 7A), when extraction was performed first, staining within the nucleus almost completely disappeared (Fig. 7C), indicating a very loose association with the nuclear compartment. Late-stage staining indicated that there was no specific localization of the entire pathway of proteins for either DSB or stalled replication fork distress within the viral replication centers, rather only a weak relocalization and sometimes a complete exclusion of many of the components. This is a likely explanation for the lack of a functional damage response in these cells.
FIG. 7.
Localization of the damage response proteins varies at late times p.i. (A) IFA of mock- and virus-infected cells for several proteins at late times p.i. One example from each of groups 1 to 4 as defined in the text is shown. Virus groupings include localization of either viral protein UL44 or UL112 in green to visualize the viral replication centers. (B) IFA in virus-infected cells for all other proteins listed in the text but not shown in panel A. Proteins are arranged according to their inclusion in groups 1 to 4. (C) IFA for ATR protein in virus-infected cells using fixation-first (left three panels) and extraction-first (right three panels) conditions. Hoechst stain is used for all cells to visualize the nucleus. Bar, 5 μm.
DNA damage proteins were not activated by HCMV infection in T98G cells.
T98G cells are glioblastoma cells that harbor a mutant p53 protein (89). This cell line is semipermissive for HCMV infection (40), as would be expected given our previous results showing that p53 plays an important role in HCMV replication (11). Despite their mutagenic profile, T98G cells are capable of mounting a DSB damage response (81). G0-arrested T98G cells infected at an MOI of 5 with BrdU-labeled HCMV displayed labeled HCMV genomes in approximately 90% of the cell nuclei by 6 h p.i. (Fig. 8A, top panels), indicating no block to viral deposition within the nucleus. HCMV IE1 gene expression was somewhat delayed in these cells, but approximately 30% of the cells showed IE1 staining by 72 h p.i. (Fig. 8A, bottom panels). We assayed several of the DSB DNA damage response proteins by immunoblotting in these cells. Nbs1/pNbs1, Mre11, γH2AX, 53BP1, and ATM/pATM levels showed essentially no differences over the duration of the infection compared with the mock-infected cells (Fig. 8B). At the later time points assayed (72 and 96 h p.i.), p53 and Ku70/80 total protein levels increased slightly compared to the mock-infected group, but there was no pp53 signal in either the HCMV-infected or the mock-infected group (data not shown). This suggested that no damage response was initiated in cells that were not actively replicating HCMV, although they were fully capable of mounting a DSB response (81).
FIG. 8.
Activation of damage proteins at late times p.i. requires active viral replication. (A) Top panels show localization of BrdU-labeled genomes inside the nucleus of T98G cells at early times p.i. Bottom panels show expression of viral antigen IE1 in approximately 30% of cells at late times p.i. (B) Immunoblot analyses of several damage response proteins in the semipermissive T98G cells. An actin loading control is included. For definitions of abbreviations, see the Fig. 1 legend.
Key DSB-sensing and transducing proteins were not needed for HCMV replication.
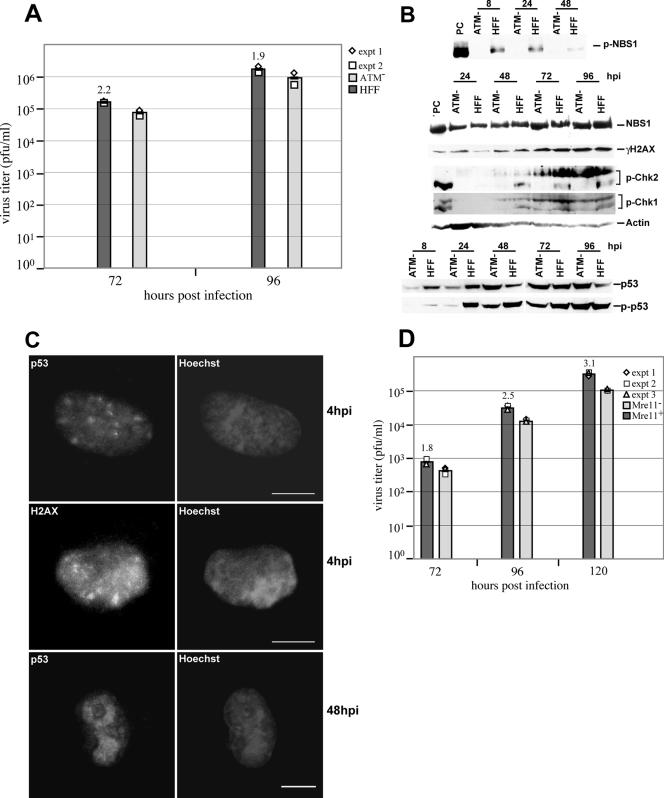
In order to investigate the role of ATM in the initiation of a DNA damage response, several different ATM− cell lines (GM02530, GM02052, and GM03487) were used. All lines behaved similarly, and data for one (GM02530) are shown. G0-synchronized ATM− and HFF cells were infected at an MOI of 5, and supernatants were harvested at 72 and 96 h p.i. for determination of virus yield by plaque assay. The virus yields in HFF cells were slightly higher than those in the several ATM− cell lines tested. Results for two separate experiments between GM02530 and HFFs are shown in Fig. 9A. The opaque bars in the figure represent the average titers for these two experiments at key time points. Average differences were not more than 2.5-fold and were therefore considered equivalent. It should be noted that experiments performed at a low MOI (0.25) gave very similar results with respect to average differences between the two cell types (data not shown).
FIG. 9.
ATM and Mre11 are not required for fully permissive infection of fibroblasts at a high MOI. (A) Cells were infected at an MOI of 5, and viral titers were calculated for ATM− and wt fibroblasts (HFF) at 72 and 96 h p.i. (as described in Materials and Methods). Average differences (n-fold) between cell types are shown above opaque bars. (B) Western blot analysis of key proteins in ATM− and HFF. Late-stage increases in Nbs1, γH2AX, and pChk1 still occur in ATM− cells. However, increases in p53 and pp53 are slightly delayed, changes in phosphorylation of Chk2 are only partial, and Nbs1 is not phosphorylated in the absence of ATM. (C) Relocalization of both p53 and γH2AX still occurs in the absence of ATM at both early and late times p.i. (D) Mre11+ and Mre11− cells infected at an MOI of 5 produced equivalent amounts of viral particles at three different times p.i. For the definition of PC, please see the Fig. 1 legend.
To investigate if the early- and late-stage changes observed in the HFFs could be due to activation via ATM, we looked at steady-state protein levels by immunoblot assay for several key proteins in the ATM− cells (Fig. 9B). There were no differences between virus-infected ATM− and HFF cells in their levels of total Nbs1. However, in the absence of ATM, Nbs1 was not phosphorylated at any time p.i., indicating that the basal level of pATM was sufficient to cause phosphorylation in wt HFFs. Both p53 and pp53 levels were higher in virus-infected HFF cells than in ATM− cells at early infection times (8 to 24 h p.i.). After 24 h p.i. the two cell types showed equivalent levels of these proteins (Fig. 9B). The delay in the phosphorylation of Ser15 is noteworthy, as this is thought to be ATM dependent. Early-stage phosphorylation of p53, as with Nbs1, can also occur via the basal level of pATM in these HFFs. Late-stage phosphorylation events involving p53 were clearly dependent upon another kinase in these cells, possibly ATR. γH2AX and pChk1 levels did not vary in the absence of ATM. This indicated that an alternate kinase was responsible for these phosphorylation events (see below). Interestingly, only the slower-migrating form of Chk2 was present in the absence of ATM, indicating the action of multiple kinases within the wt cells.
Localizations of the DNA damage proteins in ATM− cells were similar to those in HFF cells. Both γH2AX and p53 showed similar staining patterns in HFF and ATM− cells at early times p.i., with both proteins showing punctate localization patterns in the infected ATM− nuclei at 4 h p.i. (Fig. 9C). Later during infection (48 h p.i.) in ATM− cells, p53 colocalized with the viral replication protein UL44 within the replication centers, similarly to HFFs (Fig. 9C). Also, in ATM− cells all components of the MRN complex were sequestered into the viral replication centers at late times (data not shown).
To investigate the influence of damage-sensing proteins on the HCMV life cycle, G0-synchronized A-TLD1 (Mre11-knockout cells) and A-TLD1/Mre11+ cells were infected with HCMV at an MOI of 5 and viral yields were determined at 72, 96, and 120 h p.i. The difference in virus yield was no higher than threefold between the Mre11+ and Mre11− cells at all three time points (Fig. 9D). Our results indicate that unlike an HSV infection, neither ATM nor Mre11 was needed for a productive HCMV infection.
DISCUSSION
Many viruses interact with the DNA damage machinery of their host cells after entry. We have investigated the interaction of HCMV with its permissive host cell at two distinct times p.i. We have looked at early times (between 4 and 12 h p.i.) to investigate whether the large linear dsDNA viral genome invoked a DSB response upon entry into the host. We have also investigated later times p.i. in order to see whether viral replication would elicit a damage response due to the presence of DSBs or unusual recombination intermediates. We have observed many changes in steady-state protein levels, phosphorylation patterns, and localization of proteins. However, it appears that a fully functional damage response is not mounted due to insufficient relocalization of all of the components necessary to inhibit replication to both the sites of early deposition and viral replication centers.
Early events: initiation of a DSB response is disrupted.
We have not observed any relocalization of the primary DSB-sensing molecules, the MRN complex of proteins, to the sites of viral genomes within infected cell nuclei (Fig. 1C and E). Our representative images were taken between 4 and 6 h p.i.; however, we also looked at both earlier (1 to 4 h p.i., Fig. 1E) and later (13 to 17 h p.i.) times. We also varied our experimental conditions using higher viral loads (MOI of 15) and with harsher extraction techniques (53) to try to visualize relocalization of this complex to foci (data not shown). We saw no change in patterns from the mock-infected cells. Further, when cells were costained for the MRN proteins and BrdU-labeled virus, no relocalization of the complex to the sites of viral genome deposition was seen (not shown). This is in contrast to HSV infection, in which rapid relocalization to the site of prereplication centers occurs (47). Although localization of these proteins did not change, there was a large increase in both total Nbs1 and pNbs1 during the first 12 h p.i. (starting at 4 h p.i. [Fig. 1A]), which indicated an initiation of the DSB damage response. The increases observed in total Nbs1 concentration are not consistent with the literature, where there are no reports of increases in steady-state levels of any of the MRN proteins after DNA damage. However, the timing of phosphorylation of Nbs1 that we observed (apparently via ATM) was completely within the parameters of what has been reported previously (13, 51, 53, 58, 59). Effective activation of the complex requires its relocalization to the site of damage (59), which was not observed. Perhaps a viral protein(s) is binding to this active form of Nbs1 and preventing its relocalization to the site of deposition. We intend to investigate this further in future studies.
Within the very narrow window of 4 to 6 h p.i. large and loosely formed γH2AX foci were observed in infected cell nuclei (Fig. 2C). Even though no increase in the protein level was observed at early times, there was relocalization of the basal γH2AX observed in these cells. Cells were stained at both earlier (1 to 4 h p.i.) and later (12 to 18 h p.i.) times p.i., and no γH2AX foci were observed outside this period (data not shown). Compared to the literature, this time frame for relocalization is consistent but appears to be of relatively short duration (10, 15, 25, 64, 75). These foci completely colocalized with 53BP1 foci within the infected cell nuclei at 4 to 6 h p.i. This is also consistent with the literature, which points to a codependence between 53BP1 and γH2AX with respect to localization after damage. In a prototypical response, relocalization of 53BP1 to damage sites is dependent upon γH2AX, particularly when the level of damage is low (12, 25, 91). Importantly, these 53BP1/γH2AX foci did not colocalize with either viral genomes or viral early proteins which mark the site of prereplication centers, nor with cellular p53 foci (Fig. 2C and 4E), but were juxtaposed to the p53 sites in some cases. It is tempting to speculate that the juxtaposed sites are those of active viral transcription, since this occurs only at a subset of deposited genomes (37).
The other protein that undergoes dramatic changes at early times p.i. is the downstream effector molecule p53. With this protein, we saw a very rapid induction of both total p53 and pp53 steady-state levels in our infected cells (starting between 3 and 4 h p.i.) (Fig. 4C). We saw two distinct waves of pp53 (on Ser15), the first at 4 to 10 h p.i. and the second after 21 h p.i. The first wave is apparently dependent upon ATM signaling, as it was not observed in ATM− cells (Fig. 9B). The most interesting observation regarding p53 at early times p.i. was that it seems to be the only protein within the damage cascade that did relocalize (during the narrow window of 3.5 to 5.5 h p.i.) directly to sites of viral genome deposition (Fig. 4E). During this period p53 completely colocalized with two important viral proteins (IE86 and UL112/113), which are intimately involved in establishing the viral replication sites during this time frame (61, 66). This juxtaposition of p53 foci with the input viral DNA suggests another function for p53 in the viral life cycle. There is increasing evidence that p53 may directly influence DNA repair itself, first by binding to damaged regions of DNA in a nonspecific manner with its C-terminal domain (94) and then by recruiting the repair machinery to the site via protein-protein interactions. Very relevant to our studies is a report suggesting that p53 can itself augment NHEJ activity in damaged cells in a C-terminus-dependent fashion (86). p53 may be binding to the ends of the input virus through its ability to bind nonspecifically to DNA ends, thereby aiding in early circularization of the viral genome. Noteworthy is that the time at which p53 foci were observed corresponds with the reported timing of circularization of the input genomes (52). We have studies currently under way to explore this possible role for p53 at the early stages of infection.
Interestingly, despite levels of pATM (Ser1981) that did not increase above basal levels during infection (Fig. 3A and 6C), we saw ATM-dependent phosphorylation of targets such as Nbs1 and p53. Apparently, in our cells enough endogenous pATM exists to carry out these important activation functions of ATM. Alternatively, there is another phosphorylation site (as has been suggested in the literature [79]) that has been activated in response to infection. In fact, a very recent report suggests that, at least in mice, phosphorylation of ATM is not essential for its activation, relocalization, and subsequent phosphorylation of downstream targets (65). Normal DSB induction leads to rapid increases in phosphorylation at Ser1981 (3, 13) and movement of ATM and its downstream target Chk2 to the site of damage (90; reviewed also in reference 79). We did not see relocalization of either ATM or Chk2 at early times p.i. (Fig. 3B and 4B). Relocalization of both p53 and γH2AX was not ATM dependent, since both proteins relocalized normally in the ATM− cells (Fig. 9C). This points to relocalization by a different mechanism at early times p.i.
Late events: although many damage proteins displayed changes at late times p.i., completion of a damage response did not occur.
All of the changes that we observed at late times p.i. appeared to require replication of the viral genome, as they were not observed in T98G cells, which are semipermissive for viral infection (Fig. 8). All three proteins of the MRN complex showed changes in their steady-state protein profiles at later times p.i. (Fig. 6A). During these late times, levels of total Nbs1 and pNbs1 remained elevated, although pNbs1 declined slightly by 48 h p.i. High levels of total Nbs1 and Rad50 could be due to the strong sequestration of this entire complex into viral replication centers beginning between 24 and 48 h p.i. (Fig. 7A and B). Also of interest was the shift in molecular weight of Mre11 beginning at approximately 48 h p.i. (Fig. 6A). We believe this is a phosphorylation of the protein, which can be mediated either by ATM (13) or by an ATR-mediated replication fork response (see below) (69). The MRN signaling complex has been implicated in the initiation of both ATM- and ATR-mediated responses to damage (reviewed in references 60 and 79), and its close proximity to replicating viral DNA may continuously signal for a response to apparent damage within the cell. The phosphorylation of Mre11 did not aid or inhibit viral replication. This was demonstrated by the absence of differences in viral titers when Mre11− A-TLD cells were infected at a high MOI and compared to their reconstituted counterparts (Fig. 9D).
The other components of the DSB response signaling machinery also showed notable changes at late times p.i. Unlike at early times p.i., the steady-state levels of γH2AX increased in the late stages of HCMV infection (Fig. 6B). It was clear that these γH2AX increases were driven by viral replication, since they were not observed in T98G cells (Fig. 8B). In addition, they were not ATM dependent, as they occurred in the absence of ATM (Fig. 9B). The increases may have been generated by a stalled replication fork response within the viral replication centers. This response may be generated by the activation of ATR, which can phosphorylate γH2AX (32, 90). Future experiments will test the role of this alternate kinase in this and other late-stage phosphorylation events. This activation of γH2AX appears relatively protracted, and although reports in the literature vary with respect to this aspect of activation (10, 15, 64, 75), generally if the response is prolonged it is thought to signal irreparable damage (75). γH2AX localization was within the viral replication centers and was also located outside these centers, in the nucleoplasm proper (Fig. 7B). γH2AX may be signaling distress in the cellular DNA at these late times p.i., given that dramatic changes to the cellular DNA must occur for marginalization of the chromatin to the periphery of the nucleus (55) and that at late stages of infection, breaks and pulverization of the cellular DNA do occur (30).
Beginning at approximately 48 h p.i., the total level of 53BP1 began to decline and by 72 h p.i. 53BP1 was nearly absent from the infected cells (Fig. 6B). This degradation was dependent upon viral replication, as it did not occur in the T98G cells (Fig. 8B). There was no specific localization of this protein within the nucleus at late times p.i. as shown by the very diffuse staining, with little or no specific sequestration into the viral replication centers (Fig. 7B). The lack of sequestration may render this protein vulnerable to degradation. It has been reported that some ATM substrates are not phosphorylated in the absence of 53BP1 (4), which could partially explain the ineffective response mounted against viral infection. Unlike during HSV infection, there was no apparent increase in the level of phosphorylation of 53BP1 on Ser25/29 (data not shown). However, it is clear from the literature that phosphorylation is not essential for relocalization of this protein into damage-induced foci (91).
Beginning at approximately 48 h p.i., there were slight increases in steady-state levels of total ATM in infected cells (Fig. 6C). However, pATM (Ser1981) levels never increased above the endogenous background levels observed throughout the mock infection. Localization of this protein, as well as its target Chk2, at late times p.i. showed a partial sequestration within the viral replication centers (Fig. 7A and B). This was in contrast to what has been previously reported in the literature for HCMV infection (33), where exclusively cytoplasmic sequestration for both ATM and Chk2 was reported. The infection conditions used in these previous studies were very different from ours, including the method of fixation for IFA. We used an extraction procedure prior to fixation to more clearly view nuclear architecture. The cited report stated that ATM exclusively localized in the juxtanuclear compartment (33). The extraction procedure that we used did not entirely remove viral proteins (for example the tegument protein pp65) from this compartment (data not shown). Regardless of whether the extraction procedure that we used removed ATM localized in the cytoplasm, ATM was observed in the nucleus at late times p.i. Despite these observational differences, our ATM− cell studies showed that this protein (and its activating capabilities) was not required and did not in any way impede the progress of infection (Fig. 9A).
In infected cells the increased steady-state total p53 and pp53 levels remained high throughout the later times p.i. (Fig. 6D). We know from our previously published data that p53 is highly sequestered into viral replication centers at these later times, and we have shown that p53 binds differentially to the viral genome in at least 14 different sites (73). p53 is a necessary component for a fully permissive infection in HFFs, given that we have shown decreased and delayed virion production in its absence (11). Late-stage phosphorylation events did not appear to be ATM dependent, as shown by their occurrence (with slight delays) in ATM− cells (Fig. 9B). Again, ATR may be phosphorylating this key protein in the later stages of infection. To address this issue in the future, we plan to use short interfering RNA knockdown of ATR in an ATM− background. The close proximity of the MRN complex, RPA, PCNA, and the key ATR-positioning protein, ATRIP, to the viral DNA (Fig. 7A and B) (23, 29) could provide the signaling needed for replication fork stalling in these centers. ATR's downstream kinase, Chk1, was tightly associated with the replication centers at late times (Fig. 7B), and its steady-state levels slightly increased in infected cells (Fig. 6F). Indeed, we did see increases in ATR at late times p.i. (Fig. 6F) and an appearance of more slowly migrating forms. This indicated a phosphorylation event, followed by phosphorylations of ATRIP and Chk1. Despite this typical cascade, the exclusion of ATR from the viral replication centers prevented a fully functional replication stress response (Fig. 7B and C).
We feel it is noteworthy that at late times in HCMV infection there were distinct changes in the damage repair machinery itself. While steady-state levels of Rad51 were dramatically increased over mock levels at late stages of infection (Fig. 6E), unlike with HSV (87, 92), there was no clear sequestration of this protein, indicating that it was not active on viral genomes (Fig. 7A). The literature is clear that although foci are formed in response to damage, steady-state levels of Rad51 do not generally increase in normal cells (34, 53). Raderschall et al. reported seeing increases in Rad51 levels in tumor cells and indicated that this may lead to increases in unscheduled HR and genetic instability (67). The high levels of Rad51 over extended periods of time in the region of cellular DNA may be signaling the presence of extensive damage in this DNA at late times p.i.
There were small increases in steady-state levels of both Ku proteins in infected cells during late times p.i.; however, they were not dramatic and were on par with what has been observed for Ku70 after irradiation (9). No changes were observed for the catalytic subunit of the complex, in contrast to what is sometimes observed with HSV infection (46). The notable observation that we made of this complex was that unlike HSV (87), HCMV completely excludes all three subunits from the viral replication centers (Fig. 7A and B). If the virus were replicating via a rolling-circle mechanism and cleaving single length units, it would, in essence, be generating DSBs. The NHEJ machinery would reanneal these if it were present and would dampen viral processing and production, which is what is observed in HSV infection (87). We propose that HCMV changes the localization of the NHEJ proteins to prevent this from occurring.
DNA damage responses are disrupted by HCMV.
Although the entrance of the HCMV genome into the infected cell nucleus initiates a DSB response, the virus is able to inhibit the full completion of this response. Whether there is active avoidance via viral/cellular protein-protein interactions to prevent colocalization of the sensing machinery to the site of viral genome deposition must be determined. It is clear that degradation of this machinery (as occurs with adenovirus infection [84]) does not occur. Also, there is not strong relocalization of all of the necessary components to the site of replication at later times, but rather a sieving process occurs. This allows only certain proteins to be specifically sequestered and allows others to be particularly excluded. Many other viruses (HSV, polyomavirus, and human immunodeficiency virus type 1) appear to require the activation of ATM and/or Mre11 for a fully permissive infection to ensue. HCMV differs in that the initiation of a damage response does not appear to be necessary nor is it detrimental to infection at a high MOI, since neither ATM nor Mre11 is needed for full virus replication and production. We feel that by manipulating localization, HCMV subverts the full activation of an S-phase checkpoint and arrest of all replication within the cell, which would be disastrous for the virus.
Acknowledgments
This work was supported by NIH grants RO1-AI51463 and P20 RR015587 (COBRE program) to E.A.F.
We thank John O'Dowd and Rick Wood for critical reading of the manuscript.
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Albrecht, T., M. P. Fons, C. Z. Deng, and I. Boldogh. 1997. Increased frequency of specific locus mutation following human cytomegalovirus infection. Virology 230:48-61. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, L., C. Henderson, and Y. Adachi. 2001. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell. Biol. 21:1719-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499-506. [DOI] [PubMed] [Google Scholar]
- 4.Bekker-Jensen, S., C. Lukas, F. Melander, J. Bartek, and J. Lukas. 2005. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 170:201-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boldogh, I., E. S. Huang, J. F. Baskar, L. Gergely, and T. Albrecht. 1992. Oncogenic transformation by cellular DNA isolated from human cytomegalovirus-infected cells. Intervirology 34:62-73. [DOI] [PubMed] [Google Scholar]
- 6.Boppana, S. B., K. B. Fowler, W. J. Britt, S. Stagno, and R. F. Pass. 1999. Symptomatic congenital cytomegalovirus infection in infants born to mothers with preexisting immunity to cytomegalovirus. Pediatrics 104:55-60. [DOI] [PubMed] [Google Scholar]
- 7.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150-160. [DOI] [PubMed] [Google Scholar]
- 8.Britt, W., and C. Alford. 1996. Cytomegalovirus, p. 2493-2523. In B. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 9.Brown, K. D., T. A. Lataxes, S. Shangary, J. L. Mannino, J. F. Giardina, J. Chen, and R. Baskaran. 2000. Ionizing radiation exposure results in up-regulation of Ku70 via a p53/ataxia-telangiectasia-mutated protein-dependent mechanism. J. Biol. Chem. 275:6651-6656. [DOI] [PubMed] [Google Scholar]
- 10.Burma, S., B. P. Chen, M. Murphy, A. Kurimasa, and D. J. Chen. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462-42467. [DOI] [PubMed] [Google Scholar]
- 11.Casavant, N. C., M. H. Luo, K. Rosenke, T. Winegardner, A. Zurawska, and E. A. Fortunato. 2006. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J. Virol. 80:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celeste, A., O. Fernandez-Capetillo, M. J. Kruhlak, D. R. Pilch, D. W. Staudt, A. Lee, R. F. Bonner, W. M. Bonner, and A. Nussenzweig. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675-679. [DOI] [PubMed] [Google Scholar]
- 13.Cerosaletti, K., and P. Concannon. 2004. Independent roles for nibrin and Mre11-Rad50 in the activation and function of Atm. J. Biol. Chem. 279:38813-38819. [DOI] [PubMed] [Google Scholar]
- 14.Cerosaletti, K., J. Wright, and P. Concannon. 2006. Active role for nibrin in the kinetics of Atm activation. Mol. Cell. Biol. 26:1691-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhury, D., M. C. Keogh, H. Ishii, C. L. Peterson, S. Buratowski, and J. Lieberman. 2005. Gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20:801-809. [DOI] [PubMed] [Google Scholar]
- 16.Cinque, P., R. Marenzi, and D. Ceresa. 1997. Cytomegalovirus infections of the nervous system. Intervirology 40:85-97. [DOI] [PubMed] [Google Scholar]
- 17.Cobbs, C. S., L. Harkins, M. Samanta, G. Y. Gillespie, S. Bharara, P. H. King, L. B. Nabors, C. G. Cobbs, and W. J. Britt. 2002. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 62:3347-3350. [PubMed] [Google Scholar]
- 18.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 19.Courcelle, C. T., J. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Requirement for uracil-DNA glycosylase during the transition to late-phase cytomegalovirus DNA replication. J. Virol. 75:7592-7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahl, J., J. You, and T. L. Benjamin. 2005. Induction and utilization of an ATM signaling pathway by polyomavirus. J. Virol. 79:13007-13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daniel, R., G. Kao, K. Taganov, J. G. Greger, O. Favorova, G. Merkel, T. J. Yen, R. A. Katz, and A. M. Skalka. 2003. Evidence that the retroviral DNA integration process triggers an ATR-dependent DNA damage response. Proc. Natl. Acad. Sci. USA 100:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jager, M., J. van Noort, D. C. van Gent, C. Dekker, R. Kanaar, and C. Wyman. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8:1129-1135. [DOI] [PubMed] [Google Scholar]
- 23.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupre, A., L. Boyer-Chatenet, and J. Gautier. 2006. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat. Struct. Mol. Biol. 13:451-457. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Capetillo, O., H. T. Chen, A. Celeste, I. Ward, P. J. Romanienko, J. C. Morales, K. Naka, Z. Xia, R. D. Camerini-Otero, N. Motoyama, P. B. Carpenter, W. M. Bonner, J. Chen, and A. Nussenzweig. 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 4:993-997. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Capetillo, O., A. Lee, M. Nussenzweig, and A. Nussenzweig. 2004. H2AX: the histone guardian of the genome. DNA Repair (Amsterdam) 3:959-967. [DOI] [PubMed] [Google Scholar]
- 27.Flygare, J., F. Benson, and D. Hellgren. 1996. Expression of the human RAD51 gene during the cell cycle in primary human peripheral blood lymphocytes. Biochim. Biophys. Acta 1312:231-236. [DOI] [PubMed] [Google Scholar]
- 28.Fortunato, E. A., M. L. Dell'Aquila, and D. H. Spector. 2000. Specific chromosome 1 breaks induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 97:853-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fortunato, E. A., and D. H. Spector. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 72:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fortunato, E. A., and D. H. Spector. 2003. Viral induction of site-specific chromosome damage. Rev. Med. Virol. 13:21-37. [DOI] [PubMed] [Google Scholar]
- 31.Fowler, K. B., F. P. McCollister, A. J. Dahle, S. Boppana, W. J. Britt, and R. F. Pass. 1997. Progressive and fluctuating sensorineural hearing loss in children with asymptomatic congenital cytomegalovirus infection. J. Pediatr. 130:624-630. [DOI] [PubMed] [Google Scholar]
- 32.Furuta, T., H. Takemura, Z. Y. Liao, G. J. Aune, C. Redon, O. A. Sedelnikova, D. R. Pilch, E. P. Rogakou, A. Celeste, H. T. Chen, A. Nussenzweig, M. I. Aladjem, W. M. Bonner, and Y. Pommier. 2003. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 278:20303-20312. [DOI] [PubMed] [Google Scholar]
- 33.Gaspar, M., and T. Shenk. 2006. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc. Natl. Acad. Sci. USA 103:2821-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haaf, T., E. I. Golub, G. Reddy, C. M. Radding, and D. C. Ward. 1995. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 92:2298-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harkins, L., A. L. Volk, M. Samanta, I. Mikolaenko, W. J. Britt, K. I. Bland, and C. S. Cobbs. 2002. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet 360:1557-1563. [DOI] [PubMed] [Google Scholar]
- 36.Hopfner, K. P., L. Craig, G. Moncalian, R. A. Zinkel, T. Usui, B. A. Owen, A. Karcher, B. Henderson, J. L. Bodmer, C. T. McMurray, J. P. Carney, J. H. Petrini, and J. A. Tainer. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418:562-566. [DOI] [PubMed] [Google Scholar]
- 37.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwabuchi, K., B. P. Basu, B. Kysela, T. Kurihara, M. Shibata, D. Guan, Y. Cao, T. Hamada, K. Imamura, P. A. Jeggo, T. Date, and A. J. Doherty. 2003. Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J. Biol. Chem. 278:36487-36495. [DOI] [PubMed] [Google Scholar]
- 39.Jault, F. M., J.-M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated RB, and p53, leading to cell cycle arrest. J. Virol. 69:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jault, F. M., S. A. Spector, and D. H. Spector. 1994. The effects of cytomegalovirus on human immunodeficiency virus replication in brain-derived cells correlate with permissiveness of the cells for each virus. J. Virol. 68:959-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi, J. 2004. Molecular mechanism of the recruitment of NBS1/hMRE11/hRAD50 complex to DNA double-strand breaks: NBS1 binds to gamma-H2AX through FHA/BRCT domain. J. Radiat. Res. (Tokyo) 45:473-478. [DOI] [PubMed] [Google Scholar]
- 42.Kovacs, A., M. L. Weber, L. J. Burns, H. S. Jacob, and G. M. Vercellotti. 1996. Cytoplasmic sequestration of p53 in cytomegalovirus-infected human endothelial cells. Am. J. Pathol. 149:1531-1539. [PMC free article] [PubMed] [Google Scholar]
- 43.Kudoh, A., M. Fujita, L. Zhang, N. Shirata, T. Daikoku, Y. Sugaya, H. Isomura, Y. Nishiyama, and T. Tsurumi. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 280:8156-8163. [DOI] [PubMed] [Google Scholar]
- 44.Kurz, E. U., P. Douglas, and S. P. Lees-Miller. 2004. Doxorubicin activates ATM-dependent phosphorylation of multiple downstream targets in part through the generation of reactive oxygen species. J. Biol. Chem. 279:53272-53281. [DOI] [PubMed] [Google Scholar]
- 45.Lau, A., K. M. Swinbank, P. S. Ahmed, D. L. Taylor, S. P. Jackson, G. C. Smith, and M. J. O'Connor. 2005. Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat. Cell Biol. 7:493-500. [DOI] [PubMed] [Google Scholar]
- 46.Lees-Miller, S. P., M. C. Long, M. A. Kilvert, V. Lam, S. A. Rice, and C. A. Spencer. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 70:7471-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lilley, C. E., C. T. Carson, A. R. Muotri, F. H. Gage, and M. D. Weitzman. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 102:5844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin, Y., T. Lukacsovich, and A. S. Waldman. 1999. Multiple pathways for repair of DNA double-strand breaks in mammalian chromosomes. Mol. Cell. Biol. 19:8353-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lokensgard, J. R., M. C. Cheeran, G. Gekker, S. Hu, C. C. Chao, and P. K. Peterson. 1999. Human cytomegalovirus replication and modulation of apoptosis in astrocytes. J. Hum. Virol. 2:91-101. [PubMed] [Google Scholar]
- 50.Lombard, D. B., and L. Guarente. 2000. Nijmegen breakage syndrome disease protein and MRE11 at PML nuclear bodies and meiotic telomeres. Cancer Res. 60:2331-2334. [PubMed] [Google Scholar]
- 51.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McVoy, M. A., and S. P. Adler. 1994. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J. Virol. 68:1040-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mirzoeva, O. K., and J. H. Petrini. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mocarski, E. S. 1996. Cytomegaloviruses and their replication, p. 2447-2492. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 55.Monier, K., J. C. Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2:661-665. [DOI] [PubMed] [Google Scholar]
- 56.Muganda, P., O. Mendoza, J. Hernandez, and Q. Qian. 1994. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J. Virol. 68:8028-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy, E. A., D. N. Streblow, J. A. Nelson, and M. F. Stinski. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakanishi, K., T. Taniguchi, V. Ranganathan, H. V. New, L. A. Moreau, M. Stotsky, C. G. Mathew, M. B. Kastan, D. T. Weaver, and A. D. D'Andrea. 2002. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat. Cell Biol. 4:913-920. [DOI] [PubMed] [Google Scholar]
- 59.Nelms, B. E., R. S. Maser, J. F. MacKay, M. G. Lagally, and J. H. Petrini. 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280:590-592. [DOI] [PubMed] [Google Scholar]
- 60.O'Connell, M. J., and K. A. Cimprich. 2005. G2 damage checkpoints: what is the turn-on? J. Cell Sci. 118:1-6. [DOI] [PubMed] [Google Scholar]
- 61.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paull, T. T., and M. Gellert. 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1:969-979. [DOI] [PubMed] [Google Scholar]
- 64.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 65.Pellegrini, M., A. Celeste, S. Difilippantonio, R. Guo, W. Wang, L. Feigenbaum, and A. Nussenzweig. 2006. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature 443:222-225. [DOI] [PubMed] [Google Scholar]
- 66.Penfold, M. E., and E. S. Mocarski. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46-61. [DOI] [PubMed] [Google Scholar]
- 67.Raderschall, E., K. Stout, S. Freier, V. Suckow, S. Schweiger, and T. Haaf. 2002. Elevated levels of Rad51 recombination protein in tumor cells. Cancer Res. 62:219-225. [PubMed] [Google Scholar]
- 68.Richie, C. T., C. Peterson, T. Lu, W. N. Hittelman, P. B. Carpenter, and R. J. Legerski. 2002. hSnm1 colocalizes and physically associates with 53BP1 before and after DNA damage. Mol. Cell. Biol. 22:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robison, J. G., J. Elliott, K. Dixon, and G. G. Oakley. 2004. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem. 279:34802-34810. [DOI] [PubMed] [Google Scholar]
- 70.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 72.Rosenke, K., and E. A. Fortunato. 2004. Bromodeoxyuridine-labeled viral particles as a tool for visualization of the immediate-early events of human cytomegalovirus infection. J. Virol. 78:7818-7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosenke, K., M. A. Samuel, E. T. McDowell, M. A. Toerne, and E. A. Fortunato. 2006. An intact sequence-specific DNA-binding domain is required for human cytomegalovirus-mediated sequestration of p53 and may promote in vivo binding to the viral genome during infection. Virology 348:19-34. [DOI] [PubMed] [Google Scholar]
- 74.Samanta, M., L. Harkins, K. Klemm, W. J. Britt, and C. S. Cobbs. 2003. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J. Urol. 170:998-1002. [DOI] [PubMed] [Google Scholar]
- 75.Sedelnikova, O. A., I. Horikawa, D. B. Zimonjic, N. C. Popescu, W. M. Bonner, and J. C. Barrett. 2004. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat. Cell Biol. 6:168-170. [DOI] [PubMed] [Google Scholar]
- 76.Severini, A., D. G. Scraba, and D. L. Tyrrell. 1996. Branched structures in the intracellular DNA of herpes simplex virus type 1. J. Virol. 70:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shechter, D., V. Costanzo, and J. Gautier. 2004. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair (Amsterdam) 3:901-908. [DOI] [PubMed] [Google Scholar]
- 78.Shen, Y., H. Zhu, and T. Shenk. 1997. Human cytomegalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc. Natl. Acad. Sci. USA 94:3341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shiloh, Y. 2006. The ATM-mediated DNA-damage response: taking shape. Trends Biochem. Sci. 31:402-410. [DOI] [PubMed] [Google Scholar]
- 80.Shirata, N., A. Kudoh, T. Daikoku, Y. Tatsumi, M. Fujita, T. Kiyono, Y. Sugaya, H. Isomura, K. Ishizaki, and T. Tsurumi. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 280:30336-30341. [DOI] [PubMed] [Google Scholar]
- 81.Short, S. C., S. Bourne, C. Martindale, M. Woodcock, and S. P. Jackson. 2005. DNA damage responses at low radiation doses. Radiat. Res. 164:292-302. [DOI] [PubMed] [Google Scholar]
- 82.Smith, G. C., and S. P. Jackson. 1999. The DNA-dependent protein kinase. Genes Dev. 13:916-934. [DOI] [PubMed] [Google Scholar]
- 83.Speir, E., R. Modali, E.-S. Huang, M. B. Leon, F. Sahwl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391-394. [DOI] [PubMed] [Google Scholar]
- 84.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 85.Strang, B. L., and N. D. Stow. 2005. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J. Virol. 79:12487-12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tang, W., H. Willers, and S. N. Powell. 1999. p53 directly enhances rejoining of DNA double-strand breaks with cohesive ends in gamma-irradiated mouse fibroblasts. Cancer Res. 59:2562-2565. [PubMed] [Google Scholar]
- 87.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trujillo, K. M., S. S. Yuan, E. Y. Lee, and P. Sung. 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273:21447-21450. [DOI] [PubMed] [Google Scholar]
- 89.Van Meir, E. G., T. Kikuchi, M. Tada, H. Li, A. C. Diserens, B. E. Wojcik, H. J. Huang, T. Friedmann, N. de Tribolet, and W. K. Cavenee. 1994. Analysis of the p53 gene and its expression in human glioblastoma cells. Cancer Res. 54:649-652. [PubMed] [Google Scholar]
- 90.Ward, I. M., and J. Chen. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759-47762. [DOI] [PubMed] [Google Scholar]
- 91.Ward, I. M., K. Minn, K. G. Jorda, and J. Chen. 2003. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 278:19579-19582. [DOI] [PubMed] [Google Scholar]
- 92.Wilkinson, D. E., and S. K. Weller. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.You, Z., C. Chahwan, J. Bailis, T. Hunter, and P. Russell. 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 25:5363-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zotchev, S. B., M. Protopopova, and G. Selivanova. 2000. p53 C-terminal interaction with DNA ends and gaps has opposing effect on specific DNA binding by the core. Nucleic Acids Res. 28:4005-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zou, L., D. Liu, and S. J. Elledge. 2003. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA 100:13827-13832. [DOI] [PMC free article] [PubMed] [Google Scholar]