Abstract
The complex human T-cell leukemia virus type 1 (HTLV-1) retrovirus encodes several proteins that are unique to the virus within its 3′-end region. Among them, the viral transactivator Tax and posttranscriptional regulator Rex are well characterized, and both positively regulate HTLV-1 viral expression. Less is known about the other regulatory proteins encoded in this region of the provirus, including the recently discovered HBZ protein. HBZ has been shown to negatively regulate basal and Tax-dependent HTLV-1 transcription through its ability to interact with specific basic-leucine zipper (bZIP) proteins. In the present study, we found that HBZ reduces HTLV-1 transcription and virion production. We then characterized the interaction between HBZ and the cellular transcription factor CREB. CREB plays a critical role in Tax-mediated HTLV-1 transcription by forming a complex with Tax that binds to viral cyclic AMP-response elements (CREs) located within the viral promoter. We found that HBZ and CREB interact in vivo and directly in vitro, and this interaction occurs through the bZIP domain of each protein. We also found that CREM-Ia and ATF-1, which share significant homology in their bZIP domains with the bZIP domain of CREB, interact with HBZ-bZIP. The interaction between CREB and HBZ prevents CREB binding to the viral CRE elements in vitro and in vivo, suggesting that the reduction in HTLV-1 transcription by HBZ is partly due to the loss of CREB at the promoter. We also found that HBZ displaces CREB from a cellular CRE, suggesting that HBZ may deregulate CREB-dependent cellular gene expression.
Human T-cell leukemia virus type 1 (HTLV-1) is a human retrovirus that is associated with two distinct diseases: adult T-cell leukemia (ATL), an abnormal proliferation of infected CD4+ T lymphocytes, and HTLV-1-associated myelopathy and/or tropical spastic paraparesis, a neurodegenerative disorder (19, 48, 49). The molecular mechanisms leading to the development of both diseases are unclear, although the viral protein Tax is postulated to play an important role in these processes. Tax functions as a transcription factor and is essential for strong HTLV-1 transcription. Tax activates transcription through three 21-bp repeats that contain imperfect cyclic AMP responsive elements (called viral CREs) situated within the long terminal repeat of the HTLV-1 genome (1, 6, 14, 15, 22, 27). Tax does not bind DNA alone but interacts with cellular transcription factors from the ATF/CREB family to form complexes that associate with the DNA. Within these complexes, Tax contacts the GC-rich sequences flanking the CRE core (31, 37, 38, 41). The formation of Tax/CREB/DNA complexes is critical for the recruitment of the cellular coactivators CBP/p300 and subsequent high transcriptional activation of the virus (18, 21, 32, 39, 58).
A number of cellular factors containing basic leucine zipper (bZIP) motifs have been shown to bind the viral CREs in HTLV-1-infected T cells. These factors included the ATF/CREB family members (ATF-1, ATF-2, CREB, CREB-2, and CREM) and the AP-1 family members (c-Jun and c-Fos) (34; reviewed in reference 23). Of these factors, CREB has been shown to play a critical role in Tax-mediated HTLV-1 transcription (8), although the other ATF/CREB members also form a complex with Tax and activate viral transcription (14, 16). In contrast, c-Jun and c-Fos have been shown to participate in HTLV-1 basal transcription (28).
Recently, we characterized a novel HTLV-1 protein encoded by the complementary strand of the proviral genome, termed HTLV-1 bZIP factor or HBZ (17). This protein possesses a putative bZIP domain. Among the bZIP proteins that participate in HTLV-1 transcription, CREB-2 and c-Jun have been found to interact with HBZ (5, 17, 42). These interactions occur through the bZIP domain of each protein. The interaction with CREB-2 suppresses Tax-dependent viral transcription (17), whereas the interaction with c-Jun reduces AP-1 transcription and also HTLV-1 basal transcription (5, 42). However, the molecular mechanisms involved in the downregulation of HTLV-1 transcription by HBZ have not been completely elucidated. For this reason, we were interested in studying the effect of HBZ on CREB, since CREB is a major player in Tax-mediated HTLV-1 transcription (8). We first confirmed that HBZ represses HTLV-1 virion production. We then demonstrated that HBZ binds to CREB in vivo and in vitro, and this interaction is mediated by the bZIP domain of each protein. Interestingly, CREM-Ia and ATF-1, which share significant homology in their bZIP domain with CREB (53), are also found to interact with HBZ-bZIP. A truncated form of HBZ, containing the bZIP domain, inhibits CREB binding to a viral CRE in vitro, whereas HBZ with the bZIP domain deleted fails to inhibit CREB binding. Using the chromatin immunoprecipitation (ChIP) assay, we show that Tax and CREB binding at the HTLV-1 promoter are reduced in cells expressing HBZ. These results indicate that HTLV-1 transcriptional repression by HBZ is, in part, mediated by the loss of Tax and CREB binding at the viral promoter. Finally, we found that HBZ also represses CREB transcription from a cellular CRE, extending the repressive role of the HBZ protein to CREB-dependent transcription of cellular genes.
MATERIALS AND METHODS
Cell culture, transient-cotransfection assays, and expression plasmids.
CEM, MT-4, and C8166-45 cells were cultured in Iscove modified Dulbecco medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and penicillin-streptomycin. The CHOK1-Luc hamster ovary cells (47) and 293T cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Geneticin (500 μg/ml; Invitrogen) was added to CHOK1-Luc cells. For transient-transfection assays of 293T cells with HTLV-1 proviral DNA clones, the CalPhos mammalian transfection kit was used according to the manufacturer's instructions (Clontech BD Biosciences). Quantification of HTLV-1 p19 antigen was performed by using an enzyme-linked immunosorbent assay (ELISA) HTLV-1 p19 antigen kit (Zeptometrix, Buffalo, NY) as described by the manufacturer. For transient-transfection assays of CEM cells with HTLV-1 proviral DNA clones, plasmids were transfected with a Nucleofector according to the manufacturer's instructions (Amaxa Biosystems). For the transient-cotransfection assays shown in Fig. 6A, the cells were grown to a density of 106 cells/ml and transfected with DMRIE-C (Invitrogen) and a constant amount of DNA for 5 h. The cells were allowed to recover for 24 h before harvest in the presence of 20 μM forskolin (Sigma) or dimethyl sulfoxide (carrier). The cells were lysed, and the luciferase activity was measured by using a dual-luciferase reporter assay system with a Turner Designs model TD 20-e luminometer. The luciferase activity was normalized to pRL-TK (Promega), which encodes the Renilla luciferase from the HSV-TK promoter, as an internal control. Expression plasmids for HBZ (pcDNA-HBZ-MycHis [55]) and Tax (pSG-Tax [51]) have been described elsewhere. To construct the plasmid pcDNA-HBZΔATG, both HBZ ATGs in pcDNA-HBZ-MycHis were replaced by TAG with a QuikChange site-directed mutagenesis kit (Stratagene). To construct the plasmid pcDNA-HBZ-ΔbZIP, HBZ (amino acids [aa] 1 to 132) was amplified by PCR and cloned into the EcoRI and HindIII sites of pcDNA-MycHis (Invitrogen). The luciferase reporter plasmids K30-Luc (33), pminLuc-cellular CRE (20), and the HTLV-1 molecular clones ACH (30) have also been described. The Western blots in Fig. 1 were probed with an anti-Myc antibody to detect HBZ (9E10; Sigma) and an anti-nucleolin antibody (Santa-Cruz).
FIG. 6.
HBZ disrupts CREB binding at a cellular CRE. (A) HBZ represses CREB transactivation from a minimal promoter carrying only the cellular CREs. The pminLuc-cellular CRE reporter plasmid (400 ng) was transfected into CEM T cells that were subsequently treated with forskolin (20 μM for 4 h) as indicated or dimethyl sulfoxide (carrier). pcDNA-HBZ-MycHis (400 ng) and pcDNA-HBZΔATG (400 ng) were cotransfected with the reporter plasmid as indicated. Reported values are the average luminescence from two experiments performed in duplicate and are representative of four independent experiments. The standard error is indicated. (B) HBZ-bZIP displaces CREB-bZIP from a cellular CRE. Binding reactions in lanes 1 to 14 contained 6 fmol of 32P-labeled cellular CRE probe with 0.12 pmol of CREB-bZIP (lanes 2 to 5 and lanes 7 to 14). Reactions in lanes 3 to 5 also contained increasing amounts of HBZ-bZIP (0.6, 1.2, and 1.8 pmol). Reactions in lane 8 also contained 1.2 pmol of HBZ-bZIP, and lanes 9 to 11 also contained increasing amounts of HBZ-ΔbZIP (1.2, 1.8, and 2.4 pmol). An antibody directed against the bZIP domain of CREB (500 ng, lane 13) or an antibody against histone H3 (500 ng, lane 14) were also added in the reactions. The positions of the free probe and relevant protein-DNA complexes are labeled.
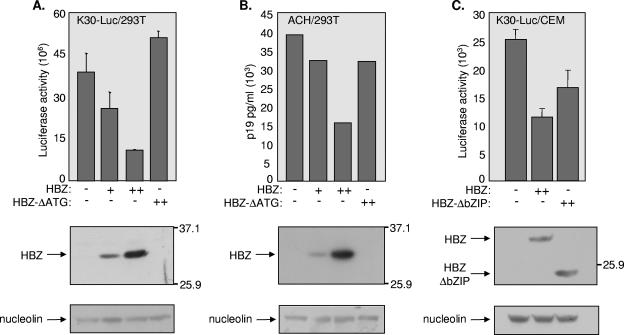
FIG. 1.
Repression of HTLV-1 proviral gene expression by HBZ. (A) HBZ abrogates proviral genome expression. 293T cells were cotransfected with K30-Luc (2 μg), pcDNA-HBZ-MycHis (1 and 5 μg), or pcDNA-HBZΔATG (5 μg) and the β-galactosidase-containing reference plasmid pcDNA-lacZ (2 μg). Luciferase assays were performed 48 h after transfection and luciferase activity was normalized to β-galactosidase activity. Reported values are the average luminescence from three experiments and the standard error is indicated. Expression of HBZ was visualized by Western blot analysis with an anti-Myc antibody (middle panel). Protein standards (in kilodaltons) are indicated at the right. Nucleolin was probed as a loading control (lower panel). (B) HBZ represses virion production. The ACH molecular clone (5 μg), pcDNA-HBZ-MycHis (1 and 5 μg) or pcDNA-HBZΔATG (5 μg), and pcDNA-lacZ (2 μg) were cotransfected into 293T cells. Expression of p19gag was measured 24 h after transfection. The results are representative of three experiments. Expression of HBZ was visualized by Western blot analysis with an anti-Myc antibody (middle panel). Protein standards (in kilodaltons) are indicated at the right. Nucleolin was probed as a loading control (lower panel). (C) HBZ abrogates proviral genome expression in T cells. CEM T cells were cotransfected with K30-Luc (1 μg), pcDNA-HBZ-MycHis (3 μg) or pcDNA-HBZΔbZIP (3 μg), and the β-galactosidase-containing reference plasmid pcDNA-lacZ (1 μg). Luciferase assays were performed 24 h after transfection and luciferase activity was normalized to β-galactosidase activity. Reported values are the average luminescence from three experiments and the standard error is indicated. Expression of HBZ was visualized by Western blot analysis with an anti-Myc antibody (middle panel). Protein standards (in kilodaltons) are indicated at the right. Nucleolin was probed as a loading control (lower panel).
Expression and purification of recombinant proteins.
The expression plasmid pGST-CREB-His6, provided by C.-Z. Giam (24), and the expression plasmid for CREM-Ia-bZIP, provided by A. E. Keating (44), were transformed in Escherichia coli BL21(DE3)/pLysS (Stratagene), and the proteins were expressed and purified by Ni2+ chelate chromatography as described by the manufacturer (QIAGEN). The pRSET-HBZ-ΔbZIP (aa 1 to 122) plasmid was constructed by cloning the HBZ truncation mutant, amplified by PCR, into the pRSET-A vector (Invitrogen). This plasmid was transformed in the E. coli BL21 codon plus (DE3) (Stratagene), and the protein was expressed and purified by Ni2+ chromatography. pGST-HBZ-bZIP (aa 123 to 209) (17) was transformed into E. coli BL21(DE3)/pLysS, expressed, and purified by glutathione-agarose affinity chromatography. Bacterial expression plasmids for human CREB (14), CREB-bZIP (aa 254 to 327) (26), HBZ-bZIP (aa 123 to 209) (17), and Tax (59) were transformed into E. coli BL21(DE3)/pLysS, and the proteins were expressed and purified as described previously (21, 26). The final HBZ-bZIP purified protein product was composed of two bands, both recognized by an anti-histidine antibody. Protein identification analysis via trypsin digest and tandem mass spectrometry positively matched the central portions of both protein bands to the HBZ amino acid sequence and identified the lower band as a C-terminal truncation of the upper band. All proteins were purified to near homogeneity, dialyzed against 0.1 M TM (50 mM Tris-HCl [pH 7.9], 100 mM KCl, 12.5 mM MgCl2, 1 mM EDTA [pH 8.0], 20% glycerol, 0.025% [vol/vol] Tween 20, 1 mM dithiothreitol [DTT]), divided into aliquots, and stored at −70°C.
GST pull-down assays.
All GST pull-down experiments were performed with 20 μl of glutathione-agarose beads equilibrated in 0.5× Superdex buffer (1× Superdex buffer is 25 mM HEPES [pH 7.9], 12.5 mM MgCl2, 10 μM ZnSO4, 150 mM KCl, 20% [vol/vol] glycerol, 0.1% Nonidet P-40, 1 mM EDTA, 2 mM DTT, and 2 mM phenylmethylsulfonyl fluoride [PMSF]), except for those shown in Fig. 3B (20 mM HEPES [pH 7.9], 2.5 mM MgCl2, 5 μM ZnSO4, 25 mM KCl, 10% [vol/vol] glycerol, 0.05% Nonidet P-40, 1 mM DTT, and 1 mM PMSF) and Fig. 3D and G, left panel (0.5× TM: 25 mM Tris, 50 mM KCl, 7.25 mM MgCl2, 10% [vol/vol] glycerol, 0.05% Tween 20, 1 mM DTT, and 1 mM PMSF). The purified glutathione S-transferase (GST) fusion protein was incubated with the glutathione agarose beads for 1 h at 4°C and then washed with binding buffer. The second protein was then added to the washed beads and incubated for 1 h (or overnight) at 4°C. Purified bovine serum albumin (BSA) used in Fig. 3H was purchased from New England Biolabs. The beads were washed with binding buffer, and bound proteins were eluted with sodium dodecyl sulfate (SDS) sample dyes. Bound proteins were separated by electrophoresis on SDS-PAGE gels, transferred to nitrocellulose, and probed with the appropriate antibody. The antibodies anti-CREB (C-21 and X-12; Santa Cruz Biotechnology) and anti-His (H-15; Santa Cruz Biotechnology) were used in the present study. For GST pull-down assays with 35S-labeled proteins (Fig. 3F, right portion of panel G, and H), plasmids pcDNA-HBZ-MycHis, pRSET-HBZ-ΔbZIP, and ATF-1 (2) were used to produce HBZ, HBZ-ΔbZIP, and ATF-1, respectively, using the TNT-Quick-Coupled transcription-translation kit (Promega).
FIG. 3.
HBZ and CREB interact through their bZIP domains. (A) Purified GST HBZ-bZIP. Purified GST HBZ-bZIP is shown by Coomassie blue staining after SDS-PAGE (lane 2). Protein standards (in kilodaltons) are indicated in lane 1. (B) CREB interacts with HBZ-bZIP. Purified CREB (10 pmol) was incubated with either 50 pmol of GST alone (lane 2) or 50 pmol of GST-HBZ-bZIP (lane 3). Bound CREB protein was detected by Western blot analysis. A fraction of the input protein (8%) is shown in lane 1, and protein standards (in kilodaltons) are indicated at the left. (C) Purified HBZ-bZIP. Purified HBZ-bZIP is shown by Coomassie blue staining after SDS-PAGE (lane 2). Protein standards (in kilodaltons) are indicated in lane 1. (D) HBZ-bZIP interacts with CREB. HBZ-bZIP (10 pmol) was incubated with either 10 pmol of GST alone (lane 2) or 10 pmol of GST-CREB (lane 3). Bound CREB-bZIP protein was detected by Western blot analysis. A fraction of the input protein is shown in lane 1, and protein standards (in kilodaltons) are indicated at the left. (E) CREB-bZIP interacts with HBZ-bZIP. Purified CREB-bZIP (50 pmol) was incubated with either 10 pmol of GST alone (lane 2) or 10 pmol of GST-HBZ-bZIP (lane 3). Bound CREB-bZIP protein was detected by Western blot analysis. A fraction of the input protein (10%) is shown in lane 1, and protein standards (in kilodaltons) are indicated at the left. (F) HBZ-ΔbZIP domain does not interact with CREB. 35S-labeled HBZ and HBZ-ΔbZIP were incubated with either GST (10 pmol, lanes 3 and 4, respectively) or GST-CREB (10 pmol, lanes 5 and 6, respectively). Bound proteins were detected by PhosphorImager analysis. A fraction of the input proteins (15%) is shown in lanes 1 and 2. (G) CREM-Ia-bZIP and ATF-1 interact with HBZ-bZIP. Purified CREM-Ia-bZIP (50 pmol) was incubated with either 50 pmol of GST alone (lane 2) or 50 pmol of GST-HBZ-bZIP (lane 3). Bound CREB-bZIP protein was detected by Western blot analysis. A fraction of the input protein (8%) is shown in lane 1, and protein standards (in kilodaltons) are indicated at the left. 35S-labeled ATF-1 was incubated with either GST (10 pmol, lane 5) or GST-HBZ-bZIP (10 pmol, lane 6). Bound ATF-1 was detected by PhosphorImager analysis. A fraction of the input protein (20%) is shown in lane 4. (H) HBZ-bZIP disrupts CREB homodimerization. In the left panel, GST (10 pmol) and GST-CREB (10 pmol) were incubated with 5 μl of 35S-labeled CREB (lanes 2 and 3, respectively). In lanes 4 to 6, 10 pmol of GST-CREB was incubated with 5 μl of 35S-labeled CREB and increasing amounts of 35S-labeled HBZ (5, 10 and 20 μl). In lanes 7 to 8, 10 pmol of GST-CREB was incubated with 5 μl of 35S-labeled CREB and increasing amounts of BSA (200, 400, and 800 ng). Complexes were separated by SDS-PAGE, and bound CREB and HBZ were detected by PhosphorImager analysis. A fraction of the input protein (10%) is shown in lane 1. In the right panel, GST (10 pmol, lane 2) and GST-CREB (10 pmol, lanes 3 to 5) were incubated with 5 μl of 35S-labeled CREB and 15 μl of 35S-labeled HBZ (lane 4) or 35S-labeled HBZ-ΔbZIP (lane 5). Complexes were separated by SDS-PAGE, and bound CREB and HBZ were detected by PhosphorImager analysis. Fractions of the input of CREB (10%), HBZ (5%), and HBZ-ΔbZIP (5%) proteins are shown in lanes 10, 15, and 16, respectively.
Coimmunoprecipitation assays.
Lysates from HTLV-1-infected T cells (MT-4 and C8166-45) and CHOK1-Luc cells transfected with pcDNA3.1 or pcDNA-HBZ-MycHis were prepared in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8.0], 1% Triton X-100, 100 mM NaCl, 1 mM MgCl2, 2 mM benzamidine, 2 μg of leupeptin/ml, and 1 mM PMSF). Antibody-bound beads (preimmune serum, HBZ serum [17], or Myc antibody [Abcam]) were washed in RIPA buffer, and cell lysates (500 μg) were added to each antibody-bead suspension, incubated overnight, and washed several times in RIPA buffer. The bound proteins were analyzed on a 10% SDS-polyacrylamide gel and detected by Western blotting with a mouse anti-CREB-1 antibody (X-12 for Fig. 2A and C-21 for Fig. 2B; Santa Cruz Biotechnology) and Myc antibody (Abcam).
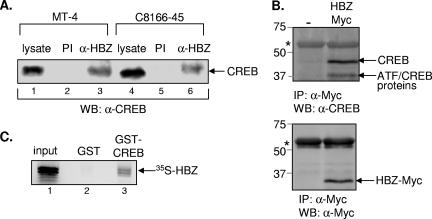
FIG. 2.
HBZ and CREB interact in vivo and in vitro. (A) HBZ and CREB interact in HTLV-1-infected cells. Cell extracts from the infected cell lines MT-4 (lanes 1 to 3) and C8166-45 (lanes 4 to 6) were immunoprecipitated with a preimmune serum (PI) or an HBZ antiserum (α-HBZ). The complexes were separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to a nitrocellulose membrane, and probed with an anti-CREB antibody. A fraction of the whole-cell extract is shown in lane 1. (B) HBZ and CREB interact in uninfected cells. Cell extracts from CHOK1-Luc cells transfected with empty vector (−) or an expression plasmid for HBZ tagged with Myc (HBZ-Myc) were immunoprecipitated with an anti-Myc antibody. The complexes were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with an anti-CREB antibody (upper panel) or an anti-Myc antibody (lower panel). Protein standards (in kilodaltons) are indicated at the left. The asterisk indicates the heavy chain of immunoglobulin G. (C) HBZ and CREB interact in vitro. 35S-labeled HBZ was incubated with either GST (10 pmol, lane 2) or GST-CREB (10 pmol, lane 3). Bound HBZ protein was detected by PhosphorImager analysis. A fraction of the input protein (35%) is shown in lane 1.
Gel shift assays.
Gel shift assays with recombinant proteins were performed as previously described (21). Briefly, the indicated amounts of purified DNA-binding proteins and HBZ-bZIP were incubated with 2 to 10 fmol of 32P-end-labeled double-stranded DNA probe and 5 ng of poly(dA)·poly(dT) DNA per reaction in 0.5× TM 0.1 M. Binding reactions were incubated on ice for 30 min and resolved on 5% nondenaturating polyacrylamide gels. Gels were dried, and complexes were visualized by PhosphorImager analysis. The DNA probes have been described elsewhere (2, 21). The viral CRE is from the promoter-proximal 21-bp repeat, and the cellular CRE is from the human chorionic gonadotropin gene. Antibodies used for supershift experiments were directed against CREB-1 (X-12; Santa Cruz Biotechnology) and histone H3 (06-755; Upstate).
ChIP assay.
For the ChIP experiments the cells were electroporated as described previously (35). Briefly, 2 × 107 cells were electroporated with the GenePulser Xcell (Bio-Rad) in the presence of 20 μg of total DNA. The cells were harvested 24 h later for luciferase and ChIP analyses. The ChIP assay was performed essentially as described previously (34). CREB antibody was purchased from Upstate (antibody 06-863). The Tax monoclonal antibody (168B17-46-34) was obtained from the NIH AIDS Research and Reference Reagent Program.
Real-time PCR.
Real-time PCR and data analysis were performed with an iCycler and the optical assembly unit (Bio-Rad). Reactions were done by using the iQ SYBR green Supermix (Bio-Rad) as described previously (34). The PCR primers for the HTLV-1 promoter were AATGACCATGAGCCCCA and GTGAGGGGTTGTCGTCA. Standard curves were generated for primer sets using fivefold serial dilutions of CHOK1-Luc input DNA from the ChIP and were included on each experimental plate. PCR efficiencies ranged from 95 to 105%, with correlation coefficients greater than 0.99. Quantitation was done by comparing threshold cycle values for coimmunoprecipitated DNA to the threshold cycle value for the input DNA in each ChIP experiment as described previously (13).
RESULTS
HBZ represses HTLV-1 virion production.
We have previously shown that HBZ represses basal and Tax-dependent transcription from the HTLV-1 promoter and that these effects occur specifically through the viral CREs within the promoter (5, 17). We were interested in extending these studies by analyzing the effects of HBZ on virion production from the complete HTLV-1 proviral genome. The effect of HBZ was first tested in the context of a proviral DNA clone containing a luciferase reporter gene inserted in frame with the envelope amino acid sequence (33). This construct was derived from the K30 proviral DNA clone without altering the splice donor site for multiply spliced mRNA. This proviral DNA was transfected into 293T cells in the presence of increasing amounts of the HBZ expression vector pcDNA-HBZ-MycHis. As shown in Fig. 1A, cotransfection of 293T cells with K30-Luc and the HBZ expression plasmid led to a dose-dependent reduction in luciferase activity. In contrast, the vector pcDNA-HBZ-ΔATG, in which the ATGs are mutated to prevent production of the HBZ protein, did not reduce the level of luciferase activity. This result demonstrates that HBZ negatively regulates the expression of K30-Luc.
The effect of HBZ on virion production was also evaluated by cotransfecting 293T cells with the HTLV-1 molecular clone ACH (30) and pcDNA-HBZ-MycHis. Cell culture supernatants were harvested 24 h after transfection, and virion production was evaluated by quantifying HTLV-1 p19 antigen production. As shown in Fig. 1B, HBZ reduced the amount of p19gag in a dose-dependent manner. In contrast, we observed only a slight decrease in virion production when the HBZ expression plasmid was substituted with pcDNA-HBZ-ΔATG. Similar results were observed with another HTLV-1 molecular clone, K30 (data not shown). Altogether, our data show that HBZ represses HTLV-1 virion production.
To determine whether HBZ inhibits viral transcription in T cells, we cotransfected CEM cells with pcDNA-HBZ-MycHis and K30-Luc. As in 293T cells, HBZ expression led to a reduction in luciferase activity in the T cells (Fig. 1C). We found that the bZIP domain of HBZ contributed to repression, since expression of an HBZ mutant lacking the last 74 aa (encompassing the bZIP domain) caused less of a reduction in viral transcription (Fig. 1C). However, the fact that this mutant was still able to decrease viral transcription suggests that other regions of HBZ are also potentially involved in regulating this process. This result contrasts with a previous study showing that an HBZ mutant with a part of the leucine zipper domain deleted does not repress HTLV-1 transcription (3). This discrepancy may be explained by the different cell lines used for each experiment (293T were used in the previous study).
HBZ interacts with CREB in vivo and in vitro.
Based on the ability of HBZ to repress Tax-dependent viral transcription and interact with CREB-2, we were interested in determining whether HBZ also binds to CREB. Like CREB-2, CREB is a member of the ATF/CREB family of transcription factors and functions in Tax transactivation from the HTLV-1 promoter. We first performed coimmunoprecipitation experiments to determine whether endogenous HBZ and CREB interact in T cells chronically infected with HTLV-1. HBZ was immunoprecipitated from whole-cell extracts of HTLV-1-infected cells (MT-4 and C8166-45) using either rabbit anti-HBZ antiserum or preimmune serum from the same rabbit (17). Both infected cell lines have been shown to express HBZ (17). Figure 2A shows CREB present in complexes immunoprecipitated with HBZ, but not preimmune serum, from both HTLV-1-infected cell lines.
Since the viral protein Tax also interacts with CREB, we were interested in determining whether Tax or other viral proteins are required for the interaction between HBZ and CREB. We therefore performed coimmunoprecipitation experiments in an uninfected cell line (CHOK1). Cells were transfected with a control plasmid or the HBZ expression plasmid pcDNA-HBZ-MycHis. Whole-cell extracts prepared from transfected CHOK1 cells were immunoprecipitated using a Myc antibody and subsequently probed for CREB. As in HTLV-1-infected cells, a CREB-HBZ interaction was detected in CHOK1 cells expressing HBZ-Myc (Fig. 2B), demonstrating that additional viral proteins are not required for the interaction between HBZ and CREB. Interestingly, a 37-kDa protein was also detected with the 42-kDa CREB protein. This lower-molecular-mass protein is likely to be either ATF-1 or CREM, since both of these proteins cross-react with the CREB antibody, directed against the C terminus of CREB, and have similar apparent molecular masses. In vitro binding assays show that both ATF-1 and CREM-Ia interact with HBZ (see below).
Recently, we and others identified a novel alternative spliced isoform of HBZ (7, 43, 52). In this isoform, HBZ SP1, the first amino acids are replaced by four different amino acids. We transfected CHOK1 cells with a control plasmid or an expression plasmid for the newly identified isoform (pcDNA-HBZ SP1-Myc-His) (7) and found by coimmunoprecipitation that HBZ SP1 also interacts with endogenous CREB (data not shown).
To determine whether the interaction between HBZ and CREB is direct, we performed GST pull-down assays. Full-length HBZ was labeled with 35S in an in vitro transcription-translation reaction. This protein was combined with either recombinant GST or CREB carrying an N-terminal GST tag. Figure 2C shows that HBZ was detected with GST-CREB, but not GST alone, indicating that CREB interacts directly with HBZ in vitro (lane 3).
The interaction between HBZ and CREB is mediated through their bZIP domains.
To better map the interaction between HBZ and CREB, we performed GST pull-down assays using a truncated form of HBZ containing the bZIP domain (aa 123 to 209; the bZIP domain is located between aa 140 and 209) fused to GST (GST-HBZ-bZIP). The purified recombinant protein is shown in Fig. 3A. We first tested the binding of purified full-length CREB to GST-HBZ-bZIP or to GST alone. As shown in Fig. 3B, we observed specific binding of CREB to the bZIP domain of HBZ (lane 3). HBZ-bZIP carrying a hexahistidine tag was also expressed and purified from bacteria to perform the inverse experiment. After purification, we obtained a mixture of two products (Fig. 3C). As shown in Fig. 3D, both polypeptides interacted with GST-tagged recombinant CREB. These results support the idea that the HBZ interaction with CREB occurs through the bZIP domain of each protein. To test this hypothesis, we analyzed the binding of GST-HBZ-bZIP with the bZIP domain of CREB (CREB-bZIP) in a GST pull-down assay. As shown in Fig. 3E, GST-HBZ-bZIP was found to interact specifically with CREB-bZIP. To ensure that the N-terminal domain of HBZ does not interact with CREB, full-length HBZ and HBZ-ΔbZIP (aa 1 to 122) were labeled with 35S in an in vitro transcription-translation reaction and combined with either recombinant GST or GST-CREB. Figure 3F shows that, whereas full-length HBZ bound to GST-CREB (lane 5), the HBZ mutant lacking the bZIP domain did not (lane 6), confirming that HBZ interacts directly with CREB through its bZIP domain.
We used GST-HBZ-bZIP to determine whether HBZ interacts directly with CREM and ATF-1, as indicated by coimmunoprecipitation experiments (see Fig. 2B). Both proteins have bZIP domains similar to that of CREB (53), and interactions with HBZ should, therefore, occur through the bZIP domains. As expected, CREM-Ia-bZIP and ATF-1 both interacted with GST-HBZ-bZIP (Fig. 3G, lanes 3 and 6), but not GST alone (Fig. 3G, lanes 2 and 5).
To then determine whether HBZ binding disrupts the CREB homodimer, we tested the ability of HBZ to compete with CREB for binding to GST-CREB. GST-CREB was combined with 35S-labeled CREB alone (Fig. 3H, lane 3), or in the presence of increasing amounts of 35S-labeled full-length HBZ (Fig. 3H, lanes 4 to 6). The formation of the HBZ/GST-CREB complex reduced the amount of CREB homodimer in a dose-dependent manner. In contrast, addition of increasing amounts of BSA did not affect CREB binding (Fig. 3H, lanes 7 to 9). Furthermore, the HBZ-ΔbZIP did not disrupt the CREB homodimer (lane 14), suggesting that the bZIP domain of HBZ is required for this effect.
HBZ inhibits CREB binding on viral CRE in vitro.
Since HBZ interacts with the CREB-bZIP domain, we were interested in determining whether HBZ inhibits the ability of CREB to bind the viral CRE DNA. We performed gel shift assays to analyze protein-DNA complexes using the third viral CRE from the HTLV-1 promoter as the DNA probe. The recombinant proteins used for these assays are shown in Fig. 4D (HBZ-bZIP is shown in Fig. 3C). As expected, CREB formed a complex with the viral CRE (Fig. 4A, lane 7). However, when increasing amounts of HBZ-bZIP were incorporated into the binding reaction, the DNA/CREB complex was diminished in a dose-dependent manner (Fig. 4A, lanes 8 to 11). We performed parallel experiments with CREB-bZIP since CREB and HBZ interact via their bZIP domains (see Fig. 3E). Similar to full-length CREB, the bZIP domain formed a complex with the viral CRE (Fig. 4A, lane 2). Again, titration of HBZ-bZIP into the binding reaction led to a reduction of DNA/CREB-bZIP complexes (Fig. 4A, lanes 3 to 6). To confirm that the effect of HBZ-bZIP on CREB binding was specific, we attempted to disrupt the complex formed between the GAL4-VP16 protein and its UAS DNA-binding site using HBZ-bZIP (Fig. 4A, lane 13). The addition of HBZ did not affect GAL4-VP16 binding to the UAS site (Fig. 4A, lanes 14 to 17).
FIG. 4.
HBZ displaces CREB from the viral CRE in vitro. (A) HBZ-bZIP reduces CREB and CREB-bZIP binding to the viral CRE. Gel shift assays were used to analyze protein-DNA interactions. Binding reactions in lanes 1 to 11 contained 2.3 fmol of 32P-labeled viral CRE 3 probe with 0.16 pmol of CREB-bZIP (lanes 2 to 6) or 0.16 pmol of full-length CREB (lanes 7 to 11). Binding reactions in lanes 12 to 17 contained 3 fmol of 32P-labeled UAS probe (lanes 12 to 17) with 0.35 pmol of Gal4-VP16 (lanes 13 to 17). The reactions shown in lanes 3 to 6 and 8 to 11 also contained increasing amounts of HBZ-bZIP (0.02, 0.04, 0.08, and 0.16 pmol) and in lanes 14 to 17 (0.044, 0.09, 0.175, and 0.35 pmol). Positions of the free probe and relevant protein-DNA complexes are labeled. The minor bands that migrated below the major CREB homodimer are truncated forms of CREB present in the purified preparation. The slower-migrating complexes in CREB-bZIP lanes are CREB-bZIP aggregates typically observed with high concentrations of protein (represented by asterisks). (B) HBZ-ΔbZIP does not reduce CREB and CREB-bZIP binding to the viral CRE. Binding reactions contained 6.8 fmol of 32P-labeled viral CRE 3 probe (lanes 1 to 12) with 0.08 pmol of CREB (lanes 2 to 6) and 0.12 pmol of CREB-bZIP (lanes 8 to 12). The reaction shown in lane 3 also contained 0.8 pmol of HBZ-bZIP, and the reactions shown in lanes 4 to 6 contained increasing amounts of HBZ-ΔbZIP (0.8, 1.6, and 2.4 pmol). The reaction shown in lane 9 also contained 1.2 pmol of HBZ-bZIP, and the reactions shown in lanes 10 to 12 contained increasing amounts of HBZ-ΔbZIP (1.2, 1.8, and 2.4 pmol). (C) Specificity of CREB and CREB-bZIP binding to the viral CRE. Binding reactions contained 6.8 fmol of 32P-labeled viral CRE 3 probe (lanes 1 to 8) with 0.2 pmol of CREB (lanes 2 and 4) and 0.5 pmol of CREB-bZIP (lanes 5 to 7). An antibody directed against the bZIP domain of CREB (500 ng, lanes 3 and 6) or an antibody against histone H3 (500 ng, lanes 4 and 7) were added in the reactions. (D) Purified proteins used in gel shift assays. Purified CREB (lane 2), CREB-bZIP (lane 3), and HBZ-ΔbZIP (lane 4) are shown by Coomassie blue staining after SDS-PAGE. Protein standards (in kilodaltons) are indicated in lane 1. (E) HBZ-bZIP does not displace CREB-bZIP prebound to the viral CRE. Binding reactions contained 6 fmol of 32P-labeled viral CRE 3 probe with 0.16 pmol of the CREB-bZIP (lanes 2 to 13). Increasing amounts of HBZ-bZIP (0.04, 0.08, 0.16, 0.32, and 0.64 pmol) were added at the same time as CREB and DNA (lanes 3 to 7) or 30 min after CREB and DNA (lanes 9 to 13). The positions of the free probe and relevant protein-DNA complexes are labeled. The slower-migrating complexes in CREB-bZIP lanes are CREB-bZIP aggregates typically observed with high concentrations of CREB-bZIP (represented by asterisks).
The specificity of HBZ-bZIP in inhibiting the formation of the DNA/CREB and DNA/CREB-bZIP complexes was tested by substituting HBZ-bZIP with other proteins in gel shift assays. Figure 4B shows that, unlike HBZ-bZIP, HBZ-ΔbZIP did not affect the binding of CREB (lanes 4 to 6) or CREB-bZIP (lanes 10 to 12) to the viral CRE. Similar results were obtained using BSA in place of HBZ-bZIP (data not shown). These results demonstrate that HBZ-bZIP specifically inhibits the ability of CREB to bind to an HTLV-1 viral CRE sequence. In agreement with this result, we also found that the DNA binding of the Tax-CREB complex was inhibited in the presence of HBZ-bZIP (data not shown).
Gel shift complexes consisting of full-length CREB or its bZIP domain and the viral CRE have been characterized (21). However, we confirmed the protein composition of these complexes by supershift using an antibody directed against the bZIP domain of CREB (Fig. 4C, lanes 3 and 6). An antibody that does not recognize CREB (anti-histone H3) did not supershift the complexes (lanes 4 and 7).
In addition, we were interested in testing whether HBZ-bZIP displaces CREB from a preformed protein-DNA complex, since complexes formed between c-Jun and the AP-1 binding site are not disrupted by the subsequent addition of HBZ (5). In the experiments shown in lanes 2 to 7 of Fig. 4E (and in experiments above), CREB and HBZ were added simultaneously to the DNA. However, when CREB-bZIP was prebound to the viral CRE DNA, the complex was refractory to disruption by HBZ (Fig. 4E, lanes 9 to 13). These results indicate that once CREB is bound to the DNA, the complex is resistant to HBZ inhibition.
HBZ displaces Tax and CREB from the HTLV-1 promoter in vivo.
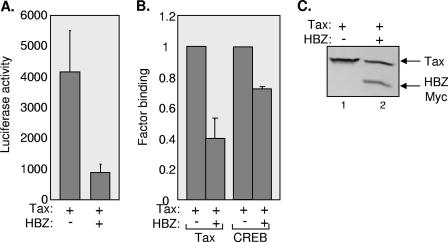
We were interested in correlating HBZ inhibition of CREB binding to the viral CRE in vitro with the ability of HBZ to repress Tax-mediated HTLV-1 transcription in living cells. To perform these experiments, we analyzed protein binding at the viral promoter in CHOK1-Luc cells that carry two to four genomically integrated copies of the HTLV-1 promoter (U3 region) cloned upstream of the luciferase gene (47). Cells were transfected with the Tax expression plasmid or cotransfected with both the Tax and the HBZ expression plasmids. Luciferase assays confirmed that HBZ significantly reduced Tax transactivation from the integrated promoter (Fig. 5A). Using this system we have previously shown by ChIP assays that Tax, when expressed in the absence of HBZ, bound to the integrated HTLV-1 long terminal repeat and promoted the recruitment of CREB (36). In Fig. 5B, we show that the coexpression of HBZ led to a reduction in Tax binding and, to a lesser extent, CREB binding (Fig. 5B). Overall, HBZ expression displaced more than 50% of Tax and ca. 25% of CREB from the HTLV-1 promoter. These results correlate the HBZ repression of Tax-dependent HTLV-1 transcription with a reduction in Tax and CREB from the viral promoter. Western blot analysis confirmed that Tax and HBZ were expressed in transfected cells (Fig. 5C).
FIG. 5.
HBZ displaces Tax and CREB from the HTLV-1 promoter in vivo. (A) HBZ represses Tax-mediated transcriptional activation from the HTLV-1 promoter. CHOK1-Luc cells were electroporated with pSG-Tax (10 μg) in the absence or presence of pcDNA-HBZ-MycHis (10 μg). Luciferase assays were performed 24 h after transfection. The reported values are the average luminescence from three independent experiments, and the standard error is indicated. (B) HBZ disrupts Tax and CREB binding at the HTLV-1 promoter. ChIP analysis was performed at 24 h posttransfection on the same transfected cells used in panel A. Real-time PCR was used to quantify the amount of viral promoter DNA in each immunoprecipitation. The levels of Tax and CREB binding in the presence of Tax and HBZ were normalized to binding in the presence of Tax alone (set to 1). The graph is the average ± the standard error from three independent ChIP experiments. (C) Tax and HBZ are expressed after transfection. Western blot analysis was performed at 24 h posttransfection with whole-cell extracts from one experiment used in panels A and B. The membrane was probed with anti-Tax and anti-Myc antibodies.
HBZ inhibits CREB binding on cellular CREs.
We wanted to determine whether HBZ also represses CREB transactivation from a cellular CRE. To address this question, we used a reporter plasmid containing three tandem copies of the consensus cellular CRE cloned upstream of a core promoter (the pminLuc-cellular CRE) (20). This reporter plasmid was transfected into CEM cells that were subsequently treated with forskolin to activate CREB (or a related factor) (20). As shown in Fig. 6A, the addition of forskolin to the culture medium produced a 20-fold increase in luciferase activity. However, cotransfection of pcDNA-HBZ-MycHis reduced the activation by fourfold, demonstrating that HBZ represses CREB activation from a cellular CRE. As expected, cotransfection of pcDNA-HBZΔATG did not reduce forskolin-mediated activation.
We then examined the effect of HBZ-bZIP on CREB-bZIP binding to a cellular CRE by using a gel shift assay. Unlike the viral CRE, the cellular CRE probe used in these experiments consists of the complete octanucleotide consensus CRE sequence. As expected, CREB-bZIP was able to bind to the cellular CRE in the absence of HBZ-bZIP (Fig. 6B, lane 2). However, the addition of increasing amounts of HBZ-bZIP to the binding reaction inhibited CREB binding to the cellular CRE (Fig. 6B, lanes 3 to 5). In contrast, increasing amounts of HBZ-ΔbZIP did not affect binding (lanes 9 to 11).
DISCUSSION
This study shows that the HBZ protein, encoded by the HTLV-1 retrovirus, abrogates virion production by repressing viral transcription. These data corroborate our previous report demonstrating that HBZ represses transcription driven from the HTLV-1 promoter and, more specifically, represses transcription mediated through the viral CREs (17). We have provided further insight into the mechanism of HBZ-mediated repression by characterizing an interaction between HBZ and the cellular transcription factor CREB. This finding is of particular interest since CREB appears to be the primary ATF/CREB member involved in Tax-mediated HTLV-1 transcription (8). We demonstrated that HBZ binds directly to CREB and that the interaction is mediated via the bZIP domain of each protein.
The consequence of this interaction is the inhibition of CREB binding to a viral CRE sequence from the HTLV-1 promoter in vitro. However, once formed, the CREB/viral CRE complex displayed resistance to subsequent challenge with HBZ. HBZ also displaced CREB and Tax from the HTLV-1 promoter in cells containing genomically integrated copies of the HTLV-1 promoter. Intriguingly, the apparent decrease in binding at the viral promoter was greater for Tax than for CREB, even though HBZ does not directly interact with Tax (17). This phenomenon could be due to the potential for HBZ to disrupt additional bZIP proteins from forming a complex with Tax and the viral CREs. In this scenario, more molecules of Tax than CREB would be prevented from binding the promoter. Indeed, several ATF/CREB members have been shown to participate in Tax-mediated transcription and form a complex with Tax at the promoter. Among these, we showed that CREB-2 is inhibited by HBZ expression (17). Furthermore, results from immunoprecipitation experiments (shown in Fig. 2B) and GST pull-down assays (shown in Fig. 3G) indicate that HBZ can additionally interact with either ATF-1 or CREM, both of which have been shown to function in Tax-mediated HTLV-1 transcription (23).
An important common denominator in the pathogenesis of the various HTLV-1-induced clinical syndromes appears to be HTLV-1 gene expression, especially Tax expression (56). Although the majority of infected individuals remain asymptomatic throughout their lifetime, ca. 5% develop inflammatory diseases and another 2 to 3% develop ATL (4). It is well established that transcription of the proviral genome is highly regulated in vivo (50). Indeed, HTLV-1 RNA is expressed in only 10% of HTLV-1-infected lymphocytes (46). An early induction of latency in HTLV-1-bearing T cells was also observed in experimentally infected squirrel monkeys (29). The precise mechanisms responsible for HTLV-1 latency remain unclear, but transcription of the viral genome can be induced in lymphocytes from ATL patients, HAM/TSP patients, and asymptomatic carriers by mitogenic stimulation of the cells in vitro (9), suggesting that the viral promoter remains functional. Based on our data, we propose that HBZ may play a role in viral latency by suppressing HTLV-1 transcription and Tax expression, which is supported by a recent report (3). HBZ expression is not the only mechanism involved in this tight control of HTLV-1 transcription. This control must require a combination of events that can act in concert, including the action of other HTLV-1-encoded proteins (45, 57), host factors (10, 12, 25, 35, 40), and epigenetic modifications (54). The repression of viral transcription, and therefore HTLV-1-antigen expression, may be a significant advantage to the virus in the infected cell by preventing its detection through a cytotoxic-T-lymphocyte response.
In addition to disrupting CREB recruitment to the HTLV-1 promoter, HBZ may also repress cellular genes that are activated by CREB. In vitro, we found that HBZ inhibited the binding of CREB to a consensus cellular CRE. In cells, HBZ also repressed luciferase expression mediated by three tandem cellular CREs upstream of a core promoter. These results are of importance since CREB has been shown to be involved in numerous cellular processes, including cell survival. However, based on the inability of HBZ to displace CREB from preformed CREB/DNA complexes, we suspect that HBZ only represses the expression of cellular genes that utilize a mechanism involving CREB recruitment. Genes that have CREB bound to the promoter would not be affected by HBZ, although the off-rate of CREB from these sites may dictate the actual degree of HBZ inhibition. Therefore, it is possible that HBZ will inhibit the expression of only a subset of genes that recruit CREB for activation, since the results of recent studies show that CREB is constitutively bound to many sites within the genome (11).
In conclusion, the HBZ protein represses HTLV-1 transcription and is potentially involved in the deregulation of cellular gene transcription. We and others recently demonstrated that another isoform of HBZ is expressed in HTLV-1-infected cells, HTLV-1 carriers, ATL cells lines, and fresh ATL cell samples, and some of these samples lack Tax expression (7, 43, 52). Furthermore, a recent study showed that the HBZ transcript is able to promote T-cell proliferation (52). Therefore, the HBZ gene possesses unique functions at both the protein and the transcript level that may contribute to leukemogenesis.
Acknowledgments
We thank K.-T. Jeang for the CHOK1-Luc cells and A. E. Keating for the CREM-Ia-bZIP plasmid.
This study was supported by a grant from the National Institutes of Health (CA55035) for J.K.N., by grants from the Centre National de la Recherche Scientifique (CNRS)-Université Montpellier I and from the Association pour la Recherche sur le Cancer (ARC no. 3606) to J.-M.M., and by The Cancer Research Society, Inc., to B.B. S.T. and P.H. were supported by fellowships from the Ligue Nationale Contre le Cancer and the Comité Régional (Gard and Hérault) de la Ligue Nationale Contre le Cancer, respectively.
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Adya, N., L. J. Zhao, W. Huang, I. Boros, and C. Z. Giam. 1994. Expansion of CREB's DNA recognition specificity by Tax results from interaction with Ala-Ala-Arg at positions 282 to 284 near the conserved DNA-binding domain of CREB. Proc. Natl. Acad. Sci. USA 91:5642-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, A., A. Franklin, M. Uittenbogaard, H. Giebler, and J. Nyborg. 1993. Pleiotropic effect of the human T-cell leukemia virus Tax protein on the DNA binding activity of eucaryotic transcription factors. Proc. Natl. Acad. Sci. USA 90:7303-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, J., B. Yamamoto, M. Li, A. J. Phipps, I. Younis, M. D. Lairmore, and P. L. Green. 2006. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood 107:3976-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barmak, K., E. W. Harhaj, C. Grant, T. Alefantis, and B. Wigdahl. 2003. Human T-cell leukemia virus type I-induced disease: pathways to cancer and neurodegeneration. Virology 9:522-529. [DOI] [PubMed] [Google Scholar]
- 5.Basbous, J., C. Arpin, G. Gaudray, M. Piechaczyk, C. Devaux, and J. M. Mesnard. 2003. The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J. Biol. Chem. 278:43620-46327. [DOI] [PubMed] [Google Scholar]
- 6.Brady, J., K.-T. Jeang, J. Duvall, and G. Khoury. 1987. Identification of p40x-responsive regulatory sequences within the human T-cell leukemia virus type I long terminal repeat. J. Virol. 61:2175-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavanagh, M., S. Landry, B. Audet, C. Arpin-Andre, P. Hivin, M.-E. Pare, J. Thete, E. Wattel, S. J. Marriott, B. Barbeau, and J.-M. Mesnard. 2006. HTLV-I antisense transcripts initiate in the 3′LTR and are alternatively spliced and polyadenylated. Retrovirology 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ching, Y. P., A. C. Chun, K. T. Chin, Z. Q. Zhang, K. T. Jeang, and D. Y. Jin. 2004. Specific TATAA and bZIP requirements suggest that HTLV-1 Tax has transcriptional activity subsequent to the assembly of an initiation complex. Retrovirology 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumais, N., M. E. Pare, S. Mercier, S. Bounou, S. J. Marriot, B. Barbeau, and M. J. Tremblay. 2003. T-cell receptor/CD28 engagement when combined with prostaglandin E2 treatment leads to potent activation of human T-cell leukemia virus type 1. J. Virol. 77:11170-11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ego, T., Y. Ariumi, and K. Shimotohno. 2002. The interaction of HTLV-1 Tax with HDAC1 negatively regulates the viral gene expression. Oncogene 21:7241-7246. [DOI] [PubMed] [Google Scholar]
- 11.Euskirchen, G., T. E. Royce, P. Bertone, R. Martone, J. L. Rinn, F. K. Nelson, F. Sayward, N. M. Luscombe, P. Miller, M. Gerstein, S. Weissman, and M. Snyder. 2004. CREB binds to multiple loci on human chromosome 22. Mol. Cell. Biol. 24:3804-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forgacs, E., S. K. Gupta, J. A. Kerry, and O. J. Semmes. 2005. The bZIP transcription factor ATFx binds human T-cell leukemia virus type 1 (HTLV-1) Tax and represses HTLV-1 long terminal repeat-mediated transcription. J. Virol. 79:6932-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin, A. A., M. F. Kubik, M. N. Uittenbogaard, A. Brauweiler, P. Utaisincharoen, M. A. Matthews, W. S. Dynan, J. P. Hoeffler, and J. K. Nyborg. 1993. Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2) ATF-2 response and cAMP element-binding protein (CREB). J. Biol. Chem. 268:21225-21231. [PubMed] [Google Scholar]
- 15.Fujisawa, J. I., M. Seiki, M. Sato, and M. Yoshida. 1986. A transcriptional enhancer sequence of HTLV-1 is responsible for transactivation mediated by p40x of HTLV-1. EMBO J. 5:713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gachon, F., A. Peleraux, S. Thebault, J. Dick, I. Lemasson, C. Devaux, and J. M. Mesnard. 1998. CREB-2, a cellular CRE-dependent transcription repressor, functions in association with Tax as an activator of the human T-cell leukemia virus type 1 promoter. J. Virol. 72:8332-8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudray, G., F. Gachon, J. Basbous, M. Biard-Piechaczyk, C. Devaux, and J. M. Mesnard. 2002. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 76:12813-12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georges, S. A., W. L. Kraus, K. Luger, J. K. Nyborg, and P. J. Laybourn. 2002. p300-mediated Tax transactivation from recombinant chromatin: histone tail deletion mimics coactivator function. Mol. Cell. Biol. 22:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calender, and G. de The. 1985. Antibodies to human T-lymphotropic virus type I in patients with tropical spastic paraparesis. Lancet ii:407-410. [DOI] [PubMed] [Google Scholar]
- 20.Giebler, H. A., I. Lemasson, and J. K. Nyborg. 2000. p53 recruitment of CREB binding protein mediated through phosphorylated CREB: a novel pathway of tumor suppressor regulation. Mol. Cell. Biol. 20:4849-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giebler, H. A., J. E. Loring, K. Van Orden, M. A. Colgin, J. E. Garrus, K. W. Escudero, A. Brauweiler, and J. K. Nyborg. 1997. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol. Cell. Biol. 17:5156-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goren, I., O. J. Semmes, K. T. Jeang, and K. Moelling. 1995. The amino terminus of Tax is required for interaction with the cyclic AMP response element binding protein. J. Virol. 69:5806-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant, C., K. Barmak, T. Alefantis, J. Yao, S. Jacobson, and B. Wigdahl. 2002. Human T-cell leukemia virus type I and neurologic disease: events in bone marrow, peripheral blood, and central nervous system during normal immune surveillance and neuroinflammation. J. Cell Physiol. 190:133-159. [DOI] [PubMed] [Google Scholar]
- 24.Harrod, R., Y. Tang, C. Nicot, H. S. Lu, A. Vassilev, Y. Nakatani, and C. Z. Giam. 1998. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol. Cell. Biol. 18:5052-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hivin, P., G. Gaudray, C. Devaux, and J. M. Mesnard. 2004. Interaction between C/EBPβ and Tax downregulates human T-cell leukemia virus type I transcription. Virology 318:556-565. [DOI] [PubMed] [Google Scholar]
- 26.Hoeffler, J. P., J. W. Lustbader and C.-Y. Chen. 1991. Identification of multiple nuclear factors that interact with cyclic adenosine 3′,5′-monophosphate response element-binding protein and activating transcription factor-2 by protein-protein interactions. Mol. Endocrinol. 5:256-266. [DOI] [PubMed] [Google Scholar]
- 27.Jeang, K. T., I. Boros, J. Brady, M. Radonovich, and G. Khoury. 1988. Characterization of cellular factors that interact with the human T-cell leukemia virus type I p40x-responsive 21-base-pair sequence. J. Virol. 62:4499-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeang, K. T., R. Chiu, E. Santos, and S. G. Kim. 1991. Induction of the HTLV-I LTR by Jun occurs through the Tax-responsive 21-bp elements. Virology 181:218-227. [DOI] [PubMed] [Google Scholar]
- 29.Kazanji, M., A. Ureta-Vidal, S. Ozden, F. Tangy, B. de Thoisy, L. Fiette, A. Talarmin, A. Gessain, and G. de The. 2000. Lymphoid organs as a major reservoir for human T-cell leukemia virus type 1 in experimentally infected squirrel monkeys (Saimiri sciureus): provirus expression, persistence, and humoral and cellular immune responses. J. Virol. 74:4860-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimata, J. T., F. H. Wong, J. J. Wang, and L. Ratner. 1994. Construction and characterization of infectious human T-cell leukemia virus type 1 molecular clones. Virology 204:656-664. [DOI] [PubMed] [Google Scholar]
- 31.Kimzey, A. L., and W. S. Dynan. 1998. Specific regions of contact between human T-cell leukemia virus type I Tax protein and DNA identified by photocross-linking. J. Biol. Chem. 273:13768-13775. [DOI] [PubMed] [Google Scholar]
- 32.Kwok, R. P., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the coactivator CBP. Nature 380:642-646. [DOI] [PubMed] [Google Scholar]
- 33.Langlois, M., B. Audet, E. Legault, M. E. Pare, M. Ouellet, J. Roy, N. Dumais, J. M. Mesnard, D. M. Rothstein, S. J. Marriott, M. J. Tremblay, and B. Barbeau. 2004. Activation of HTLV-I gene transcription by protein tyrosine phosphatase inhibitors. Virology 329:395-411. [DOI] [PubMed] [Google Scholar]
- 34.Lemasson, I., N. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2002. Transcription factor binding and histone modifications on the integrated proviral promoter in HTLV-I-infected T cells. J. Biol. Chem. 277:49459-49465. [DOI] [PubMed] [Google Scholar]
- 35.Lemasson, I., N. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2004. Transcription regulatory complexes bind the human T-cell leukemia virus 5′ and 3′ long terminal repeats to control gene expression. Mol. Cell. Biol. 24:6117-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemasson, I., N. J. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2006. Tax-dependent displacement of nucleosomes during transcriptional activation of human T-cell leukemia virus type 1. J. Biol. Chem. 281:13075-13082. [DOI] [PubMed] [Google Scholar]
- 37.Lenzmeier, B. A., E. E. Baird, P. B. Dervan, and J. K. Nyborg. 1999. The tax protein-DNA interaction is essential for HTLV-I transactivation in vitro. J. Mol. Biol. 291:731-744. [DOI] [PubMed] [Google Scholar]
- 38.Lenzmeier, B. A., H. A. Giebler, and J. K. Nyborg. 1998. Human T-cell leukemia virus type 1 Tax requires direct access to DNA for recruitment of CREB binding protein to the viral promoter. Mol. Cell. Biol. 18:721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu, H., C. A. Pise-Masison, T. M. Fletcher, R. L. Schiltz, A. K. Nagaich, M. Radonovich, G. Hager, P. A. Cole, and J. N. Brady. 2002. Acetylation of nucleosomal histones by p300 facilitates transcription from tax-responsive human T-cell leukemia virus type 1 chromatin template. Mol. Cell. Biol. 22:4450-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu, H., C. A. Pise-Masison, R. Linton, H. U. Park, R. L. Schiltz, V. Sartorelli, and J. N. Brady. 2004. Tax relieves transcriptional repression by promoting histone deacetylase 1 release from the human T-cell leukemia virus type 1 long terminal repeat. J. Virol. 78:6735-6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundblad, J. R., R. P. Kwok, M. E. Laurance, M. S. Huang, J. P. Richards, R. G. Brennan, and R. H. Goodman. 1998. The human T-cell leukemia virus-1 transcriptional activator Tax enhances cAMP-responsive element-binding protein (CREB) binding activity through interactions with the DNA minor groove. J. Biol. Chem. 273:19251-19259. [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto, J., T. Oshima, O. Isono, and K. Shimotohno. 2005. HTLV-1 HBZ suppresses AP-1 activity by impairing both the DNA-binding ability and the stability of c-Jun protein. Oncogene 24:1001-1010. [DOI] [PubMed] [Google Scholar]
- 43.Murata, K., T. Hayashibara, K. Sugahara, A. Uemura, T. Yamaguchi, H. Harasawa, H. Hasegawa, K. Tsuruda, T. Okazaki, T. Koji, T. Miyanishi, Y. Yamada, and S. Kamihira. 2006. A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization. J. Virol. 80:2495-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newman, J. R., and A. E. Keating. 2003. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300:2097-2101. [DOI] [PubMed] [Google Scholar]
- 45.Nicot, C., M. Dundr, J. Johnson, J. R. Fullen, N. Alonzo, R. Fukumoto, G. L. Princler, D. Derse, T. Misteli, and G. Franchini. 2004. HTLV-1-encoded p30II is a posttranscriptional negative regulator of viral replication. Nat. Med. 10:197-201. [DOI] [PubMed] [Google Scholar]
- 46.Ohshima, K., K. Hashimoto, S. Izumo, J. Suzumiya, and M. Kikuchi. 1996. Detection of human T lymphotrophic virus type I (HTLV-1) DNA and mRNA in individual cells by polymerase chain reaction (PCR) in situ hybridization (ISH) and reverse transcription (RT)-PCR ISH. Hematol. Oncol. 14:91-100. [DOI] [PubMed] [Google Scholar]
- 47.Okada, M., and K. T. Jeang. 2002. Differential requirements for activation of integrated and transiently transfected human T-cell leukemia virus type 1 long terminal repeat. J. Virol. 76:12564-12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osame, M., K. Usuku, S. Izumo, N. Ijichi, H. Amitani, A. Igata, M. Matsumoto, and M. Tara. 1986. HTLV-1-associated myelopathy: a new clinical entity. Lancet ii:1031-1032. [DOI] [PubMed] [Google Scholar]
- 49.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particle from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richardson, J. H., T. A. Waldmann, J. G. Sodroski, and W. A. Marasco. 1997. Inducible knockout of the interleukin-2 receptor alpha chain: expression of the high-affinity IL-2 receptor is not required for the in vitro growth of HTLV-I-transformed cell lines. Virology 237:209-216. [DOI] [PubMed] [Google Scholar]
- 51.Rousset, R., C. Desbois, F. Bantignies, and P. Jalinot. 1996. Effects on NF-κB1/p105 processing of the interaction between the HTLV-1 transactivator Tax and the proteasome. Nature 381:328-331. [DOI] [PubMed] [Google Scholar]
- 52.Satou, Y., J. Yasunaga, M. Yoshida, and M. Matsuoka. 2006. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T-cell leukemia cells. Proc. Natl. Acad. Sci. USA 103:720-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaywitz, A. J., and M. E. Greenberg. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68:821-861. [DOI] [PubMed] [Google Scholar]
- 54.Taniguchi, Y., K. Nosaka, J. Yasunaga, M. Maeda, N. Mueller, A. Okayama, and M. Matsuoka. 2005. Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology 2:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thebault, S., J. Basbous, P. Hivin, C. Devaux, and J. M. Mesnard. 2004. HBZ interacts with JunD and stimulates its transcriptional activity. FEBS Lett. 562:165-170. [DOI] [PubMed] [Google Scholar]
- 56.Yamano, Y., M. Nagai, M. Brennan, C. A. Mora, S. S. Soldan, U. Tomaru, N. Takenouchi, S. Izumo, M. Osame, and S. Jacobson. 2002. Correlation of human T-cell lymphotropic virus type 1 (HTLV-1) mRNA with proviral DNA load, virus-specific CD8+ T cells, and disease severity in HTLV-1-associated myelopathy (HAM/TSP). Blood 99:88-94. [DOI] [PubMed] [Google Scholar]
- 57.Ye, J., L. Silverman, M. Lairmore, and P. Green. 2003. HTLV-1 Rex is required for viral spread and persistence in vivo but is dispensable for cellular immortalization in vitro. Blood 102:3963-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin, M. J., and R. B. Gaynor. 1996. HTLV-1 21 bp repeat sequences facilitate stable association between Tax and CREB to increase CREB binding affinity. J. Mol. Biol. 264:20-31. [DOI] [PubMed] [Google Scholar]
- 59.Zhao, L. J., and C. Z. Giam. 1991. Interaction of the human T-cell lymphotrophic virus type I (HTLV-I) transcriptional activator Tax with cellular factors that bind specifically to the 21-base-pair repeats in the HTLV-I enhancer. Proc. Natl. Acad. Sci. USA 88:11445-11449. [DOI] [PMC free article] [PubMed] [Google Scholar]