Abstract
Herpes simplex virus type 1 is a human pathogen responsible for a range of illnesses from cold sores to encephalitis. The icosahedral capsid has a portal at one fivefold vertex which, by analogy to portal-containing phages, is believed to mediate genome entry and exit. We used electron cryotomography to determine the structure of capsids lacking pentons. The portal vertex appears different from pentons, being located partially inside the capsid shell, a position equivalent to that of bacteriophage portals. Such similarity in portal organization supports the idea of the evolutionary relatedness of these viruses.
Herpes simplex virus type 1 (HSV-1) is a large virus with a diameter of >200 nm. The first step in herpesvirus particle assembly is the formation of an icosahedral capsid 125 nm in diameter (13). The viral genome is packaged into the capsid through a specialized structure known as the portal, which is located at one of the fivefold vertices (9, 15). The other 11 vertices are occupied by pentons. The portal is believed to be organized as a dodecameric ring similar to those found in tailed double-stranded DNA bacteriophages such as Epsilon15 and P22 (2, 3, 6). A single portal subunit has a mass of 74 kDa, which makes the entire portal ∼888 kDa (9). This is similar to the mass of one penton (745 kDa) (16). This size equivalence at all vertices presents a challenge to efforts to determine the structure of the portal in the native capsid by any reconstruction method used in electron cryomicroscopy.
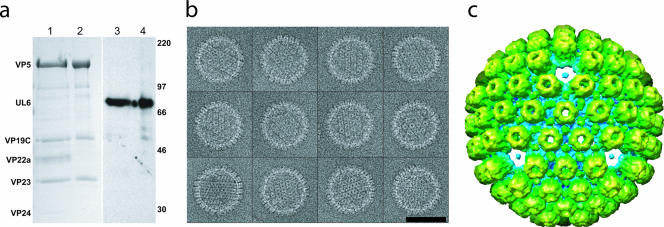
To generate a mass difference between the vertex containing the portal and the other 11 vertices, we treated the capsids with 6 M urea. This has been shown to specifically remove pentons, peripentonal triplexes, small hexon-associated protein, protease, and scaffold from capsids without disrupting the hexon-triplex shell (10). As shown in Fig. 1a, this treatment does not remove the portal protein (UL6), which is present in equivalent amounts in urea-treated and untreated capsids (11).
FIG. 1.
Urea-extracted capsids. (a) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis results for control (lanes 1 and 3) and 6 M urea-treated (lanes 2 and 4) capsids. Coomassie blue staining (lanes 1 and 2) shows that the urea-treated capsid sample contains the hexon (VP5) and triplex (VP19C, VP23) proteins but has lost the scaffolding protein (VP22a) and protease (VP24). The reduction in VP5 levels due to the loss of pentons is too small (6%) to be evident on this gel. Western blotting with MAb175 (lanes 3 and 4) (14) confirms that the portal protein (UL6) is not removed by urea extraction. Size markers are indicated in kilodaltons on the right. (b) A gallery of boxed capsids treated with 6 M urea. Bars, 100 nm. (c) A 3-nm resolution single-particle icosahedral reconstruction looking along a threefold orientation shows the capsid lacking pentons.
The urea-extracted capsids were applied to Quantifoil grids and flash frozen in liquid ethane by use of a Vitrobot. The specimens were imaged at −180°C in a JEM2010F electron microscope at 200 kV on a Gatan 4kx4k charge-coupled device camera at ×55,360 effective magnification, 15 to 20 e/Å2 dose, and 0.5 to 2.0 μm defocus (Fig. 1b). Particle boxing and contrast transfer function parameters were determined with EMAN software (7). SAVR was used to reconstruct a three-dimensional map with icosahedral symmetry imposed from randomly oriented capsid particles (4). This map shows holes at each of the fivefold vertices (Fig. 1c), confirming removal of most if not all of the pentons. The mass of the single portal vertex has been averaged over all 12 icosahedral fivefold positions, contributing insignificant density (nominally 1/12) to the reconstruction.
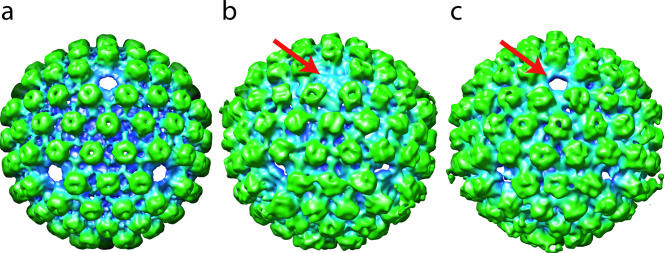
In order to visualize the unique vertex which contains the portal, we used tomographic reconstruction, which does not impose any symmetry. The same urea-extracted preparation was mixed with 15-nm colloidal gold and frozen as before. Tomographic tilt series were obtained using Mr T software (unpublished data) in a JEM2010F electron microscope operated at 200 kV. Three tilt series were imaged on a charge-coupled device camera at ×27,680 effective magnification, with a tilt range of ±70° in 2° increments. The total electron dose for each series was 47 to 50 e/Å2. Each tilt series was aligned using the gold to generate fiducial points and reconstructed using IMOD software (5). Thirty-eight subvolumes containing capsids were computationally extracted from three tomograms and spherically masked. The orientations of 33 of them were determined relative to the map shown in Fig. 1c (12), and they were rotated so that each particle had a fivefold vertex oriented along the z axis. These 33 were then icosahedrally averaged to yield a map (Fig. 2a) that resembles the single-particle-derived map (Fig. 1c) and that, as with the single-particle-derived map, displays no density at the fivefold positions. These two maps agree at up to 5.7-nm resolution according to 0.5 Fourier shell correlation criterion. The similarity of the two maps validates our three-dimensional alignment and icosahedral averaging procedures as used for the tomographic data. However, like the single-particle icosahedral reconstruction used to produce Fig. 1c, this icosahedral averaging procedure cannot reveal the portals even though they are present (Fig. 1a).
FIG. 2.
Tomographic reconstructions. (a) The icosahedrally averaged map looking along a threefold orientation is similar to that derived from single-particle reconstruction (Fig. 1c). (b) The fivefold-averaged map after orientation of the portal vertex along the positive z axis. The arrow points to the portal density. (c) Fig. 2b is rotated to show the empty vertex opposite the portal (arrow).
Next, we had to search for the vertex that contained the portal in each of the 33 tomographically reconstructed capsids. The obvious choice as the object of the search would be the vertex with the highest mass density. However, due to the effects of the limited tilt (±70°), the density map of tomographically reconstructed capsids gives differing results, with the highest density generally perpendicular to the original z axis of the tomographic volume. As a result, vertices near the “equator” of the particle in its original orientation will have apparently higher density than those near the “poles.” Our solution was to compare the densities at opposing pairs of vertices, because these are affected equally by the limited tilt. The largest intrapair density difference among the six pairs of vertices should arise when one member of the pair contains the portal while the other member is empty.
To quantify the mass at each vertex, we masked out a cylinder (45-Å radius and 110 Å tall) centered at each vertex. We determined the percentages of filled voxels at different display thresholds for each member of the pair of vertices. We repeated this procedure using the hexon densities surrounding the vertex. These measurements allowed us to determine whether the vertex contained protein density comparable to that of the hexons or not. Using this quantitative assessment, the portals in 13 out of the 33 particles were identified (data not shown). This selection was further verified by visual inspection of each particle. In the other particles, a unique vertex was not identified, possibly because the biochemical reaction was incomplete or the data were noisy (data not shown). Finally, the map of each of the 13 particles was rotated to place the portal vertex along the positive z axis and then combined and fivefold averaged (Fig. 2b and 2c) to reduce the noise and the effect of limited tilts.
Overall, the fivefold-averaged map still has strong hexon capsomeric features, and the depression at the top of each hexon suggests the position of the channel (16). However, the hexagonal nature of the capsomere is less obvious in the fivefold-averaged tomographic map (Fig. 2b and 2c) than in the icosahedrally averaged tomographic map (Fig. 2a) because of the reduced symmetry averaging and the smaller number of particles in the averaged results. A relatively flat density plug (Fig. 2b) that we interpret to be the portal is visible at only one vertex, while the other vertices lack this feature (Fig. 2b and c).
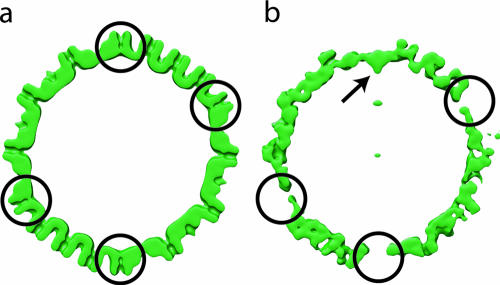
Figure 3 shows a slice through an icosahedral reconstruction of an untreated capsid (Fig. 3a) and an equivalent slice (Fig. 3b) through the reconstruction shown in Fig. 2b. Unlike the penton density results, the portal extends toward the interior of the particle. This location confirms that our density findings represent portals rather than residual pentons, since in that case the density would have been at the position expected for the penton. The other vertices of the treated capsid were empty.
FIG. 3.
Relative locations of pentons and portal in the capsid shell. (a) A slice through the icosahedral reconstruction of the native B capsids (16) shows the penton densities (circles) which are absent from panel b. (b) Tomographic reconstruction of the urea-treated capsids (Fig. 2b). The portal density is visible at only one vertex (arrow). Both maps were filtered at equivalent resolutions.
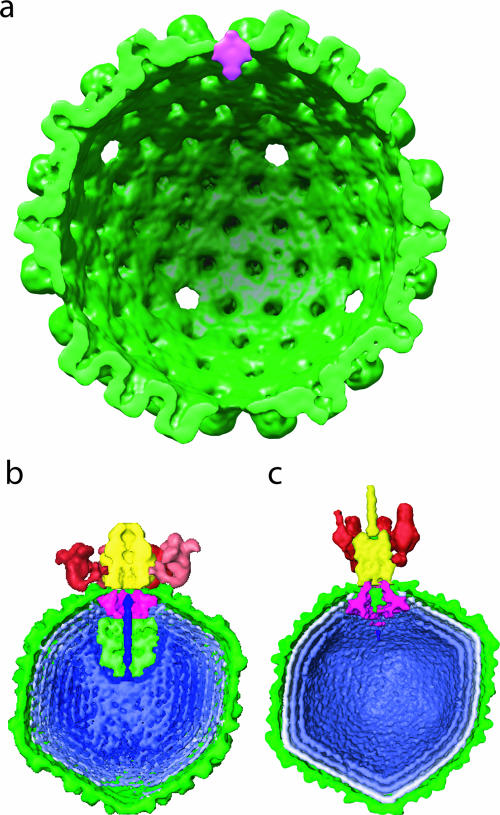
To delineate the portal as a separate mass density, we computed the difference between the tomographically derived maps with fivefold (Fig. 2b) and with icosahedral (Fig. 2a) averaging. The only significant difference density was at the portal. The portal density was cylindrically averaged to reduce the bias resulting from the previous imposition of fivefold symmetry. Figure 4a shows that much of the portal mass lies at a level that corresponds to the floor of the capsid. On the external surface of the capsid, the portal forms a low dome. On the inside, the portal extends as a conical density into the interior of the capsid.
FIG. 4.
Comparison of HSV-1 and bacteriophage portal locations. (a) The HSV-1 portal structure (magenta) was determined by rotationally averaging the results for the difference map between Fig. 2a and 2b. It is shown in the context of the icosahedral averaged tomographic map (Fig. 2a) (green). The density represented by magenta is 14 nm tall and 11 nm wide. For comparison, the ∼2-nm resolution electron cryomicroscopy maps of Epsilon15 (EBI accession number EMD-1175) (b) and P22 (EBI accession number EMD-1222) (c) are shown, with their portals also shown in magenta (2, 3). The displays of these three maps are to scale.
It is important to emphasize that Fig. 4a shows the actual location of the difference density in the map. The position of the portal as revealed by this map is at a lower radius than was suggested by the model previously obtained by manual insertion of a biochemically isolated portal structure into a capsid reconstruction (15). Despite the low resolution and the cylindrical averaging, the overall shape resembles the silhouette of the isolated portal structure (15). Interestingly, the organization at this vertex is similar to that seen in bacteriophages Epsilon15 (Fig. 4b) and P22 (Fig. 4c), whose portals also do not extend far beyond the capsid shells but do extend into the interior of the capsids (2, 3, 6). This organizational similarity between the tailed double-stranded DNA bacteriophage and herpesvirus portals provides additional evidence to support the hypothesis that these viruses share a common ancestry (1, 8).
Density map accession numbers.
The density maps determined in this work have been deposited in the European Bioinformatics Institute (EBI) database under accession numbers EMD-1305, EMD-1306, EMD-1307, and EMD-1308.
Acknowledgments
This research was supported by National Institutes of Health grants (P41RR02250 and R01AI38469) and the Robert Welch Foundation.
We thank Valerie Preston for helpful discussion and David McNab for excellent technical assistance.
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Baker, M. L., W. Jiang, F. J. Rixon, and W. Chiu. 2005. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 79:14967-14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, J., P. Weigele, J. King, W. Chiu, and W. Jiang. 2006. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14:1073-1082. [DOI] [PubMed] [Google Scholar]
- 3.Jiang, W., J. Chang, J. Jakana, P. Weigele, J. King, and W. Chiu. 2006. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439:612-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang, W., Z. Li, Z. Zhang, C. R. Booth, M. L. Baker, and W. Chiu. 2001. Semi-automated icosahedral particle reconstruction at sub-nanometer resolution. J. Struct. Biol. 136:214-225. [DOI] [PubMed] [Google Scholar]
- 5.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71-76. [DOI] [PubMed] [Google Scholar]
- 6.Lander, G. C., L. Tang, S. R. Casjens, E. B. Gilcrease, P. Prevelige, A. Poliakov, C. S. Potter, B. Carragher, and J. E. Johnson. 2006. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312:1791-1795. [DOI] [PubMed] [Google Scholar]
- 7.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 8.McGeoch, D. J., F. J. Rixon, and A. J. Davison. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90-104. [DOI] [PubMed] [Google Scholar]
- 9.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 11.Patel, A. H., and J. B. MacLean. 1995. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology 206:465-478. [DOI] [PubMed] [Google Scholar]
- 12.Schmid, M. F., A. M. Paredes, H. A. Khant, F. Soyer, W. Chiu, and J. M. Shively. 2006. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J. Mol. Biol. 364:526-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subak-Sharpe, J. H., and D. J. Dargan. 1998. HSV molecular biology: general aspects of herpes simplex virus molecular biology. Virus Genes 16:239-251. [DOI] [PubMed] [Google Scholar]
- 14.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trus, B. L., N. Cheng, W. W. Newcomb, F. L. Homa, J. C. Brown, and A. C. Steven. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 Å. Science 288:877-880. [DOI] [PubMed] [Google Scholar]