Abstract
Virus-specific neutralizing antibodies present an obstacle to the effective use of adenovirus vectors for gene therapy and vaccination. The specific sites recognized by neutralizing antibodies have not been identified for any adenovirus, but they have been proposed to reside within the hexon, in small regions of the molecule that are exposed on the capsid surface and possess sequences that vary among serotypes. We have mapped the epitopes recognized by a panel of seven hexon-specific monoclonal antibodies that neutralize the chimpanzee adenovirus 68 (AdC68). Surface plasmon resonance experiments revealed that the antibodies compete for a single hexon binding site, and experiments with synthetic peptides indicated that this site resides within just one small surface loop. Mutations within this loop (but not in other surface loops) permitted virus to escape neutralization by all seven monoclonal antibodies and to resist neutralization by polyclonal antisera obtained from animals immunized against AdC68. These results indicate that a single small surface loop defines a major neutralization site for AdC68 hexon.
Modified adenoviruses have been widely used as vehicles for gene delivery and as vaccine vectors. Most adenovirus vectors in use have been derived from the human serotype 5 (Ad5). As almost all human adults have been exposed to Ad5, they possess neutralizing antibodies to Ad5 that limit the efficiency of the virus as a delivery vector (7, 33, 42). Approaches to circumvent the problem of preexisting immunity include chemical modification of Ad5 surface proteins to mask the neutralizing epitopes (4, 19, 29) and replacement of the immunogenic capsid proteins with those of other serotypes (21, 23, 32, 41, 44). Many investigators are also exploring the use of rare human serotypes (such as human Ad48) or nonhuman adenoviruses (including those derived from dogs, fowl, and nonhuman primates) to which humans are not usually immune (2, 6, 13, 16-18, 22, 30).
An alternative approach is to identify the specific sites on adenovirus that are recognized by neutralizing antibodies and then modify those sites to generate mutants capable of escaping neutralization. One nonhuman serotype that has been proposed as an alternative vector for vaccination is the chimpanzee adenovirus 68 (AdC68) (42). AdC68 is not neutralized by most human adult sera and elicits a strong transgene product-specific immune response in animals already immune to Ad5 (7, 33, 42). However, because one immunization with an AdC68 vector will induce serotype-specific immunity, multiple-dose immunization regimens may require the availability of additional vectors. Production of antigenically modified vectors would be facilitated if the epitopes recognized by the neutralizing antibodies were well characterized.
The adenovirus capsid is an icosahedron with long fibers projecting from the vertices. Twelve copies of the trimeric major capsid protein, hexon, form each of the 20 triangular facets of the icosahedron; trimeric fibers are inserted into the pentameric penton bases at the 12 vertices (28). Each hexon trimer has a pseudo-hexagonal base, which allows for close packing within the facet, and three tower domains that are exposed on the exterior surface of the virion (Fig. 1A). Adenovirus-neutralizing antibodies can be raised against any of the major capsid proteins (9, 22, 34, 36, 37, 40). However, experiments with chimeric viruses—in which capsid components of one serotype were replaced by those of another serotype—suggest that hexon is the predominant target of serotype-specific neutralizing antibodies (10, 22, 23, 32, 44).
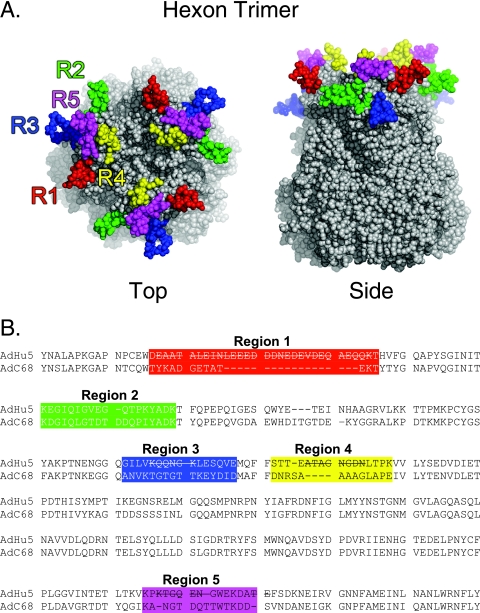
FIG. 1.
Structure and sequence of AdC68 adenovirus hexon. (A) Space-filling representation of the crystal structure of the trimeric AdC68 hexon showing the potential epitope regions (43). Potential epitope regions on hexon are located in the three tower regions at the top of the molecule. These form the exterior surface of the virion. The regions are labeled on the sequence and highlighted in the same color on the molecule: R1 (red), R2 (green), R3 (blue), R4 (yellow), and R5 (magenta). Although R3 is on the upper surface of hexon, it is buried between hexons in the intact virion and so is not accessible to antibodies. The figure was produced with PyMol v0.99. (B) Partial sequence alignment of the 932-residue AdC68 and 951-residue Ad5 hexons based on an alignment of the structures, showing residues 121 to 474 of Ad5 and 121 to 455 of AdC68. Amino acid residues in flexible regions that were not observed in the Ad5 X-ray structure are indicated by lines through the sequence. The potential epitope regions are colored as in panel A.
The typical hexon is a protein of ∼100 kDa in mass and ∼960 amino acids in length (25). Alignment of available hexon sequences and crystal structures of hexons from Ad5 and Ad2 show that all hexons share a highly conserved core structure (25). The greatest sequence variability is confined to nine hypervariable regions that map to small surface loops within the hexon towers. These form the exposed surface of the capsid and are thus likely to contain the epitopes recognized by serotype-specific neutralizing antibodies (24). The crystal structure of AdC68 hexon (43) (Fig. 1A) has more precisely defined the location of the surface loops than a homology model. In combination with the crystal structures of Ad2 (1, 25) and Ad5 (24, 25) hexons, the new structure has given an improved sequence alignment for all hexons (J. J. Rux and R. M. Burnett, unpublished). Nonetheless, the specific locations of neutralizing epitopes have not been previously identified for any adenovirus. Using a combination of immunochemical and genetic approaches, with the production of hexon-specific monoclonal antibodies neutralizing AdC68, we now have identified a single major neutralizing site on the AdC68 hexon.
MATERIALS AND METHODS
Hexon-specific neutralizing monoclonal antibodies.
Two BALB/c mice (6 to 8 weeks) (Jackson Laboratory, Bar Harbor, ME) were injected intramuscularly with 50 μl of 2 × 1011 particles/ml of CsCl-purified AdC68 virus. The mice were boosted intraperitoneally 2 weeks later with 100 μg of AdC68 hexon in phosphate-buffered saline (PBS) emulsified with TiterMax (CytRx Corp). Hexon was purified by column chromatography from the soluble structural proteins in lysates of infected cells, as described for purification of Ad5 hexon (26). A wealth of structural and biochemical evidence supports the assumption that this form of the protein is a good representation of that found in the adenovirus virion (26, 31). Three days later, the mice were sacrificed and hybridoma fusion was performed by a standard protocol (12). Hybridomas secreting hexon-specific antibodies were screened by enzyme-linked immunosorbent assay (ELISA) against 10 μg/well of purified AdC68 hexon and cloned by limiting dilution. Eighty hexon-specific clones were subsequently screened in a virus neutralization assay using AdC68 encoding green fluorescent protein (GFP) (described below); seven showed neutralizing activity (90% inhibition of GFP expression) and were subcloned (Table 1). Antibody isotypes were determined with a mouse hybridoma subtyping kit (Roche) according to the manufacturer's instructions. Antibody concentrations in hybridoma supernatants were determined with ELISA kits for both immunoglobulin G (IgG) and IgM antibodies (ZeptoMetrix).
TABLE 1.
Hexon-specific monoclonal antibodies
| MAb | Isotype | Neutralizing activity |
|---|---|---|
| 15G7 | IgM(κ) | + |
| 1B9 | IgG1(κ) | + |
| 8G10a | IgG1(κ) | + |
| 7G4a | IgG1(κ) | + |
| 10E8 | IgG2a(κ) | + |
| 4C1 | IgG2a(κ) | + |
| 6B8 | IgG2b(κ) | + |
| 4D1 | IgG2a(κ) | − |
| 11F6 | IgG2a(κ) | − |
| 12B4 | IgG2a(κ) | − |
| 6A2 | IgG2a(κ) | − |
| 3H3 | IgG2a(κ) | − |
Sequence analysis revealed that hybridomas 8G10 and 7G4 are clonally related; they most likely originated from a B-cell clone expanded in the immunized animal.
Polyclonal antisera specific for AdC68.
Rabbit antiserum was obtained from animals that had been injected intramuscularly with 1 × 1013 AdC68 particles and then boosted subcutaneously with 1 × 1013 particles in complete Freund's adjuvant. Rhesus macaque antiserum was a pooled sample from six animals, each immunized once intramuscularly with 2 × 1011 AdC68 particles.
Defining potential epitope regions.
A structural alignment of three hexon crystal structures, Ad2 (protein data bank identifier [PDB] 1P2Z), Ad5 (PDB 1P30), and AdC68 (43), was used to guide the alignment of the full-length hexon sequences using SwissPDBViewer (11) as described previously (25). PyMOL v0.99 (3) was used to generate the space-filling representation of the AdC68 hexon structure. Five regions within hexon that show serotype-specific sequence variation, and which are located on the surface of the molecule, were selected as potential sites for recognition by neutralizing antibodies (R1 [residues 136 to 148], R2 [residues 163 to 181], R3 [residues 233 to 248], R4 [residues 253 to 265], R5 [residues 412 to 425]) (Fig. 1). These correspond approximately to the positions of five of the nine hypervariable regions defined earlier from an alignment of multiple hexon sequences (25). Information from the AdC68 hexon crystal structure has led to an improved global alignment, resulting in some shifting of the AdC68 sequence relative to the sequences of other hexons (Rux and Burnett, unpublished). Based on this new alignment, the putative AdC68 neutralizing sites described above correspond approximately to hypervariable regions 1, 2, 4, 5, and 8 as previously defined.
Surface plasmon resonance.
Experiments were performed at room temperature on a Biacore X optical biosensor (Biacore AB). AdC68 hexon (10 μg/ml), diluted in 10 mM acetate buffer, pH 4.5, was directly coupled via primary amines to a CM5 chip using a standard protocol (BIA applications handbook, Biacore AB, Uppsala, Sweden) (14) in a running buffer of PBS-0.005% Tween 20. In short, an equal mix of 0.2 M EDC [N-ethyl-N′-(dimethylaminopropyl carbodiimide)] and 0.5 M NHS (N-hydroxysuccinimide) was used to activate the reactive ester groups on flow cell 2 (FC2) for coupling with hexon. The immobilization resulted in 900 resonance units of hexon on FC2. Ethanolamine was then used to block the remaining activated ester groups. As a control, FC1 was similarly activated for coupling and blocked with ethanolamine but was not coupled with hexon.
For antibody binding measurements, hybridoma supernatant was injected over both FC1 and FC2 at 5 μl/min in a buffer consisting of PBS with 0.9 mM CaCl2, 0.5 mM MgCl2, and 0.005% Tween 20. All experiments were performed with hybridoma supernatants. Antibodies with high apparent affinity (4C1, 10E8, 6B8, 4D1, and 11F6) were diluted (3 μg/ml) in running buffer prior to injection; antibodies of lower affinity (7G4 and 8G10) were used undiluted. Two neutralizing antibodies were not used in this experiment (1B9 and 15G7). The concentration of 1B9 in the supernatant was too low to achieve significant binding to hexon on the chip; 15G7 antibody is an IgM, and we were concerned that its multivalent binding would prove difficult to compete with IgGs.
Antibody binding competition assays were performed as described by Krummenacher et al. (14). Briefly, 20 μl of the first antibody (Ab) was injected for 4 min at 5 μl/min to saturate available binding sites. Once a steady state was achieved, as evidenced by a steady plateau in the binding curve, the second antibody (20 μl) was immediately injected for 4 min at 5 μl/min. Injections of 0.2 M glycine (pH 2) after the competition experiment removed bound antibody and returned the response signal to baseline. The binding of each monoclonal antibody (MAb) in the absence of a competing first MAb was defined as BA. Binding of an antibody after presaturation with itself was defined as BA−A. Binding of antibody A after saturation by a first antibody X was defined as BX−A. Competition data were calculated as a percentage of binding for each second Ab after blocking with each first Ab, using the formula [(BX−A − BA−A) × 100/(BA − BA−A)].
Antibody binding to variable region peptides, purified hexon, and whole virus.
Synthetic peptides (see Fig. 6A) representing the potential neutralizing sites (R1 to R5) on the surface of the AdC68 hexon were synthesized (Sympep). Peptides were solubilized in trifluoroacetic acid, which has been reported to enhance antibody recognition by promoting the formation of secondary structures within peptides (15). Peptides (0.5 μg/well in 100 μl of trifluoroacetic acid) or purified AdC68 hexon (0.4 μg/well in 100 μl PBS) was immobilized on tissue culture-treated polystyrene 96-well plates (Corning, Inc.) by overnight incubation at 37°C. The plates were washed four times with PBS-0.05% Tween 20 and then blocked for 1 h at room temperature in 1% bovine serum albumin-PBS-0.05% Tween 20. The immobilized peptides and hexon were incubated with each neutralizing antibody (0.1 μg Ab/well) or with an anti-AdC68 rabbit polyclonal antibody (1:1,000 dilution) for 1 h at room temperature. Bound MAb was detected with horseradish peroxidase (HRP)-conjugated goat antibody to murine IgG (Amersham Pharmacia Biotech) or IgM (Santa Cruz), and rabbit antibody was detected with HRP-conjugated goat antibody to rabbit immunoglobulin (Santa Cruz). Plates were developed with 3,5,3′,5′-tetramethylbenzidine (KPL) at room temperature for 5 min. The reaction was stopped with o-phosphoric acid, and absorbance was read at 450 nm.
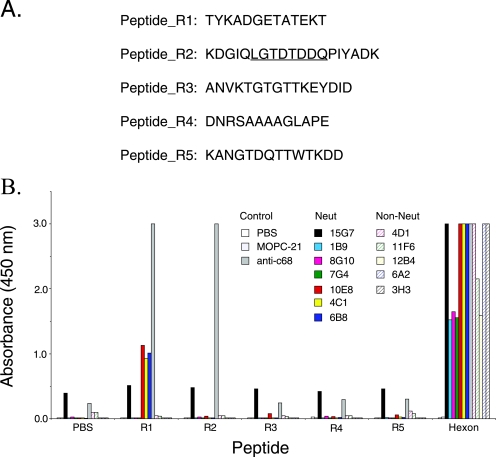
FIG. 6.
Antibody interaction with synthetic peptides. (A) Sequences of peptide_R1 to peptide_R5. Each peptide sequence represents a small exposed loop on the hexon molecule. To enhance its solubility, peptide_R2 contains additional residues that flank the corresponding loop; the sequence of the exposed loop is underlined. (B) Peptides and purified hexon (0.5 μg/well) were immobilized on 96-well plates and probed with hexon-specific monoclonal antibodies (0.1 μg Ab/well) and anti-AdC68 polyclonal serum. Hexon was plated as a positive control (0.5 μg/well). The 10E8, 4C1, and 6B8 neutralizing MAbs bound to peptide_R1. Anti-AdC68 polyclonal serum recognized both peptide_R1 and peptide_R2.
For the virus ELISA, AdC68 was diluted in 100 μl of 100 mM sodium carbonate (pH 9.5) (from 0.5 × 109 particles to 9 × 109 particles/well) and immobilized on a 96-well plate by overnight incubation at 4°C. Each neutralizing MAb and control antibody was tested against the virus, and bound antibody was detected as described above.
For immunoblot analysis (data not shown), purified AdC68 hexon was incubated for 10 min in Laemmli sample buffer containing 1% sodium dodecyl sulfate (SDS), either at room temperature or at 95°C. Heated and unheated hexon samples were electrophoresed side-by-side (10 μg/lane) on SDS-polyacrylamide gels and then transferred to a polyvinylidene difluoride membrane; membrane segments (each segment contained heated and unheated samples side-by-side) were incubated with MAbs diluted in TBST (0.15 M NaCl, 0.01 M Tris-HCl [pH 7.4], 0.05% Tween 20). The bound antibodies were detected with HRP-conjugated goat antibody to murine IgG (Amersham Pharmacia Biotech) or IgM (Santa Cruz) diluted to 1:20,000 in TBST. Western blots were analyzed by enhanced chemiluminescence with the WestDura kit (Bio-Rad).
Construction of AdC68 with mutated hexon.
A plasmid encoding an AdC68 vector with an E1 deletion (pPAN9pkGFP) (6) was obtained from the Vector Core at the University of Pennsylvania. Since the ∼38-kb adenovirus genome contains few unique restriction sites, the hexon variable region was cloned into smaller vectors that would facilitate the shuttling of an altered hexon into the full-length viral plasmid (Fig. 2).
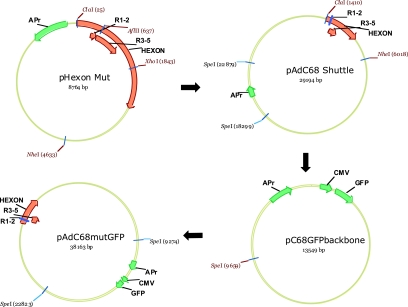
FIG. 2.
Shuttling mutated hexons into the complete AdC68 genome. The R1 and 2 and R3 to 5 variable regions are labeled and represented as arrows (orange) below a larger arrow identifying the hexon gene (orange) in the plasmid. The size of these arrows varies according to the size of the plasmid, and in the larger plasmids (i.e., pAdC68mutGFP), the R1 and 2 region is represented as a small blue box rather than an arrow. Nonunique restriction sites are labeled in black and unique sites in red. In all plasmids, f1 ORI, APr, CMV, and GFP (green) identify the origin of replication, ampicillin resistance, CMV promoter, and green fluorescent protein genes, respectively. The mutated hexon gene was excised from a mutagenesis vector (pHexon Mut) with the restriction enzymes ClaI and NheI and cloned into these same sites in a shuttle vector (pAdC68Shuttle) for final ligation into a plasmid containing the complete coding sequence for AdC68. In the last step of the cloning strategy, the 25-kb SpeI fragment from pAdC68Shuttle, containing the hexon mutation, was ligated with the SpeI-digested pC68GFP backbone to generate a pPAN9CMVGFP (AdC68mutGFP) mutant vector.
Construction of the mutagenesis vector pHexon Mut.
pPAN9pkGFP was digested with the restriction endonucleases ClaI and EcoRI, and a 7.3-kb fragment, containing the hexon, was isolated by gel electrophoresis and ligated into the ClaI/EcoRI sites of pBR322 (New England Biolabs). From the resulting plasmid, pBR7.3kb Hexon, a 4.6-kb hexon-containing fragment was isolated, by digestion with ClaI/NheI, and cloned into corresponding overhangs in pBR322 to yield the mutagenesis vector pHexon Mut (Fig. 2). pHexon Mut contained the unique restriction sites ClaI, AflII, and XhoI. Specific mutations were introduced into the hexon by splice-overlap extension PCR (38). PCR products containing mutations in variable regions R1 and R2 were ligated into the ClaI and AflI sites of pHexon Mut, and products with mutations in R3 to R5 were inserted into the AflII and XhoI sites.
Construction of the shuttle vector pAdC68 Shuttle.
A vector (pAdC68 Shuttle) was created to facilitate the transfer of mutated hexon fragments from the mutagenesis vector into the full-length AdC68 genome (Fig. 2). A fragment (pAdx-SphI) isolated from pAdenoXGFP (obtained from the Vector Core, University of Pennsylvania) contained the origin of replication, the ampicillin resistance marker, and a unique SpeI site. To isolate this fragment, pAdenoXGFP was digested with SpeI and self-ligated to create pAdx-SpeI. A 4.6-kb fragment, generated by digesting pAdx-SpeI with SphI and AflII, was then self-ligated to produce pAdx-SphI. Finally, both pAdx-SphI and pPAN9pkGFP were digested with SpeI. The 25-kb fragment from pPAN9pkGFP was ligated into the SpeI site of pAdx-SphI. The resulting vector, pAdC68 Shuttle, contained unique restriction sites ClaI and NheI flanking the hexon gene for shuttling the mutated hexon from the mutagenesis vector.
Construction of the pC68GFP backbone.
SpeI sites divide pPAN9pkGFP into two major components: a 25-kb fragment containing the hexon gene and a 14-kb fragment containing both the components for replication and restriction sites I-CeuI/PI-SceI, which can accommodate a gene insert (sites not shown but surround the GFP insert). For our experiments, GFP, under the control of a cytomegalovirus (CMV) promoter, was inserted into the I-CeuI/PI-SceI region of the pPAN9pkGFP. The resulting plasmid was called pPAN9CMVGFP. Digestion of this plasmid with SpeI resulted in a 14-kb fragment that contains the origin of replication and is sufficient for growth in bacterial cells. This 14-kb plasmid (pAdC68GFPbackbone) served as the backbone, which was ligated to the 25-kb fragment from pAdC68Shuttle, to reconstruct the complete viral genome (pAdC68mutGFP) (Fig. 2). The presence of the mutation was verified by sequencing the final cDNA clone.
Full-length mutant genomes were introduced into HEK-293 by calcium phosphate transfection (BD CalPhos transfection kit). The day before the transfections, 4 × 105 HEK-293 cells were added to each well of a six-well plate and grown overnight at 37°C with 5% CO2. Prior to transfection into HEK-293 cells, pAdC68mutGFP vector was digested with PacI to isolate the recombinant genome from the vector backbone. Transfected cells were incubated at 37°C with 5% CO2. Recombinant adenovirus was harvested when 95 to 100% cytopathic effect was observed.
Neutralization assays. (i) Screening hybridoma supernatants.
Approximately 100 fluorescent focus forming units of AdC68 encoding GFP was mixed with 100 μl of hybridoma supernatant and incubated for 1 h at 37°C in a 96-well plate in the presence of 5% CO2. The virus-Ab mixtures were transferred to a 96-well plate containing confluent monolayers of 293 cells (5 × 104 cells/well) and incubated for 48 h. After the 48 h incubation, infection was monitored using fluorescence microscopy to look for the appearance of cells expressing GFP.
(ii) Quantitative neutralization assay with MAbs.
Replication-deficient wild-type or mutant virus encoding GFP (6 × 107 particles in 500 μl Dulbecco's modified Eagle medium-10% fetal calf serum) was mixed with 500 μl MAb supernatant and incubated for 1 h at 37°C in a 24-well plate in the presence of 5% CO2. The virus-MAb mixtures were transferred to a 24-well plate containing confluent monolayers of 293 cells (2.5 × 105 cells/well) and incubated for 48 h. Cells were harvested with trypsin-EDTA and analyzed by flow cytometry to quantitate GFP expression.
(iii) Neutralization assay with anti-AdC68 polyclonal serum.
Each polyclonal serum was serially diluted in PBS, and 500 μl of each dilution was mixed with wild-type or mutant AdC68 or wild-type Ad5, all encoding GFP (6 × 107 particles in 500 μl PBS). The antibody-virus mixtures were incubated for 1 h at 37°C and then transferred to HEK-293 monolayers in 24-well plates. Monolayers were incubated for 48 h, and then the cells were washed with PBS and trypsinized. The cells were spun and resuspended in 500 μl of PBS, and GFP expression was quantified by flow cytometry. The number of GFP-positive cells was directly proportional to the number of input viral particles in the range at which these assays were performed (data not shown). The reduction in neutralization was calculated as [(1 − 1/n-fold difference in serum dilution) × 100%], where a fivefold difference would indicate that the virus was able to escape from 80% of the neutralizing antibody response.
Protein structure accession number.
Crystallographic refinement of the AdC68 hexon structure has been completed, and the coordinates have been deposited in the Protein Data Bank (identification code 2OBE).
RESULTS
Production of AdC68 hexon-specific neutralizing MAbs.
BALB/c mice were immunized with AdC68 virus and then boosted with purified AdC68 hexon, and splenocytes were isolated and used to generate hybridomas. Hexon-specific antibodies, selected by ELISA on purified hexon, were screened for the capacity to neutralize an AdC68 vector expressing enhanced GFP. Seven hexon-specific neutralizing antibodies were identified (Table 1); five hexon-specific antibodies with no neutralizing activity were selected as negative controls. Experiments were performed with supernatants of hybridomas that had been subcloned twice by limiting dilution. The AdC68 neutralizing antibodies had no neutralizing activity against Ad5.
Binding of the MAbs to AdC68 hexon and virions.
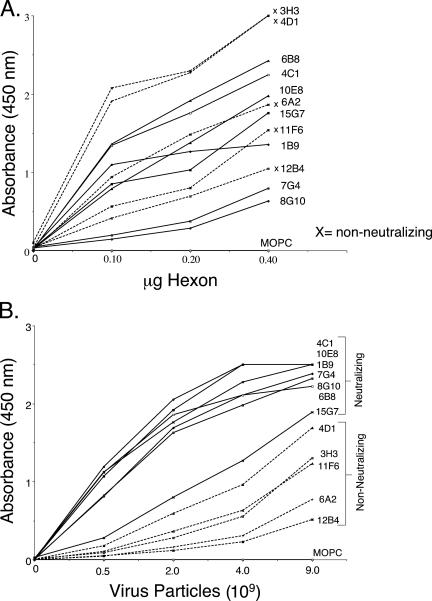
In an attempt to identify differences between the neutralizing and nonneutralizing antibodies, we tested the binding of the antibodies to hexon and to intact virions in ELISAs (Fig. 3). Because antibodies were selected for their ability to bind purified hexon, it was possible that the nonneutralizing antibodies recognized epitopes that are inaccessible in the intact virion. In ELISAs, both neutralizing and nonneutralizing antibodies bound to purified hexon (Fig. 3A) as well as to intact virus (Fig. 3B). There was no obvious relationship between neutralizing activity and avidity for purified hexon; however, the neutralizing MAbs appeared to bind with more avidity to intact virus than did the nonneutralizing MAbs. These results suggest that epitopes recognized by neutralizing antibodies may be highly exposed on the virion surface, whereas nonneutralizing antibodies may bind to sites that are accessible on the purified hexon but partially masked in the intact virion.
FIG. 3.
Antibody interactions with purified hexon and intact virus. (A) Purified hexon. Hexon (twofold serial dilutions) was immobilized and probed with MAbs. Nonneutralizing antibodies are labeled with an x and represented with dotted lines; neutralizing antibodies are shown with solid lines. (B) Intact virions. AdC68 purified by CsCl centrifugation was diluted, immobilized, and probed as performed for hexon. An IgG1 myeloma protein, MOPC21, was used as a nonspecific control.
Analysis of MAb competition by surface plasmon resonance.
We used an optical biosensor to determine if neutralizing antibodies competed for a single site within hexon. In each experiment, one antibody was injected over hexon covalently attached to the chip. When available binding sites were saturated and a plateau was reached, binding of a second antibody was measured. In a representative experiment, saturation of binding sites by the neutralizing MAb 4C1 did not inhibit subsequent binding of a nonneutralizing antibody, 11F6 (Fig. 4A). In contrast, saturation with 4C1 did prevent subsequent binding of another neutralizing antibody, 7G4 (Fig. 4B). This strongly suggests that 4C1 and 7G4 (but not 11F6) compete for the same binding site. Similar experiments were performed with each of the antibodies in combination (Fig. 5) with competition scored as described in Materials and Methods.
FIG. 4.
Representative biosensor antibody competition experiments. (A) No competition. The MAb 4C1 was permitted to bind to immobilized hexon; once a plateau was reached, MAb 11F6 was injected. The binding of 11F6 was not inhibited (did not differ from binding in the absence of 4C1). (B) Competition. MAb 4C1 was permitted to bind and then MAb 7GF was injected; 7G4 binding was inhibited by 4C1. RU, resonance units.
FIG. 5.
Summary of competition results. The chart indicates the amount of inhibition of binding of each second antibody (rows) in the presence of a first antibody (columns). Black boxes indicate >60% inhibition. Gray boxes indicate 30 to 60%. Inhibition of <30% is indicated by white boxes. The percentage of binding was calculated by the formula [(BX−A − BA−A) × 100/(BA − BA−A)], where BA represents the binding of antibody A to a chip not previously saturated. BA−A represents the residual binding of antibody A to a chip previously saturated with antibody A. BX−A represents the binding of antibody A to a chip previously saturated with antibody X. Neutralizing antibodies are indicated in black letters and are surrounded by a dotted box. The nonneutralizing antibodies are indicated in gray letters.
Among the neutralizing antibodies, competition was nearly complete. When binding sites were saturated with the neutralizing MAbs 4C1, 10E8, or 6B8, no subsequent binding was seen for any other neutralizing antibody. In contrast, binding of the nonneutralizing controls (11F6 and 4D1) was not inhibited. Saturation of the chip with neutralizing MAb 7G4 or 8G10 only partially inhibited binding of neutralizing MAb 4C1, 10E8, or 6B8; however, in the reciprocal experiments, MAb 4C1, 10E8, or 6B8 completely inhibited binding of MAbs 7G4 and 8G10. The incomplete inhibition by 7G4 and 8G10 is most likely explained by these antibodies' low apparent affinity for immobilized hexon (data not shown), consistent with their relatively poor binding to purified hexon in the ELISA (Fig. 3A).
Antibody recognition of hexon peptides.
We reasoned that epitopes for serotype-specific neutralizing antibodies would be exposed on the virion surface and would reside within regions of hexon where sequences vary among serotypes. Using the crystal structure of AdC68 hexon (Fig. 1A) and sequence alignment data (Fig. 1B), we identified five small surface loops as potential neutralizing sites (R1 to 5). Synthetic peptides were then designed to represent the loop sequences (Fig. 6A). Peptides were not made for hexon loops that would be inaccessible when hexons are assembled into a viral capsid (5, 31). When the seven neutralizing MAbs were tested against peptides in an ELISA (Fig. 6B), three of the seven neutralizing Abs recognized peptide_R1 (10E8, 4C1, and 6B8). A rabbit polyclonal anti-AdC68 serum with high-titer neutralizing activity bound to peptides_R1 and _R2. The remaining four neutralizing MAbs and the nonneutralizing MAbs did not bind any of the peptides. All antibodies bound to purified hexon.
To determine if the neutralizing and nonneutralizing antibodies recognized conformation-dependent or linear epitopes, we immunoblotted hexon that had been electrophoresed after exposure to 1% SDS at room temperature or at 95°C (data not shown). At room temperature, hexon in SDS maintains its trimeric form (8). None of the antibodies listed in Table 1 recognized hexon that had been heated to 95°C in the presence of SDS, suggesting that all the antibodies must recognize conformation-dependent epitopes.
R1 mutations reduce susceptibility to neutralization.
The biosensor experiments suggested that the neutralizing antibodies recognized a single site (composed of one or more individual epitopes), and the peptide ELISA suggested that this site was likely to be contained within R1. To map neutralizing epitopes by an independent approach, we produced viruses in which single site mutations were introduced into each small surface loop likely to contain an epitope (Table 2). (After refinement of the crystal structure, R3 was found to be involved in interhexon contacts and inaccessible from the surface of the virion; no mutations were introduced into R3.) These mutant viruses were then screened against the panel of neutralizing antibodies. Mutations in regions R2, R4, and R5 did not affect neutralization by any of the MAbs. When a single amino acid substitution was introduced within R1 (ETA to EDA), the mutant virus showed reduced susceptibility to four of the neutralizing antibodies (15G7, 1B9, 7G4, and 8G10; data not shown), consistent with the idea that these MAbs recognize epitopes within R1. A mutant with three R1 residues altered (ETA to CDQ) escaped neutralization by all seven neutralizing MAbs (Fig. 7). This mutant virus (termed CDQ) replicated to the same titers and showed the same particle/PFU ratio as did the wild-type virus (not shown), suggesting that mutations in R1 did not significantly alter the virion structure. In a virus ELISA, the CDQ mutant was recognized by nonneutralizing antibodies 4D1 and 11F6 but not by any of the neutralizing antibodies (not shown), suggesting that mutation of R1 did not perturb antibody epitopes elsewhere on the virus capsid. (Similarly, the failure of mutations in other loops to affect antibody recognition by MAbs 10E8, 4C1, and 6B8, which we found to bind directly to peptide_R1, suggests that mutations in the small surface loops have only local effects and do not prevent antibody recognition of remote epitopes.)
TABLE 2.
Mutations in small hexon surface loops
| R (loop region) | Wild-type sequencea | Mutation(s) introducedb | MAbs escaped by mutant |
|---|---|---|---|
| 1 | ...ADGETAT... | ADGATAT | None |
| ADRETAT | None | ||
| ADGEDAT | 1B9, 7G4, 8G10, 15G7 | ||
| ADGCDQT | 1B9, 4C1, 6B8, 7G4, 8G10, 10E8, 15G7 | ||
| 2 | ...DTDDQ... | DAAAQ | None |
| 4 | ...SAAAAG... | SDDDAG | None |
| SAARAG | None | ||
| 5 | ...TDQTTW... | TDQTPW | None |
| TDETTW | None |
The sequence shown represents only a small region of the loop. The dots on either side of the sequence indicate that additional residues within the loop are not represented.
Mutated residues are underlined.
FIG. 7.
The AdC68 R1 mutant (CDQ) virus resists neutralization by MAbs. Particles (6 × 107; 0.5 to 0.6 infectious units per cell) of GFP-encoding wild-type or CDQ mutant virus were incubated with culture medium or with 100 μl of each hybridoma supernatant for 1 h at 37°C and then added to HEK-293 cells. After 48 h, GFP-expressing cells were counted in a flow cytometer. Error bars indicate standard deviations for triplicate samples. Hybridoma supernatants contained the following concentrations of antibody: 8G10, 15 μg/ml; 7G4, 10 μg/ml; 6B8, 55 μg/ml; 4C1, 60 μg/ml; 1B9, 1 μg/ml; 15G7, 30 μg/ml; 10E8, 60 μg/ml; 4D1, 60 μg/ml; 11F6, 40 μg/ml.
We tested the CDQ mutant against neutralizing antisera prepared by immunizing animals with AdC68. These animals would be expected to generate virus-specific antibodies directed against multiple epitopes. High-titer rabbit serum (50% neutralization at a 2 × 106-fold dilution of the serum) was fivefold less efficient in neutralizing the CDQ mutant than it was in neutralizing wild-type virus, suggesting that the mutant escapes 80% of the neutralizing response (Fig. 8A). Serum pooled from rhesus macaques exposed to one dose of AdC68 (50% neutralization at a 5 × 103-fold dilution) showed 32-fold less activity against the mutant (Fig. 8B), suggesting escape from more than 90% of the neutralizing antibody. These results indicate that R1 contains a dominant neutralization site, which accounts for much of the neutralizing antibody response to AdC68.
FIG. 8.
The AdC68 R1 mutant resists neutralization by polyclonal antisera. Particles (6 × 107) of GFP-expressing wild-type (black circles) or CDQ mutant (black squares) virus or Ad5 control virus (white squares) (0.5 to 0.6 infectious units per cell) were incubated with serial dilutions of rabbit (A) or rhesus macaque (B) anti-AdC68 polyclonal antiserum for 1 h at 37°C then added to HEK-293 cells. After 48 h, cells expressing GFP were counted in a flow cytometer. Arrows indicate the 50% neutralization point in each experiment.
DISCUSSION
In the experiments described here, we found that the R1 region of the AdC68 hexon is a major target for neutralizing antibodies. Surface plasmon resonance experiments showed that a panel of hexon-specific neutralizing MAbs competed for a single binding site. Several of the monoclonal antibodies bound to a linear peptide derived from R1, directly confirming that they recognize epitopes within R1. Finally, mutations within R1 permitted virus to escape from neutralization by all seven monoclonal antibodies and to resist neutralization by polyclonal antisera from animals immunized with intact virus. Although it is formally possible that mutations in R1 might affect the structure of epitopes elsewhere on the viral capsid, the combination of genetic, competition, and peptide ELISA results leads us to conclude that the site for antibody recognition is the small surface loop corresponding to R1.
The monoclonal antibodies we tested were all generated by immunizing inbred mice, and we cannot exclude the possibility that the neutralizing antibody response may differ in other hosts. However, the results obtained with antisera from immunized rabbits and monkeys suggest that, in these animals, most of the polyclonal response was directed at R1. Since humans are not commonly infected by AdC68 (6), we cannot determine how closely the neutralizing responses in humans may resemble those we observed in mice, rabbits, and monkeys.
Our data are consistent with previous reports that hexon is the major target of adenovirus neutralizing antibodies (10, 20, 22, 23, 32, 44). Recently, Roberts et al. (21) replaced all of the exposed Ad5 hexon variable regions with those from human adenovirus 48 to produce a vector that escaped preexisting immunity to Ad5. The neutralizing site we mapped in AdC68 lies within the broad region swapped by these investigators and confirms the importance of the hexon region as a major target for neutralizing antibodies. Neutralizing responses against minor epitopes may become more evident after repeated exposures to virus; we observed that the CDQ mutant was less resistant to a very high titer serum obtained from rabbits given repeated doses of virus. Other investigators (20) have reported that fiber-specific neutralizing antibodies could be detected in high-titer human sera but not in sera with lower neutralizing titers. We do not know whether the additional neutralization of the CDQ mutant we see with the higher-titer rabbit serum is directed at other epitopes within the hexon or epitopes within other capsid proteins.
To our knowledge, R1 is the first neutralizing site to be identified for any adenovirus. It has been reported previously that neutralizing antibodies to Ad2 hexon targeted a 15-kDa fragment within the protein, a fragment that includes R1 (36), but the precise location of the epitope was not identified. A recent cryoelectron microscopy reconstruction of Ad5 bound by a neutralizing monoclonal antibody shows contact between the antibody and Ad5 in the region corresponding to R1 and R5 in AdC68 hexon (27, 37). It will be interesting to determine if the loop corresponding to AdC68 R1 is the major neutralization target in other adenoviruses.
Although the mechanism by which antibodies neutralize adenoviruses is unclear, some neutralizing antibodies do not prevent virus entry into cells and thus may act by inhibiting postentry events (34, 37, 39, 40). It has been proposed that an acidic cluster in the subgroup C adenoviruses may interact with histone-H1 during capsid disassembly at the nuclear pore complex (35); it has also been suggested that this acidic cluster may contribute to conformational changes in the capsid that occur after endosomal acidification (36, 39). Antibody interaction with AdC68 R1 might conceivably interfere with capsid disassembly or genome entry into the nucleus. AdC68 R1 occupies the same location in the hexon structure as do the acidic loops in Ad5 and Ad2; however, it does not contain an acidic cluster of amino acids.
Our results indicate that much of the neutralizing response to AdC68 is directed against a single site and that mutation of this site results in a vector that resists neutralization in vitro. Because neutralizing antibodies interfere with adenovirus-mediated gene delivery and immunization, capsid modifications that permit escape from neutralization may be important for successful applications of adenoviral vectors in the clinic. Structure-based immunological characterization is a powerful tool that has permitted us to focus our efforts on those hexon regions most likely to contain epitopes accessible to neutralizing antibodies.
Acknowledgments
We thank B. S. Sachais and the Optical Biosensor Shared Resource Facility at Path BioRESOURCE, University of Pennsylvania, for assistance with the optical biosensor experiments. We also thank Yang Zhu Du and Nina Luning Prak of the Department of Pathology at the University of Pennsylvania for help with hybridoma genotyping and antibody sequence interpretation.
This work was supported by NIH PAR-00093 and AI-52271 and by a grant from the Pennsylvania Department of Health.
Footnotes
Published ahead of print on 15 November 2006.
REFERENCES
- 1.Athappilly, F. K., R. Murali, J. J. Rux, Z. Cai, and R. M. Burnett. 1994. The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 A resolution. J. Mol. Biol. 242:430-455. [DOI] [PubMed] [Google Scholar]
- 2.Bangari, D. S., S. Shukla, and S. K. Mittal. 2005. Comparative transduction efficiencies of human and nonhuman adenoviral vectors in human, murine, bovine, and porcine cells in culture. Biochem. Biophys. Res. Commun. 327:960-966. [DOI] [PubMed] [Google Scholar]
- 3.DeLano, W. L. 2002. The PyMOL, user's manual. DeLano Scientific LLC, San Carlos, CA.
- 4.Eto, Y., J. Q. Gao, F. Sekiguchi, S. Kurachi, K. Katayama, H. Mizuguchi, T. Hayakawa, Y. Tsutsumi, T. Mayumi, and S. Nakagawa. 2004. Neutralizing antibody evasion ability of adenovirus vector induced by the bioconjugation of methoxypolyethylene glycol succinimidyl propionate (MPEG-SPA). Biol. Pharm. Bull 27:936-938. [DOI] [PubMed] [Google Scholar]
- 5.Fabry, C. M., M. Rosa-Calatrava, J. F. Conway, C. Zubieta, S. Cusack, R. W. Ruigrok, and G. Schoehn. 2005. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 24:1645-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farina, S. F., G. P. Gao, Z. Q. Xiang, J. J. Rux, R. M. Burnett, M. R. Alvira, J. Marsh, H. C. Ertl, and J. M. Wilson. 2001. Replication-defective vector based on a chimpanzee adenovirus. J. Virol. 75:11603-11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald, J. C., G. P. Gao, A. Reyes-Sandoval, G. N. Pavlakis, Z. Q. Xiang, A. P. Wlazlo, W. Giles-Davis, J. M. Wilson, and H. C. Ertl. 2003. A simian replication-defective adenoviral recombinant vaccine to HIV-1 gag. J. Immunol. 170:1416-1422. [DOI] [PubMed] [Google Scholar]
- 8.Fortsas, E., M. Petric, and M. Brown. 1994. Electrophoretic migration of adenovirus hexon under non-denaturing conditions. Virus Res. 31:57-65. [DOI] [PubMed] [Google Scholar]
- 9.Gahéry-Ségard, H., F. Farace, D. Godfrin, J. Gaston, R. Lengagne, T. Tursz, P. Boulanger, and J. G. Guillet. 1998. Immune response to recombinant capsid proteins of adenovirus in humans: antifiber and anti-penton base antibodies have a synergistic effect on neutralizing activity. J. Virol. 72:2388-2397. Gahéry-Ségard [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gall, J., A. Kass-Eisler, L. Leinwand, and E. Falck-Pedersen. 1996. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J. Virol. 70:2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 12.Kohler, G., and C. Milstein. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256:495-497. [DOI] [PubMed] [Google Scholar]
- 13.Kremer, E. J., S. Boutin, M. Chillon, and O. Danos. 2000. Canine adenovirus vectors: an alternative for adenovirus-mediated gene transfer. J. Virol. 74:505-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krummenacher, C., I. Baribaud, M. Ponce de Leon, J. C. Whitbeck, H. Lou, G. H. Cohen, and R. J. Eisenberg. 2000. Localization of a binding site for herpes simplex virus glycoprotein D on herpesvirus entry mediator C by using antireceptor monoclonal antibodies. J. Virol. 74:10863-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang, E., G. I. Szendrei, V. M. Lee, and L. Otvos, Jr. 1994. Spectroscopic evidence that monoclonal antibodies recognize the dominant conformation of medium-sized synthetic peptides. J. Immunol. Methods 170:103-115. [DOI] [PubMed] [Google Scholar]
- 16.Mack, C. A., W. R. Song, H. Carpenter, T. J. Wickham, I. Kovesdi, B. G. Harvey, C. J. Magovern, O. W. Isom, T. Rosengart, E. Falck-Pedersen, N. R. Hackett, R. G. Crystal, and A. Mastrangeli. 1997. Circumvention of anti-adenovirus neutralizing immunity by administration of an adenoviral vector of an alternate serotype. Hum. Gene Ther. 8:99-109. [DOI] [PubMed] [Google Scholar]
- 17.Mastrangeli, A., B. G. Harvey, J. Yao, G. Wolff, I. Kovesdi, R. G. Crystal, and E. Falck-Pedersen. 1996. “Sero-switch” adenovirus-mediated in vivo gene transfer: circumvention of anti-adenovirus humoral immune defenses against repeat adenovirus vector administration by changing the adenovirus serotype. Hum. Gene Ther. 7:79-87. [DOI] [PubMed] [Google Scholar]
- 18.Moffatt, S., J. Hays, H. HogenEsch, and S. K. Mittal. 2000. Circumvention of vector-specific neutralizing antibody response by alternating use of human and non-human adenoviruses: implications in gene therapy. Virology 272:159-167. [DOI] [PubMed] [Google Scholar]
- 19.Mok, H., D. J. Palmer, P. Ng, and M. A. Barry. 2005. Evaluation of polyethylene glycol modification of first-generation and helper-dependent adenoviral vectors to reduce innate immune responses. Mol. Ther. 11:66-79. [DOI] [PubMed] [Google Scholar]
- 20.Nanda, A., D. M. Lynch, J. Goudsmit, A. A. Lemckert, B. A. Ewald, S. M. Sumida, D. M. Truitt, P. Abbink, M. G. Kishko, D. A. Gorgone, M. A. Lifton, L. Shen, A. Carville, K. G. Mansfield, M. J. Havenga, and D. H. Barouch. 2005. Immunogenicity of recombinant fiber-chimeric adenovirus serotype 35 vector-based vaccines in mice and rhesus monkeys. J. Virol. 79:14161-14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts, D. M., A. Nanda, M. J. Havenga, P. Abbink, D. M. Lynch, B. A. Ewald, J. Liu, A. R. Thorner, P. E. Swanson, D. A. Gorgone, M. A. Lifton, A. A. Lemckert, L. Holterman, B. Chen, A. Dilraj, A. Carville, K. G. Mansfield, J. Goudsmit, and D. H. Barouch. 2006. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 441:239-243. [DOI] [PubMed] [Google Scholar]
- 22.Roy, S., D. S. Clawson, R. Calcedo, C. Lebherz, J. Sanmiguel, D. Wu, and J. M. Wilson. 2005. Use of chimeric adenoviral vectors to assess capsid neutralization determinants. Virology 333:207-214. [DOI] [PubMed] [Google Scholar]
- 23.Roy, S., P. S. Shirley, A. McClelland, and M. Kaleko. 1998. Circumvention of immunity to the adenovirus major coat protein hexon. J. Virol. 72:6875-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rux, J. J., and R. M. Burnett. 2000. Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol. Ther. 1:18-30. [DOI] [PubMed] [Google Scholar]
- 25.Rux, J. J., P. R. Kuser, and R. M. Burnett. 2003. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution x-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 77:9553-9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rux, J. J., D. Pascolini, and R. M. Burnett. 1999. Large-scale purification and crystallization of adenovirus hexon, p. 259-275. In W. S. M. Wold (ed.), Adenovirus methods and protocols—methods in molecular medicine, vol. 21. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 27.Saban, S. D., R. R. Nepomuceno, L. D. Gritton, G. R. Nemerow, and P. L. Stewart. 2005. CryoEM structure at 9A resolution of an adenovirus vector targeted to hematopoietic cells. J. Mol. Biol. 349:526-537. [DOI] [PubMed] [Google Scholar]
- 28.San Martin, C., and R. M. Burnett. 2003. Structural studies on adenoviruses. Curr. Top Microbiol. Immunol. 272:57-94. [DOI] [PubMed] [Google Scholar]
- 29.Steel, J. C., H. M. Cavanagh, M. A. Burton, and W. H. Kalle. 2004. Microsphere-liposome complexes protect adenoviral vectors from neutralising antibody without losses in transfection efficiency, in-vitro. J. Pharm. Pharmacol. 56:1371-1378. [DOI] [PubMed] [Google Scholar]
- 30.Stevenson, M., E. Boos, C. Herbert, A. Hale, N. Green, M. Lyons, L. Chandler, K. Ulbrich, N. van Rooijen, V. Mautner, K. Fisher, and L. Seymour. 2006. Chick embryo lethal orphan virus can be polymer-coated and retargeted to infect mammalian cells. Gene Ther. 13:356-368. [DOI] [PubMed] [Google Scholar]
- 31.Stewart, P. L., S. D. Fuller, and R. M. Burnett. 1993. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 12:2589-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sumida, S. M., D. M. Truitt, A. A. Lemckert, R. Vogels, J. H. Custers, M. M. Addo, S. Lockman, T. Peter, F. W. Peyerl, M. G. Kishko, S. S. Jackson, D. A. Gorgone, M. A. Lifton, M. Essex, B. D. Walker, J. Goudsmit, M. J. Havenga, and D. H. Barouch. 2005. Neutralizing antibodies to adenovirus serotype 5 vaccine vectors are directed primarily against the adenovirus hexon protein. J. Immunol. 174:7179-7185. [DOI] [PubMed] [Google Scholar]
- 33.Tatsis, N., L. Tesema, E. R. Robinson, W. Giles-Davis, K. McCoy, G. P. Gao, J. M. Wilson, and H. C. Ertl. 2006. Chimpanzee-origin adenovirus vectors as vaccine carriers. Gene Ther. 13:421-429. [DOI] [PubMed] [Google Scholar]
- 34.Toogood, C. I., J. Crompton, and R. T. Hay. 1992. Antipeptide antisera define neutralizing epitopes on the adenovirus hexon. J. Gen. Virol. 73(Pt 6):1429-1435. [DOI] [PubMed] [Google Scholar]
- 35.Trotman, L. C., N. Mosberger, M. Fornerod, R. P. Stidwill, and U. F. Greber. 2001. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 3:1092-1100. [DOI] [PubMed] [Google Scholar]
- 36.Varga, M. J., T. Bergman, and E. Everitt. 1990. Antibodies with specificities against a dispase-produced 15-kilodalton hexon fragment neutralize adenovirus type 2 infectivity. J. Virol. 64:4217-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varghese, R., Y. Mikyas, P. L. Stewart, and R. Ralston. 2004. Postentry neutralization of adenovirus type 5 by an antihexon antibody. J. Virol. 78:12320-12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warrens, A. N., M. D. Jones, and R. I. Lechler. 1997. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186:29-35. [DOI] [PubMed] [Google Scholar]
- 39.Wohlfart, C. 1988. Neutralization of adenoviruses: kinetics, stoichiometry, and mechanisms. J. Virol. 62:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wohlfart, C. E., U. K. Svensson, and E. Everitt. 1985. Interaction between HeLa cells and adenovirus type 2 virions neutralized by different antisera. J. Virol. 56:896-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, H., I. Dmitriev, E. Kashentseva, T. Seki, M. Wang, and D. T. Curiel. 2002. Construction and characterization of adenovirus serotype 5 packaged by serotype 3 hexon. J. Virol. 76:12775-12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang, Z., G. Gao, A. Reyes-Sandoval, C. J. Cohen, Y. Li, J. M. Bergelson, J. M. Wilson, and H. C. Ertl. 2002. Novel, chimpanzee serotype 68-based adenoviral vaccine carrier for induction of antibodies to a transgene product. J. Virol. 76:2667-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue, F., and R. M. Burnett. 2006. Capsid-like arrays in crystals of chimpanzee adenovirus hexon. J. Struct. Biol. 154:217-221. [DOI] [PubMed] [Google Scholar]
- 44.Youil, R., T. J. Toner, Q. Su, M. Chen, A. Tang, A. J. Bett, and D. Casimiro. 2002. Hexon gene switch strategy for the generation of chimeric recombinant adenovirus. Hum. Gene Ther. 13:311-320. [DOI] [PubMed] [Google Scholar]