Abstract
West Nile virus (WNV) can cause fatal murine and human encephalitis. The viral envelope protein interacts with host cells. A murine brain cDNA phage display library was therefore probed with WNV envelope protein, resulting in the identification of several adherent peptides. Of these, peptide 1 prevented WNV infection in vitro with a 50% inhibition concentration of 67 μM and also inhibited infection of a related flavivirus, dengue virus. Peptide 9, a derivative of peptide 1, was a particularly potent inhibitor of WNV in vitro, with a 50% inhibition concentration of 2.6 μM. Moreover, mice challenged with WNV that had been incubated with peptide 9 had reduced viremia and fatality compared with control animals. Peptide 9 penetrated the murine blood-brain barrier and was found in the brain parenchyma, implying that it may have antiviral activity in the central nervous system. These short peptides serve as the basis for developing new therapeutics for West Nile encephalitis and, potentially, other flaviviruses.
West Nile virus (WNV) is closely related to other flaviviruses of global importance, including dengue, yellow fever, and tick-borne encephalitis viruses, among others (7, 28). These pathogens are typically transmitted to vertebrates by mosquitoes or ticks and are responsible for severe morbidity and mortality in both humans and animals (28).
Since emerging in North America in 1999, WNV has rapidly spread throughout the continental United States as well as parts of Canada, Mexico, the Caribbean, and Latin America (11). Most infections are asymptomatic and remain undetected, except by later serologic evidence of exposure to the virus (9). Approximately 1 in 150 infected individuals, however, develops neurologic symptoms, such as meningitis, encephalitis, or flaccid paralysis (22). The elderly and patients with compromised immune systems are particularly at risk for the more severe forms of disease (25). The total number of human cases collectively reported in the United States from 1999 to 2005 was 19,655, with 782 fatalities (CDC [http://www.cdc.gov/ncidod/dvbid/westnile/index.htm]). Unfortunately, treatment for WNV infection currently primarily consists of supportive measures (9).
Dengue virus imposes one of the largest social and economic burdens of any mosquito-borne pathogen (10). More than half of the world's population inhabits areas with potential for dengue virus transmission, and there are 50 to 100 million cases of human infection yearly (20). The spectrum of human illness ranges from asymptomatic infection to potentially fatal dengue hemorrhagic fever, which is marked by increased vascular permeability, bleeding, and circulatory failure.
Novel approaches against flaviviral infections are essential, and blocking virus attachment or entry into host cells or inhibiting viral replication are potential strategies. Enfuvirtide, a successful peptide human immunodeficiency virus (HIV) fusion inhibitor, is the prototype for the development of a new class of antivirals (26). This viral entry inhibitor blocks structural rearrangements of viral envelope that are essential for viral infection. The flaviviruses are enveloped viruses, and their envelope (E) proteins participate in virus attachment to host cells and fusion with the host membrane (1, 8). These interactions are required for the internalization of virus particles (14).
Phage display technology allows for the rapid high-throughput screening of billion-clone peptide libraries and has become a useful approach to identify and improve peptide molecules as pharmaceuticals. This methodology has produced novel peptides that are in clinical trials as therapeutics for a variety of diseases (18). Peptides that bind and neutralize Newcastle disease virus have been discovered by using phage display technology (24). We have now screened a murine brain cDNA phage display library to identify peptides that bind to the WNV E protein and that also inhibit virus infectivity. Since WNV is neurovirulent and causes encephalitis, brain tissue cDNAs may encode a receptor(s) or ligand(s) with high affinity for WNV E proteins. Portions of these proteins may serve as peptide-based drugs or lead to the development of small-molecule antiviral agents.
MATERIALS AND METHODS
Viruses, cells, and mice.
West Nile viral isolate 2471 (2) and dengue virus 2 (strain New Guinea C) were used in these studies. Vero cells and HeLa cells were maintained in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 1% l-glutamine, 1% penicillin-streptomycin, and 10% fetal bovine serum. Neuro-2a and C6/36 cells were maintained in minimum essential medium Eagle (ATCC, Manassas, VA) supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum. Seven-week-old female BALB/c mice were purchased from the Jackson Laboratory (Bar Harbor, ME). In the murine experiments, 1,000 PFU of WNV was preincubated with 1 mM of peptides and 1 mM of dithiothreitol for 1 h at room temperature. The virus-peptide mixture was administered to the mice by intraperitoneal (i.p.) injection.
Phage display library screening.
An M13 phage library displaying protein and peptides expressed from mouse brain cDNAs was purchased from Spring Bioscience (Fremont, CA). The phages were first exposed to a plate coated with highly purified recombinant WNV E protein (19). Unbound phages were washed away. Specifically bound phages were eluted, amplified, and subjected to a second round of screening against recombinant WNV E protein. After three to four rounds of screening, phages were selected that showed significant binding to recombinant WNV E protein by an indirect enzyme-linked immunosorbent assay. The amino acid sequences of the peptides presented by bound phages were revealed by sequence analyses of phage DNA. The peptide amino acid sequences were aligned using the ClustalW alignment algorithm and searched for database homologies using BLAST. Peptides representing sequences isolated from multiple isolates of bound phage were chemically synthesized by the Yale University Keck Biotechnology Resource Laboratory or by the Tufts University Core Facility.
Q-PCR and plaque formation assay.
WNV-specific RNA was quantified using a quantitative PCR (Q-PCR) as we previously reported (4). Dengue virus gene Q-PCR primers and probe were designed according to a previous study (3). Viral copy number was expressed as a ratio to cellular β-actin cDNA copies measured by Q-PCR. The plaque formation assay and plaque reduction neutralization test (PRNT) protocols were carried out as previously described (4).
In vitro binding assay.
For preincubation experiments, WNV was incubated with serially diluted peptides for 1 h at room temperature. The virus and peptide mixture was added to cell cultures for 1 h at 4°C. The cells were washed three times with 4°C phosphate-buffered saline (PBS) and then processed for total RNA extraction and Q-PCR. In a postattachment infection assay, virus was allowed to attach to cells for 1 h at 4°C. The unattached virus was then washed away with 4°C PBS three times. Serial diluted peptides were then added into fresh cell medium, and cells were returned to an incubator for 24 h and evaluated by Q-PCR for infection.
Cytotoxicity assay.
Peptides P1 and P9 (from 1 to 500 μM final concentration) were incubated with Vero cells for 24, 48, and 72 h. Cytotoxicity was determined by a fluorescent assay that measures lactate dehydrogenase (LDH) release from cells with compromised membranes (Cytotox-One homogenous membrane integrity kit; Promega, Madison, WI).
SPR analysis.
Peptide-WNV E protein interactions were studied using a biosensor surface plasmon resonance (SPR) assay as described elsewhere (27) using a BIAcore 2000. Recombinant WNV E protein was biotinylated using a sulfo-NHS-biotinylation kit (Pierce, Rockford, IL) and then immobilized on SA sensor chips (BIAcore, Piscataway, NJ) using the immobilization protocol provided by the manufacturer. Data were analyzed and fitted using the BIAevaluation 3.0 software package.
Immunofluorescence microscopy.
Naïve mice were administered biotin-conjugated antiviral peptide P9 via an i.p. route of administration. At various time points after peptide administration, mice were sacrificed under isofluorane anesthesia and transcardially perfused with 60 ml of ice-cold PBS. Brains and spleens were rapidly isolated and immersion fixed in 4% paraformaldehyde overnight at 4°C. The following day, tissues were cryoprotected in 10% (wt/vol) sucrose in PBS, followed by 20% and then 30% sucrose in PBS, with each incubation step for 24 h at 4°C. Thereafter, tissues were frozen on dry ice in optimal cutting temperature compound (Tissue-Tek, Japan) and cryosectioned using a freezing microtome (Leica model CM1850). Four 10-μm sections, spaced 50 μm apart, were cut from each organ. Tissue sections were air dried, a PAP pen was applied, and sections were preblocked for 30 min at ambient temperature in serum-free protein block (DAKO Cytomation, Carpintera, CA). An antibody directed against biotin was then applied (1:200 in serum-free protein block; Vector Laboratories, Burlingame, CA) for 30 min at ambient temperature. Tissue sections were then rinsed three times for 5 min each in PBS in a Coplin jar. Thereafter, tyramide signal amplification was performed according to the manufacturer's instructions (Perkin-Elmer, Wellesley, MA). Sections were rinsed an additional three times for 5 min each and incubated with an antibody against streptavidin conjugated with AlexaFluor 568 (1:200 in serum-free protein block; Invitrogen, Carlsbad, CA). Following a final three rinses in PBS, sections were air dried and mounted in fluorescence mounting medium containing 4′,6′-diamidino-2-phenylindole (DAPI) nuclear counterstain (ProLong Gold; Invitrogen).
For analysis of WNV infection after i.p. injection of WNV preincubated with P9 or control (noninhibition) P2, mice were sacrificed and perfused as described above, and tissue handling was performed as above. Tissue sections were incubated overnight at 4°C with a mouse ascites fluid antibody directed against the WNV E protein (1:200 in serum-free protein block, obtained from J. F. A. Anderson's laboratory, CT Agricultural Experiment Station) and a polyclonal antibody against the neuron marker, microtubule-associated protein 2 (1:500; Chemicon, Temicula, CA). The following day, sections were rinsed three times for 5 min each in PBS at ambient temperature in a Coplin jar. Sections were then incubated with secondary antibodies against mouse immunoglobulin G conjugated with AlexaFluor 568 or against rabbit immunoglobulins conjugated with AlexaFluor 488 (1:200 in serum-free protein block; Invitrogen). Following a final three rinses in PBS, sections were air dried and mounted in fluorescence mounting medium containing DAPI (ProLong Gold; Invitrogen). All tissue sections were observed under dark field in independent fluorescence channels using an Olympus BX-61 automated microscope (Japan) with an attached QImaging QICAM charge-coupled device (CCD) camera (Canada).
Statistical analysis.
All statistical analyses were performed using GraphPad Prism (version 4.0; GraphPad Software).
RESULTS
Identification of peptides that bind the WNV E protein by using a murine brain cDNA phage display library.
WNV can infect many vertebrates, including humans, horses, and mice. The murine model of WNV encephalitis partially mimics severe human infection, as the virus penetrates the blood-brain barrier and causes death within several weeks. We therefore used a murine brain cDNA phage display library to identify peptides that bound recombinant WNV E protein. Phages were amplified and enriched in four rounds of screening and then eluted. DNAs of 177 phages that showed significant binding to the E protein in an indirect enzyme-linked immunosorbent assay were sequenced. Significant homologies with putative receptor proteins were not identified based on the ClustalW alignment algorithm and BLAST. Many of the phages, however, displayed similar 19- to 94-amino-acid sequences encoded by the cDNAs. The three most frequently encountered peptide (P) sequences, P1, P2, and P3 (Table 1), were chemically synthesized and examined for antiviral activity.
TABLE 1.
Peptides synthesized and tested for inhibitory activitya
| Peptide | Amino acid sequence | No. of amino acid residues | MW | pI | GRAVY |
|---|---|---|---|---|---|
| P1 | DTRACDVIALLCHLNT | 16 | 1,758.0 | 5.21 | 0.569 |
| P2 | TGPEFPGRPTRP | 12 | 1,311.4 | 9.26 | −1.525 |
| P3 | NTTHYRVIRLTIG | 13 | 1,543.7 | 10.84 | −0.192 |
| P4 | DTRACDVIALL | 11 | 1,189.3 | 4.21 | 0.927 |
| P5 | DTRACDVIPLL | 11 | 1,215.4 | 4.21 | 0.618 |
| P6 | CDVIALL | 7 | 745.9 | 3.80 | 2.443 |
| P7 | DTRAPLAI | 8 | 855.9 | 5.84 | 0.200 |
| P8 | CDVIALLCHLNT | 12 | 1,314.5 | 5.08 | 1.333 |
| P9 | CDVIALLACHLNT | 13 | 1,385.6 | 5.08 | 1.369 |
| P10 | CDVIALLCHLNTPSF | 15 | 1,645.9 | 5.08 | 1.093 |
| P11 | CDVIALLCHLNTPSFNTTHYRESWY | 25 | 2,984.3 | 5.99 | −0.160 |
| P12 | CDVIALLACHLNTPSF | 16 | 1,717.0 | 5.08 | 1.137 |
| P13 | CDVIALLACHLNTPSFNTTHYRESWY | 26 | 3,055.4 | 5.99 | −0.085 |
| P14 | CDVIALLECHLNT | 13 | 1,443.7 | 4.35 | 0.962 |
| P15 | TRACDVIALLECHLNT | 16 | 1,772.0 | 5.29 | 0.569 |
| P16 | DTRACDVIALLECHLNT | 17 | 1,887.1 | 4.54 | 0.329 |
The molecular weight (MW), pI, and grand average of hydropathicity (GRAVY) were predicted with the ProtParam algorithm.
Peptide inhibition of WNV infectivity in vitro.
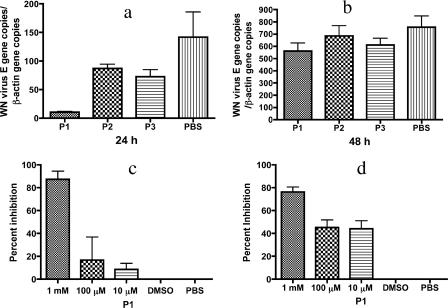
The ability of synthetic peptides to inhibit WNV was determined by reverse transcription-Q-PCR. P1, P2, and P3 were incubated with virus at a concentration of 1 mM at room temperature for 1 h. One hundred μl of the virus (multiplicity of infection, 0.2) and peptide mixture was then added to 900 μl of cell medium prior to adding to African green monkey kidney (Vero) cells and incubated for selected intervals. Q-PCR demonstrated that P1 significantly reduced the WNV load (P < 0.03) at 24 h, while P2 and P3 did not show significant inhibition (P > 0.05) (Fig. 1a). At 48 h, the viral RNA levels increased in all cultures (Fig. 1b).
FIG. 1.
(a and b) Peptide (P1, P2, and P3) inhibition of WNV infection in Vero cells at 24 h (a) and 48 h (b). P1 (1 mM) significantly (P < 0.03) inhibited WNV infection at 24 h, as measured by Q-PCR. (c and d) Q-PCR (c) and PRNT (d) demonstrated that DMSO does not influence the inhibition of WNV. DMSO groups contained the same concentration of DMSO as the 1 mM P1 sample. Statistical significance for Q-PCR: overall, P < 0.02; 1 mM versus PBS, P < 0.05; DMSO versus PBS, P > 0.05. For PRNT: overall, P < 0.0001; 1 mM, 100 μM, and 10 μM versus PBS, P < 0.001; DMSO versus PBS, P > 0.05. Any average value below 0 is considered no inhibition. All the experiments were repeated at least twice, and all the measurements were made in triplicate, with the mean ± standard deviation shown. Statistical analysis was done by one-way analysis of variance, in some cases followed by Tukey's test.
P1 is hydrophobic and was solubilized in Dulbecco's PBS with 2% dimethyl sulfoxide (DMSO). To determine whether DMSO influenced the experimental results, WNV and P1, or the same concentration of DMSO vehicle, were preincubated for 1 h at room temperature and then added to cells and examined at 24 h by both Q-PCR and a PRNT. Q-PCR and PRNT showed that P1 significantly inhibited WNV infectivity (P < 0.05) at a concentration of 1 mM (Fig. 1c and d). DMSO did not show any inhibition (P > 0.05) compared with the PBS control (Fig. 1c and d). These data indicate that P1 inhibits WNV infectivity in vitro.
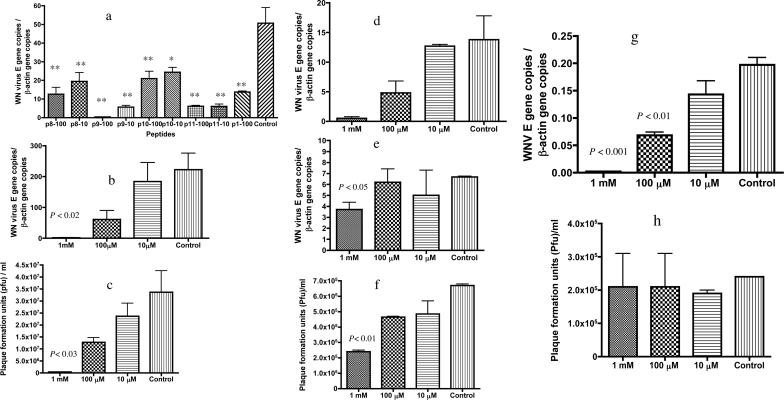
Thirteen additional peptides, P4 to P16, with amino acid insertions, deletions, or substitutions based on P1, were then synthesized to delineate the residues that are critical for antiviral activity. Several peptides, P8, P9, P10, and P11, significantly reduced viral RNA levels in the host cells as measured by Q-PCR, with P9 exhibiting the greatest degree of inhibition (96.3 ± 2.8%; P < 0.001) at 100 μM (Fig. 2a). As P9 was the most potent inhibitor, we determined whether P9 influenced WNV infectivity by blocking virus entry, replication, or viral release. The viral RNA levels in host cells were measured by Q-PCR (Fig. 2b), and a plaque formation assay was used to determine WNV titers in culture medium (Fig. 2c). The plaque formation assay was consistent with the Q-PCR data, indicating that P9 may interfere with viral entry or replication, rather than block the release of virus particles from host cells. Q-PCR assays were, therefore, performed to test this hypothesis. Serially diluted P9 was preincubated with WNV at room temperature. Virus binding to cells and washing with PBS were performed at 4°C. The amount of viruses that bound to the cell surface was evaluated by using Q-PCR. The results demonstrated that P9 reduced 96.3 ± 2.7% and 65.2 ± 14.5% of WNV attachment and entry to Vero cells at 1 mM and 100 μM concentrations, respectively (Fig. 2d). To test whether P9 inhibited WNV entry after the virus attached to the host cell surface receptor, postattachment experiments were performed with P9 at final concentrations of 200 μM, 20 μM, and 2 μM. The results suggested that P9 failed to inhibit WNV infection in Vero cell cultures (data not shown). As WNV is transmitted by mosquitoes and can infect humans and mice, we also examined whether P9 inhibited WNV infection in HeLa (human), Neuro-2a (mouse), and C6/36 (mosquito) cells. P9 inhibited WNV in HeLa and Neuro-2a cell cultures but not in C6/36 cell cultures (Fig. 2e, f, g, and h), suggesting that the mechanism of attachment and entry for WNV in mammalians and in the arthropod vector may differ. It cannot be excluded, however, that during preincubation the binding of P9 to WNV induces conformation changes in the E protein that may specifically interfere with the attachment to certain cell lines.
FIG. 2.
Evaluation of peptides derived from P1 against WNV infection of cultured Vero cells. (a) Selected peptides (P8, P9, P10, and P11) inhibited viral infectivity in Vero cell cultures. Two concentrations (100 μM and 10 μM) of each peptide were tested, and 100 μM P1 was included as a positive control. WNV RNA levels 24 h postinfection were determined by Q-PCR. All the groups significantly inhibited WNV infectivity compared with the DMSO vehicle-only control. *, P < 0.01; **, P < 0.001. (b and c) P9 inhibited WNV infection in Vero cell cultures in a dose-dependent manner as determined by both Q-PCR (b) and by a plaque formation assay (c) for viral titers in the cell supernatant 24 h postinfection. (d) P9 reduced WNV attachment or entry into Vero cells after 1 h of binding at 4°C (P < 0.001). (e and f) P9 inhibited WNV infection in HeLa cell cultures as measured both by Q-PCR (e) and a plaque formation assay (f) for viral titer in the cell supernatant. (g) P9 inhibited WNV infection in cultured Neuro 2a cells as measured by Q-PCR. (h) P9 did not inhibit WNV in C6/36 cells in the plaque formation assay (P = 0.96). All the experiments were repeated at least twice, and all the measurements were made in triplicate, with the mean ± standard deviation shown. Data were analyzed by one-way analysis of variance followed by Tukey's test.
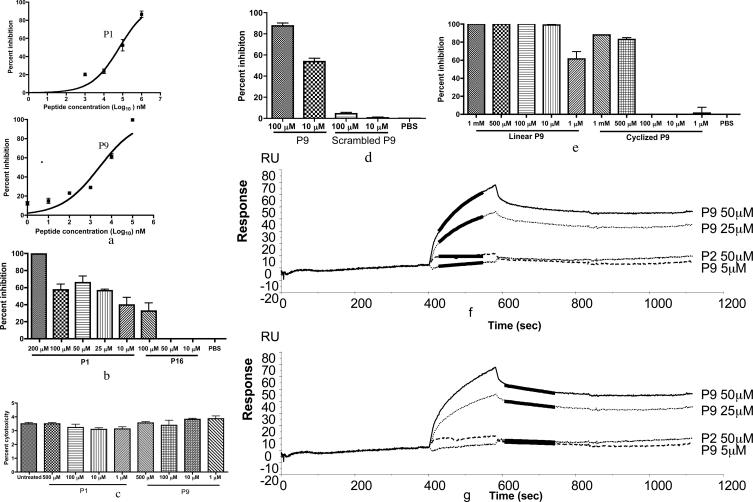
To determine the 50% inhibitory concentrations (IC50) of P1 and P9 against WNV, dose-response curves were performed using serially diluted peptides in PRNT experiments. P1 showed a maximum inhibitory activity against WNV of 86.7 ± 4.9% at 1 mM, with an IC50 of 67.0 ± 0.1 μM (Fig. 3a). P9 demonstrated 100% inhibition at 100 μM, and the IC50 was 2.60 ± 0.01 μM (Fig. 3a).
FIG. 3.
P1 and P9 inhibition of WNV or dengue virus infection of Vero cells. (a) Dose-response curves of P1 (IC50 of 67 μM) and P9 (IC50 of 2.6 μM) against WNV infection in vitro. Serially diluted P1 and P9 were preincubated with 100 PFU of WNV for 1 h at room temperature and then examined in a PRNT. Plots were made using GraphPad Prism. (b) P1 inhibited dengue virus infection in Vero cells, as measured by Q-PCR 24 h postinfection. All the measurements were made in triplicate, with the mean ± standard deviation shown. (c) P1 and P9 are not cytotoxic. Vero cells were exposed to peptide (1 μM to 500 μM) for 72 h, and LDH levels in cell supernatants were monitored. Each sample was assayed in duplicate, and data represent the mean ± standard deviation. No statistical difference was observed by one-way analysis of variance (P > 0.05). (d) Scrambled P9 showed no inhibition compared with P9, at either 100 μM or 10 μM, as measured by PRNT. (e) Cyclized P9 (at ≤100 μM) did not demonstrate inhibitory activity. Serially diluted peptides were tested and compared between P9 (linear) and P9 cyclized with an intramolecular disulfide bond. Any average value below zero in panels b and e is considered no inhibition. P9 binds to WNV E protein. (f and g) P9 binds to WNV E protein with a Kd of approximately 6 μM. P2 did not show affinity for the WNV E protein. The black line indicates the theoretical fit. All the experiments were repeated at least twice.
Since flavivirus E proteins have a significant amount of homology, we tested whether selected peptides inhibited the infectivity of dengue virus. Dengue virus serotype 2 was chosen because it is widespread and results in significant human morbidity and mortality. We preincubated peptides with dengue virus (multiplicity of infection, 0.3) for 1 h at room temperature and performed studies in an identical fashion as with WNV. Experimental results showed that 200 μM of P1 significantly inhibited dengue virus infection of Vero cells (99.3 ± 0.7%) at 24 h (Fig. 3b). Peptides P9 and P16, which were derived from P1, were not active against dengue virus.
Specificity of peptide inhibitory activity.
To address whether the peptides were cytotoxic and thereby nonspecifically inhibited flavivirus entry or replication, Vero cells were exposed to from 1 μM to 500 μM of P1 or P9 for 24, 48, and 72 h. Cytotoxicity was measured by monitoring the release of LDH. The LDH levels of untreated and peptide-treated cells were not statistically different at any time points examined (data not shown). Results of the 72-h time point are shown in Fig. 3c, and these data demonstrate that the peptides are not cytotoxic.
P1 had an IC50 of 67.0 μM against WNV (Fig. 3a) and also inhibited dengue virus replication (99.3 ± 0.7%) at a concentration of 200 μM (Fig. 3b). The insertion, however, of a glutamate into the P1 sequence (P16) eliminated the inhibitory effect against dengue virus (Fig. 3b). On the other hand, P9 was effective at inhibiting WNV but was relatively ineffective at blocking dengue virus infectivity. These results suggest that the activity of P1 against dengue virus is highly dependent on the peptide sequence. To rule out the possibility that P9 inhibited WNV in a nonspecific manner, a scrambled peptide was tested for its ability to modulate viral infectivity. This peptide has the same amino acid residues as P9 but a randomly altered sequence, AHTCADILVLCLN. PRNT experiments demonstrated that the scrambled P9 did not significantly inhibit WNV infection compared with P9 (Fig. 3d). These results further support the notion that P1 and P9 inhibited virus infectivity in a sequence-specific manner.
P9 has two cysteine residues that could form disulfide bonds in solution. We tested whether cyclic P9, in which the two cysteines form a disulfide bond, inhibited WNV. Cyclic P9 inhibited WNV plaque formation only in concentrations higher than 100 μM, whereas linear P9, in which the two cysteines are free, was able to inhibit WNV in concentrations as low as 1 μM (Fig. 3e).
Inhibitory peptides bind to WNV E protein.
Since phages were screened for binding to E protein, we determined whether the chemically synthesized peptides bound to recombinant WNV E protein by using SPR. SPR experiments showed that P9 bound to E protein with a Kd of 6 μM (Fig. 3f and g). Noninhibitory peptide P2 and scrambled P9 did not bind to the E protein (data of the scrambled P9 are not shown). Cyclized P9 did not bind to E protein unless 1 mM dithiothreitol, which reduces disulfide bonds, was present in the peptide solution (data not shown).
Peptide inhibition of WNV infection in vivo.
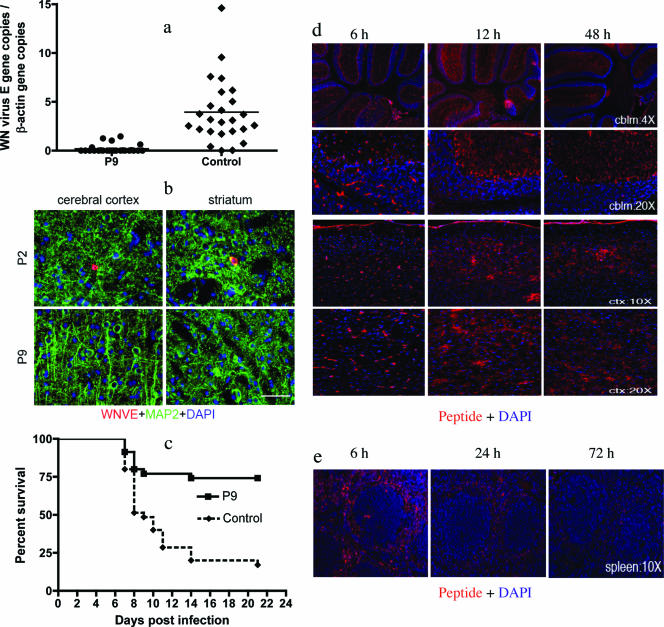
As the peptides were not cytotoxic and significantly inhibited WNV in vitro, we next determined whether select peptides could protect mice from a lethal dose of WNV. WNV-infected mice develop viremia that peaks after several days. We therefore challenged mice with WNV and peptide and evaluated the viral load in the blood at 3 days. Q-PCR showed that P9 significantly reduced the viremia compared with a noninhibitory peptide, P2 (Fig. 4a). WNV then enters the murine brain at about day 4 to 5, and animals begin to die at about 1 week. In selected experiments, we randomly sacrificed half the number of mice at day 6 to collect brains for histological analysis. Indirect immunofluorescence assays showed that the P9-treated group had little or no evidence of virus in the cerebral cortex and striatum, where WNV was detected in P2-treated mice (Fig. 4b). Similar results were observed in other brain regions, including the olfactory bulb, hippocampal formation, cerebellum, and brainstem (data not shown), brain areas in which we previously detected WNV (29). The remaining mice were observed for 3 weeks. Eighty percent of mice administered P9-treated virus survived, compared with 10% of mice that received control peptide-treated virus (P < 0.005) (Fig. 4c). These data suggest that preincubation of WNV with P9 protected mice from death. We also tested whether P9 could protect mice after WNV infection. The same dose of P9 used for preincubation was i.p. injected 3 h after challenge with WNV. Results showed that P9 failed to protect mice from viral infection in this experiment (P > 0.05) (data not shown).
FIG. 4.
P9 inhibited WNV infectivity in vivo. (a) Mixing P9 and WNV prior to challenge significantly reduced murine viremia at day 3 (P < 0.0001; Student's t test). (b) Indirect immunofluorescence revealed a reduced viral burden in brains of P9-treated mice. The red signal represents the WNV E protein, and the green signal indicates microtubule-associated protein 2 (MAP2; a neuron marker). Bar, 100 μm. (c) Preincubation of P9 with WNV significantly increased murine survival (n = 35; P < 0.0001, log rank test). P2 was used as a noninhibitory control peptide. Data were combined from seven independent experiments. Control mice were treated with noninhibitory P2. (d) P9 entered the brain after i.p. injection. Two hundred μl of 1 mM biotin-labeled P9 was injected via the i.p. route of administration. Mice were sacrificed at various time points postinjection, and brain and spleen tissues were collected after perfusion. Biotin-labeled P9 was detected using an enzymatic tyramide signal amplification method. Biotin-labeled P9 was detected in the cerebellum (cblm) and in the cerebral cortex (ctx). At 6 h, most of the signal was associated with cerebral blood vessels, but thereafter signal relocalized into the brain parenchyma. (e) Biotin-labeled P9 was detected in the spleen as early as 6 h postinjection and declined in intensity thereafter.
To monitor the distribution of the peptide, naïve mice were administered two 100-μl doses of 1 mM biotin-labeled P9 at 0 and 6 h. Spleens and brains were collected after perfusion at 0, 6, 12, 24, 48, and 72 h. Biotin-conjugated peptide in the tissues was detected using an enzymatic tyramide signal amplification method. In the brain, the biotin-conjugated peptide was identified as early as 6 h postinjection; at this time point it was concentrated in cerebral vessels. A relocalization of biotin-conjugated peptide was evident at later time points, where a punctuate pattern of staining was evident in brain parenchyma and little or no cerebral vascular signal was observed. The peak of signal intensity was noted at 12 h in the cerebellum and the cerebral cortex (Fig. 4d). The signal was still present at 24, 48, and 72 h (Fig. 4d and data not shown). A similar staining pattern was noted in all of the brain regions surveyed, including the olfactory bulb, hippocampal formation, cerebellum, and brainstem (data not shown). In the spleen, peptide was detected as early as 6 h postinjection, when the signal peaked (Fig. 4e). The signal remained detectable at 24 h and mostly disappeared at 72 h, suggesting that peripheral clearance of the peptide had ensued by this time point.
DISCUSSION
WNV has become a significant public health concern in North America, and effective therapies are lacking. Identifying novel compounds with antiflavivirus activity may, therefore, lead to new prevention or treatment strategies. In this study, we screened a murine brain cDNA phage display library for amino acid sequences that bind WNV E protein. Several peptides, most notably P1 and P9, inhibited WNV infection of cells. P9 also protected mice from lethal infection with WNV, and P1 demonstrated in vitro inhibition against dengue virus.
Flaviviral E proteins mediate attachment and entry to host cells (1, 8). Virions first attach to the cell surface and then enter the cell by receptor-mediated endocytosis. Acidification of the endosomal vesicle triggers conformational changes in the virion, fusion of the virus and cell membranes, particle disassembly, and release of the viral genome into the cytoplasm (14, 17, 23). The inhibitory peptides had weak to moderate affinity with the target E protein in SPR studies: as an example, the Kd of P9 binding to the WNV E protein was 6 μM, while noninhibitory peptides showed no affinity for the E protein. This suggests that the peptides' inhibitory effects depend on their capacity to bind the target E protein. In vitro experiments also provided direct evidence that P9 blocked virus attachment (Fig. 2b, c, and d) but not viral RNA replication or the release of progeny virions from the host cells.
Previous studies have developed rationally designed antiviral peptides based on viral envelope protein sequences. Enfuvirtide, a 36-amino-acid peptide based on the stem region of the HIV gp41, which is a viral envelope protein that mediates fusion with host cells, provides significant clinical anti-HIV activity (26). Advantages of enfuvirtide are clinical responses in patients who have failed other anti-HIV therapies and that the inhibitor is generally well tolerated. Disadvantages include a twice-daily subcutaneous route of administration and a relatively high cost. Small molecules with a similar mode of action are being developed and are expected to overcome some of the limitations of enfuvirtide peptide therapy (15). In a recent study, investigators designed several peptide inhibitors of dengue virus infectivity by targeting the viral E protein based on its structure (13). Viral inhibition assays suggested that some of these peptides inhibit dengue virus infectivity in cell cultures with an IC50 in the 10 μM range, and one of them cross-inhibits WNV infection in Vero cells (13). Of the WNV inhibitory peptides that were identified in the current study, P9, inhibited WNV infection with an IC50 of about 2.6 μM, and P1 cross-inhibited dengue virus infection. Inhibitory activity (Vero and Neuro-2a > HeLa > C6/36) suggests host cell specificity in antiviral activity. We therefore performed animal experiments to determine if P9 could reduce WNV infection in a vertebrate animal model. Moreover, the incubation of an otherwise fatal dose of WNV with P9 could protect animals from lethality by reducing WNV viremia and viral load in the brain. The peptide may block viral attachment in the periphery and subsequently reduce viremia, thereby enabling the murine immune system to clear the virus. It is noteworthy that peptide-based therapeutics are generally unable to penetrate the blood-brain barrier due to their size and hydrophilicity (5). Interestingly, we found that biotin-labeled P9 was able to penetrate the mouse blood-brain barrier and could be detected in the brain parenchyma. Although the transport mechanism for this 1.4-kDa hydrophobic peptide is not clear, this observation suggests that short peptide drugs could potentially treat WNV encephalitis.
There are several possibilities for why P9 was not effective when given to mice after WNV infection was established. P9 binds WNV E protein and interferes with virus attachment in a concentration-dependent manner. The peptide, however, may have failed to block the viral progeny, for P9 was significantly diluted in mice after the virus challenge. Second, efficacy of peptide therapeutics can be limited by fast renal clearance or degradation caused by enzymes occurring in the blood, liver, and kidney (30). Peptides and proteins often display half-lives in the range of a few minutes to a few hours (30). P9, however, reached brain blood vessels and was distributed in brain tissue 12 h later. Inhibitory effects of P9 and P1 could be potentially improved by modifying the amino acid residue composition by using computational modeling analysis. In addition, previous successful strategies will be tested to improve peptide plasma half-life without decreasing inhibition, e.g., modification of N and C terminus, head, and tail cyclization (21) or using sustained delivery systems (6, 12, 16).
In conclusion, noncytotoxic inhibitory peptides that inhibit WNV and dengue virus infections have been identified. Moreover, P9 could penetrate the mouse blood-brain barrier and increase survival in lethal WNV challenges after preincubation with virus. With some modifications, these inhibitory peptides may serve as peptide-based drugs or represent lead compounds for high-throughput screening assays to identify novel antiviral chemical compounds against WNV and other flaviviruses.
Acknowledgments
We thank Deborah Beck, Nathalie Bonafé, and James Elliott for assistance and Haibei Liu for help with statistical analysis.
F. Bai was supported by a career development fellowship from the Northeast Biodefense Center. T. Town was supported by an NIH/NRSA/NIA postdoctoral fellowship and an Alzheimer's Association grant. Ashish was supported by the National Science Foundation. This project was supported by grants from the National Institutes for Health.
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. F., T. G. Andreadis, C. R. Vossbrinck, S. Tirrell, E. M. Wakem, R. A. French, A. E. Garmendia, and H. J. Van Kruiningen. 1999. Isolation of West Nile virus from mosquitoes, crows, and a Cooper's hawk in Connecticut. Science 286:2331-2333. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong, P. M., and R. Rico-Hesse. 2001. Differential susceptibility of Aedes aegypti to infection by the American and Southeast Asian genotypes of dengue type 2 virus. Vector Borne Zoonotic Dis. 1:159-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai, F., T. Wang, U. Pal, F. Bao, L. H. Gould, and E. Fikrig. 2005. Use of RNA interference to prevent lethal murine West Nile virus infection. J. Infect. Dis. 191:1148-1154. [DOI] [PubMed] [Google Scholar]
- 5.Bickel, U., T. Yoshikawa, and W. M. Pardridge. 2001. Delivery of peptides and proteins through the blood-brain barrier. Adv. Drug Deliv. Rev. 46:247-279. [DOI] [PubMed] [Google Scholar]
- 6.Bjerregaard, S., H. Pedersen, H. Vedstesen, C. Vermehren, I. Soderberg, and S. Frokjaer. 2001. Parenteral water/oil emulsions containing hydrophilic compounds with enhanced in vivo retention: formulation, rheological characterisation and study of in vivo fate using whole body gamma-scintigraphy. Int. J. Pharm. 215:13-27. [DOI] [PubMed] [Google Scholar]
- 7.Brinton, M. A. 2002. The molecular biology of West Nile virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 56:371-402. [DOI] [PubMed] [Google Scholar]
- 8.Crill, W. D., and J. T. Roehrig. 2001. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 75:7769-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gould, L. H., and E. Fikrig. 2004. West Nile virus: a growing concern? J. Clin. Investig. 113:1102-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubler, D. J. 2002. The global emergence/resurgence of arboviral diseases as public health problems. Arch. Med. Res. 33:330-342. [DOI] [PubMed] [Google Scholar]
- 11.Hayes, E. B., N. Komar, R. S. Nasci, S. P. Montgomery, D. R. O'Leary, and G. L. Campbell. 2005. Epidemiology and transmission dynamics of West Nile virus disease. Emerg. Infect. Dis. 11:1167-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heya, T., Y. Mikura, A. Nagai, Y. Miura, T. Futo, Y. Tomida, H. Shimizu, and H. Toguchi. 1994. Controlled release of thyrotropin releasing hormone from microspheres: evaluation of release profiles and pharmacokinetics after subcutaneous administration. J. Pharm. Sci. 83:798-801. [DOI] [PubMed] [Google Scholar]
- 13.Hrobowski, Y. M., R. F. Garry, and S. F. Michael. 2005. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahn, R., T. Lang, and T. C. Sudhof. 2003. Membrane fusion. Cell 112:519-533. [DOI] [PubMed] [Google Scholar]
- 15.Jin, B. S., W. K. Lee, K. Ahn, M. K. Lee, and Y. G. Yu. 2005. High-throughput screening method of inhibitors that block the interaction between 2 helical regions of HIV-1 gp41. J. Biomol. Screen. 10:13-19. [DOI] [PubMed] [Google Scholar]
- 16.Kim, I. S., H. G. Choi, H. S. Choi, B. K. Kim, and C. K. Kim. 1998. Prolonged systemic delivery of streptokinase using liposome. Arch. Pharm. Res. 21:248-252. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landon, L. A., J. Zou, and S. L. Deutscher. 2004. Is phage display technology on target for developing peptide-based cancer drugs? Curr. Drug Discov. Technol. 1:113-132. [DOI] [PubMed] [Google Scholar]
- 19.Ledizet, M., K. Kar, H. G. Foellmer, T. Wang, S. L. Bushmich, J. F. Anderson, E. Fikrig, and R. A. Koski. 2005. A recombinant envelope protein vaccine against West Nile virus. Vaccine 23:3915-3924. [DOI] [PubMed] [Google Scholar]
- 20.Mairuhu, A. T., J. Wagenaar, D. P. Brandjes, and E. C. van Gorp. 2004. Dengue: an arthropod-borne disease of global importance. Eur. J. Clin. Microbiol. Infect. Dis. 23:425-433. [DOI] [PubMed] [Google Scholar]
- 21.Marastoni, M., S. Salvadori, V. Scaranari, S. Spisani, E. Reali, S. Traniello, and A. Tomatis. 1994. Synthesis and activity of new linear and cyclic peptide T derivatives. Arzneimittel-Forschung 44:1073-1076. [PubMed] [Google Scholar]
- 22.Mostashari, F., M. L. Bunning, P. T. Kitsutani, D. A. Singer, D. Nash, M. J. Cooper, N. Katz, K. A. Liljebjelke, B. J. Biggerstaff, A. D. Fine, M. C. Layton, S. M. Mullin, A. J. Johnson, D. A. Martin, E. B. Hayes, and G. L. Campbell. 2001. Epidemic West Nile encephalitis, New York, 1999: results of a household-based seroepidemiological survey. Lancet 358:261-264. [DOI] [PubMed] [Google Scholar]
- 23.Mukhopadhyay, S., R. J. Kuhn, and M. G. Rossmann. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13-22. [DOI] [PubMed] [Google Scholar]
- 24.Ozawa, M., K. Ohashi, and M. Onuma. 2005. Identification and characterization of peptides binding to Newcastle disease virus by phage display. J. Vet. Med. Sci. 67:1237-1241. [DOI] [PubMed] [Google Scholar]
- 25.Petersen, L. R., and A. A. Marfin. 2002. West Nile virus: a primer for the clinician. Ann. Intern. Med. 137:173-179. [DOI] [PubMed] [Google Scholar]
- 26.Root, M. J., and H. K. Steger. 2004. HIV-1 gp41 as a target for viral entry inhibition. Curr. Pharm. Des. 10:1805-1825. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz, M. F., J. K. Duong, Z. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9:1055-1065. [DOI] [PubMed] [Google Scholar]
- 28.Tsai, T. 2000. Flaviviruses, p. 1196-1206. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Philadelphia, Pa.
- 29.Wang, T., T. Town, L. Alexopoulou, J. F. Anderson, E. Fikrig, and R. A. Flavell. 2004. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat. Med. 10:1366-1373. [DOI] [PubMed] [Google Scholar]
- 30.Werle, M., and A. Bernkop-Schnurch. 2006. Strategies to improve plasma half life time of peptide and protein drugs. Amino Acids 30:351-367. [DOI] [PubMed] [Google Scholar]