Abstract
Phosphorylation plays a key role in regulating many signaling pathways. Although studies investigating the phosphorylated forms of signaling pathways are now commonplace, global analysis of protein phosphorylation and kinase activity has lagged behind genomics and proteomics. We have used a kinomics approach to study the effect of virus infection on host cell signaling in infected guinea pigs. Delineating the host responses which lead to clearance of a pathogen requires the use of a matched, comparative model system. We have used two passage variants of the arenavirus Pichinde, used as a biosafety level 2 model of Lassa fever virus as it produces similar pathologies in guinea pigs and humans, to compare the host cell responses between infections which lead to either a mild, self-limiting infection or lethal disease. Using this model, we can begin to understand the differences in signaling events which give rise to these markedly different outcomes. By contextualizing these data using pathway analysis, we have identified key differences in cellular signaling matrices. By comparing these differentially involved networks, we have identified a number of key signaling “nodes” which show differential phosphorylations between mild and lethal infections. We believe that these nodes provide potential targets for the development of antiviral therapies by acting at the level of the host response rather than by directly targeting viral proteins.
The family Arenaviridae includes a number of hemorrhagic fever viruses, including Lassa fever virus and Junin virus. Previous findings suggest that arenavirus pathogenesis could well involve the dysregulation of cytokines and immune signaling (5, 14, 15, 41); however, we do not understand how arenaviruses induce cell-signaling changes, which lead to this dysregulation and potentially to clinical disease. A characteristic of hemorrhagic arenavirus disease is a perturbation of host cytokine responses. Modulation of the host innate immune response as a countermeasure against infectious agents is a significant area of research (2), with current approaches focusing predominantly on “boosting” the immune response. However, treating a number of infections in this way may lead to an exacerbation of clinical disease, rather than to protection, due to the limited understanding of the molecular basis of how these viruses cause disease. Many viruses have evolved mechanisms to interfere with host cell signaling, either to evade the immune response (21, 32, 42) or to activate signaling pathways required for viral replication (3, 9). A detailed knowledge of how viruses and the host interact, and how these interactions change over time, would allow researchers to more rationally target the design of immunomodulatory compounds.
Lassa fever virus is endemic in West Africa, where it causes significant morbidity and mortality and is responsible for a large proportion of hospitalizations (40). In addition to its threat as an emerging infectious disease, Lassa fever virus and other hemorrhagic fever viruses are CDC and NIAID category A biothreat agents (6, 11, 20). While there is an effective vaccine available for Junin virus and ring vaccination could play an important role in outbreak control, vaccination may not be effective within a suitable time frame for treatment of primary cases during an epidemic or in response to intentional release. For these reasons, expansion of the therapeutic armamentarium is urgently required.
Phosphorylation events are critical in the understanding of cell-signaling pathways (25). A number of signaling pathways are controlled in the initial response to activation, not at the transcriptional or translational level but by a series of phosphorylations and other modifications, such as acetylation, ubiquitination, methylation, citrullination, and sumoylation (47). While a number of methodologies are available for the study of protein phosphorylation (27), there have been relatively few investigations of the broad changes in the cellular kinome in response to a stimulus. While genomic and proteomic studies have allowed us to further understand the transcriptional and translational changes induced by activating cell-signaling pathways, the activities of any differentially expressed proteins have often remained undefined due to incomplete characterization of relevant posttranslational modifications which may act to modulate their activities.
Unraveling the complex cell-signaling events which lead to clearance of a pathogen or to clinical disease requires a comparative model system in which attenuated and virulent pathogens can be directly compared. While attenuated or vaccine strains of viruses such as Japanese encephalitis virus (38), rabies virus (45), and Junin virus (1) provide good comparative models, they are limited by the fact that they require higher levels of containment for study. We have used Pichinde virus infection of guinea pigs, which produces a similar pathology to Lassa fever in humans (26), to study these differences at biosafety level 2. We have used an attenuated virus variant, P2, and a variant which causes lethal disease, P18, to dissect the differences in cell-signaling networks which lead to differing outcomes of infection. We hypothesized that infection of guinea pigs with P2 virus would lead to the activation of signaling pathways which produce a protective immune response, viral clearance, and recovery, while infection with P18 virus would induce a differential network of signaling pathways which would lead to an inappropriate response and to lethal disease. We have previously shown that infection of cells with P2 or P18 virus induces differential responses of host transcription factors and signaling intermediates (12, 19). We sought to expand these findings by investigating global cell-signaling changes in response to Pichinde virus infection.
Genomic arrays have previously been used to investigate global changes in transcription in response to pathogens. While gene arrays have provided a valuable insight into virally induced changes in the host cell, they are limited by the fact that they do not address translational or posttranslational effects. We have shown that infection with Pichinde virus causes changes in the total levels of a number of proteins compared to mock infection (12). A key regulator of signal transduction is phosphorylation of specific signaling proteins at one or more residues, which can cause activation or inhibition of the protein. The role of phosphorylation in regulating signaling is reviewed extensively in reference 25. A number of viruses have been shown to interfere with the phosphorylation of cellular proteins. For example, herpes simplex virus and measles virus have been shown to interfere with interferon signaling by inhibiting STAT phosphorylation (43, 49). The nucleocapsid protein of lymphocytic choriomeningitis virus, the prototype arenavirus, was recently shown to inhibit type I interferon signaling by preventing nuclear translocation of interferon regulatory factor 3 (35).
Analysis of signaling pathways in the guinea pig model system has been hampered by the lack of available reagents which cross-react with guinea pig proteins. This, combined with the fact that the genome was only recently sequenced, has hindered the use of traditional genomic and proteomic approaches in this model. For this reason, we utilized a substrate-based kinomics approach to investigate the effect of virus infection on signaling pathways in guinea pig macrophages, the primary target cell of Pichinde infection, and key mediators of the immune response. We have utilized the PepChip kinase assay system to assay the ability of cytoplasmic extracts from infected macrophages from infected guinea pigs to phosphorylate synthetic peptide kinase substrates ex vivo. This technology has previously been used to assay the effect of lipopolysaccharide on signal transduction in human peripheral blood mononuclear cells (17) and to investigate protein phosphorylation in big mitogen-activated protein kinase 1 (MAPK1) knockout cells (23). A kinomics-based approach using an array of phosphorylation state-specific antibodies has also been used to investigate the host response to infection with mycobacteria (24). To our knowledge, this study represents the first use of a broad kinomics approach to investigate global changes in protein phosphorylation and kinase activity in response to viral infection.
MATERIALS AND METHODS
Cell lines and culture conditions.
P388D1 (murine monocyte-like) cells were maintained in RPMI medium supplemented with 5% fetal bovine serum and 2 mM glutamine. Cells were infected with purified P2 or P18 Pichinde virus at a multiplicity of infection of 1 or mock infected with an equivalent fraction of virus purification medium. Cells were harvested at various times postinfection (2 to 16 h) and cytoplasmic and nuclear extracts prepared.
Virus stocks.
Viruses were obtained from serial spleen-passaged stocks from inbred strain 13 guinea pigs infected with Pichinde Munchique strain CoAn 4763 (53). Virus was quantified in a standard plaque assay on Vero cells as described previously (4). Viruses used for in vitro experiments were purified using polyethylene glycol (PEG; Sigma, St. Louis, MO) gradients to remove potential contamination from cytokines and other soluble factors.
Cytoplasmic and nuclear extract preparation.
Cytoplasmic and nuclear extracts were prepared from primary guinea pig macrophages and P388D1 cells as previously described (18), with the addition of a nuclear purification step using OptiPrep (Axis-Shield, Oslo, Norway) gradients. Briefly, lysates were underlayered with 10 ml 30% OptiPrep and 5 ml 35% OptiPrep and centrifuged at 4°C for 30 min at 4,300 × g. The interface was removed and placed in a fresh tube, which was filled with sucrose buffer I (described in reference 18) plus 1.5 mM CaCl2. Following centrifugation at 4°C for 15 min at 1,900 × g, the pellet was resuspended in sucrose buffer I and the centrifugation repeated. Nuclear lysis was completed following the referenced protocol.
Gel electrophoresis and immunoblotting.
Five micrograms of cytoplasmic (approximately 140,000 cell equivalents) or nuclear (approximately 370,000 cell equivalents) extracts was electrophoresed on standard 10% sodium dodecyl sulfate-polyacrylamide gels and transferred to nitrocellulose (Hybond ECL; Amersham Biosciences, Piscataway, NJ). Phosphotyrosine residues were detected by immunodetection following standard methods. Anti-phosphotyrosine PY20 was purchased from BD Biosciences (Rockville, MD). Protein A-horseradish peroxidase conjugate was purchased from Abcam (Cambridge, MA).
Animal protocols.
All animal experiments were conducted following approved institutional animal guidelines and protocols. Male outbred Hartley strain guinea pigs, 400 to 500 g, were inoculated intraperitoneally with 1 ml of phosphate-buffered saline (PBS) containing 1,000 PFU of Pichinde virus (P2 or P18 strain) or with mock spleen homogenate diluted in PBS. Weights and rectal temperatures were recorded daily. Animals were sacrificed at 1 day or 6 days postinfection. Peritoneal cells were harvested immediately following death by aseptic lavage with 100 ml PBS (calcium and magnesium free), collected by centrifugation, and washed in 10 ml PBS. Cells were counted and used to prepare cytoplasmic extracts following the procedure used for cultured cells.
PepChip kinase assay.
Arrays of kinase substrates on glass slides (PepChip kinase v1; PepScan Systems, The Netherlands) were incubated with gamma-32P-ATP, kinase buffer, and cell extracts according to the manufacturer's instructions. Cytoplasmic extracts from four guinea pigs per treatment/time point were used to phosphorylate two PepChip slides (4 arrays), giving a data set of 16 arrays per treatment/time point. Arrays were used to expose X-ray film, which was then analyzed by densitometry using Gel-Pro v.4.5 software (Media Cybernetics, Silver Spring, MD). Background was subtracted on a per spot basis by subtracting the optical density at each spot edge.
Array data analysis.
Data from arrays which showed high background were excluded from analysis. Data were normalized and scaled using the spotting control peptides throughout each array. Following production of a scaled data set, data were analyzed using S-Plus and Spotfire software packages. Student's t test was run on all possible data combinations; analysis of variance (ANOVA) was also performed for each time point and each condition as well as the combination of all treatments in one analysis. Hierarchical clustering was performed on the significant peptides (P ≤ 0.05) resulting from the ANOVA to produce heat maps for P2 versus mock infection and P18 versus mock infection at day 1 and day 6 postinfection. Spots which showed a significant difference in intensity following pairwise comparison (P < 0.05 by Student's t test) were used for the pathway analysis.
Construction of the Ingenuity Knowledge Base.
The Ingenuity Systems Pathways Analysis (IPA) Knowledge Base (Ingenuity Systems, Redwood City, CA) has been described in detail previously (13). Briefly, functions of, and interactions between, cellular proteins are mined from peer-reviewed literature and encoded into an ontology by postdoctoral level scientists. A network analysis of the knowledge base is used to construct interaction-based relationships, both direct and indirect, between proteins in the knowledge base.
A data set containing Swiss-Prot protein identifiers and their corresponding phosphorylation status values was uploaded as an Excel spreadsheet using the template provided in the application. Each protein identifier was mapped to its corresponding object in the Ingenuity Pathways Knowledge Base. Change cutoffs of 1.3-fold for day 1 postinfection data and 2-fold for day 6 postinfection data were set to identify proteins whose phosphorylation status was significantly differentially regulated. The genes were then used as the starting point for generating biological networks. Networks were constructed using direct interactions only.
Biological functions were assigned to each protein network by using the findings that have been extracted from the scientific literature and stored in the Ingenuity Pathways Knowledge Base. The biological functions assigned to each network are ranked according to the significance of that biological function to the network. Fisher's exact test was used to calculate a P value determining the probability that the biological function assigned to that network is explained by chance alone.
Transcription factor binding enzyme-linked immunosorbent assay (ELISA).
Transcription factor binding assays were performed using the TransAM assay system (Active Motif, Carlsbad, CA) following the manufacturer's instructions. Briefly, P388D1 cells were infected at a multiplicity of infection of 5 with P2 or P18 Pichinde virus or mock infected with PEG purification medium and nuclear extracts prepared following harvesting at various times postinfection. Time course infections were performed in triplicate and pooled and protein concentrations determined using a bicinchoninic acid assay (Pierce, Aalst, Belgium). Equivalent amounts of protein were added in quadruplicate wells in a 96-well plate per treatment/time point and allowed to bind to the consensus transcription factor binding site oligonucleotide bound to the well. Following washing, bound transcription factor was detected by antibody binding and colorimetric absorbance.
Phosphoprotein cell-based ELISA.
A cell-based ELISA was performed using the FACE assay system (Active Motif) following the manufacturer's instructions. Briefly, a 96-well plate was seeded with P388D1 cells and a quadruplicate time course of infection with P2 or P18 Pichinde virus was performed. Control cells were mock infected with PEG purification medium. Cells were formaldehyde fixed and assayed for the presence of the specific phosphoprotein using antibody binding and detection by colorimetric absorbance.
RESULTS
Phosphorylation changes following arenavirus infection.
Although we had previously found that total protein levels of specific transcription factors are similar following P2 and P18 infections, we found that their phosphorylation statuses are different between P2 and P18 infections (S.M.F. and N.K.H., unpublished data). These findings suggested that while pathways leading to a generalized increase in specific protein level may be up-regulated as a consequence of viral infection, activation of these proteins may be differentially affected between attenuated and lethal infections. We expanded on these specific findings by examining nuclear extracts from Pichinde-infected murine macrophage cells for phosphorylated proteins by immunoblotting against phosphotyrosine residues. We observed a difference in the quantitative and temporal expression of tyrosine-phosphorylated proteins in the nucleus between P2- and P18-infected cells (data not shown). Following infection with both virus variants, we observed reduced nuclear levels of phosphoproteins and a more rapid reduction in these levels following P18 infection than following infection with P2.
Analysis of high-throughput kinomic data.
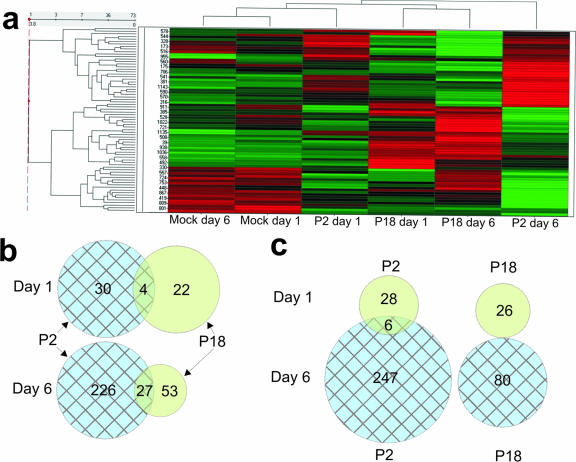
We used a high-throughput kinase substrate array to investigate the host cell response to mild and lethal arenavirus infections. Hierarchical cluster analysis was used to visualize host cell phosphorylation changes in response to attenuated and lethal virus infections. Figure 1a shows the results of hierarchical cluster analysis following ANOVA of substrate phosphorylation by macrophage extracts from P2- and P18-infected guinea pigs compared to mock infection at 1 day and 6 days postinfection. Further analysis of these data by pairwise comparison using Student's t test revealed a number of peptides which were differentially phosphorylated between mock and P2 infections, between mock and P18 infections, and between P2 and P18 infections at 1 day and 6 days postinfection (see Table S1 in the supplemental material). Figure 1b and c show the commonality in protein phosphorylation between P2 and P18 infections. We observed a considerably smaller number of changes at 1 day postinfection than at 6 days postinfection. We believe that as fewer peritoneal cells are infected at this time point, the majority of the smaller changes (n-fold) will be undetectable due to the presence of a majority of uninfected cells in the sample. We have previously shown that infection does not significantly alter the resident cell population and that monocytic cells were the dominant cell types in the samples (19). We also observed that P2 infection induced a greater number of differences in peptide phosphorylation than P18 infection. This correlates with our observations using other experimental systems and is consistent with our hypothesis that P18 actively suppresses the host responses that lead to viral clearance. A number of our findings correlated with our previous data and results from other investigators. For example, we observed that the virulent P18 variant caused a greater reduction in fibrinogen α phosphorylation than P2 at 6 days postinfection. This is consistent with a previous finding which showed increased fibrinogen α dephosphorylation in patients infected with more highly virulent variants of Junin virus compared to that in patients infected with isolates which cause a milder clinical disease (31).
FIG. 1.
Analysis of PepChip kinase data. Peptide phosphorylation was assayed by densitometry, scaled, and normalized using Spotfire software, analyzed by ANOVA, and displayed as a heat map following hierarchical clustering. Panel a shows the heat map produced following ANOVA of spots, which showed differential phosphorylations between treatments. The scaled and normalized data sets were then analyzed in a pairwise fashion using Student's t test. Spots which showed a significant (P < 0.05) differential phosphorylation between P2 and mock infections and P18 and mock infections were compiled into a data set. Commonality between spot phosphorylation is represented as Venn diagrams showing the number of changes compared to that of mock-infected samples. Panel b shows the degree of overlap in peptide phosphorylation between P2 and P18 infections at each time point. Panel c shows the degree of overlap within an infection between time points.
Network analysis of phosphorylation changes.
We sought to integrate these data into functional signaling networks in an attempt to better understand the host cell pathways involved in the response to viral infection. A recent study used a network-based analysis to model the leukocyte response to systemic inflammation by using a genomic array approach (13). Using a web-based knowledge base, the authors of that study modeled the cellular pathways involved in the leukocyte response to inflammation. We used a similar approach to contextualize our data into functional upstream-to-downstream signaling pathways.
We have used IPA (Ingenuity Systems, Redwood City, CA) to model the signaling networks which, based on our identification of differential phosphorylation states, are likely to be involved in the cellular response to attenuated and virulent arenavirus infections. IPA has traditionally been used to analyze genomic data sets, whereby proteins in networks are represented as being up- or down-regulated based on their mRNA level following microarray analysis. We have previously used IPA to construct networks from the results of a high-throughput immunoblot analysis (12), in which total protein levels are directly represented in the network. In the present study, we assayed the changes (n-fold) in peptide phosphorylation following attenuated or virulent Pichinde infection relative to changes in phosphopeptide levels induced by mock infection. Representing these data as simple up- and down-regulation in a network may be misleading due to the different effects of phosphorylation on the activity of the protein. Also, since some proteins have multiple phosphorylation sites represented in the data set, not all of the data could be analyzed and represented by the software.
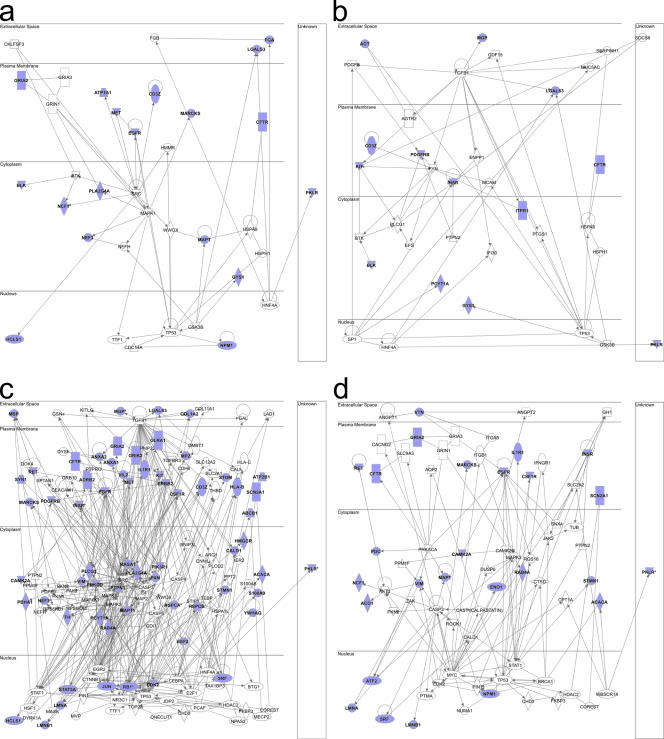
For these reasons, our initial analysis disregarded changes (n-fold) and differential phosphorylations at multiple sites. All proteins, represented by individual phosphorylation site-containing peptides, which showed a significant difference in phosphorylation (P < 0.05), were compiled into a data set and used to construct signaling networks. Figure 2 shows signaling networks produced following IPA after pairwise comparison between P2 versus mock infection and P18 versus mock infection at 1 day and 6 days postinfection. Identified networks with scores of >5 (P = 10−5) were merged and displayed according to subcellular location. Figure 2a shows the network created from differential phosphorylations between mock and P2 infections at 1 day postinfection; Fig. 2b shows the differences between mock and P18 infections. Of interest is the identification of pathways leading to p53 following infection with both viruses. Figure 2c and d show the networks identified at 6 days postinfection following P2 (c) or P18 (d) infection. Of note is the greatly increased complexity of the network induced by P2 infection compared to that induced by P18 at 6 days postinfection.
FIG. 2.
Construction of integrated signaling networks. Data sets compiled from pairwise comparisons of spot data were uploaded to the Ingenuity Pathways Analysis application. The software was used to create functional signaling networks of known interactions based around the differentially phosphorylated proteins we identified using direct interactions only. Pathways with scores of >5 were merged to create one global network for each treatment/time point. (a) P2 on day 1. (b) P18 on day 1. (c) P2 on day 6. (d) P18 on day 6. The proteins shown in blue are those in which we observed significant differential phosphorylation at one or more sites between the infected and control samples.
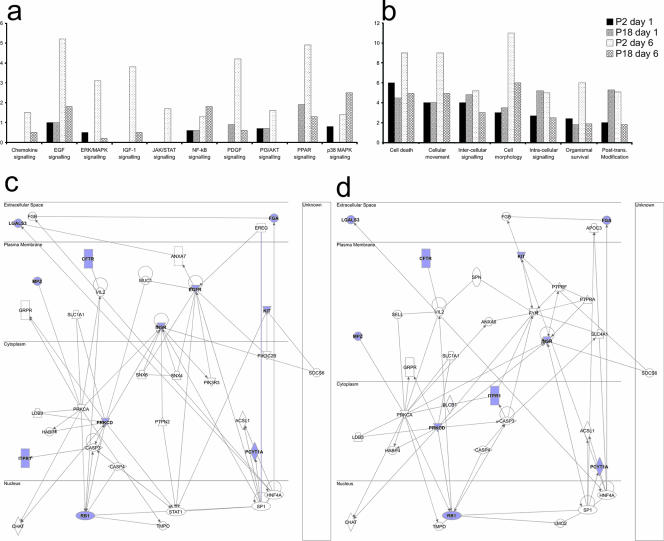
As the focus of our study was to identify the differences in host cell-signaling events between attenuated and lethal infections, we used the IPA application to perform a comparison analysis between our data sets. Figure 3a shows the significance of the involvement of a number of canonical signaling pathways between infections at both time points. As can be seen, P2 induced a higher significance of pathway involvement in the cases of most canonical signaling pathways. Interestingly, we observed a higher significance of involvement of the NF-κB and p38 MAPK pathways following P18 infection at 6 days postinfection. Comparing P2- and P18-induced phosphorylation changes in the context of functional roles of the modified proteins, we observed that at 6 days postinfection, P2 infection induces a higher significance of involvement of proteins in all studied categories. However, at 1 day postinfection, P18 infection alters the phosphorylation status of more proteins involved in intracellular signaling and posttranslational modification than infection with P2. We next performed pathway analysis on the proteins which showed significant differential phosphorylations between P2 and P18 infections at 1 day (Fig. 3c) and 6 days (Fig. 3d) postinfection. The networks show that insulin receptor, epidermal growth factor receptor (EGFR), protein kinase Cδ, and retinoblastoma protein may all be key factors in regulating the differential responses to these virus variants.
FIG. 3.
Comparison analysis of P2- and P18-induced phosphorylation changes. We used the IPA software comparison function to compare the significances of pathway involvement between attenuated and virulent infections at 1 day and 6 days postinfection. We compared the significances of the involvement of canonical signaling pathways (a) and the functional roles of proteins (b). The y axis represents increasing significance of the pathway or process as determined by the number of differentially phosphorylated proteins; it does not represent a level of activity. We compared P2- and P18-induced changes with each other by pairwise comparison, and we created a data set of proteins which are differentially phosphorylated between P2 and P18 infections. We uploaded these data to IPA and used this data set to create signaling networks around the proteins which showed differential phosphorylations between P2 and P18 infections at 1 day (c) and 6 days (d) postinfection. The proteins shown in blue are those in which we observed significant differential phosphorylation at one or more sites between the infected and control samples. ERK, extracellular signal-related kinase; IGF-1, insulin-like growth factor 1; PDGF, platelet-derived growth factor; PI3, phosphatidylinositol-3; PPAR, peroxisome proliferator-activated receptor; Post-trans., posttranslational.
Functional analysis of phosphorylation changes.
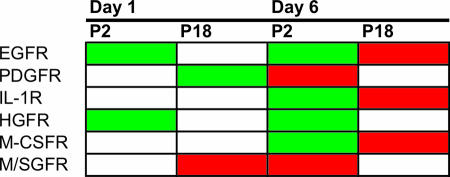
In order to further investigate the role of these phosphorylations and to improve our understanding of the intercellular/intracellular signaling interface, we mined the literature to determine the functional effects of phosphorylation changes on cellular receptors. The identified differential phosphorylation changes in receptors were categorized as being indicative of receptor activation or inhibition. Where receptors were represented by multiple phosphorylations in our array, all phosphorylation sites were investigated and we did not observe any conflicting changes, i.e., increased phosphorylation events in both inhibitory and activation domains at a given treatment and time point. A summary of these changes is shown in Fig. 4. We observed differential receptor activities in a number of proteins, including EGFR, platelet-derived growth factor receptor, interleukin-1 receptor, and macrophage colony-stimulating factor receptor.
FIG. 4.
Functional characterization of receptor activation. In order to correlate our phosphorylation data with functional consequences, we mined the literature for the roles of various phosphorylation sites of receptors to understand how our identified differential phosphorylations affected the activation of a number of cellular receptors. Phosphorylation sites were categorized as inhibitory or as reflecting the activated receptor. The effects of phosphorylation of multiple sites were consistent with increased phosphorylation in activation domains correlating with decreased phosphorylation in inhibitory domains, and decreased phosphorylation in activation domains correlating with increased phosphorylation in inhibitory domains. A number of differential receptor activities were identified at 1 day and 6 days postinfection between infection with P2 and P18 viruses. PDGFR, platelet-derived growth factor receptor; IL-1R, interleukin-1 receptor; HGFR, hepatocyte growth factor receptor; M-CSFR, macrophage colony-stimulating factor receptor; M/SGFR, mast/stem cell growth factor receptor; red, upregulation compared to mock infection; green, downregulation compared to mock infection.
Validation of involvement of identified signaling pathways.
In order to confirm the involvement of the pathways and the identification of specific proteins following pathway analysis, we investigated the effects on specific signaling intermediates and transcription factors implicated in the signaling networks.
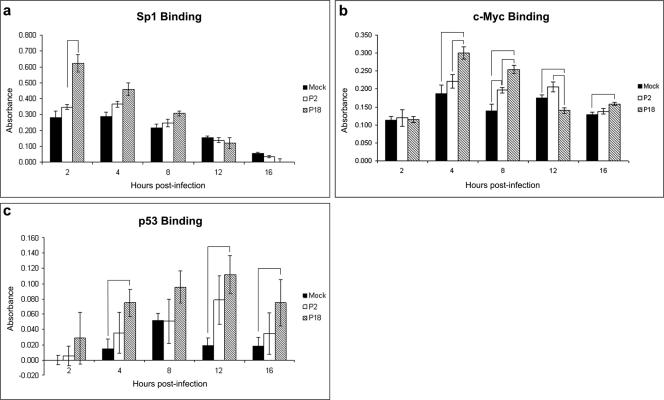
The transcription factor SP1 was implicated in a signaling network following infection with P18 at 1 day postinfection (Fig. 2b). We performed a transcription factor binding ELISA to confirm the involvement of this protein in Pichinde infection. Figure 5a shows a 1.8-fold increase in SP1 binding following P18 infection compared to that following P2 infection at 2 h postinfection (P = 0.0029). No significant differences were observed at other time points. This is consistent with the lack of SP1 involvement in signaling networks constructed at 6 days postinfection.
FIG. 5.
Transcription factor binding assays. Murine P388D1 monocyte-like cells were mock infected or infected with P2 or P18 Pichinde virus for the times indicated and nuclear extracts prepared and protein levels quantified. Five micrograms of protein was assayed for transcription factor binding to its consensus sequence. Bound transcription factor was detected by an indirect immunoassay. Error bars show the standard errors of the means. The binding abilities of Sp1 (a), c-Myc (b), and p53 (c) were assayed. Bracketed bars are those in which the change (n-fold) was statistically significant (P < 0.05); P values are given in the text.
The transcription factor c-Myc was a node in a signaling network induced by P18 infection at 6 days postinfection (Fig. 2d). The ability of c-Myc to bind to its DNA consensus sequence was assayed by a transcription factor binding ELISA. Figure 5b shows a number of small, but significant, changes in c-Myc binding between times and infections. c-Myc binding activities were equivalent in mock-, P2-, and P18-infected cells at 2 h postinfection. At 4 h postinfection, c-Myc binding activity in P18-infected cells had increased 1.6-fold over that in mock-infected cells (P = 0.0078), 1.4-fold over that in P2-infected cells (P = 0.0212), and 2.6-fold over that in P18-infected cells at 2 h postinfection (P < 0.0001). At 8 h postinfection, c-Myc binding in P18-infected cells was increased 1.8-fold compared to that in mock-infected cells (P = 0.0002). Levels of c-Myc binding were 1.3-fold higher in P18-infected cells than in P2-infected cells (P = 0.0069) and 1.4-fold higher in P2-infected cells than in mock-infected cells (P = 0.0287). At 12 h postinfection, binding was 1.3-fold lower in P18-infected cells than in mock-infected cells (P = 0.0147) and 1.5-fold lower than in P2-infected cells (P = 0.0042). At 16 h postinfection, binding was 1.2-fold higher following P18 infection than following mock infection (P = 0.013).
The transcription factor p53 was implicated in all analyses in which infected cells were compared to mock-infected cells (Fig. 2a to d); it was not incorporated in networks in which P2-induced kinome changes were compared with changes induced by P18 infection (Fig. 3c and d). We investigated the binding activity of p53 throughout a time course of infection (Fig. 5c). There were no significant differences in p53 binding activities between P2 and P18 infections. There were significant increases in p53 binding induced by P18 infection compared to that induced by mock infection at 4 (P = 0.0195), 12 (P = 0.0136), and 16 (P = 0.0027) hours postinfection.
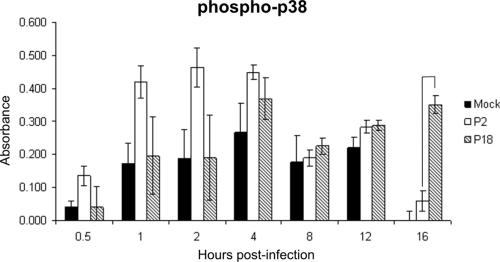
Comparison analysis of P2- versus P18-induced signaling networks revealed a higher significance of involvement of the p38 MAPK pathway following P2 infection at 1 day postinfection and a higher significance following P18 infection at 6 days postinfection (Fig. 3a). We used a cell-based ELISA to investigate the degree of p38 phosphorylation through a time course of infection (Fig. 6). While the early times postinfection show a trend for increased phosphorylation of p38 following infection with P2, these differences were not statistically significant. At 16 h postinfection, however, we observed a significant increase in p38 phosphorylation induced by P18 infection: a sevenfold increase compared to p38 phosphorylation induced by P2 infection (P < 0.0001).
FIG. 6.
Phosphorylation of p38 MAPK. Murine P388D1 monocyte-like cells were mock infected or infected with P2 or P18 Pichinde virus in quadruplicate for the times indicated in a 96-well plate. Cells were fixed, and phospho-p38 was detected by an indirect immunoassay. Error bars show the standard errors of the means. Bracketed bars are those in which the change (n-fold) was statistically significant (P < 0.05); P values are given in the text.
DISCUSSION
These studies show the complexity of virus-induced cellular signaling networks when investigated at the level of protein phosphorylation. We have used a comparative model system to reveal differentially involved signaling networks between attenuated and virulent arenavirus infections. Of particular interest is the small amount of overlap between the differential phosphorylation events between mock and P2 infections and those which differed between mock and P18 infections. This contrasts with our previous investigations in which we observed a significant amount of commonality in total protein expression changes following P2 and P18 infections (12). This finding reveals the importance of understanding the virus-induced host response at a number of levels, from genomic to postproteomic.
Our observation that P18 infection induces a much more limited host response than infection with P2 is consistent with our hypothesis that P18 actively suppresses host cell signaling. These networks reveal a number of differentially phosphorylated targets at all levels of signal transduction pathways, from the plasma membrane to the nucleus, which are targets for further investigation and possible therapeutic intervention. For example, STAT-5A, Jun, and activating transcription factor 2 are all differentially phosphorylated between P2 and P18 infections and modulation of their activities could be used to “push” a P18-induced response towards that induced by P2 infection in an attempt to elicit a protective immune response. This result is consistent with our hypothesis that P18 infection activates signaling pathways early in infection but is able to actively inhibit host cell signaling following replication, leading to the suppression of the immune response.
Of particular interest is the observed difference in the receptor activity of EGFR. EGFR is a receptor tyrosine kinase which can activate a number of downstream signaling pathways (reviewed in reference 10). It has been shown that inhibition of EGFR increases the expression of proinflammatory and apoptotic genes (46). Our finding that EGFR shows decreased activity during P2 infection and increased activity at 6 days following P18 infection is consistent with our hypothesis that P2 facilitates a normal, proinflammatory response early in infection, allowing clearance of the virus. However, it has also been shown that infection of lung epithelial cells with respiratory syncytial virus induced EGFR activation, which leads to increased inflammation (36). This apparent contradiction reveals the importance of studying signaling networks in a global, integrated context, as cellular responses to pathogens are likely to be a delicate balance between pro- and anti-inflammatory pathways.
Analysis of posttranslational modification is likely to be a key next step in our understanding of biological systems. Genomic and proteomic methods have played a crucial role in furthering this understanding but do not reveal the complete story with respect to biological functionality. Assimilating data from large data sets, from the genomic to the metabonomic, will be fundamental in fully understanding the complexity of the regulation of intra- and intercellular signaling. As data analysis tools continue to evolve in parallel with new methodologies, we are in a position where researchers can begin to integrate data from a number of different viral pathogens and from diverse biological disciplines.
Our finding that infection with the lethal P18 passage variant of Pichinde viruses induces a more limited host response than infection with the attenuated P2 variant is consistent with observations of other arenavirus systems. It has been shown that the apathogenic arenavirus Mopeia virus activates human macrophages, whereas the pathogenic Lassa virus does not (7, 39); Lassa virus infection of macrophages reduces tumor necrosis factor alpha and interleukin-8 production following stimulation of cells with lipopolysaccharide, whereas Mopeia virus infection does not (33). It has also been shown that monocyte-derived dendritic cells infected with Ebola and Lassa viruses do not up-regulate costimulatory molecules or secrete proinflammatory cytokines and are inhibited in their ability to stimulate T cells (34). These results suggest that virulent arenaviruses are able to actively suppress host-signaling events which lead to the development of a protective immune response. We have previously shown a potential mechanism for these observations: infection of macrophages with the P2 variant of Pichinde virus causes an increase in DNA binding of the activating RelA/p50 dimer of the NF-κB transcription factor in contrast to P18 virus infection, which causes an increase in binding of the repressive p50/p50 homodimeric form (19).
The guinea pig/Pichinde model is a good system for studying the pathology of arenavirus infection. However, there are few reagents available for use in this system, and there is not an available guinea pig monocyte/macrophage cell line in which to confirm in vivo results using an in vitro system. This made validating our array findings problematic. By following canonical signaling pathways from upstream to downstream, we found that the phosphorylation changes we observed made sense in a biological context. We also investigated the effect on downstream transcription factors and a signaling intermediate by using conventional in vitro assays with murine monocyte-like cells. Our findings with the transcription factor binding and p38 phosphorylation assays were consistent with the networks and comparisons produced following network analysis of the kinomic data, in terms of both infection-based and temporal observations. These data, while validating the conclusions drawn from the kinome array results, also provide interesting starting points to focused investigations aimed at understanding the role these pathways play in virus replication and pathogenesis.
The pathways we targeted for verification have previously been shown to be implicated in virus infection. Infection with cytomegalovirus causes an increase in SP1 mRNA and protein levels, which results in increased NF-κB expression (50). Virus infection has been shown to activate c-Myc, with a 50- to 100-fold increase in c-Myc transcription reported following Rous sarcoma virus infection (30). A number of viruses have been shown to activate p38 MAPK, including herpes simplex virus type 1 (51, 52) and infectious bursal disease virus (28). Viruses from several families have been shown to modulate p53 activity at a number of different levels. African swine fever virus induces up-regulation, stabilization, and nuclear accumulation of p53 (22), adenovirus inhibits p53-mediated transactivation to facilitate replication (48), and cells infected with influenza virus show up-regulated p53 activity, which may play a key role in influenza virus-induced cell death (44).
The c-Myc binding activities observed do not follow the pathway observations as closely as the SP1 data, with significant differences observed at 4 h postinfection. However, no significant changes were observed at the earliest time point and the P18 versus mock changes then persisted through intermediate to late times postinfection. However, the changes (n-fold) observed are small and may not be relevant in the host response to infection, although this could be an artifact of using the murine system. The use of the guinea pig system could reveal larger differences which may be important in infection. It has been shown that EBNA3 of Epstein-Barr virus association with RBP-Jκ causes down-regulation of c-Myc, suggesting a role for RBP-Jκ in c-Myc regulation (16). Differences in c-Myc activity could perhaps be explained by the difference in RBP-Jκ complex size caused by infection with P2 virus, but not by P18, observed in our previous study (19).
This study, combined with our previous proteomic data, our focused transcription factor results, and the observations of other investigators into the functional effects of arenavirus infection, reveals a clear trend which suggests that interference with cellular signaling, potentially at a number of levels of regulation, is involved in arenavirus pathogenesis. Attenuated and apathogenic arenaviruses induce appropriate signaling events which lead to the development of a protective immune response and viral clearance; pathogenic viruses result in a limited response, likely an active process mediated by viral proteins, to inhibit the activation of these pathways or activate inhibitory factors. As yet, we do not know the viral proteins responsible for these effects, but with an increased understanding of the cellular pathways involved, we can rationally target our future investigations to those pathways and proteins which are likely to be key players in mediating pathogenesis.
We have used a novel kinomic assay to investigate the host cell response to attenuated and virulent arenavirus infection. By integrating our results using pathway analysis, we have shown differentially involved signaling networks between these two infections. By employing this approach, we can begin to understand the complex host response to pathogens and start to unravel the cross talk between signaling pathways and how these act to cause the cell to respond in one way or another. By studying the phosphorylation state of proteins, we can dissect their roles in cellular signaling at a more physiological level than by investigating mRNA or total protein levels. A recent article has highlighted the importance of understanding the host response to infection as an alternative approach to antimicrobial drug design (37). By employing a high-throughput assay and integrated signaling network analysis approach, investigators can quickly identify candidate proteins for further investigation and rational targeting of novel antiviral and antibacterial therapeutics.
Many viruses modulate the signaling pathways of the host cell to escape activation of key innate immune mechanisms and establish a productive infection. It is clear that infection with the P18 variant of Pichinde virus suppresses host cell signaling compared to infection with an attenuated variant. By understanding molecular events responsible for the clinical disease observed at the tissue, organ, and organism levels, we can attempt to elucidate the determinants of pathogenesis at the level of signaling pathways. A number of small-molecule agonists and antagonists to receptors and signaling intermediates are already available due to their use as anticancer therapies. By combining these classical treatments with novel drug design concepts such as decoy thioaptamers to transcription factors (8, 29), we may be able to inhibit microbial pathogenesis at the level of the host response rather than directly target viral or bacterial proteins. This approach could have important consequences with regard to the current problem of microbial resistance to existing therapies.
Supplementary Material
Acknowledgments
We thank Alan Barrett for critical reading of the manuscript.
This work was supported by grants from DARPA (DAAD19011037), DTRA (DAAD17-01-D0001), and the NIH (N01-HV-28184, U01 AI054827, and R01 A127744) and by a grant from the NIAID to N.K.H. through the Western Regional Center of Excellence for Biodefense and Emerging Infectious Disease Research, NIH grant number U54 AI057156.
We declare no competing financial interests.
Footnotes
Published ahead of print on 6 December 2006.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Albarino, C. G., P. D. Ghiringhelli, D. M. Posik, M. E. Lozano, A. M. Ambrosio, A. Sanchez, and V. Romanowski. 1997. Molecular characterization of attenuated Junin virus strains. J. Gen. Virol. 78:1605-1610. [DOI] [PubMed] [Google Scholar]
- 2.Amlie-Lefond, C., D. A. Paz, M. P. Connelly, G. B. Huffnagle, K. S. Dunn, N. T. Whelan, and H. T. Whelan. 2005. Innate immunity for biodefense: a strategy whose time has come. J. Allergy Clin. Immunol. 116:1334-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrade, A. A., P. N. Silva, A. C. Pereira, L. P. De Sousa, P. C. Ferreira, R. T. Gazzinelli, E. G. Kroon, C. Ropert, and C. A. Bonjardim. 2004. The vaccinia virus-stimulated mitogen-activated protein kinase (MAPK) pathway is required for virus multiplication. Biochem. J. 381:437-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aronson, J. F., N. K. Herzog, and T. R. Jerrells. 1994. Pathological and virological features of arenavirus disease in guinea pigs. Comparison of 2 Pichinde virus strains. Am. J. Pathol. 145:228-235. [PMC free article] [PubMed] [Google Scholar]
- 5.Aronson, J. F., N. K. Herzog, and T. R. Jerrells. 1995. Tumor necrosis factor and the pathogenesis of Pichinde virus infection in guinea pigs. Am. J. Trop. Med. Hyg. 52:262-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atlas, R. M. 2003. Bioterrorism and biodefence research: changing the focus of microbiology. Nat. Rev. Microbiol. 1:70-74. [DOI] [PubMed] [Google Scholar]
- 7.Baize, S., J. Kaplon, C. Faure, D. Pannetier, M. C. Georges-Courbot, and V. Deubel. 2004. Lassa virus infection of human dendritic cells and macrophages is productive but fails to activate cells. J. Immunol. 172:2861-2869. [DOI] [PubMed] [Google Scholar]
- 8.Bassett, S. E., S. M. Fennewald, D. J. King, X. Li, N. K. Herzog, R. Shope, J. F. Aronson, B. A. Luxon, and D. G. Gorenstein. 2004. Combinatorial selection and edited combinatorial selection of phosphorothioate aptamers targeting human nuclear factor-kappaB RelA/p50 and RelA/RelA. Biochemistry 43:9105-9115. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Israel, H., and T. Kleinberger. 2002. Adenovirus and cell cycle control. Front. Biosci. 7:d1369-d1395. [DOI] [PubMed] [Google Scholar]
- 10.Bogdan, S., and C. Klambt. 2001. Epidermal growth factor receptor signaling. Curr. Biol. 11:R292-R295. [DOI] [PubMed] [Google Scholar]
- 11.Borio, L., T. Inglesby, C. J. Peters, A. L. Schmaljohn, J. M. Hughes, P. B. Jahrling, T. Ksiazek, K. M. Johnson, A. Meyerhoff, T. O'Toole, M. S. Ascher, J. Bartlett, J. G. Breman, E. M. Eitzen, Jr., M. Hamburg, J. Hauer, D. A. Henderson, R. T. Johnson, G. Kwik, M. Layton, S. Lillibridge, G. J. Nabel, M. T. Osterholm, T. M. Perl, P. Russell, and K. Tonat. 2002. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA 287:2391-2405. [DOI] [PubMed] [Google Scholar]
- 12.Bowick, G. C., S. A. Fennewald, B. A. Elsom, J. F. Aronson, B. A. Luxon, D. G. Gorenstein, and N. K. Herzog. 2006. Differential signaling networks induced by mild and lethal hemorrhagic fever virus infections. J. Virol. 80:10248-10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvano, S. E., W. Z. Xiao, D. R. Richards, R. M. Felciano, H. V. Baker, R. J. Cho, R. O. Chen, B. H. Brownstein, J. P. Cobb, S. K. Tschoeke, C. Miller-Graziano, L. L. Moldawer, M. N. Mindrinos, R. W. Davis, R. G. Tompkins, and S. F. Lowry. 2005. A network-based analysis of systemic inflammation in humans. Nature 437:1032-1037. [DOI] [PubMed] [Google Scholar]
- 14.Colle, J. H., M. F. Saron, B. Shidani, M. P. Lembezat, and P. Truffa-Bachi. 1993. High frequency of T lymphocytes committed to interferon-gamma transcription upon polyclonal activation in spleen from lymphocytic choriomeningitis virus-infected mice. Int. Immunol. 5:435-441. [DOI] [PubMed] [Google Scholar]
- 15.Colle, J. H., M. F. Saron, and P. Truffa-Bachi. 1993. Altered cytokine genes expression by conA-activated spleen cells from mice infected by lymphocytic choriomeningitis virus. Immunol. Lett. 35:247-253. [DOI] [PubMed] [Google Scholar]
- 16.Cooper, A., E. Johannsen, S. Maruo, E. Cahir-McFarland, D. Illanes, D. Davidson, and E. Kieff. 2003. EBNA3A association with RBP-Jκ down-regulates c-myc and Epstein-Barr virus-transformed lymphoblast growth. J. Virol. 77:999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diks, S. H., K. Kok, T. O'Toole, D. W. Hommes, P. van Dijken, J. Joore, and M. P. Peppelenbosch. 2004. Kinome profiling for studying lipopolysaccharide signal transduction in human peripheral blood mononuclear cells. J. Biol. Chem. 279:49206-49213. [DOI] [PubMed] [Google Scholar]
- 18.Dyer, R. B., and N. K. Herzog. 1995. Isolation of intact nuclei for nuclear extract preparation from a fragile B-lymphocyte cell line. BioTechniques 19:192-195. [PubMed] [Google Scholar]
- 19.Fennewald, S. M., J. F. Aronson, L. Zhang, and N. K. Herzog. 2002. Alterations in NF-κB and RBP-Jκ by arenavirus infection of macrophages in vitro and in vivo. J. Virol. 76:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisbert, T. W., and P. B. Jahrling. 2004. Exotic emerging viral diseases: progress and challenges. Nat. Med. 10:S110-S121. [DOI] [PubMed] [Google Scholar]
- 21.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 22.Granja, A. G., M. L. Nogal, C. Hurtado, J. Salas, M. L. Salas, A. L. Carrascosa, and Y. Revilla. 2004. Modulation of p53 cellular function and cell death by African swine fever virus. J. Virol. 78:7165-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi, M., C. Fearns, B. Eliceiri, Y. Yang, and J. D. Lee. 2005. Big mitogen-activated protein kinase 1/extracellular signal-regulated kinase 5 signaling pathway is essential for tumor-associated angiogenesis. Cancer Res. 65:7699-7706. [DOI] [PubMed] [Google Scholar]
- 24.Hestvik, A. L. K., Z. Hmama, and Y. Av-Gay. 2003. Kinome analysis of host response to mycobacterial infection: a novel technique in proteomics. Infect. Immun. 71:5514-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter, T. 2000. Signaling—2000 and beyond. Cell 100:113-127. [DOI] [PubMed] [Google Scholar]
- 26.Jahrling, P. B., R. A. Hesse, J. B. Rhoderick, M. A. Elwell, and J. B. Moe. 1981. Pathogenesis of a Pichinde virus strain adapted to produce lethal infections in guinea pigs. Infect. Immun. 32:872-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, S. A., and T. Hunter. 2005. Kinomics: methods for deciphering the kinome. Nat. Methods 2:17-25. [DOI] [PubMed] [Google Scholar]
- 28.Khatri, M., and J. M. Sharma. 2006. Infectious bursal disease virus infection induces macrophage activation via p38 MAPK and NF-kappaB pathways. Virus Res. 118:70-77. [DOI] [PubMed] [Google Scholar]
- 29.King, D. J., S. E. Bassett, X. Li, S. A. Fennewald, N. K. Herzog, B. A. Luxon, R. Shope, and D. G. Gorenstein. 2002. Combinatorial selection and binding of phosphorothioate aptamers targeting human NF-kappa B RelA(p65) and p50. Biochemistry 41:9696-9706. [DOI] [PubMed] [Google Scholar]
- 30.Kuchino, Y., K. Nemoto, S. Kawai, and S. Nishimura. 1985. Activation of c-myc gene transcription by Rous sarcoma virus infection. Jpn. J. Cancer Res. 76:75-78. [PubMed] [Google Scholar]
- 31.Lerer, G. D., M. C. Saavedra, R. Falcoff, J. I. Maiztegui, and F. C. Molinas. 1991. Activity of a platelet protein kinase that phosphorylates fibrinogen and histone in Argentine hemorrhagic fever. Acta Physiol. Pharmacol. Ther. Latinoam. 41:377-386. [PubMed] [Google Scholar]
- 32.Lisowska, K., and J. M. Witkowski. 2003. Viral strategies in modulation of NF-kappaB activity. Arch. Immunol. Ther. Exp. 51:367-375. [PubMed] [Google Scholar]
- 33.Lukashevich, I. S., R. Maryankova, A. S. Vladyko, N. Nashkevich, S. Koleda, M. Djavani, D. Horejsh, N. N. Voitenok, and M. S. Salvato. 1999. Lassa and Mopeia virus replication in human monocytes/macrophages and in endothelial cells: different effects on IL-8 and TNF-alpha gene expression. J. Med. Virol. 59:552-560. [PMC free article] [PubMed] [Google Scholar]
- 34.Mahanty, S., K. Hutchinson, S. Agarwal, M. McRae, P. E. Rollin, and B. Pulendran. 2003. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J. Immunol. 170:2797-2801. [DOI] [PubMed] [Google Scholar]
- 35.Martínez-Sobrido, L., E. I. Zúñiga, D. Rosario, A. García-Sastre, and J. C. de la Torre. 2006. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 80:9192-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monick, M. M., K. Cameron, J. Staber, L. S. Powers, T. O. Yarovinsky, J. G. Koland, and G. W. Hunninghake. 2005. Activation of the epidermal growth factor receptor by respiratory syncytial virus results in increased inflammation and delayed apoptosis. J. Biol. Chem. 280:2147-2158. [DOI] [PubMed] [Google Scholar]
- 37.Nature. 2006. Infection biology. Nature 441:255-256. [DOI] [PubMed] [Google Scholar]
- 38.Ni, H. L., G. J. J. Chang, H. Xie, D. W. Trent, and A. D. T. Barrett. 1995. Molecular basis of attenuation of neurovirulence of wild-type Japanese encephalitis virus strain Sa14. J. Gen. Virol. 76:409-413. [DOI] [PubMed] [Google Scholar]
- 39.Pannetier, D., C. Faure, M. C. Georges-Courbot, V. Deubel, and S. Baize. 2004. Human macrophages, but not dendritic cells, are activated and produce alpha/beta interferons in response to Mopeia virus infection. J. Virol. 78:10516-10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richmond, J. K., and D. J. Baglole. 2003. Lassa fever: epidemiology, clinical features, and social consequences. BMJ 327:1271-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saron, M. F., J. H. Colle, A. Dautry-Varsat, and P. Truffa-Bachi. 1991. Activated T lymphocytes from mice infected by lymphocytic choriomeningitis virus display high affinity IL-2 receptors but do not proliferate in response to IL-2. J. Immunol. 147:4333-4337. [PubMed] [Google Scholar]
- 42.Seow, H. F. 1998. Pathogen interactions with cytokines and host defence: an overview. Vet. Immunol. Immunopathol. 63:139-148. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi, K., S. Kadota, M. Takeda, N. Miyajima, and K. Nagata. 2003. Measles virus V protein blocks interferon (IFN)-alpha/beta but not IFN-gamma signaling by inhibiting STAT1 and STAT2 phosphorylation. FEBS Lett. 545:177-182. [DOI] [PubMed] [Google Scholar]
- 44.Turpin, E., K. Luke, J. Jones, T. Tumpey, K. Konan, and S. Schultz-Cherry. 2005. Influenza virus infection increases p53 activity: role of p53 in cell death and viral replication. J. Virol. 79:8802-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Z. W., L. Sarmento, Y. Wang, X.-q. Li, V. Dhingra, T. Tseggai, B. Jiang, and Z. F. Fu. 2005. Attenuated rabies virus activates, while pathogenic rabies virus evades, the host innate immune responses in the central nervous system. J. Virol. 79:12554-12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woodworth, C. D., E. Michael, D. Marker, S. Allen, L. Smith, and M. Nees. 2005. Inhibition of the epidermal growth factor receptor increases expression of genes that stimulate inflammation, apoptosis, and cell attachment. Mol. Cancer Ther. 4:650-658. [DOI] [PubMed] [Google Scholar]
- 47.Yang, X. J. 2005. Multisite protein modification and intramolecular signaling. Oncogene 24:1653-1662. [DOI] [PubMed] [Google Scholar]
- 48.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 357:82-85. [DOI] [PubMed] [Google Scholar]
- 49.Yokota, S., N. Yokosawa, T. Kubota, T. Suzutani, I. Yoshida, S. Miura, K. Jimbow, and N. Fujii. 2001. Herpes simplex virus type 1 suppresses the interferon signaling pathway by inhibiting phosphorylation of STATs and janus kinases during an early infection stage. Virology 286:119-124. [DOI] [PubMed] [Google Scholar]
- 50.Yurochko, A. D., M. W. Mayo, E. E. Poma, A. S. Baldwin, Jr., and E. S. Huang. 1997. Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-κB promoters. J. Virol. 71:4638-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zachos, G., B. Clements, and J. Conner. 1999. Herpes simplex virus type 1 infection stimulates p38/c-Jun N-terminal mitogen-activated protein kinase pathways and activates transcription factor AP-1. J. Biol. Chem. 274:5097-5103. [DOI] [PubMed] [Google Scholar]
- 52.Zachos, G., M. Koffa, C. M. Preston, J. B. Clements, and J. Conner. 2001. Herpes simplex virus type 1 blocks the apoptotic host cell defense mechanisms that target Bcl-2 and manipulates activation of p38 mitogen-activated protein kinase to improve viral replication. J. Virol. 75:2710-2728. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Zhang, L. H., K. Marriott, and J. F. Aronson. 1999. Sequence analysis of the small RNA segment of guinea pig-passaged Pichinde virus variants. Am. J. Trop. Med. Hyg. 61:220-225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.