Abstract
Cryphonectria parasitica strain EP721 is infected with a strain of hypovirus CHV1, CHV1-EP721, and exhibits typical hypovirulence-associated traits such as reduced pigmentation and reduced asexual sporulation. However, the accumulation of the viral double-stranded RNA (dsRNA) in this hypovirus-infected C. parasitica strain is atypically low. We now report the complete nucleotide sequence and construction of a full-length infectious cDNA clone for hypovirus CHV1-EP721. The genome sequence of CHV1-EP721 was determined to be 12,724 bp in length and to share extensive homology with two other hypovirus strains, CHV1-Euro7 and CHV1-EP713, with an average of 99% and 90% identities at the nucleotide level and 99% and 92% identities at the amino acid level, respectively. CHV1-EP721 was successfully introduced into virus-free fungal host strain EP721(-v) by transfection with transcripts derived from a full-length viral cDNA. The transfected strain had a phenotype indistinguishable from that of EP721, and the accumulation of CHV1-EP721 dsRNA in the transfectant was lower than those transfected by CHV1-Euro7 and CHV1-EP713 transcripts. Through the construction of chimeric viruses by domain swapping using infectious cDNA clones of CHV1-EP721, CHV1-EP713, and CHV1-Euro7 hypoviruses, the determinant for the low level of viral dsRNA accumulation in CHV1-EP721 was mapped to the second of two CHV1-EP721 open reading frames (ORFs), ORF B. Further refined swapping of domains within ORF B identified a 2.5-kb coding region between p48 and the polymerase domain of CHV1-EP721 as being responsible for the low viral dsRNA accumulation. Evidence is also provided that low rates of hypovirus transmission through conidial spores correlates with low viral dsRNA accumulation.
Hypoviruses are a group of unencapsidated, cytoplasmically replicating RNA viruses of the chestnut blight fungus Cryphonectria parasitica that can attenuate virulence (hypovirulence) and alter host biological processes, which results in suppressed pigmentation, reduced asexual production, and loss of female fertility (3, 5, 15, 24). These phenotypic changes are accompanied by the modulation of approximately 13.4% of the C. parasitica transcriptome (1, 2). Four virus species have been identified as belonging to the genus Hypovirus in the family Hypoviridae: CHV1 (3, 29), CHV2 (18, 20), CHV3 (23, 31), and CHV4 (16, 22). Interest in research on this group of viruses stemmed initially from their capability to attenuate the virulence of their phytopathogenic fungal host as biocontrol agents (3, 15, 24) and, more recently, as a model system to explore fungal pathogenic mechanisms (15, 25). In these regards, the development of infectious cDNA clones and transformation and transfection systems for hypoviruses has greatly advanced our understanding of the nature of these viruses and provided the ability to manipulate hypoviruses for both fundamental and practical applications (6-9, 13, 15).
Hypoviruses do not encode a capsid protein, and hypovirus RNA replication occurs in association with host membrane vesicles that accumulate viral double-stranded RNA (dsRNA). Like other mycoviruses, hypoviruses lack an extracellular route of infection. Natural virus transmission is restricted to cytoplasmic mixing as a result of fusion (anastomosis) between vegetatively compatible strains or to various degrees through asexual spores (conidia). The genome of the prototypic hypovirus CHV1-EP713 consists of a 12.7-kb coding strand that contains two open reading frames (ORFs), ORF A and ORF B, separated by a UAAUG pentanucleotide (29). ORF A encodes a polypeptide, p69, which is autocatalytically processed into two products, p29 and p40, by the action of a papain-like protease domain located in the p29 coding region. Cleavage occurs between p69 Gly248 and Gly249, and the residues Cys162 and His215 are essential for autolytic cleavage (12). Expression of ORF A in C. parasitica tranformants in the absence of virus infection resulted in a hypovirus-associated loss of orange pigment production (white phenotype) and a significant reduction in asexual sporulation but not in virulence attenuation (10). Craven et al. subsequently mapped the ORF A suppressive function to p29 at the N-terminal portion of p69 (14).
The CHV1-EP713 ORF A coding domain has been found to contribute to viral replication competency and RNA accumulation levels. Suzuki et al. (32) previously reported that the N-terminal 24 codons of p29 were required for virus replication, while the remaining 598 codons of ORF A were dispensable. Deletion of the dispensable portion of p29 resulted in the reduced accumulation of viral RNA and reduced virus transmission through asexual spores. It was also shown that p29 could function in trans to elevate viral genomic RNA accumulation and vertical transmission of p29 deletion mutant virus to the level of wild-type virus (33). Mutation of p29 residues Cys70 and Cys72 resulted in the loss of both p29-mediated suppressive activity in virus-free transgenic C. parasitica and in trans enhancement of RNA accumulation and transmission (33). Segers et al. (28) recently reported that p29 functions as a suppressor of RNA silencing both in the fungal host and in a heterologous plant system.
Deletion of the C-terminal portion of p69, which encodes the highly basic protein p40, resulted in a replication-competent mutant virus that was significantly reduced in RNA accumulation (34). C. parasitica colonies infected with the Δp40 mutant virus exhibited an increased growth rate, increased pigment production, and more conidial formation relative to those of colonies infected with wild-type virus. However, the reduced virus RNA accumulation resulting from the deletion of p40 had little effect on virus-mediated hypovirulence. Through gain-of-function analysis, the p40 symptom determinant was mapped to the N-terminal domain, encompassing p69 residues Thr288 to Arg312. Moreover, the restoration of symptoms correlated with an increased accumulation of viral RNA (34). However, p40 could not function in trans to elevate the viral dsRNA accumulation or restore virus-mediated symptom expression to the level of wild-type virus.
Considerable variation can be observed in the level of hypovirus RNA accumulation in infected C. parasitica field isolates. A surprisingly low level of viral dsRNA accumulation was observed in European C. parasitica strain EP721. While the dsRNA in EP721 is only about 1/10 of the level observed in CHV1-EP713-infected strain EP713 in terms of amount, the infected strain shows hypovirulence-associated traits such as reduced orange pigmentation and reduced asexual sporulation levels. Since EP721 appears not to belong to the same vegetative compatibility group as EP155 (virus free and isogenic to EP713) or Euro7(-v) (virus free and isogenic to Euro7) as judged by the failure of transmission of the virus via anastomosis, this low level of viral dsRNA accumulation has not been assigned either to the nature of the host fungus or to the property of the virus. Given the fact that little is known about the replication mechanism of the hypovirus, a species or strain that exhibits a significantly low level of viral dsRNA accumulation in the host cell would provide opportunities for comparative approaches to investigate mechanisms underlying hypovirus replication. We now report the cloning, whole-genome sequence analysis, construction of a full-length cDNA clone for hypovirus CHV1-EP721, and mapping of determinants in the viral genome responsible for the extremely low level of viral dsRNA accumulation.
MATERIALS AND METHODS
Fungal strains and growth conditions.
C. parasitica strain EP721 (ATCC 66024) was generated by the anastomosis-mediated transfer of hypovirus RNA from the Italian hypovirulent strain HI2 to American strain EP60 (ATCC 38765) that was isolated from Michigan (William McDonald, personal communication). Strain EP721(-v) is a virus-free single conidial isolate derived from EP721. EP155 (ATCC 38755) and Euro7(-v), a virus-free single conidial isolate derived from virus-containing strain Euro7 (ATCC 66021), were obtained from our laboratory stock. The fungal strains were cultured on potato dextrose agar (PDA; Difco) under laboratory bench-top conditions or in EP complete liquid medium (27) at room temperature.
Isolation of viral RNAs.
For the purpose of cDNA library construction, viral dsRNA was extracted from EP721 mycelia cultured in liquid EP medium for 5 days at 28°C as described previously (19). The partially purified dsRNA was further digested with S1 nuclease. Following phenol-chloroform extraction, the viral dsRNA was passaged through a Spin Column-1000 (Sigma) to remove small nucleotide fragments. For the purpose of semiquantitative determination of viral dsRNA and single-stranded RNA (ssRNA) concentrations, total nucleic acids were extracted sequentially with phenol, phenol-chloroform, and chloroform-isoamyl alcohol. The nucleic acid samples were exposed to RQ1-DNase (Promega) digestion for 2 h at 37°C. The quality and quantity of the RNA were examined by agarose gel (0.8%) electrophoresis in 1× Tris-acetate-EDTA (TAE) buffer.
Generation of viral cDNA library.
Oligo(dT) and a series of coding-strand primers spaced about 2 kb apart along the CHV1-Euro7 genome were designed to synthesize cDNA of CHV1-EP721 dsRNA with a Timesaver cDNA synthesis kit (Ampharmacia). The resulting cDNA fragments were cloned into the plasmid pCR-Script SK(+) (Stratagene). The library was screened for larger inserts, which were subsequently sequenced. Gaps were filled by amplifying fragments generated by PCR with Pfu polymerase (8).
Construction of full-length infectious CHV1-EP721 cDNA clone.
The construction of a CHV1-EP721 infectious cDNA clone followed the general protocol used for CHV1-Euro7 dsRNA (8). cDNA clones of the terminal domains were modified by PCR to incorporate a unique NotI site followed by a T7 polymerase promoter fused to the 5′ terminus of the coding strand and to add a unique SpeI site following the 3′-terminal poly(A). Several large intermediate clones were generated from overlapped partial cDNA clones by the use of common endonuclease restriction sites contained within the neighboring clones. The full-length cDNA was obtained by ligating three large cDNA clones through two endonuclease restriction sites, SacI (coordinate 5220) and MluI (coordinate 7843), and was then cloned into the plasmid vector pCR-BluntII-TOPO (Invitrogen) to generate plasmid p721. Transcripts corresponding to the CHV1-EP721 coding strand were synthesized from SpeI-digested p721 in a T7 polymerase reaction and used to transfect C. parasitica spheroplasts as described previously by Chen et al. (6).
Construction of chimeric viruses.
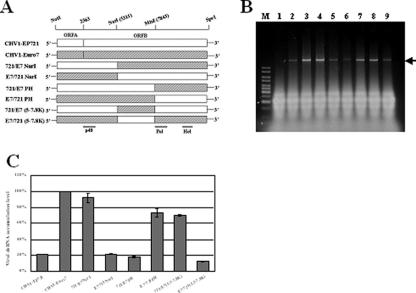
The ORF A or ORF B domain of CHV1-EP721 was interchanged with those of CHV1-EP713 or CHV1-Euro7 with the aid of corresponding infectious cDNA clones of CHV1-EP713 (9) and CHV1-Euro7 (8) as described previously by Chen and Nuss (8). The first group of chimeric viruses was configured with complete ORF A or ORF B domains swapped among CHV1-EP721, CHV1-Euro7, and CHV1-EP713. Briefly, between CHV1-EP721 and CHV1-EP713, the chimeric ORF A and ORF B junctions were first constructed by PCR so that the sequences of the domains representing target viruses were precisely configured. For chimera 721A713B, the junction contained the restriction sites SspI (position 2357) and SacI (position 3395), and for 713A721B, the junction contained the restriction sites HindIII (position 2284) and XhoI (position 3581). These restriction sites allowed the joining of the rest of the sequence of the 5′- and 3′-end portions of the target virus. Since the genome sequence from the SspI site in position 2357 to the end of ORF A is exactly the same between CHV1-EP721 and CHV1-Euro7, this restriction site was used to swap ORF A (NotI/SspI fragment) or ORF B (SspI/SpeI fragment) for making chimeric viruses. The second group of chimeric viruses was made by swapping specific domains within ORF B between CHV1-EP721 and CHV1-Euro7 by relying on common NarI (position 5311) and MluI (position 7843) restriction sites as illustrated in Fig. 4A. The integrity of each chimera was examined by restriction pattern and sequence analysis of the junction of interchanged domains. The chimeric viral dsRNA from the corresponding fungal transfectants was further confirmed by reverse transcription-PCR (RT-PCR) and restriction analysis.
FIG. 4.
Quantification of viral dsRNA levels in transfectants containing different chimeric hypoviruses. (A) Schematic diagram of parental and chimeric hypoviruses used to dissect the viral dsRNA accumulation determinant in ORF B. The coding regions of CHV1-EP721 and CHV1-Euro7 are presented as a white box and a gray box, respectively. The unique restriction sites NotI and SpeI were introduced into 5′ and 3′ ends of viral cDNA, respectively, to facilitate large-fragment swapping. Internal restriction sites NarI at genome position 5311 and MluI at position 7843 are both located within ORF B and were used for the construction of chimeric hypoviruses. The 107 nucleotides that extend from the MluI site upstream to the putative cleavage site of the 5′ portion of Pol (position 7736) are identical between CHV1-EP721 and CHV1-Euro7. The approximate positions of p48 and putative Pol and Hel coding domains are indicated at the bottom of the figure. (B) Agarose gel electrophoretic analysis of viral dsRNA of chimeric hypoviruses. Ten micrograms of total RNA from virus-infected hosts was loaded into each lane in a 0.8% agarose gel. Lane 1, virus-free strain EP721(-v); lane 2 to lane 8, transfectants with wild-type CHV1-EP721 and CHV1-Euro7 and chimeric virus 721/E7NarI, E7/721NarI, 721/E7PH, E7/721PH, 721/E7(5.3-7.8K), and E7/721(5.3-7.8K). The lane marked with an M contains 100 ng of a 1-kb DNA ladder. The arrow indicates the position of viral dsRNA. (C) Viral dsRNA quantification of transfectants of chimeric hypoviruses with the ORF B internal domain swapped. All transfectants were in the EP721(-v) genetic background. The viral dsRNA level of CHV1-Euro7 was set at 100%, and the levels of other viruses were expressed as percentages of that of CHV1-Euro7. Values were calculated from three independent experiments, and bars indicate mean deviations.
Transfection.
Synthetic full-length plus-sense viral RNAs were used to transfect C. parasitica spheroplasts as described previously (6, 8).
Viral dsRNA semiquantification.
After separation in a 0.8% agarose gel in TAE buffer, the ethidium bromide-stained gels were scanned with a Typhoon 9410 phosphorimager (Amersham). The scanned gel images were then analyzed by densitometry with the aid of ImageQuant TL-1D gel analysis software to quantify the relative amount of large and medium dsRNA in each sample. The florescence intensities of the viral dsRNA bands were normalized against the 18S rRNA in the samples.
Viral ssRNA quantification.
The relative accumulation levels of viral ssRNA in fungal colonies infected with wild-type or chimeric viruses were examined by semiquantitative real-time PCR as described previously by Suzuki and Nuss (34). cDNAs specific for viral plus- and minus-strand RNA were generated with primers RTQ-2 (positions 12545 to 12526 of CHV1-EP721) and RTQ-1 (positions 12376 to 12396 of CHV1-EP721), respectively, using a ReverAid First Strand cDNA synthesis kit (Fermentas). Nucleotides in the primer regions are identical for CHV1-EP721, CHV1-EP713, and CHV1-Euro7. Each cDNA synthesis reaction mixture also contained primer CP18SR (5′-CAGCACGACAGAGTTTCACAAG-3′), which was complementary to the 3′ portion of C. parasitica 18S rRNA. The primer sets used for real-time PCR were RTQ-1 and RTQ-2 for the viral RNA and CP18SR and CP18SF (5′-ACGCTGGCTTCTTAGAGGGACT-3′) for 18S rRNA. The reaction was performed using a volume of 20 μl containing 5 μl of cDNA template, 5 pmol of primers, 2.5 μmol of deoxynucleoside triphosphate, 2.0 mM of MgCl2, and 0.5 U of ExTag polymerase (Takaha) in 0.5× SGB1 mix (Shanghai Ope Technology Development Company, Shanghai, People's Republic of China). The reaction was performed using DNA Engine OPTICON 2 (MJ Research Incorporated). Viral cDNA values were normalized against the cDNA value of 18S rRNA. For comparisons among experiments, cloned target DNA sequences, a 596-bp cDNA fragment of CHV1-EP721 dsRNA from positions 12130 to 12724 and a 1,178-bp cDNA fragment of 18S rRNA generated with primer sets CP18SR and 18SF2 (5′-TCTCGAATCGCATGGCCT-3′), were used as references in each experiments.
Assay of virus transmission through conidial spores.
Fungal colonies were cultured in an incubator maintained at 23 to 25°C with 12 h light at 600 to 1,000 lx on PDA for 2 weeks to support conidiation. Conidia were harvested from the plate by washing with 0.15% Tween 80 as described previously by Hillman et al. (19). The spores were spread onto PDA plates at a density of 100 spores per plate. Colonies derived from single spores were transferred onto new PDA plates and cultured for 1 to 2 weeks in order to record white (virus-containing) and orange (virus-free) phenotypes.
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequence of the full-length cDNA of CHV1-EP721 genomic RNA is DQ861913.
RESULTS
Accumulation of CHV1-EP721 dsRNA in host fungus strain EP721.
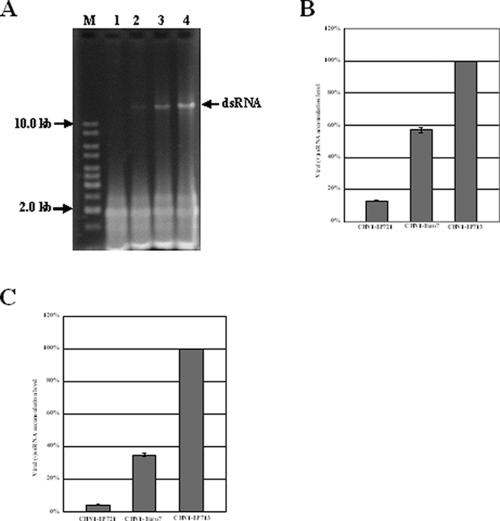
When cultured on PDA under laboratory bench-top conditions, EP721 showed a typical hypovirus-infected phenotype: a white colony and reduced asexual sporulation. Compared with strain EP713 that harbors prototypic hypovirus CHV1-EP713, EP721 grows more quickly and has a higher level of asexual sporulation. Since the fungal phenotype can be virus-host specific, hypoviruses CHV1-EP713 and CHV1-Euro7 were introduced into the virus-free strain EP721(-v) by transfection. The resulting transfectants, EP721(-v)/CHV1-EP713 and EP721(-v)/CHV1-Euro7, showed phenotypes and dsRNA accumulation levels very similar to those of EP713 and Euro7, respectively. An examination of viral dsRNA in the infected fungal strains revealed that viral dsRNA in strain EP721 was at a significantly lower level than were CHV1-EP713 and CHV1-Euro7 in the same host fungal strain (Fig. 1A). The viral dsRNA accumulation for CHV1-EP713 was about twice that for CHV1-Euro7 and 10 times higher than that for CHV1-EP721. Quantification of viral plus-strand ssRNA [(+)ssRNA] showed that the ratio of CHV1-EP713, CHV1-Euro7, and CHV1-EP721 was 100:57:13, a trend very similar to that shown by viral dsRNAs (Fig. 1B). An investigation into minus-strand ssRNA [(−)ssRNA] revealed a ratio of 100:35:5 for CHV1-EP713, CHV1-Euro7, and CHV1-EP721, a general trend similar to that of viral (+)ssRNA, but the ratio for CHV1-Euro7 and CHV1-EP721 appeared to drop by half (Fig. 1C). It was apparent from Fig. 1 that viral dsRNA levels can serve as an indicator of the relative levels of the viral (+)ssRNA level in the cell.
FIG. 1.
Viral dsRNA accumulation level and ssRNA semiquantification of hypovirus strains in the fungal host EP721(-v). (A) Agarose gel electrophoretic analysis of viral dsRNA accumulation. Equal amounts (10 μg) of total RNA from uninfected strain EP721(-v) (lane1), native CHV1-EP721-infected strain EP721 (lane 2), and transfectants EP721(-v)/CHV1-Euro7 (lane 3) and EP721(-v)/CHV1-EP713 (lane 4) were electrophoresed on a 0.8% agarose gel in 1× TAE buffer. Lanes marked with M contained 100 ng of 1-kb DNA ladder (Fermentas) as a size marker. (B) Real-time RT-PCR quantification of (+)ssRNA of hypovirus strains. The viral ssRNA level of CHV1-EP713 was set at 100%, and the levels of CHV1-EP721 and CHV1-Euro7 were expressed as percentages of that of CHV1-EP713. Values were calculated from three independent experiments. Bars indicate mean deviations. (C) Real-time RT-PCR quantification of (−)ssRNA of hypovirus strains. The viral ssRNA level of CHV1-EP713 was set at 100%, and the levels of CHV1-EP721 and CHV1-Euro7 were expressed as percentages of that of CHV1-EP713. Values were calculated from three independent experiments. Bars indicate mean deviations.
Genome organization of CHV1-EP721.
Since CHV1-EP721 and CHV1-Euro7 were both isolated in Italy, we considered the possibility that they might share certain similarities in their genome organizations and sequences. The CHV1-EP721 genome was determined to be 12,724 bp in length, with a typical CHV1-type organization. A comparison with the genome sequences of CHV1-Euro7 and CHV1-EP713 showed that CHV1-EP721 shared an average of 99% and 90% identities at the nucleotide level. The surprisingly high level of similarity between CHV1-EP721 and CHV1-Euro7 was observed throughout the whole genome: at the 5′ noncoding region, there was only one base difference, a guanine at position 329 in CHV1-Euro7 was substituted with an adenine in CHV1-EP721, and only three base differences were found at the entire 3′ noncoding region.
At the amino acid level, CHV1-EP721 shares 99% and 92% identities with CHV1-Euro7 and CHV1-EP713, respectively. All the predominant features identified in CHV1-Euro7 and CHV1-EP713 were found in CHV1-EP721. These include amino acid residues Gly248 and Gly249, the cleavage site between p29 and p40 in ORF A (12); Cys162 and His215, essential for p29 cleavage (11); Cys70 and Cys72, important for p29 enhancement of viral dsRNA accumulation and vertical transmission in trans (33); and the portion from Thr288 to Arg312, responsible for p40-mediated RNA accumulation (34). Strikingly, only 46 amino acid residue differences between CHV1-EP721 and CHV1-Euro7 were found throughout the genome, suggesting a very recent divergence for these two virus strains. The two virus strains differ at 3 residues in p29, 8 in p40, 3 in p48, 4 in the polymerase domain, 7 in the helicase domain, 19 in the region between p48 and the polymerase (from ORF B Gly419 to Ser1791), and 2 in the region between the polymerase and the helicase domains (ORF B positions 2208 to 2656).
Infectious cDNA clone of CHV1-EP721.
CHV1-EP721 RNA accumulates in strain EP721 to a significantly lower level than CHV1-Euro7 and CHV1-EP713 do in the same host, suggesting that the low level of CHV1-EP721 RNA accumulation is a property of the virus and not due to preference of the host fungus. The availability of a full-length CHV1-EP721 infectious cDNA clone would provide the means to conclusively test this possibility and to further probe the mechanism underlying the regulation of hypoviral RNA replication and accumulation.
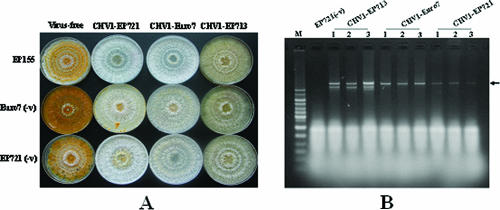
When RNA transcripts synthesized from the full-length CHV1-EP721 cDNA template were used to transfect the virus-free fungal strain EP721(-v), colonies with traits indistinguishable from those of CHV1-EP721-containing strain EP721, including a white phenotype, suppressed sporulation, and the presence of cytoplasmically replicative viral dsRNA, were obtained, demonstrating a successful infection. To further examine whether different fungal hosts would have an impact on the accumulation of viral dsRNA, hypoviruses CHV1-EP713, CHV1-Euro7, and CHV1-EP721 were introduced into virus-free and genetically different fungal strains EP155, Euro7(-v), and EP721(-v), respectively, via transfection. A change of colony pigmentation from orange to white indicated that a productive infection had been established for all these viruses. All three fungal hosts exhibited similar symptoms (white pigmentation and suppression of conidial spore production) upon infection by the same hypovirus, and different virus strains incited subtle yet distinguishable symptoms in these hosts (Fig. 2A). These results demonstrate that the phenotype of a transfectant is determined predominantly by the virus and not by the host. It is evident from Fig. 2A that morphologies of transfectants infected by CHV1-EP721 are more similar to those of transfectants infected by CHV1-Euro7 than by CHV1-EP713. It was also observed that at a later stage of infection (7 to 10 days), the colony morphology and pigmentation exhibited some differences for the three fungal hosts infected with CHV1-EP721, with hosts of Euro7(-v) and EP721(-v) being whiter and making some stroma-like structures, two characteristics found in native EP721. This could be regarded as the contribution of the host genome to symptom expression (8). An examination of viral dsRNA in the infected cells revealed that the lowest dsRNA accumulation was found in the CHV1-EP721-infected strains, and the highest level was found in the CHV1-EP713-infected strains, with CHV1-Euro7 RNA accumulating to a level about half of that of CHV1-EP713 (Fig. 2B). These results clearly demonstrated that the viral dsRNA accumulation level was determined solely by the input virus.
FIG. 2.
Colony morphology and viral dsRNA accumulation in hypovirus-infected fungal hosts. Hypovirus strains CHV1-EP721, CHV1-Euro7, and CHV1-EP713 were introduced into virus-free C. parasitica host strains EP155, Euro7(-v), and EP721(-v) by transfection. Viral dsRNA was extracted from a 5-day-old culture in liquid EP complete medium. (A) Colony morphology of virus-free and virus-infected host fungal strains on PDA. The photograph was taken on day 7. (B) Agarose gel electrophoretic analysis of viral dsRNA accumulation in different host-virus combinations. Ten micrograms of total RNA from virus-infected hosts was loaded into each lane in a 0.8% agarose gel in 1× TAE buffer. The numbers 1, 2, and 3 represent hosts C. parasitica strain EP155, Euro7(-v), and EP721(-v), respectively. Viruses that infected the hosts are shown above the numbers. Lanes marked with M contain 100 ng of 1-kb DNA ladder. The arrow indicates the position of viral dsRNA.
ORF B of CHV1-EP721 is responsible for the low level of viral RNA accumulation.
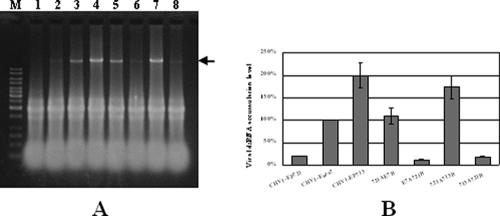
The high level of sequence identity shared by CHV1-EP713, CHV1-Euro7, and CHV1-EP721 suggested the possibility that viral coding domains responsible for differences in viral dsRNA accumulation in a given host fungus could be mapped through the construction of chimeric viruses by genome domain swapping. Four chimeric viruses, 721A713B, 713A721B, 721AE7B, and E7A721B, with ORF A and ORF B from different hypovirus strains were obtained through swapping of the polypeptide coding domains between CHV1-EP721 and CHV1-EP713 and between CHV1-EP721 and CHV1-Euro7. All these chimeric viruses were able to establish a productive infection in the host fungus EP721(-v) by transfection. An examination of the viral dsRNA by agarose gel electrophoresis revealed that the accumulation of viral dsRNA varied considerably among the transfectants: 721A713B dsRNA accumulated to the highest level, which was comparable to that of CHV1-EP713; 721AE7B RNA accumulated to a moderate level, which was comparable to that of CHV1-Euro7; and both 713A721B and E7A721B dsRNA accumulated to the lowest level, which was comparable to that of CHV1-EP721 (Fig. 3A). A semiquantification of the viral dsRNAs was performed by scanning the gel with a Typhoon 9410 (Amersham) scanner and analyzing the gel with ImageQuant TL-1D gel analysis software. With viral dsRNA from the transfectant infected with CHV1-Euro7 set at 100%, the highest amount of viral dsRNA accumulation was found in the transfectant infected with CHV1-EP713 (200%), followed by the transfectants infected with 721A713B (175%), 721AE7B (109%), CHV1-EP721 (20%), 713A721B (18%), and E7A721B (12%) (Fig. 3B). These data clearly showed a correlation between the origin of ORF B and the viral dsRNA accumulation level; i.e., a chimeric virus with ORF B derived from a donor virus with high dsRNA accumulation has a high dsRNA level, while a chimeric virus with ORF B derived from a donor virus with low dsRNA accumulation has a low dsRNA level.
FIG. 3.
Viral dsRNA quantification of transfectants containing wild-type or chimeric hypoviruses. All transfectants were in the EP721(-v) genetic background. (A) Agarose gel electrophoretic analysis of viral dsRNA. Ten micrograms of total RNA from virus-infected hosts was loaded into each lane in a 0.8% agarose gel. Lane 1, virus-free strain EP721(-v); lane 2 to lane 8, transfectants with wild-type CHV1-EP721, CHV1-Euro7, and CHV1-EP713 and chimeric viruses 721AE7B, E7A721B, 721A713B, and 713A721B. The lane marked with an M contains 100 ng of a 1-kb DNA ladder. The arrow indicates the position of viral dsRNA. (B) Semiquantification of viral dsRNA from different transfectants. The viral dsRNA level of CHV1-Euro7 was set at 100%, and levels of CHV1-EP721, CHV1-EP713, and chimeric viruses were expressed as percentages of that of CHV1-Euro7. Values were calculated from three independent experiments, and bars indicate mean deviations.
Virus accumulation level influences virus transmission efficiency.
It has been observed that the CHV1-EP721-carrying native fungal strain EP721 has a much lower virus transmission rate through conidia than the CHV1-EP713-carrying native strain EP713 (data not shown). To investigate whether this is due to the function of a specific viral protein or is related to the general load of virus in the cell, we compared fungal strains infected with wild-type virus strains and chimeric viruses for their efficiencies of transmission into the asexual conidial spores. Under high-light conditions, the sporulation levels for transfectants containing CHV1-EP721 or CHV1-Euro7 were of the same magnitude (109 conidia per plate), while the transfectant containing CHV1-EP713 consistently produced a much lower number of conidia (106 conidia per plate). The same pattern of sporulation was observed for transfectants containing chimeric viruses, with the origin of ORF B determining the sporulation level. Transfectants containing CHV1-EP713 and 721A713B had the highest virus transmission efficiency (100%), CHV1-Euro7 and 721AE7B were transmitted at rates of 85% and 86.4%, and CHV1-EP721, E7A721B, and 713A721B had the lowest efficiency (∼75%) (Table 1). These results suggest that virus transmission efficiency correlates with the virus accumulation level in the host.
TABLE 1.
Efficiency of hypovirus transmission through conidia in the host strain EP721(-v)
| Virus strain | No. of infected spore germinates/No. of tested spore germinates
|
Efficiency of transmission (%) | |||
|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Total | ||
| CHV1-EP713 | 100/100 | 65/65 | 100/100 | 265/265 | 100 |
| CHV1-EP721 | 71/85 | 44/64 | 46/60 | 161/209 | 77.0 |
| CHV1-Euro7 | 79/100 | 89/95 | 82/99 | 250/294 | 85.0 |
| 721A713B | 100/100 | 100/100 | 95/95 | 295/295 | 100 |
| 713A721B | 54/90 | 76/100 | 52/60 | 182/250 | 72.8 |
| 721AE7B | 92/100 | 79/90 | 71/90 | 242/280 | 86.4 |
| E7A721B | 34/50 | 69/85 | 65/100 | 168/235 | 71.5 |
Fine mapping of the determinant for low accumulation of viral dsRNA in CHV1-EP721.
In order to map the determinant for low viral dsRNA accumulation in CHV1-EP721 to a more defined domain within ORF B, we took advantage of the high level of sequence identity (99%) shared by the CHV1-EP721 and CHV1-Euro7 infectious clones to construct a series of additional chimeric viruses (Fig. 4A). All resulting chimeras were infectious upon transfection of the fungal host EP721(-v), and all transfectants showed a white phenotype. An examination of the dsRNA from the transfectants revealed a significant difference in viral dsRNA accumulation among these chimeric viruses (Fig. 4B and C). The viral dsRNA of 721/E7NarI, a chimera with a portion of ORF B (from the NarI site at position 5311 through the 3′ end) from CHV1-Euro7 in a CHV1-EP721 background, accumulated to a level similar to that observed for CHV1-Euro7, while the reciprocal chimera E7/721NarI with the NarI fragment from CHV1-EP721 in the CHV1-Euro7 background showed dsRNA accumulation similar to that observed for CHV1-EP721. However, when the MluI fragment (from position 7843 through the 3′ end) of CHV1-EP721 was replaced with the counterpart from CHV1-Euro7, the dsRNA of chimera 721/E7PH accumulated to a level comparable to that observed for CHV1-EP721 (22%). When the MluI fragment of CHV1-Euro7 was exchanged with the fragment from CHV1-EP721, the dsRNA of E7/721PH accumulated to a level about 74% of the wild-type CHV1-Euro7 dsRNA level. Chimera 721/E7 (5.3-7.8K) was constructed by replacing the NarI/MluI fragment (positions 5311 to 7843) of CHV1-EP721 with the fragment of the same position from CHV1-Euro7. This chimera resulted in a dsRNA level of 70% of that of wild-type CHV1-Euro7. Its reciprocal counterpart, E7/721(5.3-7.8K), accumulated dsRNA at a level of 14%, close to that for CHV1-EP721. Combined, these results indicate that the determinant for low viral dsRNA accumulation in CHV1-EP721 is located within the NarI/MluI domain of ORF B.
DISCUSSION
Comparative studies with infectious cDNA clones of hypoviruses CHV1-EP713 and CHV1-Euro7 have provided insight into the relative contribution of hypovirus and fungal host genomes to symptom expression (8), identified differences in the effects of hypoviruses on host signaling pathways (26), identified hypovirus-specific and common alterations of host transcript accumulation (2), and mapped viral determinants responsible for these differences (7, 8, 26). Whole-genome sequencing and the development of an infectious cDNA clone for a closely related, but low-accumulating hypovirus, CHV1-EP721, reported here, have allowed similar comparative approaches to evaluate virus-host interactions that contribute to viral RNA accumulation in the infected fungal host.
The observation that CHV1-EP713, CHV1-Euro7, and CHV1-EP721 viral dsRNAs accumulated to the same relative levels in several different C. parasitica strains (Fig. 2) clearly indicates that viral dsRNA accumulation in the infected fungal hosts is determined by intrinsic properties of the virus. The correlation observed between relative hypovirus dsRNA accumulation and efficiency of transmission through asexual spores for these three CHV1 isolates (Table 1) is consistent with similar correlations between viral dsRNA accumulation and vertical transmission with laboratory-generated mutant strains of CHV1-EP713 (33). The extensive sequence homology between CHV1-EP721, CHV1-Euro7, and CHV1-EP713 also allowed the construction of chimeric viruses and mapping of the determinant responsible for low viral dsRNA accumulation and rates of transmission to the NarI/MluI fragment located within ORF B of CHV1-EP721 (extending from nucleotide residues 5311 to 7843 and corresponding to ORF B amino acid residue positions 983 to 1791) (Fig. 4).
ORF B of hypovirus CHV1 encodes a large polypeptide of 3,164 amino acid residues. Except for the N terminus of p48, a protease that autocatalytically cleaves itself from the rest of ORF B-encoded peptide (30), the nature of the mature protein products derived from the ORF B-encoded polyprotein has not been determined. While RNA polymerase and helicase motifs have been found within the C-terminal region (21), functions associated with the large domain residing between p48 and the RNA polymerase remain ill defined. There is a difference of 19 amino acid residues between CHV1-EP721 and CHV1-Euro7 in this region, and 7 of these amino acid residues reside within the NarI/MluI fragment. Of these seven amino acid variations, three do not change the general property of the amino acid, and two (ORF B amino acid residue positions 1495 and 1502) are a change from glycine in CHV1-Euro7 to glutamine in CHV1-EP721 and from glutamate in CHV1-Euro7 to glycine in CHV1-EP721. Changes from glutamate to lysine at position 1439 and from asparagine to aspartate at position 1492 are also found in CHV1-EP721. Overall, these changes appear to be subtle, and a search of the current databases failed to find any high-scored match to this putative protein. Replacement of the NarI/MluI fragment of CHV1-Euro7 with that of the CHV1-EP721 effectively reduced the dsRNA accumulation of chimera E7/721(5.3-7.8K) to the level of CHV1-EP721, demonstrating that this NarI/MluI fragment is necessary for the low dsRNA accumulation. However, replacement of the NarI/MluI fragment of CHV1-EP721 with that of CHV1-Euro7 could restore the dsRNA accumulation of chimera 721/E7(5.3-7.8K) to only 70% of the level reached by CHV1-Euro7, indicating that the NarI/MluI fragment from CHV1-Euro7 alone is not sufficient for the accumulation of viral dsRNA to the CHV1-Euro7 level. This suggests that there may be a second domain in the 3′ portion of ORF B that interacts or coordinates with the protein encoded within the NarI/MluI fragment. This hypothesis is further supported by the observation that E7/721PH, a chimera in the CHV1-Euro7 background with the polymerase and helicase domains replaced by those of CHV1-EP721, also accumulated at 74% of the CHV1-Euro7 level. Since polymerase and helicase are the major domains that follow the NarI/MluI domain at the 3′ portion of ORF B and since these two proteins are implicated in virus replication, it is hypothesized that the product of the NarI/MluI domain may interact with the viral polymerase or helicase to exert its function on viral dsRNA replication. It is difficult to correlate the point mutation(s) of any of the seven amino acids in the NarI/MluI region of CHV1-EP721 to the low level of viral replication at this stage, partially because of the lack of knowledge of the mature form of this protein. The NarI/MluI domain also overlaps with the portion of ORF B extending from positions 5310 to 9897, which Parsley et al. (26) previously reported as being responsible for the modulation of cyclic AMP-dependent signaling of the host fungus. Whether these two functions are determined by the same domain also remains to be clarified.
Previous studies have implicated both p29 and p40 as contributing to viral dsRNA accumulation (33, 34). The contribution of p29 to viral dsRNA accumulation is likely related to the function of this protein as a suppressor of RNA silencing (28), while p40 appears to provide an in cis accessory role in viral RNA amplification (33). Suzuki et al. (33) previously reported that the 70% to 80% reduction in viral dsRNA accumulation observed for the CHV1-EP713 p40 deletion mutant correlated with a significant relief in virus-mediated suppression of pigmentation, conidiation, and mycelial growth while having no effect on virus-mediated hypovirulence. These observations led to the hypothesis that the magnitude and range of hypovirus-mediated symptom expression can be influenced by the level of virus dsRNA accumulation (33). The observations reported here, that CHV1-EP721 and chimeric viruses harboring the low-viral RNA determinant from CHV1-EP721 caused a phenotype very similar to that of the closely related CHV1-Euro7, even though levels of accumulated RNA for the former viruses were as much as fivefold lower than those of the latter virus, are not consistent with this hypothesis. However, interpretations of the combined results are complicated by the comparison of a mutant and wild-type virus in one case (CHV1-EP713 and the p40 deletion mutant) and two different hypovirus isolates (CHV1-EP721 and CHV1-Euro7) in the second. Additional systematic experimentation will be required to determine the influence of virus RNA accumulation on symptom expression.
The hypovirus replication cycle has not been studied extensively. The fact that CHV1 hypovirus coding-strand RNA is infectious when electroporated into C. parasitica spheroplasts (6) is consistent with a plus-strand RNA virus replication strategy and the close phylogenetic relationship to the ssRNA of plant potyviruses (21). Membrane vesicles isolated from the infected fungus contain hypovirus dsRNA and support the synthesis of viral RNAs of both polarities (17). Since hypoviruses do not encode a coat protein, viral ssRNA is not sequestered in a virus particle. The absence of a capsid protein has been postulated to be a contributing factor to explain why hypovirus positive-strand RNA is not found in large excess over viral minus-strand RNA as reported for capsid-encoding positive-strand RNA viruses such as poliovirus (4) and why hypovirus dsRNA is found to be present in relatively high abundances in infected mycelia (17, 34). A clearer understanding of the mechanisms that regulate hypovirus dsRNA accumulation and the corresponding consequences for hypovirus transmission and the spectrum of virus-mediated symptom expression will benefit ongoing efforts to engineer hypoviruses for enhanced biological control (15, 25). The identification of a determinant for low viral dsRNA accumulation within the CHV1-EP721 ORF B NarI/MluI domain should significantly facilitate future studies on hypovirus RNA replication.
Acknowledgments
This work was supported in part by grants from the National Key Basic Research Project of China (2006CB101906), National Natural Science Foundation of China (30130020 and 39925003), National Hightech Program of China (2001AA223111 and 2004AA223100), and International Collaboration Key Project (2001CB711104) to B.C. and by Public Health Service grant GM55981 to D.L.N. H.L. is a recipient of Guangxi Graduate Education Creativity Program grant 2006105930710D01, and T.B.P. was a postdoctoral trainee on Public Health Service grant AI07510-3.
Footnotes
Published ahead of print on 29 November 2006.
REFERENCES
- 1.Allen, T. D., A. L. Dawe, and D. L. Nuss. 2003. Use of cDNA microarrays to monitor transcriptional responses of the chestnut blight fungus Cryphonectria parasitica to infection by virulence-attenuating hypoviruses. Eukaryot. Cell 2:1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. D., and D. L. Nuss. 2004. Specific and common alterations in host gene transcript accumulation following infection of the chestnut blight fungus by mild and severe hypoviruses. J. Virol. 78:4145-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anagnostakis, S. L. 1982. Biological control of chestnut blight. Science 215:466-471. [DOI] [PubMed] [Google Scholar]
- 4.Baltimore, D. 1969. The replication of picornaviruses, p. 101-176. In H. B. Levy (ed.), The biochemistry of viruses. Marcel Dekker, New York, NY.
- 5.Chen, B., C. Chen, B. H. Bowman, and D. L. Nuss. 1996. Phenotypic changes associated with wild-type and mutant hypovirus RNA transfection of plant pathogenic fungi phylogenetically related to Cryphonectria parasitica. Phytopathology 86:301-310. [Google Scholar]
- 6.Chen, B., G. H. Choi, and D. L. Nuss. 1994. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264:1762-1764. [DOI] [PubMed] [Google Scholar]
- 7.Chen, B., L. M. Geletka, and D. L. Nuss. 2000. Using chimeric hypoviruses to fine-tune the interaction between a pathogenic fungus and its plant host. J. Virol. 74:7562-7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, B., and D. L. Nuss. 1999. Infectious cDNA clone of hypovirus CHV1-Euro7: a comparative virology approach to investigate virus-mediated hypovirulence of the chestnut blight fungus Cryphonectria parasitica. J. Virol. 73:985-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, G. H., and D. L. Nuss. 1992. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257:800-803. [DOI] [PubMed] [Google Scholar]
- 10.Choi, G. H., and D. L. Nuss. 1992. A viral gene confers hypovirulence-associated traits to the chestnut blight fungus. EMBO J. 11:473-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, G. H., D. M. Pawlyk, and D. L. Nuss. 1991. The autocatalytic protease p29 encoded by a hypovirulence-associated virus of the chestnut blight fungus resembles the potyvirus-encoded protease HC-Pro. Virology 183:747-752. [DOI] [PubMed] [Google Scholar]
- 12.Choi, G. H., R. Shapira, and D. L. Nuss. 1991. Cotranslational autoproteolysis involved in gene expression from a double-stranded RNA genetic element associated with hypovirulence of the chestnut blight fungus. Proc. Natl. Acad. Sci. USA 88:1167-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Churchill, A. C. L., L. M. Ciuffetti, D. R. Hansen, H. D. van Etten, and N. K. van Alfen. 1990. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17:25-31. [Google Scholar]
- 14.Craven, M. G., D. M. Pawlyk, G. H. Choi, and D. L. Nuss. 1993. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67:6513-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawe, A. L., and D. L. Nuss. 2001. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annu. Rev. Genet. 35:1-29. [DOI] [PubMed] [Google Scholar]
- 16.Enebak, S. A., W. L. MacDonald, and B. I. Hillman. 1994. Effect of dsRNA associated with isolates of Cryphonectria parasitica from the Central Appalachians and their relatedness to other dsRNAs from North America and Europe. Phytopathology 84:528-534. [Google Scholar]
- 17.Fahima, T., P. Kazmierczak, D. R. Hansen, P. Pfeiffer, and N. K. Van Alfen. 1993. Membrane-associated replication of an unencapsidated double-strand RNA of the fungus, Cryphonectria parasitica. Virology 195:81-89. [DOI] [PubMed] [Google Scholar]
- 18.Hillman, B. I., B. T. Halpern, and M. P. Brown. 1994. A viral dsRNA element of the chestnut blight fungus with a distinct genetic organization. Virology 201:241-250. [DOI] [PubMed] [Google Scholar]
- 19.Hillman, B. I., R. Shapira, and D. L. Nuss. 1990. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80:950-956. [Google Scholar]
- 20.Hillman, B. I., Y. Tian, P. J. Bedker, and M. P. Brown. 1992. A North American hypovirulent isolate of the chestnut blight fungus with European isolate-related dsRNA. J. Gen. Virol. 73:681-686. [DOI] [PubMed] [Google Scholar]
- 21.Koonin, E. V., G. H. Choi, D. L. Nuss, R. Shapira, and J. C. Carrington. 1991. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. USA 88:10647-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linder-Basso, D., J. N. Dynek, and B. I. Hillman. 2005. Genome analysis of Cryphonectria hypovirus 4, the most common hypovirus species in North America. Virology 337:192-203. [DOI] [PubMed] [Google Scholar]
- 23.Melzer, M. S., and G. J. Boland. 1999. CHV3-type dsRNAs and the GH2 genotype in a population of Cryphonectria parasitica in Ontario. Can. J. Plant Pathol. 21:248-255. [Google Scholar]
- 24.Nuss, D. L. 1992. Biological control of chestnut blight: an example of virus-mediated attenuation of fungal pathogenesis. Microbiol. Rev. 56:561-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuss, D. L. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3:632-642. [DOI] [PubMed] [Google Scholar]
- 26.Parsley, T. B., B. Chen, L. M. Geletka, and D. L. Nuss. 2002. Differential modulation of cellular signaling pathways by mild and severe hypovirus strains. Eukaryot. Cell 1:401-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puhalla, J. E., and S. L. Anagnostakis. 1971. Genetics and nutritional requirements of Endothia parasitica. Phytopathology 61:169-173. [Google Scholar]
- 28.Segers, G. C., R. van Wezel, X. Zhang, Y. Hong, and D. L. Nuss. 2006. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell 5:896-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapira, R., G. H. Choi, and D. L. Nuss. 1991. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 10:731-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapira, R., and D. L. Nuss. 1991. Gene expression by a hypovirulence-associated virus of the chestnut blight fungus involves two papain-like protease activities. J. Biol. Chem. 266:19419-19425. [PubMed] [Google Scholar]
- 31.Smart, C. D., W. Yuan, R. Foglia, D. L. Nuss, D. W. Fulbright, and B. I. Hillman. 1999. Cryphonectria hypovirus 3, a virus species in the family Hypoviridae with a single open reading frame. Virology 265:66-73. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, N., L. M. Geletka, and D. L. Nuss. 2000. Essential and dispensable virus-encoded replication elements revealed by efforts to develop hypoviruses as gene expression vectors. J. Virol. 74:7568-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki, N., K. Maruyama, M. Moriyama, and D. L. Nuss. 2003. Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J. Virol. 77:11697-11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, N., and D. L. Nuss. 2002. Contribution of protein p40 to hypovirus-mediated modulation of fungal host phenotype and viral RNA accumulation. J. Virol. 76:7747-7759. [DOI] [PMC free article] [PubMed] [Google Scholar]