Abstract
MKlp2 is a kinesin-like motor protein of the central mitotic spindle required for completion of cytokinesis. Papillomavirus E2 is a sequence specific DNA binding protein that regulates viral transcription and replication and is responsible for partitioning viral episomes into daughter cells during cell division. We demonstrate that MKlp2 specifically associates with the E2 protein during mitosis. Using chromatin immunoprecipitation, we show viral genomes are in complex with MKlp2 only within this stage of cell cycle. By immunofluorescence, a subpopulation of papillomavirus E2 colocalizes with MKlp2 in the midbody/midplate during late mitosis. We conclude that during specific stages of mitosis, the papillomavirus E2 protein binds to MKlp2, and infer that association with this motor protein ensures viral genome partitioning during cytokinesis.
Rather than causing lytic infection of host epithelial cells, papillomaviruses (PVs) induce slowly growing lesions. For viral genome persistence, the virus must access the basal epithelial cells and be maintained in these cells through cytokinesis, as the daughter cells migrate outward and undergo terminal differentiation. The approximately 8,000-bp PV genome replicates as autonomous episomes at a relatively stable low copy number in the nuclei of these infected basal epithelial cells. To establish persistent infection, the replicated viral episomes must be retained within the nucleus as it reforms during late mitosis and partition into the two cells. This mode of replication is reproduced in monolayer cultures of human and murine cell lines that stably harbor viral episomes. Bovine papillomavirus type 1 (BPV-1) genomes replicate in synchrony with cellular DNA as multicopy nuclear plasmids that do not integrate into the host genome (19). The ID13 cell line originated from a clone of transformed mouse C127 cells and maintains on average about 25 copies of the viral episome.
The virus-encoded E2 protein is necessary for long-term episomal maintenance of viral genomes within replicating cells. The C-terminal domain of E2 binds with high affinity and specificity to sites within the viral DNA, including the viral origin of replication. Its N-terminal domain binds to the viral DNA helicase E1. Together, E1 and E2 are sufficient to trigger initiation of transient viral DNA replication from this origin sequence, but for long-term viral persistence, a cis-acting region with multiple E2 binding sites is necessary (34). The observation that E2 colocalizes with cellular DNA through mitosis implies its role in tethering the viral genome to the cellular proteins, organelles, or chromosomes, directly or indirectly driven by the cellular motor proteins to move along either dynein or microtubule networks into the dividing cells (1, 20, 31). Several groups using immunofluorescence have observed dots of E2 protein randomly associated with chromosomes during mitosis (14, 15, 20). It was recently reported that human papillomavirus type 11 (HPV-11) E2 colocalized to mitotic spindles, especially the middle of spindle and midbody/midplate (5, 35). It has also been proposed that a subpopulation of E2 proteins are in close proximity to the kinetochore (5, 35), the macromolecular complex that coordinates attachment of mitotic spindles to the centromeric region on replicated chromatids.
In mammalian cells, alignment and accurate segregation of chromosomes during mitosis occur on a highly ordered apparatus called the mitotic spindle, which is composed of a bipolar array of microtubules. A constant dissipation of energy is required for assembly and maintenance of the spindle as well the cargo proteins moving along the microtubules. The kinesin and kinesin-like proteins (Klps) superfamily are the motor proteins that actuate movement along the microtubules (24). A subset of motor proteins from this family are known as mitotic or chromosomal kinesins, transporting different cellular components, including chromosomes, transcription complexes, the Golgi apparatus and other cellular organelles, and even viral particles into daughter cells or positioning them to specific sites during mitosis (9, 36).
How the E2 protein and the associated viral genome are maintained, tethered, and transported during all of the stages of mitosis is unresolved. Brd4 has been reported as the tethering bridge between PV E2 and mitotic chromosomes (38). Brd4 interaction has also been shown to be necessary for E2-mediated transcriptional activation (15, 23, 30). Therefore, the cellular protein that attaches E2 and the viral genomes to mitotic chromosomes and the mechanism by which these are segregated remain unclear. Utilizing yeast two-hybrid screens to identify cellular proteins that bind to bovine papillomavirus E2, we identified human kinesin-like protein MKlp2 (mitotic kinesin-like protein 2; Rab6KIFL/KIF20) as an E2-interacting protein. We show MKlp2 binds BPV and HPV E2 proteins and suggest its involvement in transport of E2-associated viral genomes to the mitotic apparatus.
MATERIALS AND METHODS
Yeast two-hybrid assays.
All two-hybrid experiments were performed with Saccharomyces cerevisiae AH109 using the Clontech Matchmaker 3 system (Clontech, Mountain View, CA). A human brain cDNA library fused with the GAL4 activation domain (GAL4-AD) was screened with a hybrid protein consisting of the DNA binding domain (DBD) of the GAL4 transcription factor fused to the “bait” E2 E39G (3, 4).
In vivo protein association assay.
The 3xFlag epitope-MKlp2 fusion was cloned onto the full-length MKlp2 cDNA sequence and transferred into pCMV 3xFlag 7.1 (Sigma, Saint Louis, MO). For coimmunoprecipitation experiments, pCMV 3xFlag-MKlp2 and pCG E2, pCG E2R, or pCG 1-215 plasmids (3, 4) were transfected into C33A cells using calcium phosphate. Thirty-six hours posttransfection, cells were harvested and pelleted in ice-cold phosphate-buffered saline (PBS). Cell lysis and immunoprecipitation were performed as described previously (4). Proteins in the immunoprecipitate were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted to BioTrace polyvinylidene difluoride (Pall Life Science, Pensacola, FL). The blot was probed sequentially by anti-Flag M2 monoclonal antibody (Sigma, Saint Louis, MO) and goat anti-mouse immunoglobulin G (IgG) conjugated with horseradish peroxidase (Jackson Immuno Research, West Grove, PA), then developed with a SuperSignal West PICO chemiluminescent kit (Pierce, Rockford, IL). In studies involving endogenous MKlp2 and Flag-11E2, affinity-purified anti-MKlp2 antibody was used for immunoprecipitation. Antiserum against MKlp2 was raised in rabbit using the C-terminal amino acids (aa) 496 to 886 of hMKlp2. In studies involving HPV-16 E2, anti-HPV-16 E2 antibody TVG261 was used for immunoblots and was developed with a WestDura chemiluminescent kit (Pierce, Rockford, IL).
GST fusion protein expression.
Escherichia coli BL21 pLysS cells transformed by glutathione S-transferase (GST)-MKlp2 (aa 496 to 886) were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2.5 h. Cell pellets were solubilized in 8 M urea and disrupted by sonication on ice. Triton X-100 was added to 1%, and the lysate was further diluted in NETN (100 mM NaCl, 0.1 mM EDTA, 20 mM Tris-HCl, pH 8.0, 0.1% Nonidet P-40) with 1 mM phenylmethylsulfonyl fluoride and centrifuged at 15,000 × g to remove the debris. Soluble GST fusion proteins were collected on glutathione-Sepharose beads (Amersham/GE Healthcare, Piscataway, NJ) and resuspended with NETN.
Cell synchronization.
HeLa, U2OS, and hTERT-RPE1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) or DMEM-F-12 with 10% fetal bovine serum to 40% confluence and synchronized by double-thymidine blockade (33). Half of the cells were harvested, and the other half were released for 8 to 9 h until at least 40% of the cell population appeared mitotic by microscopy. HeLa cells were arrested in complete media with nocodazole at 100 ng/ml for 5 h and harvested at 9 h after release from thymidine.
ChIP assay.
We used the chromatin immunoprecipitation (ChIP) assay kit from Upstate Biotechnology (Lake Placid, NY; catalog no. 17-295). ID13 cells were transfected with Flag human MKlp2 expression vectors using FuGene 6 (Roche, Germany) and exposed to formaldehyde added directly into the medium at final concentration of 1% for 10 min at room temperature. Cells were rinsed and lysed in 200 μl SDS cell lysis buffer per 1 × 106 cells. Sonicated lysates were cleared by centrifugation and diluted 10-fold into ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8, 167 mM NaCl). The supernatant fraction was incubated overnight at 4°C with anti-Flag (M2) or appropriate antibody, salmon sperm DNA, and protein G/A-agarose. Antibody-bound complexes were pelleted by centrifugation, and the supernatant containing unbound, nonspecific DNA was removed. The protein G/A-agarose-antibody complexes were washed once in low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl), once with high-salt wash buffer (same as low-salt buffer with 500 mM NaCl), once with LiCl wash (0.25 M LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0) and twice with Tris-EDTA (pH 8.0). To elute the DNA-protein complex from the antibody, 250 μl of 1% SDS-0.1 M NaHCO3 solution was added to the agarose pellet at room temperature for 15 min. Supernatants were collected, and elutions were repeated. Twenty microliters of 5 M NaCl was added to the eluate and heated to 65°C for 4 h to reverse the cross-link. Proteinase K was added to the eluate for 1 h at 45°C. DNA was recovered by phenol-chloroform extraction and ethanol precipitation. PCR was performed by using Ex-Taq (Takara Bio, Inc., Japan). In studies conducted with ID13 cells, the sense strand primer sequence was 5′-AAAGTTTCCATTGCGTCTGG-3′. The antisense strand primer sequence was 5′-GCTTTTTATAGTTAGCTGGCTATTTT-3′. In experiments involving the HPV-16 genome, the respective sense and antisense sequences are as follows: 5′-GTGTAACTATTGTGTCATGCAACAT-3′ and 5′-CCTTAGAAGTTTAAACCTTATGCCA-3′.
Indirect immunofluorescence.
Telomerase-immortalized human retinal epithelial RPE1cells (Clontech Laboratories, Mountain View, CA) were cultured on glass coverslips in 1:1 DMEM-Ham's F-12 medium (Invitrogen/GIBCO). After synchronization, cells were rinsed in PBS and sequentially incubated in cytoskeletal buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES, pH 6.8) for 30 s on ice, cytoskeletal buffer with 0.5% Triton X-100 for 30 s, and again in cytoskeletal buffer for another 30 s. Cells were then fixed in 3.7% paraformaldehyde in PBS for 10 min at room temperature and permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature. To visualize tubulin, methanol at −20°C was added on the coverslips and incubated for 15 min. Specimens were blocked in PBS buffer containing 0.1% bovine gelatin, 1% bovine serum albumin, and 0.1% Tween 20 before incubation with primary antibodies for 3 to 4 h at room temperature. Mouse antibodies against BPV-1 E2 (B201, IgG2b) and α-tubulin (DM1α, IgG1) were preincubated with Zenon labeling fluorophores Alexa 488 and Alexa 547, respectively, prior to use. After 2 h of incubation with affinity-purified rabbit anti-MKlp2 serum at room temperature, slides were gently rinsed in PBS and Alexa anti-rabbit 633 was added for another 2 h. Slides were rinsed again in PBS-Tween (PBST). Hoechst 34580 in PBS at a final concentration at 5 μg/ml was used to visualize genomic DNA. Slides were mounted in Fluoromount-G (Southern Biotech, Birmingham, AL), air dried, and kept at 4°C. Alexa reagents and Hoechst stain were purchased from Molecular Probes/Invitrogen (Eugene, OR). Images were acquired on Leica DM IRE2 microscopic system and scanned with a Leica True confocal scanner SP2, with postacquisition processing performed on LCS Lite (Leica Microsystems, Wetzlar GmbH).
RESULTS
Isolation of MKlp2 in yeast two-hybrid screen.
To search for cellular proteins that bind to BPV-1 E2, we performed a yeast two-hybrid screen with an E2 bait containing a missense mutation replacing glutamate 39 with glycine (E39G) fused to the GAL4 DBD. This mutant is unable to activate transcription from E2 binding sites in metazoan or yeast promoters (4). E2 E39G was selected since it retains the ability to stimulate BPV E1-induced DNA replication (10). After coexpression of a human brain cell cDNA library fused with the GAL4-AD, yeast transformants were selected under stringent conditions for adenine, histidine, leucine, and tryptophan auxotrophy and α-galactosidase expression. The cDNA from one strongly blue-stained colony on X-α-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid) was identified as MKlp2 by DNA sequencing.
The yeast two-hybrid system was also used to determine the region of MKlp2 that interacted with E2-E39G. The full-length 886-aa MKlp2 and truncated fragments were amplified by reverse transcription-PCR of MCF-7 cellular RNA and inserted in frame with the GAL4-AD. Yeast transformants expressing full-length or C-terminal (aa 541 to 886) but not other truncated portions of MKlp2 stained blue on X-α-Gal plates in the presence of Gal4 DBD-E2 E39G (Table 1). E2 E39G did not interact with N-terminal regions of MKlp2 encompassing amino acids 1 to 468 or 1 to 528 in this assay. These results imply that BPV E2 associates with the carboxyl-terminal region of MKlp2.
TABLE 1.
Yeast two-hybrid results
| pGBK insert | pGAD insert | X-α-Gal staina |
|---|---|---|
| Lamin | SV40b large T | − |
| E2-E39G | MKlp2 1-468 | − |
| E2-E39G | MKlp2 1-528 | − |
| E2-E39G | MKlp2 541-886 | ++ |
| E2-E39G | MKlp2FL 1-886 | + |
| P53 | SV40 large T | ++ |
| E2-E39G | GPS2 | + |
−, negative; +, positive; ++, strongly positive.
SV40, simian virus 40.
MKlp2 binds the BPV-1 E2 protein.
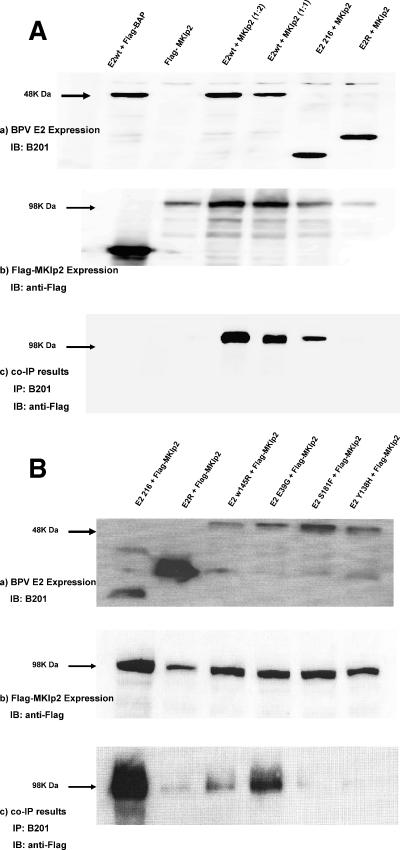
To further validate the association of MKlp2/Rab6KIFL with the BPV-1 E2 protein in vivo, we performed coimmunoprecipitation (co-IP) assays. C33A cells were cotransfected with plasmids encoding Flag-epitope-tagged MKlp2 and BPV-1 wild-type E2, E2R (aa 162 to 410), or E2 N-terminal transactivation domain (TAD; aa 1 to 216). E2 proteins were immunoprecipitated using the E2 monoclonal antibody B201, and the complexes were probed using Flag antibody. Flag-MKlp2 bound wild-type and TAD E2, while binding to E2R was negligible (Fig. 1A). These co-IP experiments were reversed using anti-Flag antibody to immunoprecipitate Flag-MKlp2, followed by Western blotting for E2, and also revealed their specific association (data not shown). Binding experiments using in vitro-translated MKlp2 incubated with GST-E2 1-216 or GST-E2 162-410 further confirmed that the N-terminal 162 amino acids of E2 are necessary for their association (data not shown).
FIG. 1.
(A) Coimmunoprecipitation of MKlp2 and BPV E2. Extracts of C33A cells cotransfected with Flag-MKlp2 and different E2 expression constructs as indicated were immunoblotted (IB) and probed with anti-E2 monoclonal B201 (panel a) or anti-Flag M2 (panel b). Immunoprecipitates with B201 were immunoblotted and probed with anti-Flag M2 (panel c). E2wt, wild type; BAP, bacterial alkaline phosphatase. (B) Coimmunoprecipitation of MKlp2 and BPV E2 mutants. Extracts of C33A cells cotransfected with Flag-MKlp2 and different expression constructs of E2 mutants as indicated were immunoblotted and probed with anti-E2 monoclonal B201 (panel a) or anti-Flag M2 (panel b). Immunoprecipitates with B201 were immunoblotted and probed with anti-Flag M2 (panel c).
Because MKlp2 interacted with the E2 TAD, we tested a series of point mutants within this domain (4) for MKlp2 association by co-IP. Studies of multiple transcription-active and -defective E2 mutations throughout the TAD showed no clear relationship to MKlp2 binding (Table 2). Three E2 mutants, W145R, Y138H, and S181F, did not coprecipitate with Flag-MKlp2 (Fig. 1B). However, other mutations in this region such as S171P and F173S retained the ability to coprecipitate. The MKlp2 binding-defective E2 S181F mutation showed a 90% reduction in transient transcriptional activation (3, 4). E2 S181F is also competent for transient replication of an origin-containing plasmid, while W145R and Y138H were defective. These results suggest that MKlp2 association is not absolutely necessary for E2 transcriptional and transient replication activities. Because MKlp2 acts as a mitotic motor protein, we therefore suspected its role in maintenance or partitioning of the viral genome during mitosis.
TABLE 2.
Summary of results for MKlp2-binding-defective E2 mutants
| Mutant | % Transient replication | Gps2/AMF1 bindingb | % Genome maintenance | % Transactivation | E1 bindingb |
|---|---|---|---|---|---|
| Y138H | <40 | − | NDa | 9 | + |
| W145R | <10 | − | 5 | <15 | + |
| S181F | <80 | + | ND | <10 | + |
ND, not done.
−, negative; +, positive.
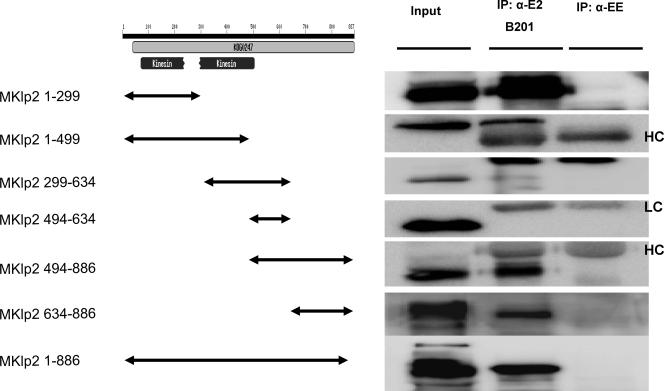
To identify the MKlp2 domain that bound to BPV-1 E2, we constructed a series of plasmids expressing Flag-tagged regions of MKlp2 (Fig. 2). Each of these plasmids was cotransfected with wild-type E2 into C33A cells. Immunoprecipitation of BPV-1 E2 with B201 and immunoblotting with Flag antibody demonstrated that two overlapping C-terminal polypeptides from MKlp2, aa 494 to 886 and aa 634 to 886, were present in the complexes, but not segments of MKlp2 spanning aa 299 to 634 and aa 494 to 634 (Fig. 2). Taken together with the yeast two-hybrid results, we infer aa 634 to 886 of MKlp2 bind E2. In these coprecipitation experiments, MKlp2 1-299 also coprecipitated with E2. This region of MKlp2 is known to bind microtubules (6, 7, 12). In contrast, E2 E39G interacted with the C-terminal but not N-terminal microtubule binding region of MKlp2 in the yeast two-hybrid experiments.
FIG. 2.
MKlp2 BPV E2-binding region mapping. Extracts of C33a cells cotransfected with BPV E2 and a series of Flag-tagged MKlp2 fragment expression constructs were immunoblotted and probed by anti-Flag M2. Immunoprecipitations were performed by using B201 or anti-EE (α-EE) monoclonal antibody as a control. HC, heavy chain; LC, light chain.
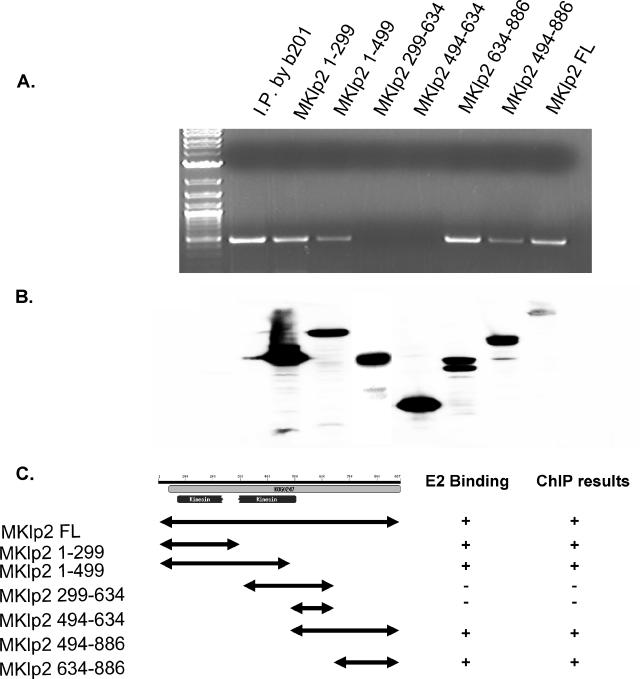
To further understand the biological significance of the E2 interaction with MKlp2, we adopted the ChIP assay to investigate MKlp2 binding to the BPV genome. BPV-1-transformed mouse ID13 cells were transfected with different segments of Flag-MKlp2, and the cells were treated with 1% formaldehyde to cross-link protein-DNA complexes. These lysates were immunoprecipitated with anti-Flag or anti-E2 antibody B201. Levels of the transfected Flag-tagged MKlp2 proteins are shown in Fig. 3B. PCR was performed after immunoprecipitation using oligonucleotides to amplify a 500-bp fragment from the BPV-1 long control region. Both the N-terminal and C-terminal regions, but not the central region of MKlp2, were found to be in association with the viral genome (Fig. 3A). These data imply the MKlp2 protein is in complex with E2 and the viral genome.
FIG. 3.
Chromatin immunoprecipitation. ID13 cells transfected with Flag-tagged full-length MKlp2 and a series of truncation constructs were used for ChIP. Anti-Flag and B201 antibodies were used as indicated. (A) ChIP PCR results. (B) Corresponding protein expression. (C) Summary of ChIP and BPV E2 binding results. −, negative; +, positive.
HPV E2 proteins bind endogenous MKlp2.
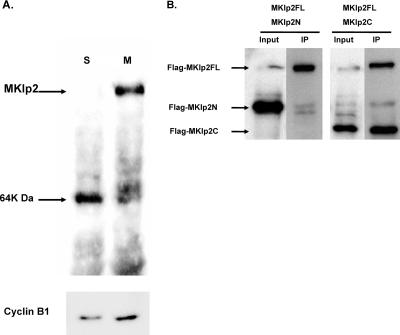
We sought to determine whether papillomavirus E2 would complex with the endogenous MKlp2 protein, which is present only during mitosis (7, 26). To demonstrate our antiserum specifically recognizes MKlp2, HeLa cells were cycled by double-thymidine block, released, and harvested, with the first time point postrelease representing S phase. Parallel cultures were treated 4 h later with nocodazole to accumulate cells in early mitosis. The serum from a rabbit immunized with the C-terminal region of human MKlp2 spanning aa 496 to 886 was found to recognize a protein at the predicted mass of 98 kDa in mitotic extracts (Fig. 4A). To demonstrate our antiserum was specific to the C-terminal region of MKlp2, C33A cells were transfected with Flag-tagged full-length N-terminal or C-terminal MKlp2. Lysates were immunoprecipitated with rabbit MKlp2 antiserum, and the washed complexes were immunoblotted with Flag antibody. As predicted, the full-length and C-terminal MKlp2 proteins were specifically recognized by both MKlp2 and Flag antibodies (Fig. 4B). Although mouse and human MKlp2 proteins share greater than 90% identity, neither sheep (7, 26) nor our rabbit antiserum recognizes mouse MKlp2 by immunoprecipitation or Western blotting (data not shown). Therefore, we could not test the association of MKlp2 with BPV E2 using murine C127-derived ID13 cells and turned to HPV E2 for further study.
FIG. 4.
Characterization of anti-MKlp2 antibody. (A) Fifty micrograms of HeLa cell extracts synchronized using a double-thymidine block were blotted and probed by affinity-purified anti-MKlp2. The same cell extracts were probed for cyclin B1 (Sc-245; Santa Cruz) on the bottom panel. S, S phase; M, mitotic cell lysate. (B) Extracts of C33A cells cotransfected with Flag-tagged full-length MKlp2 plus Flag-MKlp2N (aa 1 to 499) or MKlp2C (aa 496 to 886) followed by immunoprecipitation with anti-MKlp2 antibody were blotted and probed with anti-Flag M2.
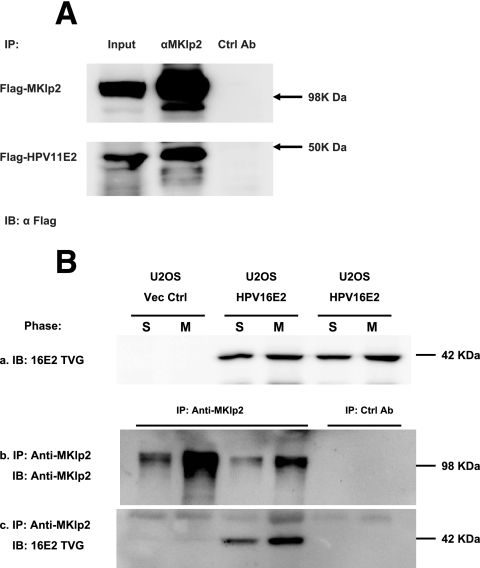
Using our MKlp2 antiserum, we therefore sought to determine whether association with MKlp2 was conserved among HPV E2 proteins by performing co-IP experiments. We used CV-1 cells expressing Flag-tagged HPV-11 E2 under the control of the metallothionein promoter (27) and found HPV-11 E2 coimmunoprecipitated with MKlp2, using the rabbit MKlp2 antiserum (Fig. 5A). We also tested whether high-risk HPV-16 E2 binds and coprecipitates the endogenous MKlp2 protein during mitosis. U2OS cells stably expressing HPV-16 E2 were synchronized using double-thymidine blockade. Half of the cells were harvested at the end of synchronization at S phase. The other half were released into complete media for 9 h before harvest. Lysates from these time points were analyzed on immunoblots probed with anti-HPV-16 E2 monoclonal antibody TVG261 (11) (Fig. 5B, subpanel a). As expected, the 42-kDa HPV-16 E2 protein was detected in S- and M-phase lysates from U2OS-16E2 but not in control U2OS cells. Lysates were incubated with affinity-purified anti-MKlp2 antiserum to immunoprecipitate the endogenous MKlp2 protein or GST-MKlp2-preabsorbed antiserum as a negative control. Washed complexes were resolved by SDS-PAGE, and immunoblots were probed with TVG261 or anti-MKlp2 antiserum (Fig. 5B, subpanels b and c). We confirmed that the endogenous MKlp2 is complexed with the HPV-16 E2 during mitosis. A smaller amount of HPV-16 E2 was detected in the S-phase MKlp2 immunoprecipitate fraction. This is likely due to a subpopulation of cells in the S-phase fraction escaping thymidine blockade and entering mitosis.
FIG. 5.
HPV E2 protein complex with MKlp2. (A) CV-1 cells with inducible Flag-HPV-11 E2 were transfected with the Flag-MKlp2 expression construct. Four hours before harvesting, expression of HPV-11 E2 was induced by 2 μM CdSO4 in complete medium. Anti-MKlp2 (αMKlp2) and antigen-preabsorbed antisera (Ctrl Ab) were used for immunoprecipitations. Immunoprecipitates were blotted and probed with anti-Flag M2 (α Flag). (B) U2OS cells with or without stably expressed HPV-16 E2 were synchronized using a double-thymidine block. Extracts were blotted and probed with TVG261 (top panel) to demonstrate E2 expression. Anti-MKlp2 and antigen-preabsorbed antisera (Ctrl Ab) were used for immunoprecipitation. Precipitates and cell extracts as input were blotted and probed with anti-MKlp2 and TVG261, respectively. S, S phase; M, mitotic cell lysate.
Endogenous MKlp2 and E2 colocalization in mitosis.
The question remained whether or where during mitosis endogenous MKlp2 would colocalize with E2. As we noted above, we could not use BPV genome-transformed ID13 cells for these studies since our antiserum does not recognize murine MKlp2. We were also unable to detect HPV-16 E2 by immunofluorescence in mitotic U2OS HPV-16 E2 cells using the TVG261 antibody. We therefore used human RPE-1 cells transfected with BPV-1 E2. The morphology of these flat cells is well suited for immunofluorescence studies.
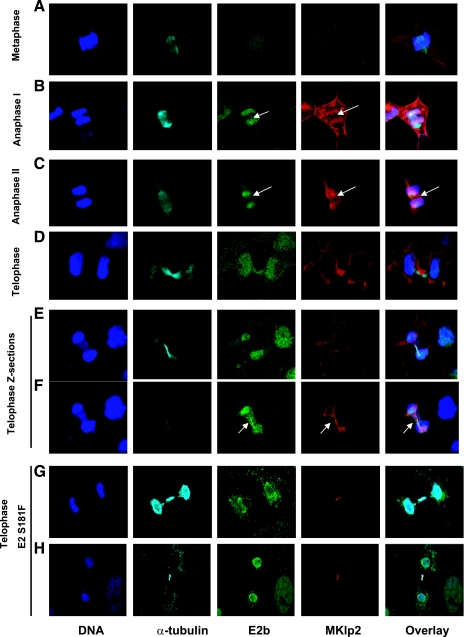
Using affinity-purified rabbit MKlp2 antibody, we determined the localization of endogenous MKlp2 throughout the cell cycle (Fig. 6) in RPE-1 cells transfected with BPV E2. Our observations were consistent with previous reports using different antisera (7, 26). MKlp2 staining was absent in interphase cells. A diffuse cytosolic pattern was observed in metaphase (Fig. 6A). By early anaphase, MKlp2 concentrated in the middle of spindle, appearing as a series of short bars running parallel with its axis (Fig. 6B and C). At late anaphase, as the cleavage furrow pinched in, MKlp2 was restricted to filament-like structures in the middle of spindle, probably overlapping the microtubules in this region (Fig. 6C). In telophase and cytokinesis, MKlp2 staining was limited to the midbody (Fig. 6D and E).
FIG. 6.
Indirect immunofluorescence of MKlp2 and E2 through cell cycles. RPE-1 cells transfected with BPV E2 expression construct were fixed and stained. Each panel represents the specific stage of the cell cycle as indicated. Each column shows a single fluorescent signal corresponding to DNA, α-tubulin, E2b, and MKlp2, with the overlays on the far-right column. Panels E and F show different Z-sections of the same field containing a cell in telophase/cytokinesis. Panels G and H show cells in telophase/cytokinesis with E2 mutant S181F.
BPV E2-transfected RPE-1 cells displayed nuclear E2 with nucleolar exclusion in interphase. In metaphase, the E2 protein was less intense and diffusely nuclear, with a fraction appearing in spots overlapping the cellular DNA (Fig. 6). In early anaphase, the majority of the E2 staining overlapped the DNA staining in the middle of the spindle. With anaphase progression and as chromosomes move from the midplate towards the poles, most of the E2 colocalized with the mitotic DNA, while a fraction remained in the middle of the spindle, appearing as faint lines across the midplate. This pattern resembles a microtubule-like pattern, similar to the distribution of MKlp2 at this stage of mitosis. By telophase, the majority of E2 overlapped the cellular DNA. Using stepwise Z series images at this stage of mitosis, we observed that E2 was also found in the midbody, colocalizing with the MKlp2 signal. Intense α-tubulin appeared in a Z-section close to but not coincident with MKlp2 and E2. Cells transfected with the MKlp2-binding-defective E2 mutant S181F did not demonstrate midbody colocalization similar to wild-type BPV E2 (Fig. 6G and H).
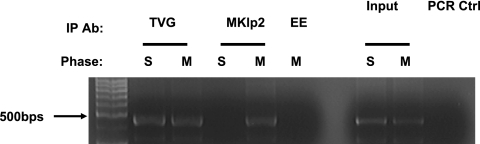
Although we observed the colocalization of endogenous MKlp2 with a subpopulation of E2, it was not direct evidence that viral genomes existed in the same fraction. We again adopted ChIP assays to explore MKlp2 protein association with viral genomes. U2OS cells were first blocked in complete medium with 2 mM thymidine for 16 h. After release from the first thymidine block, cells were transfected with the HPV-16 genome using Fugene 6 (Roche, Germany). After 8 h, cells were arrested with 2 mM thymidine in complete medium for another 16 h. After release from the second thymidine block, half of the cells were treated with formaldehyde to cross-link DNA-protein complexes and harvested. The remaining half of the cells were incubated in complete medium for about 9 h until at least 40% of cell population appeared rounded and were judged mitotic. These cells were then processed for ChIP assays. For immunoprecipitation of the protein-DNA complexes, we used TVG261 for HPV-16 E2 and affinity-purified rabbit anti-MKlp2. By specific PCR of the HPV DNA present in the complexes, anti-MKlp2 antibody was found to precipitate the viral genome specifically during mitosis but not in S phase, while this PCR product was not detected with a negative control antibody (Fig. 7). Control PCR was performed in both U2OS cells transfected by HPV-16 genomes to confirm successful transfection of viral genomes. These data strongly indicate association between MKlp2 and viral genomes during mitosis.
FIG. 7.
PCR results of ChIP in U2OS cells with the HPV-16 genome. U2OS cells were transfected with HPV-16 genomes and synchronized. Extracts from S-phase (S) and mitotic (M) cells were immunoprecipitated with TVG261, anti-MKlp2, and anti-EE as a control antibody (Ab).
DISCUSSION
Forty-four putative members of the kinesin superfamily proteins (KIFs) function as microtubule-associated motor proteins. Together with actin-associated myosin, these proteins are responsible for organelle, protein vesicle, DNA, and RNA transport and hence are critical for a numerous cellular processes (25). KIFs can be subdivided into three classes according to the position of their motor domain: the N-terminal motor domain type (N-kinesins), middle motor domain type (M-kinesins), and C-terminal motor domain type (C-kinesins). Among these, N-terminal KIFs are the most abundant. They can be further grouped according to their functions. MKlp2, also called KIF20A or Rab6KIFL (32), belongs to the proposed N-6 kinesins (13). The other kinesin protein in this N-6 class is MKlp1, also known as CHO1 or KIF23, which is involved in central spindle organization and cytokinesis (16). The CENP-E/KIF10 family, comprising N-7 kinesins, was identified as a centromere-associated protein responsible for chromosome segregation (21). KIF2C, an M-kinesin, was reported as a mitotic centromere-associated kinesin (37).
The human MKlp2 gene has been mapped to 5q31 (17, 18), a segment commonly deleted in myeloid leukemia. Interestingly, this 1.5-Mbp genomic sequence (D5s479 to D5s500) contains another gene, CDC23, a component of the anaphase-promoting complex and substrate for Plk1 (17, 18). High levels of MKlp2 are found in mitotically active tissues, including skin, testis, and spleen. Expression of MKlp2 is cell cycle dependent and limited to mitosis (7). MKlp2 has also been visualized in the Golgi apparatus (6, 12).
Structurally, the MKlp2 protein has a tubulin binding site within its N-terminal kinesin domain and another within a carboxyl end. This is posited to allow binding and contraction between two parallel microtubules in the midbody that occurs in telophase. It is well documented that MKlp2 is essential for completion of cytokinesis (6, 7, 12). Depletion of MKlp2 by RNA interference (RNAi) results in failure of cytokinesis (26, 39). Microinjection of MKlp2-specific antibodies induces binucleate cells that arrest in telophase (12), further demonstrating the function of MKlp2 at this stage of mitosis.
Using both yeast two-hybrid and in vivo coimmunoprecipitation experiments, we show E2 binds the C-terminal region of MKlp2. N-terminal segments (amino acids 1 to 299) of MKlp2 expressed in C33A cells were also immunoprecipitated in complex with BPV E2. We suspect this may be indirect since this “head” region of MKlp2 binds microtubules. One group recently referred to as unpublished data the detection of β-tubulin by analysis of HPV-11 E2-bound proteins using mass spectrometry, although, in vitro-translated β-tubulin did not bind E2 (5). Moreover, HPV-11 E2 has been reported to localize to the mitotic spindle, which is composed of polymerized α- and β-tubulin (5, 35). These observations likely explain the coimmunoprecipitation of the microtubule-binding N-terminal domain of MKlp2 with E2. We suspect this interaction of MKlp2 is relatively weak, since in our attempts at a direct binding assay, bacterially synthesized MKlp2 and BPV E2 showed slight association above background. We infer that tubulin or another spindle component stabilizes the interaction of E2 with MKlp2 during a specific stage of mitosis.
We have demonstrated that the association between endogenous MKlp2 and papillomavirus HPV-16 E2 occurs in mitosis by immunoprecipitation of synchronized cell extracts. In ChIP experiments, we consistently detected viral genomes in immunoprecipitates from cells in mitosis using anti-MKlp2 antibody, but not in S-phase extracts. These results strongly infer that E2, together with viral genome, is in complex with MKlp2. Due to the inexact nature of cell synchronization using double-thymidine blockade, the phase of mitosis in which E2 associates with MKlp2 cannot be precisely determined.
Colocalization studies of MKlp2 with BPV E2 were performed to determine when in mitosis the proteins associate. After transfection and synchronization of human RPE1 cells, BPV E2 decorated the mitotic chromosomes, consistent with earlier reports (22). A fraction of E2 protein was detected in the spindle midzone after metaphase and through anaphase (Fig. 6B and C). E2 remained in the midbody with formation of the cleavage furrow, although the majority was associated with the host chromosomes. GFP-tagged full-length and TAD HPV-18 E2 may also be seen in this midbody pattern (2). GFP-tagged HPV-11 E2 has also been reported to associate with mitotic spindle; however, BPV E2 was not observed in this locale (35). In this study, we found a minor fraction of BPV E2 in the spindle midzone and midbody. Serial Z-section images revealed that BPV E2 was adjacent to the microtubules in the midzone and midbody, but appeared in more close proximity to MKlp2, which is microtubule associated.
What is the role of MKlp2 association with E2 during mitosis? By using the ChIP assay, we demonstrate MKlp2 is in the E2 protein-viral DNA complex in a cell-cycle-dependent manner. A previous report of ChIP experiments regarding Brd4 (38) used asynchronous cells. In contrast to Brd4, endogenous MKlp2 protein is only expressed in mitosis. This may be the first report of a protein-DNA complex in mitosis that contains the PV genome and represents strong supporting evidence that MKlp2 plays a role in genome partitioning.
In confocal microscopy studies, the majority of the E2 protein, whether BPV or HPV, appears as speckles that randomly coat the chromosomes throughout mitosis. This is not the pattern of MKlp2. We do not know whether the major E2 staining pattern represents the localization of the PV genomes or whether this population of E2 is partitioning the viral DNA. It is conceivable that PV genomes are associated with E2 and MKlp2 during metaphase around the midplate and then further partition towards the poles, transported on MKlp2, a positive-end kinesin that actuates movement towards the spindle poles.
The mechanism of mitotic partitioning of PV genomes likely requires the participation of multiple cellular factors during each stage of mitosis. We have found that binding of E2 with the ChlR1 protein is necessary for the initial loading of E2 proteins onto mitotic chromosomes (28). RNAi-mediated depletion of ChlR1 prevents viral genome attachment to mitotic chromatids (28). Because MKlp2 depletion induces growth arrest in telophase (26, 39), leading to cell death or apoptosis, RNAi directed at MKlp2 cannot be used to discriminate the requirement for Mklp2 in E2-mediated viral genome partitioning in dividing cells in culture. Brd4 association may represent a subsequent step in the ordered events necessary for recognition and movement of the viral DNA during the multiple steps of mitosis. Since BPV has been reported to replicate through a random choice mechanism (8, 29), distinct pathways may be needed to retain and partition replicated and nonreplicated genomes. One possibility is that MKlp2 selectively binds E2 attached to all viral genomes and mediates their movement. Another speculative scenario is that the E2 interaction with MKlp2 represents a transport mechanism to segregate viral episomes that were not randomly selected for replication.
Acknowledgments
We thank Francis Barr for providing the sheep anti-MKlp2 antibody. We are grateful to Alison McBride, Bruno Goud, Sally Roberts, and Iain Morgan for providing reagents and cell lines essential for this work. We greatly appreciate the many useful discussions with members of our laboratory.
This work was supported by NIH R01 CA58376.
Footnotes
Published ahead of print on 29 November 2006.
REFERENCES
- 1.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134. [DOI] [PubMed] [Google Scholar]
- 2.Bellanger, S., S. Blachon, F. Mechali, C. Bonne-Andrea, and F. Thierry. 2005. High-risk but not low-risk HPV E2 proteins bind to the APC activators Cdh1 and Cdc20 and cause genomic instability. Cell Cycle 4:1608-1615. [DOI] [PubMed] [Google Scholar]
- 3.Breiding, D. E., M. J. Grossel, and E. J. Androphy. 1996. Genetic analysis of the bovine papillomavirus E2 transcriptional activation domain. Virology 221:34-43. [DOI] [PubMed] [Google Scholar]
- 4.Breiding, D. E., F. Sverdrup, M. J. Grossel, N. Moscufo, W. Boonchai, and E. J. Androphy. 1997. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol. Cell. Biol. 17:7208-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dao, L. D., A. Duffy, B. A. Van Tine, S.-Y. Wu, C.-M. Chiang, T. R. Broker, and L. T. Chow. 2006. Dynamic localization of the human papillomavirus type 11 origin binding protein E2 through mitosis while in association with the spindle apparatus. J. Virol. 80:4792-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Echard, A., F. Jollivet, O. Martinez, J. J. Lacapere, A. Rousselet, I. Janoueix-Lerosey, and B. Goud. 1998. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science 279:580-585. [DOI] [PubMed] [Google Scholar]
- 7.Fontijn, R. D., B. Goud, A. Echard, F. Jollivet, J. van Marle, H. Pannekoek, and A. J. Horrevoets. 2001. The human kinesin-like protein RB6K is under tight cell cycle control and is essential for cytokinesis. Mol. Cell. Biol. 21:2944-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert, D. M., and S. N. Cohen. 1987. Bovine papilloma virus plasmids replicate randomly in mouse fibroblasts throughout S phase of the cell cycle. Cell 50:59-68. [DOI] [PubMed] [Google Scholar]
- 9.Greber, U. F., and M. Way. 2006. A superhighway to virus infection. Cell 124:741-754. [DOI] [PubMed] [Google Scholar]
- 10.Grossel, M. J., F. Sverdrup, D. E. Breiding, and E. J. Androphy. 1996. Transcriptional activation function is not required for stimulation of DNA replication by bovine papillomavirus type 1 E2. J. Virol. 70:7264-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hibma, M. H., K. Raj, S. J. Ely, M. Stanley, and L. Crawford. 1995. The interaction between human papillomavirus type 16 E1 and E2 proteins is blocked by an antibody to the N-terminal region of E2. Eur. J. Biochem. 229:517-525. [DOI] [PubMed] [Google Scholar]
- 12.Hill, E., M. Clarke, and F. A. Barr. 2000. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 19:5711-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirokawa, N., and R. Takemura. 2004. Kinesin superfamily proteins and their various functions and dynamics. Exp. Cell Res. 301:50-59. [DOI] [PubMed] [Google Scholar]
- 14.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilves, I., K. Mäemets, T. Silla, K. Janikson, and M. Ustav. 2006. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J. Virol. 80:3660-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuriyama, R., C. Gustus, Y. Terada, Y. Uetake, and J. Matuliene. 2002. CHO1, a mammalian kinesin-like protein, interacts with F-actin and is involved in the terminal phase of cytokinesis. J. Cell Biol. 156:783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai, F., A. A. Fernald, N. Zhao, and M. M. Le Beau. 2000. cDNA cloning, expression pattern, genomic structure and chromosomal location of RAB6KIFL, a human kinesin-like gene. Gene 248:117-125. [DOI] [PubMed] [Google Scholar]
- 18.Lai, F., L. A. Godley, J. Joslin, A. A. Fernald, J. Liu, R. Espinosa III, N. Zhao, L. Pamintuan, B. G. Till, R. A. Larson, Z. Qian, and M. M. Le Beau. 2001. Transcript map and comparative analysis of the 1.5-Mb commonly deleted segment of human 5q31 in malignant myeloid diseases with a del(5q). Genomics 71:235-245. [DOI] [PubMed] [Google Scholar]
- 19.Law, M.-F., D. R. Lowy, I. Dvoretzky, and P. M. Howley. 1981. Mouse cells transformed by bovine papillomavirus contain only extrachromosonal viral DNA sequences. Proc. Natl. Acad. Sci. USA 78:2727-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lombillo, V. A., C. Nislow, T. J. Yen, V. I. Gelfand, and J. R. McIntosh. 1995. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J. Cell Biol. 128:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride, A. A., M. G. McPhillips, and J. G. Oliveira. 2004. Brd4: tethering, segregation and beyond. Trends Microbiol. 12:527-529. [DOI] [PubMed] [Google Scholar]
- 23.McPhillips, M. G., K. Ozato, and A. A. McBride. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol. 79:8920-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miki, H., Y. Okada, and N. Hirokawa. 2005. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 15:467-476. [DOI] [PubMed] [Google Scholar]
- 25.Miki, H., M. Setou, K. Kaneshiro, and N. Hirokawa. 2001. All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. USA 98:7004-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neef, R., C. Preisinger, J. Sutcliffe, R. Kopajtich, E. A. Nigg, T. U. Mayer, and F. A. Barr. 2003. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 162:863-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveira, J. G., L. A. Colf, and A. A. McBride. 2006. Variations in the association of papillomavirus E2 proteins with mitotic chromosomes. Proc. Natl. Acad. Sci. USA 103:1047-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parish, J. L., A. M. Bean, R. B. Park, and E. J. Androphy. 2006. ChlR1 is required for loading papillomavirus E2 onto mitotic chromosomes and viral genome maintenance. Mol. Cell 24:867-876. [DOI] [PubMed] [Google Scholar]
- 29.Ravnan, J. B., D. M. Gilbert, K. G. Ten Hagen, and S. N. Cohen. 1992. Random-choice replication of extrachromosomal bovine papillomavirus (BPV) molecules in heterogeneous, clonally derived BPV-infected cell lines. J. Virol. 66:6946-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schweiger, M.-R., J. You, and P. M. Howley. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 80:4276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taniuchi, K., H. Nakagawa, T. Nakamura, H. Eguchi, H. Ohigashi, O. Ishikawa, T. Katagiri, and Y. Nakamura. 2005. Down-regulation of RAB6KIFL/KIF20A, a kinesin involved with membrane trafficking of discs large homologue 5, can attenuate growth of pancreatic cancer cell. Cancer Res. 65:105-112. [PubMed] [Google Scholar]
- 33.Thomas, D. B., and C. A. Lingwood. 1975. A model of cell cycle control: effects of thymidine on synchronous cell cultures. Cell 5:37-42. [DOI] [PubMed] [Google Scholar]
- 34.Ustav, E., M. Ustav, P. Szymanski, and A. Stenlund. 1993. The bovine papillomavirus origin of replication requires a binding site for the E2 transcriptional activator. Proc. Natl. Acad. Sci. USA 90:898-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Tine, B. A., L. D. Dao, S. Y. Wu, T. M. Sonbuchner, B. Y. Lin, N. Zou, C. M. Chiang, T. R. Broker, and L. T. Chow. 2004. Human papillomavirus (HPV) origin-binding protein associates with mitotic spindles to enable viral DNA partitioning. Proc. Natl. Acad. Sci. USA 101:4030-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vernos, I., and E. Karsenti. 1996. Motors involved in spindle assembly and chromosome segregation. Curr. Opin. Cell Biol. 8:4-9. [DOI] [PubMed] [Google Scholar]
- 37.Wordeman, L., and T. J. Mitchison. 1995. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 128:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349-360. [DOI] [PubMed] [Google Scholar]
- 39.Zhu, C., J. Zhao, M. Bibikova, J. D. Leverson, E. Bossy-Wetzel, J. B. Fan, R. T. Abraham, and W. Jiang. 2005. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell 16:3187-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]