Abstract
The sheep genome harbors approximately 20 endogenous retroviruses (enJSRVs) highly related to the exogenous Jaagsiekte sheep retrovirus (JSRV). One of the enJSRV loci, enJS56A1, acts as a unique restriction factor by blocking JSRV in a transdominant fashion at a late stage of the retroviral cycle. To better understand the molecular basis of this restriction (termed JLR, for JSRV late restriction), we functionally characterized JSRV and enJS56A1 Gag proteins. We identified the putative JSRV Gag membrane binding and late domains and determined their lack of involvement in JLR. In addition, by using enJS56A1 truncation mutants, we established that the entire Gag protein is necessary to restrict JSRV exit. By using differentially tagged viruses, we observed, by confocal microscopy, colocalization between JSRV and enJS56A1 Gag proteins. By coimmunoprecipitation and molecular complementation analyses, we also revealed intracellular association and likely coassembly between JSRV and enJS56A1 Gag proteins. Interestingly, JSRV and enJS56A1 Gag proteins showed distinct intracellular targeting: JSRV exhibited pericentrosomal accumulation of Gag staining, while enJS56A1 Gag did not accumulate in this region. Furthermore, the number of cells displaying pericentrosomal JSRV Gag was drastically reduced in the presence of enJS56A1. We identified amino acid residue R21 in JSRV Gag as the primary determinant of centrosome targeting. We concluded that JLR is dependent on a Gag-Gag interaction between enJS56A1 and JSRV leading to altered cellular localization of the latter.
Endogenous retroviruses (ERVs) are fixed in the genomes of virtually all vertebrates and are believed to be the result of germ line infections of ancient exogenous (i.e., horizontally transmitted) retroviruses (6, 36). The great majority of ERVs have accumulated, during evolution, mutations and/or deletions that have altered their genomic structure. As a result of these genomic changes, most ERVs are not infectious and are nonpathogenic (15).
The selection in various animal species of transcriptionally active ERVs with intact open reading frames suggests that some of these elements have provided (and could still provide) a beneficial effect to their host. One of the possible biological functions of ERVs is the protection of the host against infection by exogenous pathogenic retroviruses. For example, some mouse and chicken ERVs block infection of related exogenous retroviruses at the early phases of the viral replication cycle by receptor interference (6). Fv1, a murine gene derived from an endogenous retrovirus, restricts infection in mice by the Friend strain of murine leukemia virus at a postentry step (3).
Of particular interest is a unique mechanism of restriction displayed at a late step of the retroviral replication cycle by an endogenous betaretrovirus of sheep (19). Sheep betaretroviruses include the exogenous oncogenic Jaagsiekte sheep retrovirus (JSRV) and approximately 20 copies of endogenous viruses highly related to JSRV (hence the name enJSRVs) (23, 25, 26). JSRV is the causative agent of ovine pulmonary adenocarcinoma, a transmissible lung cancer of sheep. enJSRVs are, in turn, highly expressed in the genital tract of the ewe (23, 35) and play a role in the reproductive biology of this animal species (11, 12, 24).
Three enJSRV proviruses have been cloned and characterized to date, including enJS56A1, enJS5F16, and enJS59A1 (23). Locus enJS56A1 possesses intact open reading frames for all of its genes, although a frameshift in the last portion of pol likely results in a nonfunctional viral integrase (23). Cells transfected with an expression plasmid for enJS56A1 do not release viral particles into the supernatant, although abundant Gag is detected in cell lysates and intracytoplasmic viral particles can be observed by electron microscopy (19, 23). Notably, the defect possessed by enJS56A1 is transdominant over JSRV (19). In particular, enJS56A1 blocks JSRV at a late step of the virus replication cycle. For convenience, we will refer to the enJS56A1-induced block of JSRV particle release as JLR, for JSRV late restriction.
We previously mapped the main determinant of JLR to residue 21 of the enJS56A1 Gag polyprotein (19). Gag, the structural protein of the retroviral nucleocapsid core, plays a central role in retroviral assembly and budding (38). Interestingly, there is an arginine residue in Gag position 21 of JSRV which is conserved in all betaretroviruses but replaced by a tryptophan in enJS56A1. The mechanism and timing of JLR are not yet understood. However, the observation of viral particles by electron microscopy in cells expressing enJS56A1 (or coexpressing enJS56A1 and JSRV) suggests that JLR is dependent on a defect in Gag trafficking, possibly occurring after assembly. From studies conducted on Mason-Pfizer monkey virus (M-PMV), it is hypothesized that betaretroviruses assemble in the cytoplasm in a pericentrosomal region and then traffic to the cell membrane by incompletely characterized mechanisms involving the recycling endosomes and the viral envelope glycoprotein (32, 33).
Some of the determinants of Gag trafficking to the cell membrane and exit, such as the membrane binding (M) and late (L) domains, have been characterized for other retroviruses (38), but their involvement (if any) in JLR was not known. Most retroviruses have a bipartite M domain composed of a myristate linked to the N-terminal part of Gag and a patch of basic amino acid residues located in the N-terminal matrix (MA) domain (38). Mutation of the M domain alters the ability of retroviral Gag to reach the cell membrane (20, 27, 28, 42). L domains are short amino acid sequences required during viral exit for virus-cell separation (9, 14). Typical L domain mutants are retained at the cell membrane, as they are unable to pinch off from the cell (14).
In the present study, we functionally characterized the JSRV and enJS56A1 Gag polyproteins. We identified putative JSRV M and L domains and determined their lack of involvement in JLR. We show that truncated enJS56A1 Gag polyproteins are not able to block JSRV. Moreover, we show that JSRV and enJS56A1 Gag proteins colocalize, associate in trans, and likely coassemble. In addition, we show that the defect of enJS56A1 depends on its ability to misplace intracytoplasmic JSRV Gag localization. Indeed, JSRV Gag concentrates in the pericentrosomal area unless enJS56A1 is coexpressed in the same cell. Finally, we demonstrate that localization of JSRV Gag in the pericentrosomal area depends primarily on amino acid residue R21 in Gag. The results shown in this study argue that enJS56A1 Gag blocks JSRV in trans by hampering the progression of the latter to the centrosome.
MATERIALS AND METHODS
Plasmids.
pCMV4JS21 and pCMV2en56A1 express the full-length JSRV21 clone and the enJS56A1 locus, respectively, and have been described elsewhere (23, 25). JSRV and enJS56A1 Gag proteins were differentially tagged with the FLAG and hemagglutinin (HA) epitopes, at the carboxy-terminal position of Gag and within the matrix domain before variable region 1, by overlap PCR. pJSRVHA-MA, pJSRVFLAG-MA, penJS56A1HA-MA, and penJS56A1FLAG-MA contain the HA or FLAG epitope in the MA domain and were derived from pCMV4JS21 or from pCMV2en56A1. pJSRVHA-C was derived from pCMV4JS21. In pJSRVHA-C, the JSRV Gag gene is fused at its C terminus-encoding end with the HA coding sequence and is followed by the JSRV env gene and the 3′ long terminal repeat (LTR) of JSRV. penJS56A1HA-C was derived from pJSRVHA-C by replacing JSRV gag with the enJS56A1 gag gene. pJSRVFLAG-C was derived from pCMV4JS21 by introducing the FLAG epitope at the carboxy-terminal portion of Gag by overlap PCR. penJS56A1FLAG-C encodes the enJS56A1 Gag protein tagged at the carboxy terminus with the FLAG epitope and was derived from the plasmid pGePEx (23).
Mutants JSR21A, JSK19E, JSH20E, JSK19E/H20E, JSR21A, JSR21E, JSR21K, and JSG22A were obtained by site-directed mutagenesis, using QuickChange (Stratagene) as suggested by the manufacturer. The nomenclature of the mutants indicates the virus from which they were derived (JS, JSRV; and en56, enJS56A1), followed by a single letter indicating the amino acid mutated, a number representing its position in Gag, and a letter indicating the amino acid residue in the resulting mutant. Mutants JSR21W, JSR98C, en56W21R, and en56W21R/C98R have been described previously (19).
Single and double late domain (LD) mutants were designated as follows: pJSΔLD201 carries a mutation of the proximal LD PSAP (Gag positions 201 to 204) to AGAP, whereas in pJSΔLD207, the distal late domain PPAY is mutated to AAAY (Gag positions 207 to 210). The double LD mutant pJSΔLD201-7 carries both mutations.
Mutants penJS56A1ΔCA-NC and penJS56A1ΔNC were derived from pCMV2enJS56A1 by insertion of a stop codon at positions 258 and 483 of Gag, respectively. The deletion mutant penJS56A1ΔNC2 was derived from pCMV2enJS56A1 by an in-frame deletion of Gag amino acid residues 490 to 548 in the nucleocapsid (NC) domain. enJS56A1ΔNC2 has a functional protease. The mutant JSRVΔpro is a full-length JSRV molecular clone with a deletion in the pro gene resulting in a nonfunctional protease and has been described before (19). The truncated mutant penJS56A1ΔMHR was derived from pCMV2en56A1 by deletion of Gag amino acid residues 403 to 422, encompassing the major homology region of the capsid (CA) domain.
Cell cultures, transfections, and viral preparations.
293T and HeLa cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum at 37°C with 5% CO2 and 95% humidity. For Western blot analysis, virus was produced by transient transfection of 293T cells, using a Calphos mammalian transfection kit (Clontech) according to the manufacturer's instructions. Cell supernatants were collected at 24 and 48 h posttransfection, and viral particles were concentrated by ultracentrifugation. For analysis of intracellular proteins, cells were lysed by standard techniques as already described (19).
For reverse-phase high-pressure liquid chromatography (RP-HPLC) separation, concentrated viral pellets were purified twice by isopycnic centrifugation on a 25 to 55% (wt/wt) linear sucrose gradient. Fractions with buoyant densities between 1.146 and 1.176 g/ml were pooled, concentrated by ultracentrifugation as described above, and resuspended in 8 M sequencing-grade aqueous concentrate of high-purity guanidine-HCl (Pierce) and 2% β-mercaptoethanol. JSRV Gag proteins were then separated by RP-HPLC and analyzed by N-terminal protein sequencing at the NCI-SAIC AIDS Vaccine Program as previously described (17, 21).
Western blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as previously described (25), using concentrated viral particles and cell lysates (50 μg of protein extracts). Gag proteins were detected with rabbit polyclonal sera against JSRV CA, p23 (MA), and NC and with an antiserum raised against the first 100 N-terminal amino acid residues of the JSRV Gag protein (Proteintech). Gag proteins differentially tagged with either the HA or FLAG epitope were detected with mouse monoclonal anti-HA (Covance) or anti-FLAG (Sigma) antibodies. Membranes were exposed to the appropriate peroxidase-conjugated secondary antibodies and further developed by chemiluminescence using ECL Plus (Amersham). Each experiment was repeated independently at least three times, using two different maxipreps for each transfected plasmid.
Coimmunoprecipitation assays.
Coimmunoprecipitation assays were performed with 293T cells transfected with the appropriate plasmids, as indicated in Results. Forty-eight hours after transfection, cells were lysed with RIPA buffer (150 mM NaCl; 50 mM Tris-HCl, pH 7.4; 1 mM EDTA, pH 8; 1 mM NaF; 1% NP-40) supplemented with a protease inhibitor cocktail (Complete; Roche) and 1 mM phenylmethylsulfonyl fluoride, essentially as already described (40). Lysates were sonicated and then centrifuged for 20 min at 14,000 × g to remove insoluble material. For the evaluation of protein-protein interactions, 200 μg of whole-cell extract was rocked with 20 μl of protein A-agarose beads (Santa Cruz Biotechnology) and primary antibody (mouse monoclonal anti-FLAG or anti-HA) at 4°C for 3 h. After three washes with lysis buffer, beads were resuspended in SDS loading buffer, boiled for 5 min, and subjected to SDS-PAGE and Western blotting as described above.
Confocal microscopy.
HeLa cells were plated onto two-well chambered glass slides (Lab-Tek, Nalge Nunc International) and transfected with Lipofectamine (Invitrogen) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were washed with phosphate-buffered saline and fixed with ice-cold methanol for 5 min at −20°C. After fixation, cells were processed essentially as already described (19, 33). Gag proteins were detected by using a rabbit antiserum towards JSRV MA (p23) preadsorbed with HeLa cell extracts to minimize the background. Gag proteins differentially tagged with either the HA or FLAG epitope were detected with mouse monoclonal anti-HA (Covance) or rabbit polyclonal anti-FLAG (Sigma). A mouse monoclonal antibody against γ-tubulin (Abcam) was used as a centrosome marker. The secondary antibodies used were goat anti-mouse and anti-rabbit immunoglobulin G, conjugated with Alexa 594 and Alexa 488, respectively (Molecular Probes). Slides were mounted with medium containing DAPI (4′,6′-diamidino-2-phenylindole) (Vectashield; Vector Laboratories) and analyzed with a Leica TCS SP2 confocal microscope. In some experiments, the pattern of Gag staining was recorded as either diffuse, dispersed, or concentrated, as already described (19). In addition, concentrated Gag staining was scored as pericentrosomal or distant from the centrosome, as described in Results. Statistical analysis of the data was performed by the chi-square test.
RESULTS
JSRV Gag residues K19, H20, and R21 affect viral exit, but only the last residue is involved in JLR.
Our first aim was to determine whether known domains involved in retroviral Gag trafficking could play a role in JLR. Residue R21 in Gag is highly conserved among betaretroviruses (Fig. 1A) and is the main determinant of JLR, as shown when it is replaced by tryptophan (W) in enJS56A1. R21 is followed by a glycine residue that is also highly conserved among betaretroviruses. Thus, R21 and G22 could be part of a functional domain required for JSRV trafficking/exit, and we further evaluated the contributions of these amino acid residues to JLR by mutagenesis.
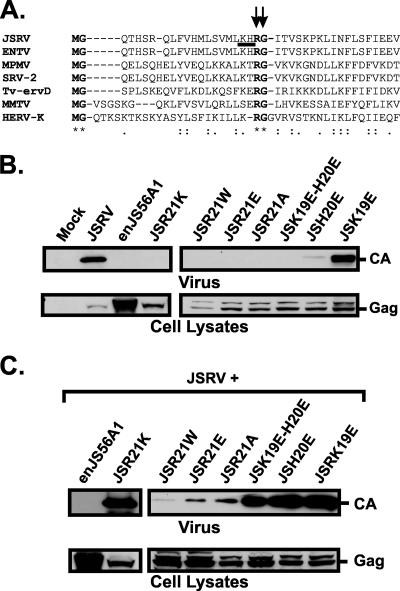
FIG. 1.
Mutational analysis of JSRV Gag amino acid residues 19, 20, and 21. (A) Alignment of the N-terminal Gag peptides from the indicated viruses, using ClustalW (39). R21 and G22 are highly conserved among betaretroviruses. Highly conserved residues are highlighted in bold. R21 and G22 are indicated with vertical arrows. K19 and H20 are underlined. Consensus symbols are displayed below the alignment, as follows: “*” indicates identical residues in all sequences in the alignment, “:” indicates conserved substitutions, and “.” indicates semiconserved substitutions. ENTV, enzootic nasal tumor virus; M-PMV, Mason-Pfizer monkey virus; SRV-2, simian retrovirus 2; TV-ervD, brushtail possum type D endogenous retrovirus; MMTV, mouse mammary tumor virus; HERV-K, human endogenous retrovirus K. (B) Analysis of JSRV mutants bearing mutations in the highly basic region of the M domain. Viral pellets (upper panel) and lysates (lower panel) of cells transfected with the indicated plasmids were resolved by SDS-PAGE and immunoblotted with a JSRV CA antiserum. (C) Western blot analysis of viral pellets (upper panel) and lysates (lower panel) of cells cotransfected with expression plasmids for JSRV and mutants, as indicated.
Previously, we showed that the single mutant JSR21W recapitulates the phenotype displayed by enJS56A1 (19). We derived JSRV mutants where the positively charged residue R21 was replaced with either glutamic acid (JSR21E), alanine (JSR21A), or lysine (JSR21K). Both the JSR21E and JSR21A mutants showed the same phenotype as that of JSR21W, while the more conservative mutant JSR21K failed to release viral particles (Fig. 1B) but did not block JSRV release (Fig. 1C). The JSG22A mutant was also incompetent for viral exit but did not block viral release when coexpressed with JSRV (data not shown).
In JSRV, R21 lies in close proximity to two other basic residues, i.e., lysine (K) 19 and histidine (H) 20, which are conserved in enJS56A1. R21 could be assumed to be part of the basic patch of amino acid residues that form the JSRV M (membrane) domain. Consequently, JLR might be the result of a transdominant defective M phenotype displayed by enJS56A1. If this was the case, disruption of the basic domain in JSRV MA would likely reproduce the interfering phenotype even in the presence of R21. In order to test this hypothesis, we derived JSRV mutants where K19 and H20 (individually or in combination) were replaced by the negatively charged amino acid glutamic acid (E). Viral release of JSH20E and JSK19E/H20E was severely impaired, while JSK19E did not display any defect in viral exit (Fig. 1B). However, when cotransfected with JSRV, neither JSH20E nor JSK19E/H20E blocked viral particle release in a transdominant fashion (Fig. 1C), suggesting that JLR is not the result of a dominant-negative M domain phenotype but is specifically dependent on residue R21.
Next, we studied cells expressing the above mutants by confocal microscopy. We previously classified cells expressing JSRV and enJS56A1 as diffuse, dispersed, and concentrated to describe the distribution of Gag staining by immunofluorescence (Fig. 2A) (19). This definition is dependent on whether the majority of Gag staining is uniformly diffuse (Fig. 2A, panel a) within the cytoplasm, dispersed in discrete dots (Fig. 2A, panel b), or concentrated (Fig. 2A, panel c) in a major location within the cell, usually the perinuclear region. Intermediate phenotypes, especially between the diffuse and dispersed phenotypes, are also observed. The phenotype of cells expressing JSRV or enJS56A1 by confocal microscopy is similar to that observed with the related betaretrovirus M-PMV (33). Available data on M-PMV (33) and the results shown below suggest that the diffuse phenotype corresponds mainly to newly synthesized Gag, the concentrated phenotype corresponds to Gag localizing to the pericentrosomal area, where it is proposed to assemble (see below), and the dispersed phenotype is Gag trafficking to the membrane or to the centrosome.
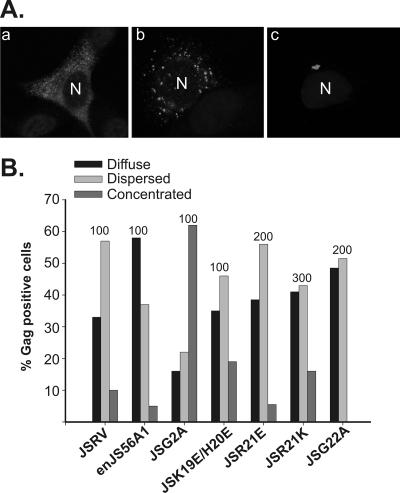
FIG. 2.
Patterns of Gag staining by confocal microscopy of cells expressing JSRV, enJS56A1, and M domain mutants. (A) By confocal microscopy, Gag staining in HeLa cells expressing JSRV and enJS56A1 is described as diffuse (panel a), dispersed (panel b), or concentrated (panel c) (19). The figure represents typical examples of enJS56A1-expressing cells; anti-MA staining is displayed in gray, and the letter “N” indicates the location of the nucleus. (B) Gag staining patterns of JSRV and enJS56A1 M domain mutants in a representative experiment. The number on top of each bar indicates the total number of cells counted for each virus/mutant.
Cells expressing the myristoylation-defective JSRV mutant JSG2A showed a significant increase in cells with concentrated perinuclear staining compared to cells expressing wild-type JSRV (P ≤ 0.001) (Fig. 2B) (19). Mutant JSK19E/H20E also showed an increase (although not as pronounced as that of JSG2A) in cells with a concentrated phenotype compared to JSRV, but in this case the difference was not statistically significant (P ≤ 0.1). In contrast, cells expressing JSR21W (19) and JSR21E showed a phenotype similar to that of JSRV and, if anything, a relative decrease in concentrated Gag staining, further reinforcing the hypothesis that the block induced by R21 mutation is distinct from trafficking defects involving the JSRV M domain. Interestingly, cells expressing JSG22A did not show concentrated Gag staining in the perinuclear region, suggesting that this mutant might induce a defect different from those caused by both R21 and H19/K20 mutations.
Overall, these results indicate that the highly conserved R21 and G22 residues are specifically required for JSRV exit, although only the former appears to be involved in JLR. JSRV K19 and H20 appear to contribute to JSRV exit and to be part of its M domain, similar to the case for other well-characterized retroviral M domains (38).
Identification of putative JSRV L domains.
We searched for putative L domains in JSRV Gag to experimentally rule out an involvement of these domains in JLR. Tandem PSAP and PPAY domains were identified at positions 201 to 204 and 207 to 210 of Gag, respectively. Both the sequences and relative positions in JSRV Gag of these domains are analogous to those of the M-PMV L domains, although the order is inverted (16, 41). To establish whether these sequences were bona fide L domains, we first transfected 293T cells with wild-type JSRV or L domain mutants and compared the amounts of virus produced by Western blotting. L domains were mutated individually (JSΔLD201 and JSΔLD207) or in combination (JSΔLD201-7), according to studies performed on M-PMV (16).
Expression of the double mutant JSΔLD201-7 resulted in reduced virus particle production and the accumulation of intracellular Gag compared to those of wild-type JSRV, as assessed by Western blotting (Fig. 3A and B). Indeed, cells expressing JSΔLD201-7 displayed variable accumulation of Gag staining at the cell membrane by confocal microscopy (Fig. 3C). Cells expressing JSRVΔLD201, in some cases, also showed an accumulation of Gag at the cell membrane, although both the number of cells displaying this phenotype and the degree of Gag accumulation were not as pronounced as those with cells expressing JSΔLD201-7. Expression of JSΔLD207 did not result in significant accumulation of Gag staining at the membrane or in cell lysates.
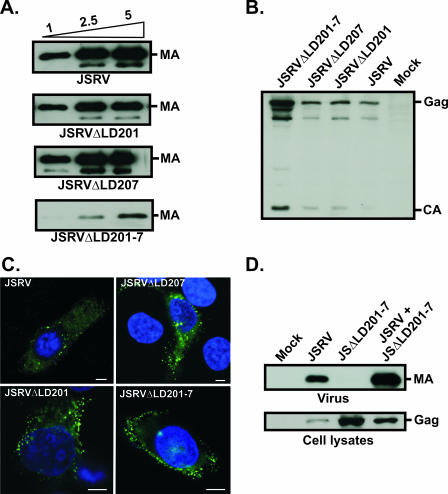
FIG. 3.
Analysis of JSRV L domains. (A) Viral pellets from cells transfected with increasing amounts of the indicated plasmids were resolved by SDS-PAGE and immunoblotted with an antiserum towards JSRV MA. The names of the transfected constructs are indicated below the panels, while the amounts of plasmid DNA transfected (in 10-cm dishes) are shown above the upper panel. (B) Lysates of cells transfected with the indicated plasmids were resolved by SDS-PAGE and immunoblotted with an antiserum towards JSRV MA. Cells expressing JSRVΔLD201-7 showed more intense bands for both immature and mature Gag, indicative of a defect in viral exit for this virus. (C) Confocal microscopy of HeLa cells expressing JSRV and JSRV L domain mutants. Anti-MA staining is displayed in green, and nuclei are shown in blue. Cells expressing JSRVΔLD201-7 show a characteristic accumulation of Gag staining at the cell membrane. Bar, 5 μm. (D) Viral pellets (upper panel) and lysates (lower panel) of cells transfected with the indicated plasmids were resolved by SDS-PAGE and immunoblotted with an antiserum towards JSRV MA. JSRVΔLD201-7 is defective for viral exit but is not transdominant over wild-type JSRV.
We ruled out the hypothesis that JLR could result from a transdominant L domain phenotype by coexpressing JSRV with JSΔLD201-7 in 293T cells. This mutant was unable to block JSRV and was further rescued by the wild-type virus (Fig. 3D). Collectively, the data presented so far suggest that the mechanisms governing JLR restriction appear to be unrelated to known signals/mechanisms of Gag trafficking.
Truncated enJS56A1 Gag mutants do not block JSRV.
Since the main determinant of JLR lies within the N-terminal region of Gag, it was of interest to determine whether enJS56A1 mutants with truncated Gag could still block JSRV release. enJS56A1 is defective and transdominant, while in general, retroviral Gag-defective mutants are rescued by homologous wild-type viruses. Therefore, a possibility was that enJS56A1 Gag could restrict JSRV without necessarily interacting with the latter but, for example, by saturating cellular factors required for Gag trafficking. If this was the case, enJS56A1 mutants with truncated Gag could still interfere with JSRV exit.
In order to design precise deletion mutants, the exact boundaries of the mature JSRV Gag protein were determined by treating sucrose gradient-purified viral preparations with 8 M guanidine-HCl, followed by RP-HPLC separation and N-terminal sequencing of the isolated peptides (L. Henderson and R. Sowder, personal communication). The results obtained suggest that JSRV Gag is cleaved into at least five products, in the following order: MA (p23)-p15-CA (p26)-NC-p4. The predicted boundaries of the mature JSRV Gag protein are indicated in Fig. 4A. These data are in agreement with previous studies of 35S-labeled JSRV virions, which revealed the presence of five polypeptides (excluding the surface and transmembrane domains of the envelope glycoprotein) with apparent molecular masses in an SDS-PAGE gel of 26, 23, 15, 10, and 5 kDa (26). We confirmed that p26 is CA, p23 is MA, and p10 is NC by Western blot analysis using a panel of specific polyclonal antisera and JSRV expression plasmids bearing HA epitopes within MA (not shown). Besides Gag, the mature surface (SU) domain of the JSRV envelope glycoprotein and cellular ubiquitin were also detected in purified viral particles. The N-terminal SU sequence started at Env amino acid residue 81 (AAFWAY…), in agreement with the previously predicted cleavage site of the leader peptide (23, 25).
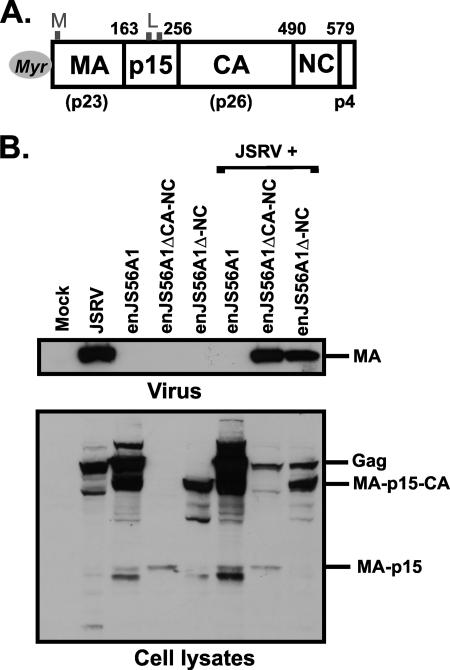
FIG. 4.
Truncated enJS56A1 Gag mutants do not block JSRV exit. (A) Organization of JSRV Gag. The names of the Gag cleavage products are displayed inside the boxes (with the exception of p4, which is indicated below). Vertical lines indicate the cleavage sites. The numbers above the bar refer to the positions of the boundaries in mature Gag of the JSRV21 infectious molecular clone (20). Myristate is represented by a gray circle. The relative positions of the L and M domains are also indicated. The apparent molecular weights of MA, CA, and NC are indicated below the bar. (B) Western blot analysis of enJS56A1 deletion mutants. Viral pellets (upper panel) and lysates (lower panel) of cells transfected with the indicated plasmids were resolved by SDS-PAGE and immunoblotted with a JSRV MA antiserum. Truncated constructs did not interfere with JSRV exit.
Truncated mutants expressing MA-p15 (enJS56A1ΔCA-NC) and MA-p15-CA (enJS56A1ΔNC) were derived by inserting a stop codon by site-directed mutagenesis at the predicted boundaries between p15 and CA and between CA and NC. As expected, cells transfected with these mutants expressed Gag proteins of lower molecular weights (Fig. 4B). Neither enJS56A1ΔCA-NC nor enJS56A1ΔNC was able to block JSRV exit, indicating that an entire enJS56A1 Gag protein is necessary for JLR. The inability of enJS56A1ΔCA-NC to block JSRV could be due to the much lower level of expression of this mutant than of enJS56A1, probably as a result of nonsense-mediated mRNA decay (2, 18). However, the expression of enJS56A1ΔNC was comparable to that of enJS56A1. To prove that the different interfering properties displayed by enJS56A1 and enJS56A1ΔNC were not due to differences in Gag expression levels, we cotransfected JSRV with either fixed amounts of enJS56A1ΔNC or serial dilutions of enJS56A1. As expected, enJS56A1 induced JLR even when its Gag expression levels were clearly lower than those of enJS56A1ΔNC (not shown).
JSRV and enJS56A1 Gag proteins colocalize.
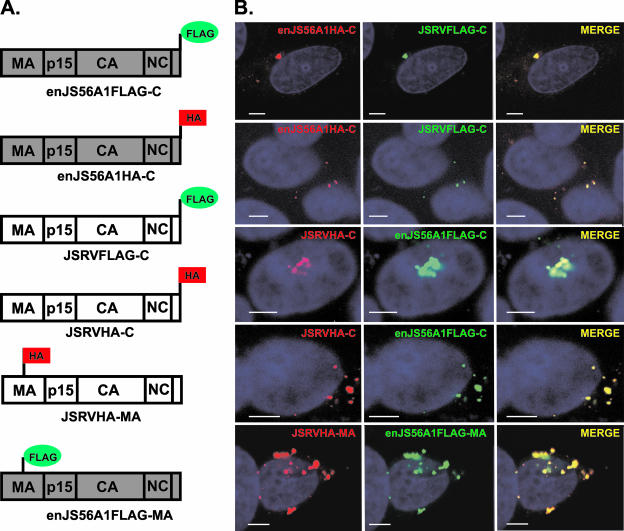
We then investigated whether enJS56A1 and JSRV Gag proteins colocalize within the cell, as this would be suggestive of a restriction mechanism occurring in trans. We differentially tagged JSRV and enJS56A1 Gag proteins by fusing the FLAG and HA epitopes at the carboxy-terminal end of Gag or within the MA domain (Fig. 5A). The phenotypes of the parental viruses were not altered by the addition of the epitopes, as all of the tagged JSRV and enJS56A1 viruses were competent for viral release and interference, respectively (not shown).
FIG. 5.
JSRV and enJS56A1 Gag proteins colocalize. (A) Schematic representation of JSRV and enJS56A1 tagged constructs. (B) Representative images of confocal microscopy of HeLa cells coexpressing either JSRVFLAG-C and enJS56A1HA-C, JSRVHA-C and enJS56A1FLAG-C, or JSRVHA-MA and enJS56A1FLAG-MA. Strong colocalization of JSRV and enJS56A1 is observed in all cases. Both dispersed and concentrated phenotypes are represented (see Results). FLAG staining is displayed in green, HA staining is shown in red, and nuclei are shown in blue. Bar, 5 μm.
By confocal microscopy, we observed strong colocalization between JSRV and enJS56A1 Gag proteins (Fig. 5B) in both cells displaying the dispersed phenotype and those displaying the concentrated phenotype. Essentially the same results were obtained with constructs where Gag was tagged at the C-terminal end or within the MA domain.
JSRV and enJS56A1 associate in trans.
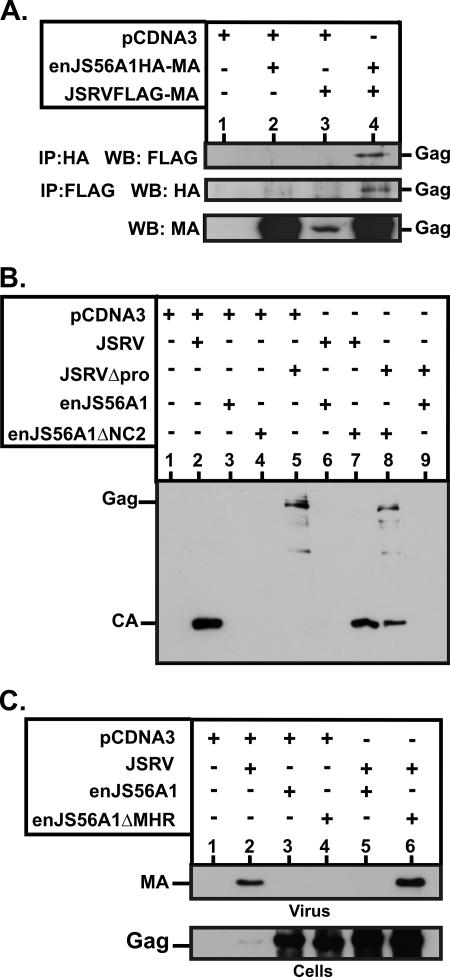
The colocalization data showed above suggest that JLR is likely due to an interaction in trans between JSRV and enJS56A1 Gag proteins. To address this point, we performed coimmunoprecipitation experiments with cells coexpressing differentially tagged JSRV and enJS56A1. As shown in Fig. 6A, JSRV and enJS56A1 Gag proteins coimmunoprecipitate, suggesting an association in trans.
FIG. 6.
Association between JSRV and enJS56A1 Gag proteins. (A) 293T cells were cotransfected with enJS56A1HA-MA and JSRVFLAG-MA. At 48 h posttransfection, cell lysates were immunoprecipitated (IP) and analyzed by SDS-PAGE/Western blotting (WB) as indicated beside each panel. Gag expression was assessed by Western blotting using a JSRV MA antiserum. enJS56A1-JSRV Gag association is evident in lysates from cells cotransfected with both viruses (lane 4). Note that the JSRV MA antiserum is a polyclonal serum and is much more sensitive in Western blotting than the anti-HA or anti-FLAG monoclonal antibody. (B) Supernatants of cells transfected with the indicated plasmids were resolved by SDS-PAGE and immunoblotted with an antiserum towards JSRV CA. Note that JSRVΔpro releases virus particles into the supernatant with an immature Gag (lane 5) because of the lack of a functional protease in this virus. Coexpression of JSRVΔpro and enJS56A1ΔNC2 (lane 8) leads to the release of viral particles with both immature and mature Gag. (C) Supernatants and lysates of cells transfected with the indicated plasmids were resolved by SDS-PAGE and immunoblotted with an antiserum towards JSRV MA. Note that enJS56A1ΔMHR is highly expressed (lane 4) but does not interfere with JSRV exit (lane 6).
Complementation assays were also performed by coexpressing JSRVΔpro and enJS56A1ΔNC2. JSRVΔpro is an exit-competent virus with a truncated, nonfunctional viral protease (19), while the defective (but not interfering) enJS56A1ΔNC2 virus contains an in-frame deletion in NC and an intact pro gene. As shown in Fig. 6B, expression of JSRVΔpro results in the release of viral particles with immature Gag only, while viral particles obtained by coexpressing JSRVΔpro with enJS56A1ΔNC2 have both immature and mature cleaved Gag proteins. Thus, JSRVΔpro rescues and coassembles with enJS56A1ΔNC2, as Gag maturation (cleavage) can happen only when the viral protease is contained within the retroviral particle (30).
Furthermore, enJS56ΔMHR, an enJS56A1 mutant deleted of the major homology region (MHR) in CA, was not able to interfere with JSRV (Fig. 6C). These data indirectly suggest that JSRV-enJS56A1 coassembly is required for JLR, given that the retroviral MHR in Gag is required for correct particle assembly (8, 29, 37).
As a whole, the data described above and our published observations of intracytoplasmic particles by electron microscopy in cells expressing enJS56A1, with or without JSRV (19), suggest that JSRV and enJS56A1 associate and, most likely, coassemble.
JSRV and enJS56A1 Gag proteins display differential intracellular targeting.
In our previous study, no major differences in Gag staining patterns were observed in cells expressing either JSRV or enJS56A1 (or both), with the exception of a higher intensity of Gag staining in enJS56A1-expressing cells (19). The staining pattern of JSRV or enJS56A1 Gag by immunofluorescence is similar to that described for cells expressing M-PMV, a betaretrovirus like JSRV (32, 33). M-PMV targets Gag to the pericentrosomal region, where it has been hypothesized to assemble, and then traffics to the cell membrane by mechanisms dependent on the recycling endosomes and the viral envelope (32, 33).
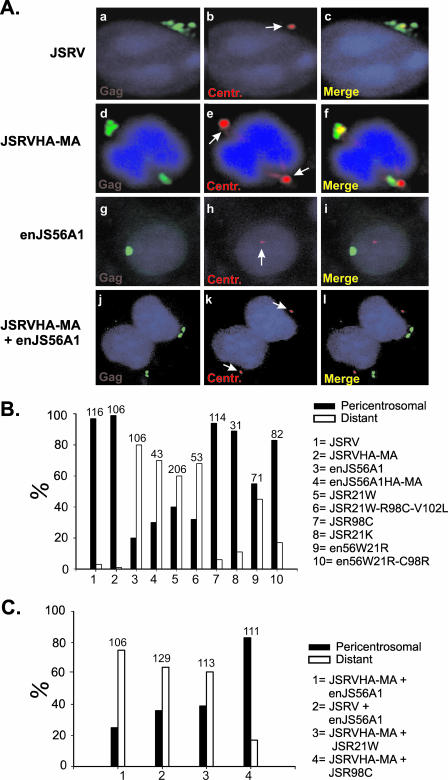
Thus, Gag targeting to the centrosome appears to be one of the first critical steps after translation in the betaretrovirus replication cycle. To begin to determine exactly the stage(s) when JLR occurs, we initially investigated whether JSRV and enJS56A1 Gag proteins concentrate in the vicinity of the centrosome. We analyzed cells transiently expressing JSRV or enJS56A1 and performed confocal microscopy using antisera towards JSRV MA and γ-tubulin (a centrosomal marker). Cells with a concentrated Gag staining pattern were then scored for the presence of Gag in the pericentrosomal area. We arbitrarily defined Gag staining within a 3-μm radius of the centrosome as “pericentrosomal,” as 97% of cells expressing concentrated JSRV Gag displayed positive staining within this area (Fig. 7A and B). In contrast, enJS56A1 showed a significantly different phenotype, with only 20% of cells having concentrated Gag staining in the pericentrosomal area (P ≤ 0.001) (Fig. 7A and B).
FIG. 7.
Intracellular targeting of JSRV and enJS56A1 Gag proteins. (A) HeLa cells were transfected with JSRV (panels a to c), JSRVHA-MA (panels d to f), or enJS56A1 (panels g to i) or cotransfected with JSRVHA-MA and enJS56A1 (panels j to l) and then stained with anti-γ-tubulin antibodies and antiserum to either JSRV MA (panels a to c and g to i) or the HA tag (panels d to f and j to l). Gag and HA staining is shown in green, while γ-tubulin staining is shown in red. Centrosomes are indicated with white arrows. (B and C) Quantification of centrosomal targeting in cells expressing concentrated Gag staining (see Results). Viruses/mutants are indicated on the right. The number on top of each bar indicates the total number of cells counted per sample.
We repeated the experiments described above, using a variety of mutants, to assess whether the primary determinants of JSRV Gag centrosome targeting corresponded to the JLR determinants. The following mutants were tested: JSR21W (defective and interfering), JSR21W-C98R-V102L (defective and interfering), JSR98C (defective but not interfering), JSR21K (defective but not interfering), and the reverse enJS56A1 mutants en56W21R and en56W21R-C98R (competent for viral exit) (19). Remarkably, only 40% of the Gag-positive cells expressing JSR21W and 32% of those expressing JSR21W-C98R-V102L displayed pericentrosomal accumulation (P ≤ 0.001), whereas the intracellular distributions of the defective-but-not-interfering mutants JSR98C and JSR21K were similar to that observed for JSRV. The revertant mutant en56A1W21R displayed 55% of cells with a concentrated phenotype in the pericentrosomal region, and the double mutant en56A1W21R/C98R displayed a phenotype more similar to that of JSRV than to that of enJS56A1 (P ≤ 0.001) (Fig. 7B). These results indicate that JSRV and enJS56A1 are targeted to different subcellular compartments: JSRV reaches the centrosome, while enJS56A1 does not appear to reach this region. The major determinant of centrosome targeting is amino acid residue R21, although residue R98 could also be an additional trafficking signal for this region.
We reasoned that the pericentrosomal localization of JSRV must be altered in the presence of enJS56A1, given the fact that both viruses colocalize when expressed within the same cell. To test this hypothesis, we used cells cotransfected with enJS56A1 and JSRV or JSRVMA-HA. By using JSRVMA-HA, we could distinguish between JSRV and enJS56A1 Gag proteins by using an anti-HA antibody. As expected, among cells cotransfected with JSRVMA-HA and enJS56A1, 75% of cells did not accumulate HA staining in the pericentrosomal area (P ≤ 0.001 compared to values obtained for cells expressing JSRVMA-HA alone) (Fig. 7A and C). Accumulation of Gag staining was also mainly distant from the centrosome in cells where wild-type JSRV (without any tag) was coexpressed with enJS56A1 (P ≤ 0.001) (Fig. 7C). Similar results were observed when JSRVMA-HA was cotransfected with the defective and interfering mutant JSR21W (P ≤ 0.001) (Fig. 7C). In contrast, when JSRVMA-HA was cotransfected with JSR98C (a defective but noninterfering mutant), the pattern of HA staining was similar to that for JSRVMA-HA alone, although there was a statistically significant increase in the number of cells with concentrated Gag staining that did not localize in the pericentrosomal area (P ≤ 0.001). Overall, these results suggest that enJS56A1 hampers the intracellular targeting of JSRV Gag to the centrosome.
DISCUSSION
Expression of enJS56A1, an endogenous sheep betaretrovirus, blocks the release of JSRV in a transdominant manner (19). The late timing of this block (referred to as JLR) is unique compared with the earlier replication steps affected by other well-characterized retroviral and cellular restriction factors (e.g., Fv1, Fv4, Trim5α, APOBEC-3G, etc.) (4, 5).
In this study, we determined that JSRV and enJS56A1 Gag proteins colocalize, associate in trans, and likely coassemble. enJS56A1 displaces the intracellular localization of JSRV Gag. More specifically, the data shown argue that JLR is due to a block in JSRV targeting to the pericentrosomal region by a mechanism independent from other previously known Gag trafficking domains.
We previously mapped the main determinant of JLR to amino acid residue R21 within JSRV Gag (19), which is a conserved residue among betaretroviruses that is mutated to tryptophan in enJS56A1. The main determinant of JSRV Gag centrosomal targeting is also residue R21. The R21W substitution is not strictly necessary for JLR, as single JSRV mutants carrying amino acid residues other than R at position 21 are incompetent for viral release and transdominant over JSRV, with the conservative mutant JSR21K being the only exception.
Thus, the block of JSRV Gag targeting to the centrosome by enJS56A1 seems to be the key event in JLR. Indeed, those JSRV mutants that are defective but still able to reach the centrosome (such as JSR98C, JSR21K, and the M and L domain mutants) do not interfere with JSRV replication. However, at the moment, we cannot rule out that the block in JSRV Gag centrosome targeting could be the consequence, rather than the cause, of JLR.
The centrosome has been hypothesized to be the site of assembly for M-PMV (another betaretrovirus). Once assembled, M-PMV particles traffic to the cell membrane by as yet uncharacterized mechanisms that require the viral envelope and recycling endosomes (32, 33). Because M-PMV appears to assemble at the centrosome, one could hypothesize that JLR results from altered assembly. Our data suggest that JSRV and enJS56A1 coassemble, considering that (i) JSRV and enJS56A1 Gag proteins colocalize and associate in trans; (ii) we observed, by electron microscopy, viral particles within the cytoplasm of cells expressing enJS56A1 or coexpressing enJS56A1 and JSRV (19); and (iii) enJS56A1 truncation mutants or mutants lacking the MHR are defective but do not block JSRV. Furthermore, in a protease complementation assay, we have shown that an enJS56A1 mutant with an in-frame deletion in NC and a functional protease (enJS56A1ΔNC2) coassembles with JSRVΔpro. It can be argued that enJS56A1ΔNC2 is not transdominant and, consequently, does not completely recapitulate the enJS56A1 phenotype. However, enJS56A1 and enJS56A1ΔNC2 possess identical CA domains and retroviral CA dictates coassembly (1). Thus, there are strong indications that JSRV and enJS56A1 Gag proteins coassemble, at least in part, and that coassembly is required for JLR.
The precise mechanisms of JLR action are not known at present. A possibility is that even a minority of enJS56A1 Gag molecules alters the overall conformation of JSRV/enJS56A1 viral particles (or multimerized Gag molecules) so that they are unable to bind cellular factors that direct them to the centrosome. In this model, enJS56A1 and JSRV Gag proteins coassemble in a noncentrosomal region where the newly formed particles are not able to reach the cell membrane.
In general, the nature of the association between any cellular protein assisting trafficking and Gag would have to be reversible or temporary. Thus, the defect of enJS56A1 could even result from its Gag protein binding cellular factors irreversibly (or more tightly than necessary). Alternatively (or additionally), enJS56A1 Gag might actively target, in a transdominant fashion, a cellular compartment where viral particles cannot reach the cell membrane and egress from the cell.
In M-PMV, a region of 18 amino acids within the MA, known as the cytoplasmic targeting-retention signal, is responsible for directing Gag molecules to the centrosome. A single mutation in this domain (R55W) abolishes centrosomal targeting, but the resulting mutant is still able to assemble at the cell membrane, like those viruses that follow the so-called C-type assembly (e.g., human immunodeficiency virus, murine leukemia viruses, etc.) (7, 31). Thus, centrosomal targeting does not appear to be absolutely necessary for betaretroviral assembly. JSRV bears a threonine in correspondence with M-PMV R55, and it has not been determined whether a cytoplasmic targeting-retention signal domain is present in this virus. JSRV R21 might be part of an equivalent domain, but if this were the case, we would expect the JSR21W mutant to display a C-type assembly and efficient exit from the cell rather than a dominant-negative phenotype.
Studies on the molecular biology of JSRV Gag are lacking, despite the vast body of knowledge of this protein in other retroviruses (9, 13). To better understand the molecular mechanisms underpinning JLR, we identified known trafficking signals and evaluated their functionality within the context of JSRV. This approach allowed us to identify putative JSRV L and M domains and to rule out their involvement in JLR.
From an evolutionary perspective, the strict requirement for R21 in JSRV release suggests that the R-to-W mutation became fixed in the sheep genome after integration. This seems feasible, since in enJS56A1 the codon TGG encodes a W, while AGG encodes an R in other enJSRV loci (23; F. Arnaud and M. Palmarini, unpublished results). Thus, a single nucleotide substitution would have been sufficient to select a provirus with dominant-negative interfering properties. enJSRVs (including enJS56A1 or similar proviruses possessing W21 in Gag) are highly expressed in the genital tract of the ewe (10, 19, 22, 23, 35). enJSRVs are also able to interfere with JSRV by receptor competition (34). enJSRVs might therefore have been selected in sheep because they protected the host against exogenous retroviral infection by a seemingly powerful two-step interference mechanism. However, interference with exogenous pathogenic retroviruses is not the only biological function of enJSRVs. Recently, we have shown that enJSRVs are absolutely required for sheep conceptus development (11, 12), further reinforcing the hypothesis that endogenous retroviruses have benefited the evolution of their host (6).
Unfortunately, there is no tissue culture system for the propagation of JSRV. Thus, our system has to rely on cells being transfected rather than infected. However, the concern related to protein overexpression is greatly reduced by our published data (19), where we have shown that enJS56A1 is still able to block JSRV exit even after transfection at a plasmid DNA ratio of 1 (enJS56A1) to 15 (JSRV). Thus, enJS56A1 blocks JSRV exit despite overexpression of the latter.
Understanding the mechanisms of JLR could inspire the design of novel antiretroviral strategies and shed light on early events in retroviral assembly and/or trafficking. In addition, this unique viral block provides additional clues to the variety of mechanisms shaping coevolution of endogenous/exogenous retroviruses and their hosts.
Acknowledgments
We thank Alan Rein, Mariana Varela, Manuela Mura, and Karen Blyth for advice and Lou Henderson and Ray Sowder II (NCI-SAIC AIDS Vaccine Program) for HPLC analysis.
This work was funded by a program grant from the Wellcome Trust. HPLC analysis was funded by the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. M.P. is a Wolfson-Royal Society research merit awardee.
Footnotes
Published ahead of print on 29 November 2006.
REFERENCES
- 1.Ako-Adjei, D., M. C. Johnson, and V. M. Vogt. 2005. The retroviral capsid domain dictates virion size, morphology, and coassembly of Gag into virus-like particles. J. Virol. 79:13463-13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amrani, N., M. S. Sachs, and A. Jacobson. 2006. Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell. Biol. 7:415-425. [DOI] [PubMed] [Google Scholar]
- 3.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109-1115. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11:286-291. [DOI] [PubMed] [Google Scholar]
- 6.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses and the evolution of retroelements, p. 343-436. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- 7.Choi, G., S. Park, B. Choi, S. Hong, J. Lee, E. Hunter, and S. S. Rhee. 1999. Identification of a cytoplasmic targeting/retention signal in a retroviral Gag polyprotein. J. Virol. 73:5431-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 10.Dunlap, K. A., M. Palmarini, D. L. Adelson, and T. E. Spencer. 2005. Sheep endogenous betaretroviruses (enJSRVs) and the hyaluronidase 2 (HYAL2) receptor in the ovine uterus and conceptus. Biol. Reprod. 73:271-279. [DOI] [PubMed] [Google Scholar]
- 11.Dunlap, K. A., M. Palmarini, and T. E. Spencer. 2006. Ovine endogenous betaretroviruses (enJSRVs) and placental morphogenesis. Placenta 27(Suppl. A):S135-S140. [DOI] [PubMed] [Google Scholar]
- 12.Dunlap, K. A., M. Palmarini, M. Varela, R. C. Burghardt, K. Hayashy, J. L. Farmer, and T. E. Spencer. 2006. Endogenous retroviruses regulate peri-implantation conceptus growth and differentiation. Proc. Natl. Acad. Sci. USA 103:14390-14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 14.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gifford, R., and M. Tristem. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291-315. [DOI] [PubMed] [Google Scholar]
- 16.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson, L. E., M. A. Bowers, R. C. Sowder II, S. A. Serabyn, D. G. Johnson, J. W. Bess, Jr., L. O. Arthur, D. K. Bryant, and C. Fenselau. 1992. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processing, and complete amino acid sequences. J. Virol. 66:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeBlanc, J. J., and K. L. Beemon. 2004. Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J. Virol. 78:5139-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mura, M., P. Murcia, M. Caporale, T. E. Spencer, K. Nagashima, A. Rein, and M. Palmarini. 2004. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc. Natl. Acad. Sci. USA 101:11117-11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ott, D. E., E. N. Chertova, L. K. Busch, L. V. Coren, T. D. Gagliardi, and D. G. Johnson. 1999. Mutational analysis of the hydrophobic tail of the human immunodeficiency virus type 1 p6(Gag) protein produces a mutant that fails to package its envelope protein. J. Virol. 73:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmarini, M., C. A. Gray, K. Carpenter, H. Fan, F. W. Bazer, and T. E. Spencer. 2001. Expression of endogenous betaretroviruses in the ovine uterus: effects of neonatal age, estrous cycle, pregnancy, and progesterone. J. Virol. 75:11319-11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmarini, M., C. Hallwirth, D. York, C. Murgia, T. de Oliveira, T. Spencer, and H. Fan. 2000. Molecular cloning and functional analysis of three type D endogenous retroviruses of sheep reveal a different cell tropism from that of the highly related exogenous Jaagsiekte sheep retrovirus. J. Virol. 74:8065-8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmarini, M., M. Mura, and T. E. Spencer. 2004. Endogenous betaretroviruses of sheep: teaching new lessons in retroviral interference and adaptation. J. Gen. Virol. 85:1-13. [DOI] [PubMed] [Google Scholar]
- 25.Palmarini, M., J. M. Sharp, M. De las Heras, and H. Fan. 1999. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J. Virol. 73:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmarini, M., J. M. Sharp, C. Lee, and H. Fan. 1999. In vitro infection of ovine cell lines by Jaagsiekte sheep retrovirus. J. Virol. 73:10070-10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parent, L. J., C. B. Wilson, M. D. Resh, and J. W. Wills. 1996. Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J. Virol. 70:1016-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Provitera, P., A. Goff, A. Harenberg, F. Bouamr, C. Carter, and S. Scarlata. 2001. Role of the major homology region in assembly of HIV-1 Gag. Biochemistry 40:5565-5572. [DOI] [PubMed] [Google Scholar]
- 30.Rabson, A. B., and B. J. Graves. 1997. Synthesis and processing of viral RNA, p. 205-261. In J. M. Coffin, S. H. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 31.Rhee, S. S., and E. Hunter. 1990. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell 63:77-86. [DOI] [PubMed] [Google Scholar]
- 32.Sfakianos, J. N., and E. Hunter. 2003. M-PMV capsid transport is mediated by Env/Gag interactions at the pericentriolar recycling endosome. Traffic 4:671-680. [DOI] [PubMed] [Google Scholar]
- 33.Sfakianos, J. N., R. A. LaCasse, and E. Hunter. 2003. The M-PMV cytoplasmic targeting-retention signal directs nascent Gag polypeptides to a pericentriolar region of the cell. Traffic 4:660-670. [DOI] [PubMed] [Google Scholar]
- 34.Spencer, T. E., M. Mura, C. A. Gray, P. J. Griebel, and M. Palmarini. 2003. Receptor usage and fetal expression of ovine endogenous betaretroviruses: implications for coevolution of endogenous and exogenous retroviruses. J. Virol. 77:749-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer, T. E., A. G. Stagg, M. M. Joyce, G. Jenster, C. G. Wood, F. W. Bazer, A. A. Wiley, and F. F. Bartol. 1999. Discovery and characterization of endometrial epithelial messenger ribonucleic acids using the ovine uterine gland knockout model. Endocrinology 140:4070-4080. [DOI] [PubMed] [Google Scholar]
- 36.Stoye, J. P. 2001. Endogenous retroviruses: still active after all these years? Curr. Biol. 11:R914-R916. [DOI] [PubMed] [Google Scholar]
- 37.Strambio-de-Castillia, C., and E. Hunter. 1992. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J. Virol. 66:7021-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-364. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varela, M., Y. H. Chow, C. Sturkie, P. Murcia, and M. Palmarini. 2006. Association of RON tyrosine kinase with the Jaagsiekte sheep retrovirus envelope glycoprotein. Virology 350:347-357. [DOI] [PubMed] [Google Scholar]
- 41.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]