Abstract
Chronic hepatitis C virus (HCV) infection affects more than 170 million persons worldwide and is responsible for the development of liver cirrhosis in many cases. Standard treatment with pegylated alpha interferon (IFN-α) in combination with the nucleoside analogue ribavirin leads to a sustained virologic response in approximately half of the patients. IFN-α is classified as an indirect treatment, as it interacts with the host's immune response. The mechanism of action of ribavirin is still unknown. The benefit of triple therapy by adding other antiviral agents, e.g., amantadine, is controversial. Currently, new direct antiviral drugs (HCV protease/polymerase inhibitors) are being evaluated in phase 1/phase 2 trials. Phenotypic resistance to antiviral therapy has been attributed to amino acid variations within distinct regions of the HCV polyprotein. While sensitivity to IFN-α-based antiviral therapy in vivo is clearly correlated with the number of mutations within the HCV NS5A protein, the underlying functional mechanisms for this association are unknown. In turn, in vitro, several mechanisms to circumvent the host immune defense or to block treatment-induced antiviral activities have been described for different HCV proteins. By the introduction of direct antiviral drugs, hepatitis C therapy now is entering a new era in which the development of resistance may become the most important parameter for treatment success or failure.
INTRODUCTION
Hepatitis C virus (HCV) is an enveloped, approximately 9.6-kb, positive-sense, single-stranded RNA virus and is classified in the family Flaviviridae (17). After translation from a large open reading frame, the polyprotein precursor is cleaved by viral and host peptidases, resulting in three structural proteins, termed core, envelope 1 (E1), and E2, and a protein named p7, and six nonstructural (NS) proteins, termed NS2, NS3, NS4A, NS4B, NS5A, and NS5B. The F protein has been described as being a result of a ribosomal frameshift of the core-encoding genomic region (149). There is a noncoding region of 324 to 341 nucleotides at the 5′ end containing the internal ribosome entry site (IRES) and a 3′ noncoding region of variable length.
According to estimations of the WHO, over 170 million individuals worldwide are infected with HCV (148). Chronic HCV infection is responsible for inflammation of the liver, and ∼20% of patients progress to liver cirrhosis with an increased risk for the development of hepatocellular carcinoma (148). The current standard treatment for patients with chronic hepatitis C consists of pegylated alpha interferon (IFN-α) (PEG-IFN) in combination with the nucleoside analogue ribavirin for 24 to 48 weeks and leads to a sustained virologic response in 54 to 56% of cases (38, 83). Sustained virologic response is defined as undetectable HCV RNA by a sensitive assay (lower detection limit of <50 IU/ml) at the end of a 24-week follow-up period after the end of treatment. Patients who do not achieve a sustained virologic response may be found to be HCV RNA negative during therapy but may relapse thereafter or may be virologic nonresponders showing detectable HCV RNA levels throughout the complete treatment period.
Virologic response rates have been shown to depend on various host and viral factors such as age, weight, sex, race, liver enzymes, stage of fibrosis, HCV genotype, and HCV RNA concentration at baseline (9, 38, 50, 62, 83, 94, 147a). To further improve sustained virologic response rates, different treatment approaches are currently under investigation. For example, individualized therapy durations on the basis of the HCV RNA concentration at baseline and early during therapy are the subjects of clinical studies. In addition, triple therapy with the addition of the antiviral drug amantadine to IFN-α and ribavirin has been evaluated in multiple studies, leading to contradictory results (7, 21, 82). Moreover, multiple new substances with different modes of action are being studied in ongoing clinical trials (e.g., albuferon, amantadine, Toll-like receptor [TLR] agonists, and viramidine). So far, none of these drugs has been established as a standard treatment for hepatitis C.
Most promising, results of phase 1/phase 2 clinical trials with direct antiviral drugs such as inhibitors of HCV-specific NS3 protease and HCV NS5B RNA-dependent RNA polymerase (RdRp) have recently been presented at international meetings (60, 112a, 115, 120a, 125a, 155a).
Resistance mechanisms that might explain how the virus may circumvent the antiviral actions of IFN-α, ribavirin, amantadine, and, most recently, new direct antiviral drugs have been proposed in vitro and in vivo (20, 36, 41, 49, 70, 74, 139). This review discusses recent achievements of HCV treatment resistance mechanisms based on in vitro experiments and clinical data.
MECHANISMS OF RESISTANCE TO IFN-α
IFN-α-Based Antiviral Therapy
Considerable improvements in the treatment of patients infected with chronic hepatitis C were achieved by the introduction of IFN-α in its pegylated form and by the combination with the nucleoside analogue ribavirin. Remarkably, the response rates of IFN-α-based therapy are influenced mainly by the HCV genotype. Due to the lack of a proofreading activity of the HCV RNA-dependent RNA polymerase, different HCV genomes have evolved and are classified into six genotypes (130). The different genotypes can be further assigned to subtypes with homologies of approximately 80% (131). Within a given individual, a large number of closely related viral variants, called HCV quasispecies, circulate. With all IFN-α-based treatment schedules, HCV genotype 1-infected patients achieve much lower sustained rates of response (42 to 52%) to antiviral therapy than those infected with HCV genotypes 2 and 3 (78 to 86%) (38, 50, 83, 153). For patients infected with HCV genotypes 4, 5, and 6, only limited data are available, but sustained response rates seem to be slightly higher than those for HCV genotype 1 (50 to 77%) (38, 83). No significantly different virologic response rates between HCV subtypes were reported (e.g., subtypes 1a and 1b).
The underlying mechanisms for the varying virologic response rates to IFN-α-based antiviral therapy between the different HCV genotypes are unknown. However, as response rates are detected independently of any host or treatment factors, many studies focused on virus-encoded mechanisms of resistance to IFN-α-based therapy.
IFN-α Signal Transduction
IFNs are a family of cytokines belonging to the host's natural immune response to various stimuli, particularly viral infection. After binding to their specific receptors on the target cell surface, an intracellular signaling cascade is activated, which leads to the upregulation of IFN-stimulated genes (ISGs), resulting in the expression of multiple antiviral effector proteins. The best-understood IFN-mediated signaling and transcriptional activation pathway is called the Jak/signal transducer and activators of transcription (STAT) pathway (Fig. 1). Thereby, two cytoplasmic protein tyrosine kinases, Tyk2 and Jak1, belonging to the Janus kinase family phosphorylate tyrosine residues of the IFN receptor that function as docking sites for STATs. The subsequent phosphorylation of the receptor-recruited STATs by the Jak kinases promotes the formation of heterodimers between STAT1 and STAT2, which further binds to a third protein, p48, to form the ISG factor 3 (ISGF3) complex. This complex is translocated to the cell nucleus, where it binds to the IFN-stimulated response element in IFN-stimulated gene promoters, leading to the expression of IFN effector proteins. HCV cell-based expression models show inhibition of IFN-α-induced signal transduction through the Jak/STAT pathway (22, 26, 55, 76). Among all ISGs, the best characterized effector proteins are the double-stranded RNA-activated protein kinase (PKR) (41), the 2′,5′-oligoadenylate synthetase (92), and the Mx proteins (134). Besides its antiviral activities, IFN-α also affects processes that regulate cell growth and the modulation of the immune response.
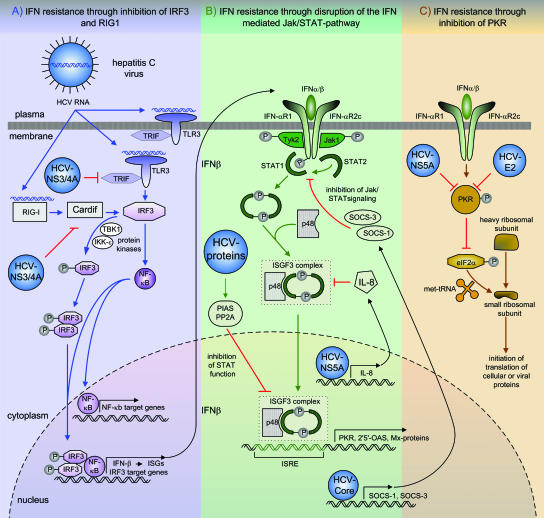
FIG. 1.
Potential strategies of the different HCV proteins to antagonize IFN therapy. (A) IFN resistance through inhibition of RIG-I and TLR3 by the HCV NS3/4A protein. Viral RNA binding to RIG-I and TLR3 leads to the activation of IRF3 and NF-κB. Both transcription factors enter the nucleus and bind to target genes, resulting in the expression of antiviral defense proteins. The NS3/4A protein disrupts the activation of IRF3 and NF-κB by the inactivation of Cardif, which is an adaptor protein in the RIG-I antiviral pathway, and through inhibition of Trif, an adaptor molecule in the TLR3 signaling pathway. (B) Disruption of the IFN-mediated Jak/STAT pathway. IFN binding to its receptor triggers the autophosphorylation of tyrosine residues through the Tyk2 and Jak1 kinases, which function as docking sites for STAT1 and STAT2. The subsequent phosphorylation of STAT1 and STAT2 promotes the formation of heterodimers between STAT1 and STAT2 and the assembly of the trimeric ISGF3 complex. After translocation of the ISGF3 complex to the cell nucleus, it binds to the IFN-stimulated response element (ISRE) in ISG promoters, leading to the expression of IFN effector proteins. The HCV core protein induces the expression of SOCs, which block Jak/STAT signaling through the IFN-α/β receptor. The HCV NS5A protein induces the expression of IL-8, which blocks the assembly and function of the ISGF3 complex. The expression of the whole HCV polyprotein inhibits STAT1 function through elevated levels of PP2A, which is associated with an increased binding of STAT1 to PIAS and a reduced activation of ISGs. (C) IFN resistance through inhibition of PKR by the HCV E2 and HCV NS5A proteins. After IFN-α binding, autophosphorylation of PKR takes place, leading to the inhibition of the phosphorylation of eIF-2α, which is responsible for the initiation of translation of cellular and viral proteins by facilitating Met-tRNA binding to the 40S ribosomal subunit. HCV E2 and HCV NS5A antagonize the effects of IFN-α by the inhibition of PKR.
Approaches to In Vitro and In Vivo Studies of IFN-α Resistance
So far, several virus-encoded factors have been suggested to be responsible for the inhibition of the antiviral effects of IFN-α (39). The in vitro studies are based mainly on the recombinant expression of the whole HCV open reading frame or on the expression of individual HCV genes. The expression of the whole HCV polyprotein has been shown to inhibit the STAT1 function in the context of elevated levels of protein phosphatase 2A (PP2A). Overexpression of the catalytic subunit of PP2A resulted in the hypomethylation of STAT1, leading to the increased binding of a protein inhibitor of activated STAT, called PIAS. Increased binding of STAT1 to PIAS leads to reduced activation of ISGs (10, 26, 55). For the HCV core, the envelope protein E2, the serine protease NS3/4A, and the NS5A protein, different mechanisms were suggested to contribute to antiviral resistance (11, 36, 41, 136, 139).
Additionally, different HCV replicon systems have been utilized to study IFN-α resistance (77, 89). The HCV replicon system is a well-defined and robust cell culture system for the replication of the nonstructural HCV genes. By definition, replicons are molecules that are capable of self-amplification. In the case of HCV, the first-generation replicons were derived from a cloned HCV genome by deleting the structural region and inserting a selectable marker (neomycin phosphotransferase, conferring G418 resistance). A second IRES element (encephalomyocarditis virus promoter) is introduced to allow the translation of the HCV nonstructural region. Transfection of synthetic RNAs derived from such a construct into the human hepatoma cell line HuH7 and G418 selection lead to cell lines carrying autonomously replicating HCV subgenomic RNA (77). By the development of HCV replicon systems, an additional tool to study interactions of the HCV proteins with the host cellular machinery is available.
However, the translation of these in vitro experiments to the situation in clinical practice has been difficult. For example, the HCV isolates used in various HCV replicon systems are not clinically characterized. So far, only HCV replicon systems harboring genotype 1a/1b genomes and a recent HCV replicon system harboring the HCV genotype 2a genome of a patient suffering from acute fulminant hepatitis C were developed. Moreover, selection of HCV replicon systems with resistance to IFN-α in vitro has not been successful so far. In this context, it would be worthwhile to construct HCV replicon systems with HCV isolates derived from patients with different sensitivities to IFN-α-based therapy.
As a consequence, the in vivo analysis of HCV treatment resistance mechanisms are restricted to cloning and sequencing approaches for HCV isolates derived from patient sera with known virologic responses to antiviral therapy. Today, the amino acid variabilities of HCV core protein and E2, NS3/4A, and NS5A proteins have been studied in correlation with the virologic treatment response.
HCV Core and IFN-α Resistance
The HCV core protein was shown to induce the expression of suppressor of cytokine signaling 3 (SOCS-3) and SOCS-1 in cultured cells, which in turn may antagonize IFN-α action by blocking the Jak/STAT pathway (11) (Fig. 1). Sequence analysis of the HCV core gene in patients with chronic hepatitis C and different sensitivities to IFN-α-based therapy has not been published so far. However, in one study, the complete open reading frame of HCV was sequenced to determine the genetic basis of resistance to IFN-α (29). A comparison of the pairs of IFN-α-resistant and IFN-α-sensitive HCV isolates from three patients infected with genotype 1b revealed only two amino acid differences within the core region in two of three patients at different codons. In conclusion, genetic variations of the highly conserved HCV core protein seem not to be relevant in determining IFN-α sensitivity in chronic hepatitis C (Table 1).
TABLE 1.
Clinical significance of HCV mutations for sensitivity/resistance to antiviral therapy
| Antiviral and HCV protein | Region analyzed | Importance of amino acid mutations | Reference(s) |
|---|---|---|---|
| IFN-α | |||
| HCV core | HCV complete open reading frame | No correlation of mutations with virologic treatment response in patients with HCV genotype 1 infection | 29 |
| HCV E2 | CD81-binding sites | No correlation of mutations with virologic treatment response in patients infected with HCV genotypes 1b and 3a | 61 |
| PePHD | No significant correlation of mutations with virologic treatment response in genotype 1- and genotype 3-infected patients | 8, 42, 125, 147, 150 | |
| For patients infected with HCV genotype 2, conflicting data about a correlation of viral PePHD variants and treatment response exist | 116, 147 | ||
| HVR1/E2 | Correlation of quasispecies heterogeneity with response to antiviral therapy | 1, 15, 32, 123 | |
| HCV NS3 | Complete NS3 gene | No correlation of mutations in patients infected with genotype 1 with sustained virologic treatment response | 120 |
| HCV NS5A | ISDR | High no. of mutations within the ISDR in patients infected with HCV genotypes 1a/1b and 2 correlates with treatment response | 16, 29, 30, 69, 102, 104, 117, 121, 122 |
| Complete NS5A gene | High no. of mutations within the complete NS5A sequence correlates with sustained virologic treatment response | 124 | |
| Ribavirin | |||
| HCV NS5B | Complete NS5B gene | F415Y mutation conferring resistance to ribavirin in HCV replicon system; larger trials are needed to assess prevalence and relevance in vivo | 151 |
| HCV NS5A | Complete NS5A gene | G404S and E442G mutations conferring resistance to ribavirin in HCV replicon system; in vivo relevance is unknown | 107 |
| Amantadine, HCV p7 | Complete p7 gene | No correlation of amino acid variations with virologic treatment response in patients with genotype 1 infection | 87 |
E2 and IFN-α Resistance
CD81-binding sites/hypervariable region 2.
The human CD81 receptor has been proposed to be a candidate receptor for HCV. The E2 glycoprotein is involved in virus entry, and two conformational CD81-binding sites located in the carboxy-terminal part of the E2 protein that overlap with parts of hypervariable region 2 (HVR2) were described (108). It was hypothesized that various capabilities for the binding of the E2 protein to CD81 may depend on amino acid substitutions within the CD81-binding sites, and HVR2 and may thus contribute to different treatment outcomes. However, sequencing of the putative CD81-binding sites within the HCV E2 gene showed no association between specific amino acid variations and the HCV RNA concentration or response to IFN-α-based therapy (61). Thus, given the uncertainty for the predicted CD81-binding sites and the negative results of that study, at present, no importance of amino acid variability within the E2 protein for the CD81 receptor interaction in correlation with the response to IFN-α-based therapy could be shown.
Hypervariable region 1.
The HCV amino acid quasispecies heterogeneity within HVR1 of the E2 protein has been correlated with the virologic response to antiviral therapy in multiple studies (Table 1). While a lower HCV quasispecies heterogeneity was associated with spontaneous recovery in patients with acute hepatitis C (31), in patients with chronic hepatitis C, conflicting data concerning the significance of HVR1 quasispecies diversity and complexity in correlation with virologic response were published. Many studies showed that a higher degree of variability in the HVR1 quasispecies correlates with lower response rates to IFN-α treatment (46, 59, 63, 68, 90, 96, 100, 105, 128). However, no such correlation was observed in other studies (72, 78, 97, 119). The discrepancies of these results may be related to (i) HCV genotype distribution in different studies, (ii) methodologies to assess quasispecies heterogeneity (i.e., band shift assays or sequencing), (iii) different assessments and quantifications of quasispecies heterogeneity, and/or (iv) different definitions of treatment outcome. However, in more recent studies using extensive cloning and sequencing approaches, in patients with chronic hepatitis C, a lower HVR1 heterogeneity of HCV quasispecies before antiviral therapy and changes in HCV E2 quasispecies early during therapy also seem to be associated with higher rates of virologic response (1, 15, 32, 123). The lower heterogeneity of HCV quasispecies in patients with a subsequent sustained virologic response to IFN-α-based therapy may indicate a more effective control of the emergence of viral variants by the immune system during successful treatment. Interestingly, this seems to be restricted to neutralizing antibodies directed towards HVR1, while within other regions (e.g., NS5A), a higher number of amino acid variations in the HCV quasispecies is associated with the virologic response to IFN-α-based therapy (see below).
PKR-eIF-2α phosphorylation homology domain.
The HCV E2 glycoprotein was reported to be involved in mediating IFN-α resistance through the inhibition of PKR (140) (Fig. 1). PKR is a primary regulator of the IFN-α-induced antiviral and antiproliferative effects of the host cell. Once activated by IFN-α, autophosphorylation of PKR takes place and inhibits the phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF-2α), which is responsible for facilitating Met-tRNA binding to the 40S ribosomal subunit (118). This results in the decreased translation of cellular or viral proteins (54). A sequence of 12 amino acids within the E2 gene that displays high homology to several autophosphorylation sites within the regulatory domain of the PKR and to its natural substrate, eIF-2α, which is called the PKR-eIF-2α phosphorylation homology domain (PePHD), was identified. Within the E2 protein of HCV subtype 1a/1b isolates, inhibition of PKR was demonstrated through interactions with the PePHD by abolishing its kinase activity and blocking its inhibitory effect on viral protein synthesis in vitro (139, 140). Interestingly, the PePHD of the most resistant HCV subtype, subtype 1a/1b, shares higher homology with the PKR autophosphorylation sites than genotypes 2 and 3, suggesting that the interaction of the PePHD with PKR specifically enhances resistance to antiviral therapy of HCV genotype 1 isolates compared with genotypes 2 and 3. Several studies that aimed to investigate whether the PePHD shows a correlation between mutation rates and treatment responses could not observe significant mutations during IFN-α treatment for isolates of HCV genotypes 1a/b and 3a (8, 42, 125, 147, 150). For HCV genotype 1, the PePHD region has turned out to be highly conserved, and no clinical relationship between the amino acid sequence of the PePHD and the outcome of IFN-α therapy could be demonstrated. For genotypes 2a and 2b, conflicting data about a putative correlation of PePHD polymorphism and IFN-α resistance exist (116, 147) (Table 1). Taken together, the potential E2 PePHD interaction with PKR may explain general differences in the IFN-α sensitivities of HCV isolates of different HCV genotypes (genotype 1 versus genotypes 2 and 3) but is highly conserved in HCV isolates from patients without an individual correlation to the outcome of IFN-α-based therapy. Investigations with the recently established full-length HCV cell culture system with E2 genes of different genotypes may further resolve the relevance of the PePHD/PKR interaction (145).
NS3/4A and IFN-α Resistance
Inhibition of the innate immune system.
Expression of the HCV NS3/4A serine protease was associated with diminished phosphorylation and nuclear translocation of IFN regulatory factor 3 (IRF3), leading to the reduced expression of IFN-induced genes (36) (Fig. 1). IRF3 functions as a key activator of the ISGs that limits virus replication at multiple points within the replicative cycle. Among these genes, ISG56 has been identified as being a suppressor of HCV translation through the disruption of viral IRES function independently of the PKR pathway (47, 135, 146). NS3/4A was shown to evade the early immune response by the disruption of the retinoic acid-inducible gene I (RIG-I) signaling pathway (35). Normally, this pathway is induced by virus-produced double-stranded RNA, resulting in the activation of type I IFN, by phosphorylation and activation of preexisting transcription factors such as NF-κB and IRF3 (151). Recently, a new adapter protein in the RIG-I pathway, called Cardif, was identified. Cardif was shown to be targeted and inactivated by the HCV NS3/4A protease (85). The other described strategy of NS3/4A, to block the expression of defense genes, is based on binding to TLR3. TLR3 functions as a signaling receptor for extracellular and intracellular double-stranded RNA (2) and transmits signals to activate the type I IFN response through its adaptor molecule, called Trif (also known as TICAM-1). Trif functions as a linkage between TLR3 and the kinases responsible for activating IRF3 and NF-κB. NS3/4A mediates the cleavage of this adaptor protein, resulting in the inhibition of the antiviral response (73) (Fig. 1).
NS3 protease sequence analysis.
Sequence analysis of the NS3 gene derived from patients infected with HCV genotype 1 revealed no significance of the NS3 protein variability in correlation with the response to IFN-α-based therapy (Table 1). Overall, the amino acid sequence within the NS3 protein as part of the NS3/4A serine protease complex was highly conserved (120). In the HCV replicon system, several mutations within the NS3 gene that lead to enhanced replication efficiency were detected. Interestingly, in HCV isolates from patients harboring identical amino acid substitutions in the NS3 protein (R1283G, P1112R, and S1496M), a slower decrease of HCV RNA during IFN-α-based therapy was observed (120). To confirm these findings and to address the overall importance of NS3 protein variability with regard to HCV replication efficiency and sensitivity to IFN-α, a phenotypic evaluation should be performed using the HCV replicon assay before any conclusions regarding the importance for the treatment of patients with chronic hepatitis C can be drawn.
NS5A and IFN-α Resistance
ISDR, complete NS5A region, and V3 region.
The first data to support a significant role for amino acid variations within the HCV NS5A protein and the response to IFN-α-based therapy came from Japan. In 1995 and 1996, a correlation of a high number of mutations in a so-called IFN-α sensitivity-determining region (ISDR) comprising 40 amino acids within the carboxy-terminal part of the NS5A protein and sustained virologic response to IFN-α therapy in HCV genotype 1b-infected patients was described by Enomoto et al. (29, 30) (Table 1). In the first report, all patients with at least four mutations within the ISDR achieved a sustained virologic response in comparison to a prototype reference sequence. In addition, an inverse correlation between an increasing number of mutations within the ISDR and the HCV RNA concentration at baseline was observed (30). Subsequently, the correlation of ISDR mutations with treatment response was investigated by different groups in Japan, Europe, and the United States. Studies from Japan were generally able to confirm the strong correlation between ISDR mutations and treatment response (16, 69). Groups from Europe and the United States were initially not able to verify the importance of ISDR mutations in IFN-α sensitivity (18, 65, 84, 99, 133, 154), and in subsequent studies, it turned out that HCV genotype 1b isolates with multiple mutations within the ISDR are rare in Europe and the United States compared to those found in Japan (104, 117, 121, 122). In further studies with larger patient cohorts and in a meta-analysis, the positive correlation of an increasing number of ISDR mutations with a sustained virologic response to IFN-α-based therapy could be confirmed (102, 104, 117, 121, 122). After reports of the existence of a PKR-binding domain within the NS5A protein comprising the ISDR in the amino-terminal part (40, 41), the clinical importance of mutations within this PKR-binding domain was investigated in additional studies. However, in several studies, no specific mutation within the carboxy-terminal part of the PKR-binding domain in correlation with IFN-α treatment response was observed (43, 93, 124). Furthermore, in in vitro studies of the expression of the complete HCV open reading frame, no colocalization of NS5A and PKR could be demonstrated (37).
To summarize the results of the different studies from Western countries, in the majority of patients with a sustained virologic response to IFN-α-based therapy, no or only a small number of mutations within the ISDR were observed. Thus, for these patients, sensitivity to IFN-α cannot be explained by the ISDR hypothesis. However, in several in vitro studies of the cellular expression of clinically characterized NS5A genes, it was shown that the NS5A protein, independently of the presence of ISDR mutations, was able to lower the protection of previously IFN-α-induced cells against infection with a surrogate virus (111, 132).
Therefore, mutational analysis by sequencing the entire NS5A gene was performed to assess the significance of NS5A mutations outside the ISDR/PKR binding domain (14, 28, 98, 124). In a study that reported the analysis of 70 full-length NS5A sequences, the overall number of mutations within the NS5A protein was highly correlated with the treatment response (124) (Table 1). Furthermore, in three studies, a local accumulation of mutations around the so-called variable region 3 (V3) within the carboxy-terminal part of the NS5A protein was found to correlate with the treatment response in HCV genotype 1a/1b isolates, which is currently under further investigation (14, 98, 124). Furthermore, it must be assumed that additional mutations outside the NS5A gene but within other genes of the HCV open reading frame are important for determinations of IFN-α sensitivity. For example, the isolated replacement of the NS5A gene in the HCV replicon system with sequences from clinical responder and nonresponder patients did not change the in vitro sensitivity to IFN-α (6).
For HCV genotype 2 isolates, a correlation between the number of mutations within the ISDR and treatment response was also observed (67, 91), while for HCV genotype 3a isolates, no such association was reported for relatively small patient cohorts reported in several studies (117, 125).
Interaction with interleukin-8 and the 2′,5′-oligoadenylate synthetase system.
Since a direct interaction of PKR with the NS5A protein has not convincingly been demonstrated so far (44), other mechanisms by which the NS5A protein could inhibit the action of IFN-α were investigated. Recently, the expression of the NS5A protein was associated with the induction of interleukin-8 (IL-8) and the subsequent inhibition of IFN-α activity in vitro (64, 109, 110) (Fig. 1). Interestingly, in patients with HCV genotype 1 infection and with a sustained virologic response to IFN-α-based therapy, levels of IL-8 in blood that were significantly lower than those of virologic nonresponders were also observed (88).
Aside from the above-described intracellular interactions of the HCV NS5A protein, multiple studies on numerous modulations of cell processes by the NS5A protein were published (4). Recently, the structure of an N-terminal zinc-binding domain of the NS5A with protein, RNA, and membrane interaction sites was described (141). Overall, the NS5A protein seems to be a promiscuous protein with the function of an important regulator of HCV replication and multiple intracellular interactions.
Furthermore, an interaction of the NS5A protein with the 2′,5′-oligoadenylate synthetase leading to the inhibition of the antiviral activity of IFN-α in vitro was described. However, the HCV core protein seems rather to activate the 2′,5′-oligoadenylate synthetase system (95, 136). Activation of 2′,5′-oligoadenylate synthetase leads to the cleavage of single-stranded UA and UU dinucleotides by RNase L, and differences in the frequencies of UA and UU dinucleotides between full-length prototype of HCV genotype 1 and those of genotypes 2 and 3, which may explain general differences in the IFN-α sensitivities between these genotypes, were described (51, 52). 2′,5′-Oligoadenylate synthetase serum levels were not associated with virologic responses in patients with chronic hepatitis C (92), whereas a specific polymorphism within the 2′,5′-oligoadenylate synthetase gene was associated with the outcome of antiviral therapy (66). Nothing is known about the frequency of UA and UU dinucleotides before and during IFN-α-based therapy in correlation with HCV RNA clearance.
Summary of NS5A and IFN-α resistance.
Taken together, the highest grade of clinical evidence suggests that mutations within the ISDR of the NS5A protein are involved in sensitivity/resistance to IFN-α-based therapy in HCV genotype 1b-infected patients. A meta-analysis of 28 studies from Japan, Europe, and the United States clearly showed the correlation between an increasing number of mutations within the ISDR and an increased probability of a sustained virologic response to IFN-α-based therapy on the basis of 1,230 isolates from single patients. Since an inverse correlation between the number of ISDR mutations with HCV RNA serum concentrations was also shown in several of these studies, it is likely that NS5A-mediated regulations of replication efficiency may explain the clinical correlation with treatment response. As for the other HCV proteins, in vitro analysis of HCV viral variants with defined mutational patterns must be performed to confirm this hypothesis.
MECHANISMS OF RESISTANCE TO RIBAVIRIN
Ribavirin (1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide) is a guanosine analogue that was synthesized more than 35 years ago and that possesses broad-spectrum antiviral activity against several RNA and DNA viruses in vitro (129). The first pilot study addressing the utility of ribavirin for oral antiviral therapy in patients with chronic hepatitis C was published in 1991 (114), which was followed by several controlled trials showing that ribavirin monotherapy led to a decline in alanine aminotransferase levels in a significant proportion of patients (23, 27). However, in the majority of patients receiving ribavirin monotherapy, no significant reduction of HCV RNA was observed, suggesting that ribavirin monotherapy has no or only marginal effects on HCV replication in vivo in some patients (103). In recent years, ribavirin was combined with PEG-IFN, leading to substantially improved sustained virologic response rates in patients with chronic hepatitis C.
Data from pivotal randomized trials suggest that the benefit of adding ribavirin to PEG-IFN treatment results predominantly from the prevention of virologic relapse. However, the distinct antiviral mechanisms of ribavirin are poorly understood, and at present, four different mechanisms of action have been proposed: (i) enhancement of the host adaptive antiviral immune response (113, 137), (ii) inhibition of host IMP dehydrogenase (IMPDH) (33), (iii) direct inhibition of HCV NS5B RNA-dependent RNA polymerase (12, 80), and, more recently, (iv) RNA virus mutagenesis and “error catastrophe” (Fig. 2). For the latter hypothesis, it has been reported that ribavirin acts as an RNA virus mutagen, thereby leading to an increased mutational frequency that exceeds the mutational threshold of viral fitness and drives RNA viruses into error catastrophe. By using a model of poliovirus polymerase, Crotty et al. showed that the misincorporation of ribavirin nonspecifically templates the incorporation of cytidine and uridine with equal efficiency, thus exerting mutagenic activity, which is strongly correlated with a decrease in poliovirus infectivity (20).
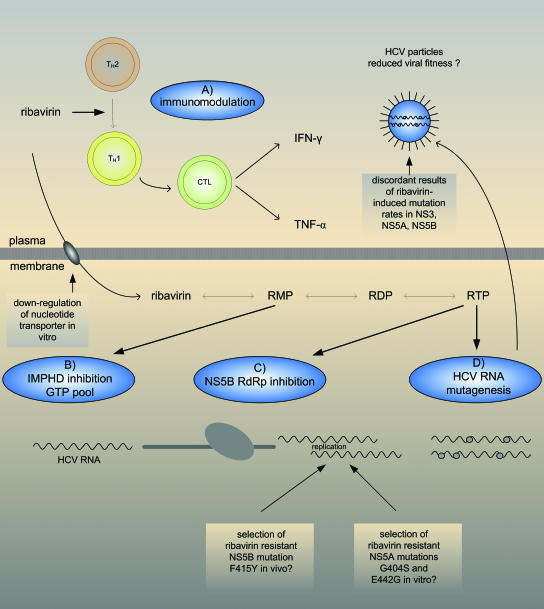
FIG. 2.
Different proposed antiviral mechanisms of ribavirin. (A) Enhancement of the host adaptive antiviral immune response. Ribavirin modulates the balance of T-helper 1 (TH1) and TH2 responses by enhancing TH1 and inhibiting TH2 cytokine production. (B) Inhibition of host IMPDH. Intracellular ribavirin is converted by cellular kinases to ribavirin monophosphate (RMP), ribavirin diphosphate (RDP), and ribavirin triphosphate (RTP). Ribavirin monophosphate is a competitive inhibitor of IMPDH, which leads to the depletion of the intracellular GTP pool necessary for viral RNA synthesis. (C) Direct inhibition of HCV NS5B RdRp. The phosphorylated form of ribavirin, ribavirin triphosphate, binds the nucleotide-binding site of polymerases, thereby competitively inhibiting viral replication. Moreover, ribavirin is utilized and incorporated by viral RNA polymerases including the HCV RdRp opposite cytosine or uridine, which can result in a significant block to RNA elongation. (D) RNA virus mutagenesis and error catastrophe. Ribavirin acts as an RNA virus mutagen, thereby leading to an increased mutational frequency that exceeds the mutational threshold of viral fitness and drives RNA viruses into lethal mutagenesis and error catastrophe. CTL, cytotoxic T lymphocyte. TNF-α, tumor necrosis factor alpha.
Ribavirin Sensitivity and Resistance In Vitro
Several in vitro studies provided evidence that ribavirin may also cause error catastrophe in HCV infection. Using a full-length binary HCV replicon system treated with increasing concentrations of ribavirin, Contreras et al. showed that the error generation rate, defined as the number of nucleotide substitutions divided by the number of nucleotides sequenced, increased significantly in both naturally variant regions such as E2 HVR1 and NS5A and invariant regions such as NS5B, underlining the presence of ribavirin-induced mutagenesis (19). These results for the NS5B region (61a) and for the NS5A ISDR region using a subgenomic HCV replicon system (138) were confirmed by others. Lanford et al. reported error-prone replication of HCV using a colony formation assay. Thus, it was shown that the colony-forming efficiency of HuH7 cells decreased after cells were transfected with total RNA from cultures harboring the HCV replicon system treated with increasing concentrations of ribavirin (71). Accordingly, Zhou et al. reported a decrease in colony formation efficiency after ribavirin treatment (156). However, in that study, the mutational frequency of the NS5A gene was found to be increased only when HCV replicon cells were treated by a combination of IMPDH inhibitors and ribavirin but not by ribavirin alone.
Pfeiffer and Kirkegaard provided in vitro evidence that resistance to ribavirin can be conferred by distinct mechanisms by using both a poliovirus cell culture system and the HCV replicon system (106, 107). By passaging HeLa cells transfected with poliovirus cDNA that were treated with ribavirin, a single amino acid replacement of glycine with serine at position 64 (G64S) located in the finger domain of poliovirus RdRp was selected. Interestingly, ribavirin-resistant poliovirus RdRp displayed an increased fidelity of RNA synthesis that may as a result reduce the frequency of ribavirin misincorporation. Accordingly, by passaging HuH7 cells harboring the HCV replicon system that were treated with increasing concentrations of ribavirin, those authors generated ribavirin-resistant HCV replicon cells. Resistance was not conferred by mutations in the HCV RdRp, and surprisingly, two other mechanisms of ribavirin resistance were reported. On the one hand, the resistant cell lines showed reduced ribavirin import in the cells, most likely by the downregulation of a nucleotide transporter. On the other hand, those authors could detect mutations within the NS5A region, G404S, and E442G that were present only in the ribavirin-resistant HCV replicon cell lines but not in the ribavirin-sensitive cell lines (107) (Table 1). The NS5A protein has been shown to interact with the HCV NS5B RdRp in vitro, and those authors suggested that the NS5A mutations may alter NS5B polymerase activity.
Ribavirin Sensitivity and Resistance In Vivo
To translate the in vitro evidence of ribavirin-induced mutagenesis to clinical settings, accumulating data from patients with chronic hepatitis C who received either ribavirin monotherapy or IFN-α-ribavirin combination therapy were recently published. Schinkel et al. were the first to evaluate nucleotide substitutions of the NS5A region by a direct sequencing approach comparing mutation rates in serum samples from patients who had not responded to IFN-α monotherapy and who were subsequently treated with IFN-α and ribavirin combination therapy (126). As the mutation rate was not increased and no significant phylogenetic sequence divergence was detectable in the NS5A region during combination treatment compared to IFN-α monotherapy, those authors concluded that ribavirin does not induce mutations, possibly rendering the virions more sensitive to IFN-α. In two other studies, the mutagenic effect of ribavirin monotherapy was studied by either direct sequencing of the NS5B region or quasispecies analyses of the NS3 and NS5A regions at several time points. In both studies, no significant increase in the mutation rate was observed (79a, 105a).
However, in a recent study, a moderate but significant increase of the NS5B and NS3 quasispecies heterogeneity was reported during the first weeks of ribavirin monotherapy (61a). In that study, an increase in the proportions of G-to-A and C-to-T transition mutations that may be specific for the misincorporation of ribavirin during RNA synthesis was observed. In concordance with those findings, Asahina et al. recently reported significantly increased mutation rates by direct sequencing of the NS5A and NS5B regions in patients who were treated with ribavirin monotherapy for 4 weeks followed by IFN-α-ribavirin combination treatment for 24 weeks when samples collected prior to treatment, at baseline, and at the end of ribavirin monotherapy were compared (5). Furthermore, the highest mutation rates within the NS5A region and nonsynonymous NS5A mutations were detected in those patients who achieved a sustained response to combination therapy.
In another study that compared NS5B quasispecies of mainly HCV subtype 1a-infected patients before and after ribavirin monotherapy, a weak but statistically not significant increase in the error generation rate was detected in samples from patients treated with ribavirin compared to controls that were treated with IFN-α or placebo (152). Interestingly, a nucleotide substitution leading to an amino acid change from phenylalanine to tyrosine at NS5B position 415 (F415Y) emerged in all five HCV subtype 1a-infected patients treated with ribavirin. By constructing HCV subgenomic replicons with both the NS5B 415F and the NS5B 415Y amino acid substitutions, those authors demonstrated in vitro that the treatment of replicon cells with ribavirin reduced the HCV RNA levels of the NS5B 415F replicon but not of the NS5B 415Y replicon (Table 1). In silico analyses showed that the NS5B 415 amino acid position maps to the thumb subdomain of NS5B and that the replacement of the phenylalanine with tyrosine may narrow the putative RNA-binding pocket in the central tunnel of the HCV RdRp, thus affecting polymerase function. Those authors concluded that the selection of the NS5B F415Y substitution during ribavirin therapy represents a mutation that confers resistance to ribavirin. In other cohorts, however, this proposed NS5B resistance mutation was less prevalent and was not associated with a lack of response to antiviral therapy (20a). Furthermore, it has to be noted that the tyrosine residue at NS5B position 415 is the consensus residue found in almost all HCV genotypes but not HCV subtype 1a.
An earlier study examined the possibility of selecting ribavirin-responsive or -resistant variants in patients receiving ribavirin monotherapy for at least 12 months. Single-stranded conformation polymorphism analysis of the HVR1 and NS5A and NS5B regions followed by direct sequencing or cloning and sequencing before treatment and at 6 and 12 months of therapy was conducted and compared to data for untreated controls (112). Phylogenetic analyses did not show any clustering of related variants in correlation with ribavirin treatment, suggesting that no selection of responsive or resistant variants occurred during ribavirin monotherapy.
Summary of Ribavirin and Treatment Resistance
Taken together, conflicting results concerning mechanisms that confer resistance to ribavirin both in vitro and in vivo have been published so far. Additional experimental and clinical data are needed to elucidate the role of ribavirin as a mutagen in hepatitis C virus infection. Furthermore, despite the apparent existence of ribavirin resistance in cell culture, the a priori presence of ribavirin resistance and possible mechanisms that confer ribavirin resistance have to be confirmed in patients undergoing antiviral therapy. As recently shown, mathematical modeling of the ribavirin effect on HCV kinetics during anti-viral therapy may help to gain more insights into ribavirin sensitivity and resistance (24, 57).
MECHANISMS OF RESISTANCE TO AMANTADINE
Amantadine, a tricyclic amine, has antiviral activity against a broad range of viruses. Used for patients with chronic hepatitis C, amantadine had no antiviral effect when it was given as monotherapy in different studies (3, 45, 127). Placebo-controlled trials with the combination treatment of IFN-α with or without amantadine have shown no significantly higher sustained virologic response rates in patients who received amantadine (56, 142, 155). Meta-analyses of all studies with IFN-α in combination with amantadine showed contradictory results (22, 81). Triple therapy with PEG-IFN, ribavirin, and amantadine showed improved sustained virologic response rates in patients who were previously nonresponders but showed no significant benefit in naïve patients or relapsers in comparison with standard combination therapy (7, 13, 22, 34, 82, 143). Additional studies with PEG-IFN, ribavirin, and higher doses of amantadine to verify a potential antiviral effect of amantadine in patients with chronic hepatitis C virus genotype 1 infection are under way.
As a potential target for amantadine, Griffin et al. identified the HCV p7 protein (48). The small, highly hydrophobic HCV p7 protein is located downstream of the HCV structural proteins and is inconsistently cleaved from the E2 protein (25). The HCV p7 protein was identified as being a calcium ion channel in artificial lipid bilayers and in membranes of mammalian cells (48, 49). Amantadine abrogates the activity of this channel, similar to its ability to inhibit the M2 protein, an integral membrane protein of influenza A virus (53). Furthermore, a study of mutated p7 variants showed that p7 is involved in the release of infectious virus particles. Mutated p7 proteins and p7 deletion mutants strongly impaired virus release in transfected HuH7 cells (134a).
In a recent study, the clinical importance of amino acid variations within the HCV p7 protein for the response to IFN-α-based antiviral therapy with and without amantadine in patients with HCV genotype 1 infection was investigated. Overall, the HCV p7 protein was highly conserved, and no significant association of amino acid variations within HCV p7 with the virologic treatment response in patients who received amantadine was observed (87) (Table 1). Interestingly, in patients with HCV subtype 1b infection and combination therapy with amantadine, a trend for a more frequent presence of amino acid substitution L20F was detected in virologic nonresponders compared with virologic responders. By in silico modeling, amino acid position 20 was located towards the p7 channel lumen, and the L20F substitution may impair the amantadine interaction by changes in the size and shape of the p7 ion channel pore. Taken together, there is no evidence for a clear additional antiviral effect of amantadine for first-line therapy of patients with chronic hepatitis C, and results of future studies, especially with higher amantadine doses, have to be awaited. Together with these studies as well as in in vitro systems (e.g., HCV replicon or patch-clamp assay), the potential importance of the L20F mutation should be addressed (87).
MECHANISMS OF RESISTANCE TO DIRECT ANTIVIRAL DRUGS
Based on the availability of an HCV replicon system and the detailed knowledge about the structure and function of HCV proteins, in recent years, a subset of new compounds comprising direct inhibitors of HCV enzymes like protease, helicase, and polymerase were developed. The most advanced new antiviral agents are directed against the HCV NS3/4A serine protease and the HCV-specific NS5B RNA-dependent RNA polymerase. Different antiviral agents for the specific inhibition of the HCV NS3/4A protease and the HCV NS5B polymerase are currently in phase 1/phase 2 trials for treatment of patients with chronic hepatitis C (Tables 2 and 3), and multiple additional direct antiviral drugs are in preclinical development. So far, results of phase 1 studies are available for three protease inhibitors (BILN2061, VX-950, and SCH503034) and three polymerase inhibitors (valopicitabine, R1479, and HCV796).
TABLE 2.
Phase 1/2 HCV NS3/4A protease inhibitors and resistance mechanisms
| Inhibitora | Company and study | Description | Resistance mechanisms | Reference(s) |
|---|---|---|---|---|
| BILN2061 | Böhringer-Ingelheim, phase 1 | Peptidomimetic inhibitor, noncovalent binding; clinical development was stopped because of cardiac toxicity observed in animal studies | In the replicon system, resistance is conferred by mutations at positions R155Q, A156S/T, and D168A/V | 74, 75, 79 |
| VX-950 (Telaprevir) | Vertex, phase 2 | Peptidomimetic inhibitor, α-ketoamide, monotherapy and combination therapy with IFN | In the replicon system, resistance is conferred by mutation at position A156S | 74, 75 |
| By sequence analysis, resistance conferred by mutations at positions V36, T54, R155, and A156 | 125a | |||
| SCH503034 | Schering-Plough, phase 2 | Peptidomimetic inhibitor, α-ketoamide, monotherapy and combination therapy with IFN | In the replicon system, low to moderate resistance is conferred by mutations at positions T54A, V170A, and A156S; longer exposure or selection led to the selection of a more resistant variant, A156T | 144 |
| By sequence analysis, only selection of HCV isolates with mutations at position T54 in single patients | 120a |
The inhibitor target is HCV NS3/4A protease.
TABLE 3.
Phase 1/2 HCV NS5B polymerase inhibitors and resistance mechanisms
| Inhibitora | Company and study | Description | Resistance mechanism(s) | Reference(s) |
|---|---|---|---|---|
| NM283 (valopicitabine) | Idenix, phase 2 | Nucleoside polymerase inhibitor NM283 (prodrug of 2′-C-methylcytosine [NM107]) | Sequence analysis of the HCV NS5B gene of several drug-resistant replicons defined a single substitution of the highly conserved S282T mutation that confers resistance to nucleosides containing 2′-methyl functionality | 86, 101 |
| R1626 | Roche, phase 1 | Nucleoside polymerase inhibitor 4′-acidocytidine (R1479) | Unknown | 115a |
| HCV796 | ViroPharma/Wyeth, phase 1 | Nonnucleoside polymerase inhibitor | One mutation, C316Y, was detected by sequence analysis of the HCV NS5B gene of patients with resistance | 144a |
The inhibitor target is HCV NS5B polymerase.
NS3/4A Protease Inhibitors
During short-term monotherapy with the protease inhibitors BILN2061 (2 days), SCH503034, and VX-950 (14 days), a decrease in HCV RNA concentrations of between 1.5 and 4 log10 HCV RNA IU/ml was observed in patients with chronic HCV genotype 1 infection. BILN2061 and VX-950 especially showed high antiviral efficacy, with HCV RNA levels below the detection limit of a highly sensitive assay (<10 IU/ml) in individual patients at the end of therapy. However, after the end of dosing, HCV RNA levels increased to concentrations at baseline in all patients (60, 70, 112b, 115, 155a). For the protease inhibitor BILN2061, studies of patients infected with genotypes 2 and 3 were published. Interestingly, a generally less pronounced antiviral activity in patients infected with genotypes 2 and 3 was observed. This shows a high probability of specificity of direct antiviral drugs for a special genotype caused by the substantial HCV sequence variability between different genotypes (60, 115). In comparison with IFN-α-based therapy, the antiviral efficacy for blocking viral production is significantly higher in patients treated with the protease inhibitor BILN2061 (>99% versus 55 to 95%) (58). Combination therapy with SCH503034 or VX-950 and PEG-IFN leads to a higher decline in the HCV RNA viral load compared with that of protease inhibitor monotherapy (120a). Restoration of the IFN signal transduction pathway, which was shown to be partially blocked by the NS3/4A protein, may be responsible for the improved effect of IFN-α under specific inhibition of the NS3/4A protease (36). Due to the error-prone nature of the HCV RNA-dependent RNA polymerase together with highly efficient replication, the selection of isolates that are resistant to compounds such as HCV NS3/4A protease and NS5B polymerase inhibitors could be a major limitation for the efficiency of direct antiviral therapies in patients with chronic hepatitis C. In the subgenomic HCV replicon system, different mutations in the HCV NS3 serine protease domain (R155Q, A156S/T, D168A/V, T54A, and V170A) that confer different levels of resistance to BILN 2061, VX-950, and SCH503034 were identified (74, 75, 79, 144) (Table 2). In patients treated with the protease inhibitor VX-950, selection of resistant variants with viral breakthrough during a 14-day treatment period was observed. By highly sensitive sequence analysis of the HCV quasispecies before treatment, at the end of treatment, and during follow-up, different amino acid positions (V36, T54, R155, and A156) with mutations that confer different levels of resistance to VX-950 were described (125a). For treatment with SCH503034, no analysis of the HCV quasispecies during therapy was performed, and by direct sequencing, only the selection of HCV isolates with mutations at position T54 in single patients was reported (120a, 155a). Whether the combination of different direct antiviral drugs as well as combination therapy with PEG-IFN could prevent the development of resistance remains to be clarified in future studies. Overall, it is clear that for HCV infection, a rapid development of resistance as in human immunodeficiency virus infection is possible and that an efficient initial blocking of HCV replication with high plasma levels of the protease inhibitor is essential for the long-term success of this treatment option.
NS5B Polymerase Inhibitors
Results of phase 1/phase 2 studies with the HCV NS5B RNA polymerase inhibitor valopicitabine (NM283) showed a decrease in the HCV RNA concentration of 1 to 2 log10 HCV RNA IU/ml in the treatment of naïve patients or IFN-α nonresponders (44a). Interim analysis of ongoing studies also showed improved antiviral efficacy for combination therapy with NM283 and PEG-IFN (98a). NM283 is the oral prodrug of 2′-C-methyl-cytidine (NM107) and is cleaved to the free nucleoside that is converted to the active triphosphate by the cellular machinery. No in vivo resistance data for NM283 are available yet, but it was shown that the 2′-C-methyl-nucleoside NM107 is susceptible to resistance development, as shown in replicon systems and isolated polymerase assays. Sequence analysis of the HCV NS5B gene of several drug-resistant replicons defined a single replacement of the highly conserved serine 282 with threonine (S282T) that confers resistance to nucleosides containing 2′-methyl functionality (86, 101). Recently, the development of resistance to NM283 was investigated using NM107 and a bovine viral diarrhea virus in vitro infection assay. Bovine viral diarrhea virus is a pestivirus related to HCV, and sequencing analysis of variants that are resistant to NM107 revealed that resistance was due to an S405T substitution, which is the homologous mutation of S282T in HCV NS5B (9a). Taken together, only limited data on HCV polymerase inhibitors are available today. So far, the antiviral activity of the drugs tested in patients with chronic hepatitis C seems to be generally weaker than that of several HCV protease inhibitors, but this may be overcome by increased specificity and higher doses of HCV polymerase inhibitors. In the future, it will be highly interesting to investigate the significance of mutations in different HCV genes of patients treated with IFN-α in combination with direct antiviral drugs (potential IFN-α resistance regions together with targets of protease and polymerase inhibitors).
CONCLUDING REMARKS
Several HCV proteins have been associated with mechanisms of resistance to different antiviral drugs in acute and chronic hepatitis C. In the vast majority of the multiple studies addressing this issue, either in vitro mechanisms together with HCV prototype isolates or amino acid sequence data for different HCV proteins obtained from patients undergoing antiviral therapies have been investigated. This restriction should be overcome in the future.
As a main viral strategy to circumvent the effects of IFN-α, the inhibition of PKR by the E2 and NS5A proteins in in vitro studies has been proposed. However, for both proteins, no concluding data in follow-up studies have been presented, and the overall importance of PKR inhibition is uncertain. More recently, the inhibition of signaling molecules of the innate immune response (Cardif and Trif) by the NS3/4A protease has been described. This mechanism may explain the high rate of HCV persistence after acute hepatitis C virus infection. However, the potential importance of the virologic response to IFN-α-based therapy in patients with chronic hepatitis C has to be investigated in future studies.
In the majority of clinical studies investigating amino acid mutations of different HCV proteins, no correlation of specific mutations or the number of mutations with the response to IFN-α-based antiviral therapy was detected. While, in all probability, the amino acid variability of HCV proteins reflects different sensitivities to IFN-α-based antiviral therapy, it became clear that simply counting mutations generally may not be sufficient for the detection of functional differences, which in turn may result in different treatment outcomes. However, multiple studies and a meta-analysis have conclusively shown that for the NS5A ISDR, indeed, just the number of mutations is significantly associated with the response to IFN-α-based therapy, and future studies should aim to elucidate the underlying functional mechanisms of this phenomenon.
None of the proposed antiviral effects of ribavirin have been conclusively proofed so far. For the error catastrophe mechanism, several supportive data exist, and also, analysis of HCV kinetics in patients with relatively low IFN-α efficacy argues in favor of this hypothesis. However, the decline of alanine aminotransferase levels despite significant changes in HCV RNA concentrations during ribavirin monotherapy is not well explained by a sole RNA virus mutagenic effect of ribavirin, and thus, additional mechanisms of action have to be assumed.
For amantadine, the interaction with the p7 protein represents an attractive model for an explanation of its mode of action. However, only preliminary in vitro studies are available so far. Furthermore, the at best weak antiviral efficacy of amantadine in patients with chronic hepatitis C will make it difficult to determine specific HCV p7 sequence variants with sensitivity/resistance to amantadine.
The new era of direct antiviral drugs for treatment of hepatitis C is just beginning, and little is known about resistance of the different HCV isolates, subtypes, and genotypes to, e.g., HCV protease and polymerase inhibitors today. However, it is already clear that different from hepatitis B and analogous to the situation in human immunodeficiency virus, depending on the drug plasma levels, clinically manifest resistance can develop very rapidly within a few days in patients with hepatitis C undergoing therapy with, for example, an NS3/4A protease inhibitor. The various possibilities of the use of direct antiviral drugs in hepatitis C together with IFN-α with or without ribavirin or combined with each other open a broad field of new treatment options with associated resistance mechanisms.
Acknowledgments
We are grateful to Stefan Zeuzem for critical reading of the manuscript.
REFERENCES
- 1.Abbate, I., I. Lo Iacono, R. Di Stefano, G. Cappiello, E. Girardi, R. Longo, D. Ferraro, G. Antonucci, V. Di Marco, M. Solmone, A. Craxi, G. Ippolito, and M. R. Capobianchi. 2004. HVR-1 quasispecies modifications occur early and are correlated to initial but not sustained response in HCV-infected patients treated with pegylated- or standard-interferon and ribavirin. J. Hepatol. 40:831-836. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 3.Andant, C., J. Lamoril, J. C. Deybach, P. Jouet, and J. C. Soule. 2000. Amantadine for chronic hepatitis C: pilot study in 14 patients. Eur. J. Gastroenterol. Hepatol. 12:1319-1322. [DOI] [PubMed] [Google Scholar]
- 4.Appel, N., T. Pietschmann, and R. Bartenschlager. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 79:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asahina, Y., N. Izumi, N. Enomoto, M. Uchihara, M. Kurosaki, Y. Onuki, Y. Nishimura, K. Ueda, K. Tsuchiya, H. Nakanishi, T. Kitamura, and S. Miyake. 2005. Mutagenic effects of ribavirin and response to interferon/ribavirin combination therapy in chronic hepatitis C. J. Hepatol. 43:623-629. [DOI] [PubMed] [Google Scholar]
- 6.aus dem Siepen, M., V. Lohmann, M. Wiese, S. Ross, M. Roggendorf, and S. Viazov. 2005. Nonstructural protein 5A does not contribute to the resistance of hepatitis C virus replication to interferon alpha in cell culture. Virology 336:131-136. [DOI] [PubMed] [Google Scholar]
- 7.Berg, T., B. Kronenberger, H. Hinrichsen, T. Gerlach, P. Buggisch, E. Herrmann, U. Spengler, T. Goeser, S. Nasser, K. Wursthorn, G. R. Pape, U. Hopf, and S. Zeuzem. 2003. Triple therapy with amantadine in treatment-naive patients with chronic hepatitis C: a placebo-controlled trial. Hepatology 37:1359-1367. [DOI] [PubMed] [Google Scholar]
- 8.Berg, T., M. A. Mas, M. Hohne, B. Wiedenmann, U. Hopf, and E. Schreier. 2000. Mutations in the E2-PePHD and NS5A region of hepatitis C virus type 1 and the dynamics of hepatitis C viremia decline during interferon alfa treatment. Hepatology 32:1386-1395. [DOI] [PubMed] [Google Scholar]
- 9.Berg, T., C. Sarrazin, E. Herrmann, H. Hinrichsen, T. Gerlach, R. Zachoval, B. Wiedenmann, U. Hopf, and S. Zeuzem. 2003. Prediction of treatment outcome in patients with chronic hepatitis C: significance of baseline parameters and viral dynamics during therapy. Hepatology 37:600-609. [DOI] [PubMed] [Google Scholar]
- 9a.Bichko, V., L. Qu, M. la Colla, M. Tausek, S. Bergelson, C. Pierra, R. Storer, G. Gosselin, J. P. Sommadossi, and D. Standring. 2005. Abstr. 56th Am. Assoc. Study Liver Dis., abstr. 860.
- 10.Blindenbacher, A., F. H. Duong, L. Hunziker, S. T. Stutvoet, X. Wang, L. Terracciano, D. Moradpour, H. E. Blum, T. Alonzi, M. Tripodi, N. La Monica, and M. H. Heim. 2003. Expression of hepatitis C virus proteins inhibits interferon alpha signaling in the liver of transgenic mice. Gastroenterology 124:1465-1475. [DOI] [PubMed] [Google Scholar]
- 11.Bode, J. G., S. Ludwig, C. Ehrhardt, U. Albrecht, A. Erhardt, F. Schaper, P. C. Heinrich, and D. Haussinger. 2003. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J. 17:488-490. [DOI] [PubMed] [Google Scholar]
- 12.Bougie I., and M. Bisaillon. 2003. Initial binding of the broad spectrum antiviral nucleoside ribavirin to the hepatitis C virus RNA polymerase. J. Biol. Chem. 278:52471-52478. [DOI] [PubMed] [Google Scholar]
- 13.Brillanti, S., F. Levantesi, L. Masi, M. Foli, and L. Bolondi. 2000. Triple antiviral therapy as a new option for patients with interferon nonresponsive chronic hepatitis C. Hepatology 32:630-634. [DOI] [PubMed] [Google Scholar]
- 14.Castelain, S., H. Khorsi, J. Roussel, C. Francois, O. Jaillon, D. Capron, F. Penin, C. Wychowski, E. Meurs, and G. Duverlie. 2002. Variability of the nonstructural 5A protein of hepatitis C virus type 3a isolates and relation to interferon sensitivity. J. Infect. Dis. 185:573-583. [DOI] [PubMed] [Google Scholar]
- 15.Chambers, T. J., X. Fan, D. A. Droll, E. Hembrador, T. Slater, M. W. Nickells, L. B. Dustin, and A. M. Dibisceglie. 2005. Quasispecies heterogeneity within the E1/E2 region as a pretreatment variable during pegylated interferon therapy of chronic hepatitis C virus infection. J. Virol. 79:3071-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chayama, K., A. Tsubota, M. Kobayashi, K. Okamoto, M. Hashimoto, Y. Miyano, H. Koike, M. Kobayashi, I. Koida, Y. Arase, S. Saitoh, Y. Suzuki, N. Murashima, K. Ikeda, and H. Kumada. 1997. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology 25:745-749. [DOI] [PubMed] [Google Scholar]
- 17.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 18.Chung, R. T., A. Monto, J. L. Dienstag, and L. M. Kaplan. 1999. Mutations in the NS5A region do not predict interferon-responsiveness in American patients infected with genotype 1b hepatitis C virus. J. Med. Virol. 58:353-358. [PubMed] [Google Scholar]
- 19.Contreras, A. M., Y. Hiasa, W. He, A. Terella, E. V. Schmidt, and R. T. Chung. 2002. Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system. J. Virol. 76:8505-8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 20a.Danehower, S. C., G. Lutchman, and M. G. Ghany. 2004. Abstr. 11th Int. Symp. Hepat. C Virus Relat. Viruses, abstr. 73.
- 21.Deltenre, P., J. Henrion, V. Canva, S. Dharancy, F. Texier, A. Louvet, S. De Maeght, J. C. Paris, and P. Mathurin. 2004. Evaluation of amantadine in chronic hepatitis C: a meta-analysis. J. Hepatol. 41:462-473. [DOI] [PubMed] [Google Scholar]
- 22.de Lucas, S., J. Bartolome, and V. Carreno. 2005. Hepatitis C virus core protein down-regulates transcription of interferon-induced antiviral genes. J. Infect. Dis. 191:93-99. [DOI] [PubMed] [Google Scholar]
- 23.Di Bisceglie, A. M., M. Shindo, T. L. Fong, M. W. Fried, M. G. Swain, N. V. Bergasa, C. A. Axiotis, J. G. Waggoner, Y. Park, and J. H. Hoofnagle. 1992. A pilot study of ribavirin therapy for chronic hepatitis C. Hepatology 16:649-654. [DOI] [PubMed] [Google Scholar]
- 24.Dixit, N. M., J. E. Layden-Almer, T. J. Layden, and A. S. Perelson. 2004. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature 432:922-924. [DOI] [PubMed] [Google Scholar]
- 25.Dubuisson, J. 2000. Folding, assembly and subcellular localization of hepatitis C virus glycoproteins. Curr. Top. Microbiol. Immunol. 242:135-148. [DOI] [PubMed] [Google Scholar]
- 26.Duong, F. H., M. Filipowicz, M. Tripodi, N. La Monica, and M. H. Heim. 2004. Hepatitis C virus inhibits interferon signaling through up-regulation of protein phosphatase 2A. Gastroenterology 126:263-277. [DOI] [PubMed] [Google Scholar]
- 27.Dusheiko, G., J. Main, H. Thomas, O. Reichard, C. Lee, A. Dhillon, S. Rassam, A. Fryden, H. Reesink, M. Bassendine, G. Norkrans, T. Cuypers, N. Lelie, P. Telfer, J. Watson, C. Weegink, P. Sillikens, and O. Weiland. 1996. Ribavirin treatment for patients with chronic hepatitis C: results of a placebo-controlled study. J. Hepatol. 25:591-598. [DOI] [PubMed] [Google Scholar]
- 28.Duverlie, G., H. Khorsi, S. Castelain, O. Jaillon, J. Izopet, F. Lunel, F. Eb, F. Penin, and C. Wychowski. 1998. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J. Gen. Virol. 79:1373-1381. [DOI] [PubMed] [Google Scholar]
- 29.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato. 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J. Clin. Investig. 96:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Marumo, and C. Sato. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 334:77-81. [DOI] [PubMed] [Google Scholar]
- 31.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 32.Farci, P., R. Strazzera, H. J. Alter, S. Farci, D. Degioannis, A. Coiana, G. Peddis, F. Usai, G. Serra, L. Chessa, G. Diaz, A. Balestrieri, and R. H. Purcell. 2002. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc. Natl. Acad. Sci. USA 99:3081-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feld, J. J., and J. H. Hoofnagle. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967-972. [DOI] [PubMed] [Google Scholar]
- 34.Ferenci, P., E. Formann, H. Laferl, M. Gschwantler, F. Hackl, H. Brunner, R. Hubmann, C. Datz, R. Stauber, P. Steindl-Munda, H. H. Kessler, A. Klingler, A. Gangl, et al. 2006. Randomized, double-blind, placebo-controlled study of peginterferon alfa-2a (40KD) plus ribavirin with or without amantadine in treatment-naive patients with chronic hepatitis C genotype 1 infection. J. Hepatol. 44:275-282. [DOI] [PubMed] [Google Scholar]
- 35.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA 102:2986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 37.Francois, C., G. Duverlie, D. Rebouillat, H. Khorsi, S. Castelain, H. E. Blum, A. Gatignol, C. Wychowski, D. Moradpour, and E. F. Meurs. 2000. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J. Virol. 74:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Gonzales, D. Häussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffmann, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 39.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436:939-945. [DOI] [PubMed] [Google Scholar]
- 40.Gale, M. J., Jr., M. J. Korth, and M. G. Katze. 1998. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin. Diagn. Virol. 10:157-162. [DOI] [PubMed] [Google Scholar]
- 41.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 42.Gaudy, C., M. Lambele, A. Moreau, P. Veillon, F. Lunel, and A. Goudeau. 2005. Mutations within the hepatitis C virus genotype 1b E2-PePHD domain do not correlate with treatment outcome. J. Clin. Microbiol. 43:750-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerotto, M., F. Dal Pero, P. Pontisso, F. Noventa, A. Gatta, and A. Alberti. 2000. Two PKR inhibitor HCV proteins correlate with early but not sustained response to interferon. Gastroenterology 119:1649-1655. [DOI] [PubMed] [Google Scholar]
- 44.Gimenez-Barcons, M., C. Wang, M. Chen, J. M. Sanchez-Tapias, J. C. Saiz, and M. Gale, Jr. 2005. The oncogenic potential of hepatitis C virus NS5A sequence variants is associated with PKR regulation. J. Interferon Cytokine Res. 25:152-164. [DOI] [PubMed] [Google Scholar]
- 44a.Godofsky, E., N. Afdhal, V. Rustgi, L. Shick, L. Duncan, X. J. Zhou, G. Chao, C. Fang, B. Fielman, M. Myers, and N. A. Brown. 2005. Abstr. 39th Eur. Assoc. Study Liver, abstr. 96.
- 45.Goff, J. S., R. M. Reveille, and J. Johnson. 2000. Treatment of chronic hepatitis C with amantadine. Dig. Dis. Sci. 45:1389-1391. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez-Peralta, R. P., K. Qian, J. Y. She, G. L. Davis, T. Ohno, M. Mizokami, and J. Y. Lau. 1996. Clinical implications of viral quasispecies heterogeneity in chronic hepatitis C. J. Med. Virol. 49:242-247. [DOI] [PubMed] [Google Scholar]
- 47.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Griffin, S. D., L. P. Beales, D. S. Clarke, O. Worsfold, S. D. Evans, J. Jaeger, M. P. Harris, and D. J. Rowlands. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 535:34-38. [DOI] [PubMed] [Google Scholar]
- 49.Griffin, S. D., R. Harvey, D. S. Clarke, W. S. Barclay, M. Harris, and D. J. Rowlands. 2004. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J. Gen. Virol. 85:451-461. [DOI] [PubMed] [Google Scholar]
- 50.Hadziyannis, S. J., H. Sette, Jr., T. R. Morgan, V. Balan, M. Diago, P. Marcellin, G. Ramadori, H. C. Bodenheimer, Jr., D. Bernstein, M. Rizzetto, S. Zeuzem, P. J. Pockros, A. Lin, and A. M. Ackrill. 2004. Peginterferon-alfa2a and ribavirin combination therapy in chronic hepatitis C. A randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346-355. [DOI] [PubMed] [Google Scholar]
- 51.Han, J. Q., and D. J. Barton. 2002. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA 8:512-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han, J. Q., G. Wroblewski, Z. Xu, R. H. Silverman, and D. J. Barton. 2004. Sensitivity of hepatitis C virus RNA to the antiviral enzyme ribonuclease L is determined by a subset of efficient cleavage sites. J. Interferon Cytokine Res. 24:664-676. [DOI] [PubMed] [Google Scholar]
- 53.Hay, A. J., A. J. Wolstenholme, J. J. Skehel, and M. H. Smith. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 4:3021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He, Y., S. L. Tan, S. U. Tareen, S. Vijaysri, J. O. Langland, B. L. Jacobs, and M. G. Katze. 2001. Regulation of mRNA translation and cellular signaling by hepatitis C virus nonstructural protein NS5A. J. Virol. 75:5090-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heim, M. H., D. Moradpour, and H. E. Blum. 1999. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J. Virol. 73:8469-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Helbling, B., I. Stamenic, F. Viani, J. J. Gonvers, J. F. Dufour, J. Reichen, G. Cathomas, M. Steuerwald, J. Borovicka, M. Sagmeister, and E. L. Renner. 2002. Interferon and amantadine in naive chronic hepatitis C: a double-blind, randomized, placebo-controlled trial. Hepatology 35:447-454. [DOI] [PubMed] [Google Scholar]
- 57.Herrmann, E., J. H. Lee, G. Marinos, M. Modi, and S. Zeuzem. 2003. Effect of ribavirin on hepatitis C viral kinetics in patients treated with pegylated interferon. Hepatology 37:1351-1358. [DOI] [PubMed] [Google Scholar]
- 58.Herrmann, E., S. Zeuzem, C. Sarrazin, H. Hinrichsen, Y. Benhamou, M. P. Manns, M. Reiser, H. Reesink, J. L. Calleja, X. Forns, G. G. Steinmann, and G. Nehmiz. 2006. Viral kinetics in patients with chronic hepatitis C treated with the serine protease inhibitor BILN 2061. Antivir. Ther. 11:371-376. [PubMed] [Google Scholar]
- 59.Hino, K., Y. Yamaguchi, D. Fujiwara, Y. Katoh, M. Korenaga, M. Okazaki, M. Okuda, and K. Okita. 2000. Hepatitis C virus quasispecies and response to interferon therapy in patients with chronic hepatitis C: a prospective study. J. Viral Hepat. 7:36-42. [DOI] [PubMed] [Google Scholar]
- 60.Hinrichsen, H., Y. Benhamou, H. Wedemeyer, M. Reiser, R. E. Sentjens, J. L. Calleja, X. Forns, A. Erhardt, J. Cronlein, R. L. Chaves, C. L. Yong, G. Nehmiz, and G. G. Steinmann. 2004. Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology 127:1347-1355. [DOI] [PubMed] [Google Scholar]
- 61.Hofmann, W. P., C. Sarrazin, B. Kronenberger, B. Schonberger, K. Bruch, and S. Zeuzem. 2003. Mutations within the CD81-binding sites and hypervariable region 2 of the envelope 2 protein: correlation with treatment response in hepatitis C virus-infected patients. J. Infect. Dis. 187:982-987. [DOI] [PubMed] [Google Scholar]
- 61a.Hofmann, W. P., A. Polta, E. Herrmann, U. Mihm, B. Kronenberger, T. Sonntag, V. Lohmann, B. Schönberger, S. Zeuzem and C. Sarrazin. Mutagenic effect of ribavirin on hepatitis C non-structural 5B quasispecies in vitro and during antiviral therapy. Gastroenterology, in press. [DOI] [PubMed]
- 62.Jeffers, L. J., W. Cassidy, C. D. Howell, S. Hu, and K. R. Reddy. 2004. Peginterferon alfa-2a (40 kd) and ribavirin for black American patients with chronic HCV genotype 1. Hepatology 39:1702-1708. [DOI] [PubMed] [Google Scholar]
- 63.Kanazawa, Y., N. Hayashi, E. Mita, T. Li, H. Hagiwara, A. Kasahara, H. Fusamoto, and T. Kamada. 1994. Influence of viral quasispecies on effectiveness of interferon therapy in chronic hepatitis C patients. Hepatology 20:1121-1130. [PubMed] [Google Scholar]
- 64.Khabar, K. S., F. Al Zoghaibi, M. N. Al Ahdal, T. Murayama, M. Dhalla, N. Mukaida, M. Taha, S. T. Al Sedairy, Y. Siddiqui, G. Kessie, and K. Matsushima. 1997. The alpha chemokine, interleukin 8, inhibits the antiviral action of interferon alpha. J. Exp. Med. 186:1077-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khorsi, H., S. Castelain, A. Wyseur, J. Izopet, V. Canva, A. Rombout, D. Capron, J. P. Capron, F. Lunel, L. Stuyver, and G. Duverlie. 1997. Mutations of hepatitis C virus 1b NS5A 2209-2248 amino acid sequence do not predict the response to recombinant interferon-alfa therapy in French patients. J. Hepatol. 27:72-77. [DOI] [PubMed] [Google Scholar]
- 66.Knapp, S., L. J. Yee, A. J. Frodsham, B. J. Hennig, S. Hellier, L. Zhang, M. Wright, M. Chiaramonte, M. Graves, H. C. Thomas, A. V. Hill, and M. R. Thursz. 2003. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun. 4:411-419. [DOI] [PubMed] [Google Scholar]
- 67.Kobayashi, M., K. Watanabe, M. Ishigami, K. Murase, H. Ito, K. Ukai, M. Yano, K. Takagi, M. Hattori, S. Kakumu, and K. Yoshioka. 2002. Amino acid substitutions in the nonstructural region 5A of hepatitis C virus genotypes 2a and 2b and its relation to viral load and response to interferon. Am. J. Gastroenterol. 97:988-998. [DOI] [PubMed] [Google Scholar]
- 68.Koizumi, K., N. Enomoto, M. Kurosaki, T. Murakami, N. Izumi, F. Marumo, and C. Sato. 1995. Diversity of quasispecies in various disease stages of chronic hepatitis C virus infection and its significance in interferon treatment. Hepatology 22:30-35. [PubMed] [Google Scholar]
- 69.Kurosaki, M., N. Enomoto, T. Murakami, I. Sakuma, Y. Asahina, C. Yamamoto, T. Ikeda, S. Tozuka, N. Izumi, F. Marumo, and C. Sato. 1997. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-beta therapy. Hepatology 25:750-753. [DOI] [PubMed] [Google Scholar]
- 70.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 71.Lanford, R. E., B. Guerra, H. Lee, D. R. Averett, B. Pfeiffer, D. Chavez, L. Notvall, and C. Bigger. 2003. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(I)-poly(C), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J. Virol. 77:1092-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Guen, B., G. Squadrito, B. Nalpas, P. Berthelot, S. Pol, and C. Brechot. 1997. Hepatitis C virus genome complexity correlates with response to interferon therapy: a study in French patients with chronic hepatitis C. Hepatology 25:1250-1254. [DOI] [PubMed] [Google Scholar]
- 73.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 102:2992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin, C., C. A. Gates, B. G. Rao, D. L. Brennan, J. R. Fulghum, Y. P. Luong, J. D. Frantz, K. Lin, S. Ma, Y. Y. Wei, R. B. Perni, and A. D. Kwong. 2005. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J. Biol. Chem. 280:36784-36791. [DOI] [PubMed] [Google Scholar]
- 75.Lin, C., K. Lin, Y. P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 76.Lin, W., W. H. Choe, Y. Hiasa, Y. Kamegaya, J. T. Blackard, E. V. Schmidt, and R. T. Chung. 2005. Hepatitis C virus expression suppresses interferon signaling by degrading STAT1. Gastroenterology 128:1034-1041. [DOI] [PubMed] [Google Scholar]
- 77.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 78.Lopez-Labrador, F. X., S. Ampurdanes, M. Gimenez-Barcons, M. Guilera, J. Costa, M. T. Jimenez de Anta, J. M. Sanchez-Tapias, J. Rodes, and J. C. Saiz. 1999. Relationship of the genomic complexity of hepatitis C virus with liver disease severity and response to interferon in patients with chronic HCV genotype 1b infection. Hepatology 29:897-903. (Erratum, 29:1915.) [DOI] [PubMed] [Google Scholar]
- 79.Lu, L., T. J. Pilot-Matias, K. D. Stewart, J. T. Randolph, R. Pithawalla, W. He, P. P. Huang, L. L. Klein, H. Mo, and A. Molla. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79a.Lutchman, G. A., S. Danehower, C. Ward, T. J. Liang, J. H. Hoofnagle, M. Z. Thomson, and M. G. Ghany. 2004. Abstr. 55th Am. Assoc. Study Liver Dis., abstr. 512.
- 80.Maag, D., C. Castro, Z. Hong, and C. E. Cameron. 2001. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J. Biol. Chem. 276:46094-46098. [DOI] [PubMed] [Google Scholar]
- 81.Mangia, A., G. Leandro, B. Helbling, E. L. Renner, M. Tabone, L. Sidoli, S. Caronia, G. R. Foster, S. Zeuzem, T. Berg, V. Di Marco, N. Cino, and A. Andriulli. 2004. Combination therapy with amantadine and interferon in naive patients with chronic hepatitis C: meta-analysis of individual patient data from six clinical trials. J. Hepatol. 40:478-483. [DOI] [PubMed] [Google Scholar]
- 82.Mangia, A., G. L. Ricci, M. Persico, N. Minerva, V. Carretta, D. Bacca, M. Cela, M. Piattelli, M. Annese, G. Maio, D. Conte, V. Guadagnino, V. Pazienza, D. Festi, F. Spirito, and A. Andriulli. 2005. A randomized controlled trial of pegylated interferon alpha-2a (40 KD) or interferon alpha-2a plus ribavirin and amantadine vs interferon alpha-2a and ribavirin in treatment-naive patients with chronic hepatitis C. J. Viral Hepat. 12:292-299. [DOI] [PubMed] [Google Scholar]
- 83.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 84.McKechnie, V. M., P. R. Mills, and E. A. McCruden. 2000. The NS5a gene of hepatitis C virus in patients treated with interferon-alpha. J. Med. Virol. 60:367-378. [PubMed] [Google Scholar]
- 85.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 86.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 87.Mihm, U., N. Grigorian, C. Welsch, E. Herrmann, B. Kronenberger, G. Teuber, M. von Wagner, W. P. Hofmann, M. Albrecht, T. Lengauer, S. Zeuzem, and C. Sarrazin. 2006. Mutations in the HCV-p7 protein and sensitivity to antiviral therapy with amantadine in chronic hepatitis C. Antivir. Ther. 11:507-519. [PubMed] [Google Scholar]
- 88.Mihm, U., W. P. Hofmann, B. Kronenberger, M. von Wagner, S. Zeuzem, and C. Sarrazin. 2004. Correlation of serum interleukin 8 with virologic response to antiviral therapy in patients with chronic hepatitis C. J. Hepatol. 40:845-852. [DOI] [PubMed] [Google Scholar]
- 89.Moradpour, D., P. Kary, C. M. Rice, and H. E. Blum. 1998. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology 28:192-201. [DOI] [PubMed] [Google Scholar]
- 90.Moribe, T., N. Hayashi, Y. Kanazawa, E. Mita, H. Fusamoto, M. Negi, T. Kaneshige, H. Igimi, T. Kamada, and K. Uchida. 1995. Hepatitis C viral complexity detected by single-strand conformation polymorphism and response to interferon therapy. Gastroenterology 108:789-795. [DOI] [PubMed] [Google Scholar]
- 91.Murakami, T., N. Enomoto, M. Kurosaki, N. Izumi, F. Marumo, and C. Sato. 1999. Mutations in nonstructural protein 5A gene and response to interferon in hepatitis C virus genotype 2 infection. Hepatology 30:1045-1053. [DOI] [PubMed] [Google Scholar]
- 92.Murashima, S., R. Kumashiro, T. Ide, I. Miyajima, T. Hino, Y. Koga, K. Ishii, T. Ueno, S. Sakisaka, and M. Sata. 2000. Effect of interferon treatment on serum 2′,5′-oligoadenylate synthetase levels in hepatitis C-infected patients. J. Med. Virol. 62:185-190. [DOI] [PubMed] [Google Scholar]
- 93.Murphy, M. D., H. R. Rosen, G. I. Marousek, and S. Chou. 2002. Analysis of sequence configurations of the ISDR, PKR-binding domain, and V3 region as predictors of response to induction interferon-alpha and ribavirin therapy in chronic hepatitis C infection. Dig. Dis. Sci. 47:1195-1205. [DOI] [PubMed] [Google Scholar]
- 94.Myers, R. P., K. Patel, S. Pianko, T. Poynard, and J. G. McHutchison. 2003. The rate of fibrosis progression is an independent predictor of the response to antiviral therapy in chronic hepatitis C. J. Viral Hepat. 10:16-22. [DOI] [PubMed] [Google Scholar]
- 95.Naganuma, A., A. Nozaki, T. Tanaka, K. Sugiyama, H. Takagi, M. Mori, K. Shimotohno, and N. Kato. 2000. Activation of the interferon-inducible 2′-5′-oligoadenylate synthetase gene by hepatitis C virus core protein. J. Virol. 74:8744-8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nagasaka, A., S. Hige, I. Tsunematsu, J. Yoshida, Y. Sasaki, T. Matsushima, and M. Asaka. 1996. Changes in hepatitis C virus quasispecies and density populations in patients before and after interferon therapy. J. Med. Virol. 50:214-220. [DOI] [PubMed] [Google Scholar]
- 97.Nakazawa, T., N. Kato, S. Ohkoshi, A. Shibuya, and K. Shimotohno. 1994. Characterization of the 5′ noncoding and structural region of the hepatitis C virus genome from patients with non-A, non-B hepatitis responding differently to interferon treatment. J. Hepatol. 20:623-629. [DOI] [PubMed] [Google Scholar]
- 98.Nousbaum, J., S. J. Polyak, S. C. Ray, D. G. Sullivan, A. M. Larson, R. L. Carithers, Jr., and D. R. Gretch. 2000. Prospective characterization of full-length hepatitis C virus NS5A quasispecies during induction and combination antiviral therapy. J. Virol. 74:9028-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98a.O'Brien, C., E. Godofsky, M. Rodriguez-Torres, N. Afdhal, S. C. Pappas, P. Pockros, E. Lawitz, N. Bzowej, V. Rustgi, M. Sulkowski, K. Sherman, I. Jacobson, G. Chao, S. Knox, K. Pietropaolo, and N. A. Brown. 2005. Abstr. 56th Am. Assoc. Study Liver Dis., abstr. 95.
- 99.Odeberg, J., Z. Yun, A. Sonnerborg, O. Weiland, and J. Lundeberg. 1998. Variation in the hepatitis C virus NS5A region in relation to hypervariable region 1 heterogeneity during interferon treatment. J. Med. Virol. 56:33-38. [DOI] [PubMed] [Google Scholar]
- 100.Okada, S., Y. Akahane, H. Suzuki, H. Okamoto, and S. Mishiro. 1992. The degree of variability in the amino terminal region of the E2/NS1 protein of hepatitis C virus correlates with responsiveness to interferon therapy in viremic patients. Hepatology 16:619-624. [DOI] [PubMed] [Google Scholar]
- 101.Olsen, D. B., A. B. Eldrup, L. Bartholomew, B. Bhat, M. R. Bosserman, A. Ceccacci, L. F. Colwell, J. F. Fay, O. A. Flores, K. L. Getty, J. A. Grobler, R. L. LaFemina, E. J. Markel, G. Migliaccio, M. Prhavc, M. W. Stahlhut, J. E. Tomassini, M. MacCoss, D. J. Hazuda, and S. S. Carroll. 2004. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob. Agents Chemother. 48:3944-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pascu, M., P. Martus, M. Hohne, B. Wiedenmann, U. Hopf, E. Schreier, and T. Berg. 2004. Sustained virological response in hepatitis C virus type 1b infected patients is predicted by the number of mutations within the NS5A-ISDR: a meta-analysis focused on geographical differences. Gut 53:1345-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pawlotsky, J. M., H. Dahari, A. U. Neumann, C. Hezode, G. Germanidis, I. Lonjon, L. Castera, and D. Dhumeaux. 2004. Antiviral action of ribavirin in chronic hepatitis C. Gastroenterology 126:703-714. [DOI] [PubMed] [Google Scholar]
- 104.Pawlotsky, J. M., G. Germanidis, A. U. Neumann, M. Pellerin, P. O. Frainais, and D. Dhumeaux. 1998. Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5A gene quasispecies mutations. J. Virol. 72:2795-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pawlotsky, J. M., M. Pellerin, M. Bouvier, F. Roudot-Thoraval, G. Germanidis, A. Bastie, F. Darthuy, J. Remire, C. J. Soussy, and D. Dhumeaux. 1998. Genetic complexity of the hypervariable region 1 (HVR1) of hepatitis C virus (HCV): influence on the characteristics of the infection and responses to interferon alfa therapy in patients with chronic hepatitis C. J. Med. Virol. 54:256-264. [PubMed] [Google Scholar]
- 105a.Pawlotsky, J. M., R. Brillet, C. Hezode, and D. Dhumeaux. 2004. Abstr. 39th Eur. Assoc. Study Liver, abstr. 62.
- 106.Pfeiffer, J. K., and K. Kirkegaard. 2003. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. USA 100:7289-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pfeiffer, J. K., and K. Kirkegaard. 2005. Ribavirin resistance in hepatitis C virus replicon-containing cell lines conferred by changes in the cell line or mutations in the replicon RNA. J. Virol. 79:2346-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 109.Polyak, S. J., K. S. Khabar, D. M. Paschal, H. J. Ezelle, G. Duverlie, G. N. Barber, D. E. Levy, N. Mukaida, and D. R. Gretch. 2001. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J. Virol. 75:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Polyak, S. J., K. S. Khabar, M. Rezeiq, and D. R. Gretch. 2001. Elevated levels of interleukin-8 in serum are associated with hepatitis C virus infection and resistance to interferon therapy. J. Virol. 75:6209-6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Polyak, S. J., D. M. Paschal, S. McArdle, M. J. Gale, Jr., D. Moradpour, and D. R. Gretch. 1999. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology 29:1262-1271. [DOI] [PubMed] [Google Scholar]
- 112.Querenghi, F., Q. Yu, G. Billaud, G. Maertens, C. Trepo, and F. Zoulim. 2001. Evolution of hepatitis C virus genome in chronically infected patients receiving ribavirin monotherapy. J. Viral Hepat. 8:120-131. [DOI] [PubMed] [Google Scholar]
- 112a.Reesink, H. W., N. Forestier, C. W. Weegink, S. Zeuzem, L. McNair, S. Purdy, H. M. Chu, and P. L. M. Jansen. 2006. Abstr. 41st Eur. Assoc. Study Liver, abstr. 737.
- 112b.Reesink, H. W., S. Zeuzem, C. J. Weegink, N. Forestier, A. van Vliet, J. van de Wetering de Rooij, L. McNair, S. Purdy, R. Kauffman, J. Adam, and P. L. M. Jansen. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology 131:997-1002. [DOI] [PubMed]
- 113.Rehermann, B., and F. V. Chisari. 2000. Cell mediated immune response to the hepatitis C virus. Curr. Top. Microbiol. Immunol. 242:299-325. [DOI] [PubMed] [Google Scholar]
- 114.Reichard, O., J. Andersson, R. Schvarcz, and O. Weiland. 1991. Ribavirin treatment for chronic hepatitis C. Lancet 337:1058-1061. [DOI] [PubMed] [Google Scholar]
- 115.Reiser, M., H. Hinrichsen, J. P. Benhamou, H. W. Reesink, H. Wedemeyer, C. Avendano, N. Riba, C. L. Yong, G. Nehmiz, and G. Steinmann. 2005. Antiviral efficacy of NS3-serine protease inhibitor BILN-2061 in patients with chronic genotype 2 and 3 hepatitis C. Hepatology 41:832-835. [DOI] [PubMed] [Google Scholar]
- 115a.Roberts, S., G. Cookdley, G. Dore, R. Robson, D. Shaw, H. Berns, M. Brandl, S. Fettner, G. Hill, D. Ipe, K. Klumpp, M. Mannino, E. O'Mara, Y. Tu, and C. Washington. 2006. Abstr. 57th Am. Assoc. Study Liver Dis., abstr. LB2.
- 116.Saito, T., T. Ito, H. Ishiko, M. Yonaha, K. Morikawa, A. Miyokawa, and K. Mitamura. 2003. Sequence analysis of PePHD within HCV E2 region and correlation with resistance of interferon therapy in Japanese patients infected with HCV genotypes 2a and 2b. Am. J. Gastroenterol. 98:1377-1383. [DOI] [PubMed] [Google Scholar]
- 117.Saiz, J. C., F. X. Lopez-Labrador, S. Ampurdanes, J. Dopazo, X. Forns, J. M. Sanchez-Tapias, and J. Rodes. 1998. The prognostic relevance of the nonstructural 5A gene interferon sensitivity determining region is different in infections with genotype 1b and 3a isolates of hepatitis C virus. J. Infect. Dis. 177:839-847. [DOI] [PubMed] [Google Scholar]
- 118.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809, table. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sandres, K., M. Dubois, C. Pasquier, J. L. Payen, L. Alric, M. Duffaut, J. P. Vinel, J. P. Pascal, J. Puel, and J. Izopet. 2000. Genetic heterogeneity of hypervariable region 1 of the hepatitis C virus (HCV) genome and sensitivity of HCV to alpha interferon therapy. J. Virol. 74:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sarrazin, C., U. Mihm, E. Herrmann, C. Welsch, M. Albrecht, U. Sarrazin, S. Traver, T. Lengauer, and S. Zeuzem. 2005. Clinical significance of in vitro replication enhancing mutations of the hepatitis C virus replicon in patients with chronic hepatitis C. J. Infect. Dis. 192:1710-1719. [DOI] [PubMed] [Google Scholar]
- 120a.Sarrazin, C., R. Rouzier, F. Wagner, N. Forestier, D. Larry, S. K. Gupta, M. Hussain, A. Shah, D. Cutler, J. Zhang, and S. Zeuzem. SCH503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alfa-2b for genotype 1 non-responders. Gastroenterology, in press. [DOI] [PubMed]
- 121.Sarrazin, C., T. Berg, J. H. Lee, B. Rüster, B. Kronenberger, W. K. Roth, and S. Zeuzem. 2000. Mutations in the protein kinase-binding domain of the NS5A protein in patients infected with hepatitis C virus type 1a are associated with treatment response. J. Infect. Dis. 181:432-441. [DOI] [PubMed] [Google Scholar]
- 122.Sarrazin, C., T. Berg, J. H. Lee, G. Teuber, C. F. Dietrich, W. K. Roth, and S. Zeuzem. 1999. Improved correlation between multiple mutations within the NS5A region and virological response in European patients chronically infected with hepatitis C virus type 1b undergoing combination therapy. J. Hepatol. 30:1004-1013. [DOI] [PubMed] [Google Scholar]
- 123.Sarrazin, C., M. Bruckner, E. Herrmann, B. Rüster, K. Bruch, W. K. Roth, and S. Zeuzem. 2001. Quasispecies heterogeneity of the carboxy-terminal part of the E2 gene including the PePHD and sensitivity of hepatitis C virus 1b isolates to antiviral therapy. Virology 289:150-163. [DOI] [PubMed] [Google Scholar]
- 124.Sarrazin, C., E. Herrmann, K. Bruch, and S. Zeuzem. 2002. Hepatitis C virus nonstructural 5A protein and interferon resistance: a new model for testing the reliability of mutational analyses. J. Virol. 76:11079-11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sarrazin, C., I. Kornetzky, B. Rüster, J. H. Lee, B. Kronenberger, K. Bruch, W. K. Roth, and S. Zeuzem. 2000. Mutations within the E2 and NS5A protein in patients infected with hepatitis C virus type 3a and correlation with treatment response. Hepatology 31:1360-1370. [DOI] [PubMed] [Google Scholar]
- 125a.Sarrazin, C., T. Kieffer, D. Bartels, B. Hanzelka, U. Mueh, M. Welker, D. Wincheringer, L. Chao, T. Grossmann, S. Purdy, C. Weegink, H. Reesink, S. Zeuzem, and A. Kwong. 2005. Abstr. 56th Am. Assoc. Study Liver Dis., abstr. LB06.
- 126.Schinkel, J., M. D. de Jong, B. Bruning, B. van Hoek, W. J. Spaan, and A. C. Kroes. 2003. The potentiating effect of ribavirin on interferon in the treatment of hepatitis C: lack of evidence for ribavirin-induced viral mutagenesis. Antivir. Ther. 8:535-540. [PubMed] [Google Scholar]
- 127.Senturk, H., A. Mert, M. Akdogan, F. Tabak, G. Basaran, S. Turkoglu, G. Ozbay, and S. Badur. 2000. Amantadine monotherapy of chronic hepatitis C patients infected with genotype lb. Scand. J. Infect. Dis. 32:575-576. [DOI] [PubMed] [Google Scholar]
- 128.Shindo, M., K. Hamada, S. Koya, K. Arai, Y. Sokawa, and T. Okuno. 1996. The clinical significance of changes in genetic heterogeneity of the hypervariable region 1 in chronic hepatitis C with interferon therapy. Hepatology 24:1018-1023. [DOI] [PubMed] [Google Scholar]
- 129.Sidwell, R. W., J. H. Huffman, G. P. Khare, L. B. Allen, J. T. Witkowski, and R. K. Robins. 1972. Broad-spectrum antiviral activity of Virazole: 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177:705-706. [DOI] [PubMed] [Google Scholar]
- 130.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 131.Simmonds, P., E. C. Holmes, T. A. Cha, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74:2391-2399. [DOI] [PubMed] [Google Scholar]
- 132.Song, J., M. Fujii, F. Wang, M. Itoh, and H. Hotta. 1999. The NS5A protein of hepatitis C virus partially inhibits the antiviral activity of interferon. J. Gen. Virol. 80:879-886. [DOI] [PubMed] [Google Scholar]
- 133.Squadrito, G., F. Leone, M. Sartori, B. Nalpas, P. Berthelot, G. Raimondo, S. Pol, and C. Brechot. 1997. Mutations in the nonstructural 5A region of hepatitis C virus and response of chronic hepatitis C to interferon alfa. Gastroenterology 113:567-572. [DOI] [PubMed] [Google Scholar]
- 134.Stark, G. R., I. M. Kerr, B. R. G. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 134a.Steinmann, E., G. Koutsoudakis, S. Kallis, F. Penin, R. Bartenschlage, and T. Pietschmann. 2005. Abstr. 12th Int. Symp. Hepat. C Virus Relat. Viruses, abstr. 31.
- 135.Sumpter, R., Jr., C. Wang, E. Foy, Y. M. Loo, and M. Gale, Jr. 2004. Viral evolution and interferon resistance of hepatitis C virus RNA replication in a cell culture model. J. Virol. 78:11591-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Taguchi, T., M. Nagano-Fujii, M. Akutsu, H. Kadoya, S. Ohgimoto, S. Ishido, and H. Hotta. 2004. Hepatitis C virus NS5A protein interacts with 2′,5′-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J. Gen. Virol. 85:959-969. [DOI] [PubMed] [Google Scholar]
- 137.Tam, R. C., B. Pai, J. Bard, C. Lim, D. R. Averett, U. T. Phan, and T. Milovanovic. 1999. Ribavirin polarizes human T cell responses towards a type 1 cytokine profile. J. Hepatol. 30:376-382. [DOI] [PubMed] [Google Scholar]
- 138.Tanabe, Y., N. Sakamoto, N. Enomoto, M. Kurosaki, E. Ueda, S. Maekawa, T. Yamashiro, M. Nakagawa, C. H. Chen, N. Kanazawa, S. Kakinuma, and M. Watanabe. 2004. Synergistic inhibition of intracellular hepatitis C virus replication by combination of ribavirin and interferon-alpha. J. Infect. Dis. 189:1129-1139. [DOI] [PubMed] [Google Scholar]
- 139.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. C. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 140.Taylor, D. R., B. Tian, P. R. Romano, A. G. Hinnebusch, M. M. Lai, and M. B. Mathews. 2001. Hepatitis C virus envelope protein E2 does not inhibit PKR by simple competition with autophosphorylation sites in the RNA-binding domain. J. Virol. 75:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tellinghuisen, T. L., J. Marcotrigiano, and C. M. Rice. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Teuber, G., T. Berg, U. Naumann, J. Raedle, S. Brinkmann, U. Hopf, and S. Zeuzem. 2001. Randomized, placebo-controlled, double-blind trial with interferon-alpha with and without amantadine sulphate in primary interferon-alpha nonresponders with chronic hepatitis C. J. Viral Hepat. 8:276-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Teuber, G., M. Pascu, T. Berg, M. Lafrenz, J. Pausch, F. Kullmann, G. Ramadori, R. Arnold, H. Weidenbach, E. Musch, U. Junge, K. H. Wiedmann, E. Herrmann, M. Zankel, and S. Zeuzem. 2003. Randomized, controlled trial with IFN-alpha combined with ribavirin with and without amantadine sulphate in non-responders with chronic hepatitis C. J. Hepatol. 39:606-613. [DOI] [PubMed] [Google Scholar]
- 144.Tong, X., R. Chase, A. Skelton, T. Chen, J. Wright-Minogue, and B. A. Malcolm. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antivir. Res. 70:28-38. [DOI] [PubMed] [Google Scholar]
- 144a.Villano, S., A. Howe, D. Raible, D. Harper, J. Speth, and G. Bichier. 2006. Abstr. 57th Am. Assoc. Study Liver Dis., abstr. 1127.
- 145.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang, C., J. Pflugheber, R. Sumpter, Jr., D. L. Sodora, D. Hui, G. C. Sen, and M. Gale, Jr. 2003. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J. Virol. 77:3898-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Watanabe, H., K. Nagayama, N. Enomoto, J. Itakura, Y. Tanabe, C. Sato, N. Izumi, and M. Watanabe. 2003. Amino acid substitutions in PKR-eIF2 phosphorylation homology domain (PePHD) of hepatitis C virus E2 protein in genotype 2a/2b and 1b in Japan and interferon efficacy. Hepatol. Res. 26:268-274. [DOI] [PubMed] [Google Scholar]
- 147a.Weich, V., E. Herrmann, C. Sarrazin, H. Hinrichsen, P. Buggisch, T. Gerlach, H. Klinker, U. Spengler, A. Bergk, B. Wiedenmann, S. Zeuzem, and T. Berg. 2004. Abstr. 55th Am. Assoc. Study Liver Dis., abstr. 389.
- 148.World Health Organization. 1999. Global surveillance and control of hepatitis C. Report of a WHO consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J. Viral Hepat. 6:35-47. [PubMed] [Google Scholar]
- 149.Xu, Z., J. Choi, T. S. B. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yang, S. S., M. Y. Lai, D. S. Chen, G. H. Chen, and J. H. Kao. 2003. Mutations in the NS5A and E2-PePHD regions of hepatitis C virus genotype 1b and response to combination therapy of interferon plus ribavirin. Liver Int. 23:426-433. [DOI] [PubMed] [Google Scholar]
- 151.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 152.Young, K. C., K. L. Lindsay, K. J. Lee, W. C. Liu, J. W. He, S. L. Milstein, and M. M. Lai. 2003. Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy. Hepatology 38:869-878. [DOI] [PubMed] [Google Scholar]
- 153.Zeuzem, S., R. Hultcrantz, M. Bourliere, T. Goeser, P. Marcellin, J. Sanchez-Tapias, C. Sarrazin, J. Harvey, C. Brass, and J. Albrecht. 2004. Peginterferon alfa-2b plus ribavirin for treatment of chronic hepatitis C in previously untreated patients infected with HCV genotypes 2 or 3. J. Hepatol. 40:993-999. [DOI] [PubMed] [Google Scholar]
- 154.Zeuzem, S., J. H. Lee, and W. K. Roth. 1997. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology 25:740-744. [DOI] [PubMed] [Google Scholar]
- 155.Zeuzem, S., G. Teuber, U. Naumann, T. Berg, J. Raedle, S. Hartmann, and U. Hopf. 2000. Randomized, double-blind, placebo-controlled trial of interferon alfa2a with and without amantadine as initial treatment for chronic hepatitis C. Hepatology 32:835-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155a.Zeuzem, S., C. Sarrazin, and J. Zhang. 2005. Abstr. 56th Am. Assoc. Study Liver Dis., abstr. 233.
- 156.Zhou, S., R. Liu, B. M. Baroudy, B. A. Malcolm, and G. R. Reyes. 2003. The effect of ribavirin and IMPDH inhibitors on hepatitis C virus subgenomic replicon RNA. Virology 310:333-342. [DOI] [PubMed] [Google Scholar]