Abstract
Trichophyton rubrum is a pathogenic filamentous fungus of increasing medical concern. Two antifungal agents, ketoconazole (KTC) and amphotericin B (AMB), have specific activity against dermatophytes. To identify the mechanisms of action of KTC and AMB against T. rubrum, a cDNA microarray was constructed from the expressed sequence tags of the cDNA library from different developmental stages, and transcriptional profiles of the responses to KTC and AMB were determined. T. rubrum was exposed to subinhibitory concentrations of KTC and AMB for 12 h, and microarray analysis was used to examine gene transcription. KTC exposure induced transcription of genes involved in lipid, fatty acid, and sterol metabolism, including ERG11, ERG3, ERG25, ERG6, ERG26, ERG24, ERG4, CPO, INO1, DW700960, CPR, DW696584, DW406350, and ATG15. KTC also increased transcription of the multidrug resistance gene ABC1. AMB exposure increased transcription of genes involved in lipid, fatty acid, and sterol metabolism (DW696584, EB801458, IVD, DW694010, DW688343, DW684992), membrane transport (Git1, DW706156, DW684040, DMT, DW406136, CCH1, DW710650), and stress-related responses (HSP70, HSP104, GSS, AOX, EB801455, EB801702, TDH1, UBI4) but reduced transcription of genes involved in maintenance of cell wall integrity and signal transduction pathways (FKS1, SUN4, DW699324, GAS1, DW681613, SPS1, DW703091, STE7, DW703091, DW695308) and some ribosomal proteins. This is the first report of the use of microarray analysis to determine the effects of drug action in T. rubrum.
Dermatophytosis is a common disease that can affect a large proportion of the population (5, 36). The incidence of dermatophytoses has increased over recent years, particularly in immunocompromised patients (8, 29, 31). The dimorphic anthrophilic fungus Trichophyton rubrum is an important cause of superficial dermatomycoses such as onychomycosis and tinea pedis (15, 39) and is known to account for as many as 69.5% of all dermatophyte infections. T. rubrum infections are often intractable, and relapse frequently occurs after cessation of antifungal therapy (20).
The azole antifungal agents such as itraconazole, clotrimazole, and ketoconazole are generally used to treat dermatomycosis (9, 22, 30). The mechanism of action of azoles involves inhibiting the cytochrome P450 enzyme lanosterol demethylase by binding to the heme in the active site of the enzyme (26). Amphotericin B (AMB) is a type of polyene antifungal drug, which binds to ergosterol in the fungal cell membrane, thereby compromising membrane integrity and ultimately leading to cell death (10). AMB is not effective in the treatment of dermatophyte infections (27) but has been shown to be very active against T. rubrum in vitro (7). The difference between the antifungal activity of AMB against T. rubrum in vivo and in vitro is a reason to determine its mechanism of action against this pathogenic fungus. cDNA microarrays are a good tool for drug target identification, as they can survey the global effects mediated by the addition of antifungal agents. Although the mechanisms of action of antifungal agents against some model fungi such as Saccharomyces cerevisiae and Candida albicans have been studied by cDNA microarrays (1, 3, 17), the inhibitory mechanisms of ketoconazole (KTC) and AMB against T. rubrum are still poorly understood.
To establish a molecular base for understanding the biological function of T. rubrum, we created a T. rubrum cDNA library derived from various developmental stages. The T. rubrum genome was estimated to be 22.05 Mb (4). We obtained 11,085 first-pass unique expressed sequence tags (ESTs) and used sequence analysis and database searching to identify known genes, define putative novel genes, and propose some possible metabolic networks in this fungus. To identify class-specific and mechanism-independent changes in gene expression, we examined changes in transcriptional profiles of T. rubrum in response to the azole and polyene classes of antifungal agents by cDNA microarray.
As mentioned above, the prevalence of T. rubrum infections and its anthrophilic nature make it a good model for the study of human pathogenic filamentous fungus, and our work in identifying ESTs in T. rubrum cDNA libraries will improve our understanding of the molecular mechanisms of its growth, metabolism, pathogenesis, and drug resistance.
MATERIALS AND METHODS
Fungus and material.
The T. rubrum clinical isolate BMU 01672 used in this study was obtained from the Research Center for Medical Mycology, Peking University. The isolate was confirmed as T. rubrum by morphological identification of both microscopic and macroscopic characteristics (6, 16) as well as by PCR amplification and sequencing of the 18S ribosomal DNA and internal transcribed spacer regions (13). The potato dextrose agar (PDA), yeast extract, peptone, and d-glucose used for the strain cultures were bought from Difco.
Antifungal agents.
KTC and AMB were obtained from Sigma (St. Louis, MO). Stock solutions of varying concentrations were made in dimethyl sulfoxide (Sigma).
Preparation of cell cultures.
Potato dextrose agar (39 g/liter) was inoculated with a few hyphae of T. rubrum and incubated at 28°C for 2 to 3 weeks until good conidiation was produced. The mixture of conidia and hyphal fragments was collected in distilled water. The conidia, sprouted conidia, and mycelia of different developmental stages to be used for construction of the cDNA library were collected as follows: (i) the conidia were harvested by Whatman's filter model 40, (ii) a group of conidia were incubated at 28°C in YPG medium (10 g/liter yeast extract, 20 g/liter peptone, 10 g/liter d-glucose) for approximately 8 h to produce sprouted conidia, and (iii) the hyphae were placed in 100-ml aliquots of YPG medium and incubated in a 28°C bath shaker for 7, 10, 14, 15, 16, 20, 22, 26, 28, 34, and 36 days. The above-mentioned collections were centrifuged, respectively, the supernatant was discarded, and the pelleted material was washed twice with phosphate-buffered saline.
Construction of cDNA libraries and sequencing.
The cDNA libraries were constructed following the protocols of the SUPERSCRIPT plasmid system with GATEWAY Technology for cDNA synthesis and cloning (Invitrogen). The cDNA plasmids were isolated by the Millipore method using MADV filter plates. Sequencing was performed with a generic T7 primer located 5′ upstream of the inserted segments, following the protocol of the PRISM Big Dye terminator kit on an ABI3700 automated sequencer.
EST processing pipeline and annotation.
Phred quality assessment and computational analysis were carried out as follows. The trace files from the sequencer were “basecalled” by Phred with the quality value set at >Q15. The cross-match software from the Phrap package (http://www.phrap.org) was used to remove vector sequences. The poly(A) tails were clipped from some of the ESTs using the Trimmest program from the EMBOSS package. Processed ESTs longer than 200 bp were clustered by comparing all base pairs using BLASTN and collecting those with >95% identity over regions longer than 40 bp and with unmatched overhangs of <20 bp. The sequences contained in each cluster were assembled using CAP3 to identify the consensus ESTs. The BLASTX program (2) was used for annotation, together with GenBank nonredundant clusters of orthologous groups and gene ontology databases. The metabolic pathway networks of T. rubrum were partially reconstructed by searching for known pathway homologs in the Kyoto Encyclopedia of Genes and Genomes database.
cDNA microarrays.
PCR fragments used for printing the microarray chip were amplified from the EST library in 96-well plates using vector-PCR amplification with T7 and SP6 universal primers. PCR products were analyzed on gels to confirm the success of the reactions and were subsequently purified using MultiScreen-PCR plates (Millipore). Purified PCR products were resuspended in 50 μl of 3× SSC (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate [pH 7.0]) to produce the microarrays. A set of microarrays containing a total of 11,232 spots (including 10,250 clones in the form of PCR products and 982 controls in each block, including blank, negative, and positive controls) were spotted in duplicate on slides (Corning) with a Cartesian arrayer. The spotted cDNA was cross-linked to the surface of the slides (at 65 mJ) using a StrataLinker instrument and washed with 1% sodium dodecyl sulfate (SDS) to minimize background signal. Slides were subsequently placed in a blocking solution containing 0.2 M succinic anhydride and 0.05 M sodium borate prepared in 1-methyl-2-pyrrolidinone for 20 min, washed for 2 min in 95°C water, and rinsed five times in 95% ethanol. Slides were spin-dried at 500 rpm for 5 min and stored for future hybridizations.
MIC determinations.
The broth microdilution assay for antifungal susceptibility testing of dermatophytes was previously developed as a modification of the National Committee for Clinical Laboratory Standards (NCCLS) M38-A method (21). RPMI 1640 medium (GIBCO BRL, Barcelona, Spain) with l-glutamine but without sodium bicarbonate and buffered at pH 7.0 with 0.165 M morpholinepropanesulfonic acid (MOPS) (Sigma, Madrid, Spain) was used for broth microdilution susceptibility testing. Test concentrations for KTC and AMB ranged from 0.01 to 64.0 μg/ml. A standardized inoculum was prepared by counting the microconidia microscopically. Cultures was subcultured onto PDA and incubated at 28°C for 7 days to produce conidia. Sterile normal saline (0.85%) was added to the agar slant, and the cultures were gently swabbed with a cotton-tipped applicator to dislodge the conidia from the hyphal mat. The mixture of conidia and hyphae fragments was filtered with a Whatman filter model 40 (pore size 8 μm), which retains hypha fragments and only allows passage of T. rubrum microconidia. The suspension was transferred to a sterile centrifuge tube, and the volume was adjusted to 5 ml with sterile normal saline. Microdilution plates were set up in accordance with the NCCLS M38-A reference method. The final inoculum size was adjusted to between 0.4 × 104 and 5.0 × 104 CFU/ml. The microdilution plates were incubated at 30°C and were assessed visually after 72 h of incubation. For azole agents, the MIC was defined as the lowest concentration at which growth was markedly inhibited (approximately 50% growth of the control). For AMB, the MIC was defined as the lowest concentration at which 100% growth was inhibited. MIC results were recorded in micrograms per milliliter. Quality control strains Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 were used to validate the susceptibility testing results.
Cell culture and drug exposure for microarray experiments.
The isolates were subcultured onto PDA plates at 28°C. Stock inoculum suspensions of each isolate were prepared for each experiment from 7-day-old cultures grown on PDA. The mixtures of conidia and hyphal fragments were collected in distilled water. These stock suspensions were diluted in RPMI medium to obtain inoculum sizes of 1.0 × 106 CFU/ml. For a single microarray experiment, a total of six 100-ml cultures were prepared (three independent 100-ml cultures were grown for each drug). All drug exposures were performed on the same day using the same starting culture to minimize experimental variations, and final inoculum sizes of 1.0 × 104 CFU/ml were reached. The cultures were incubated at 30°C and 140 rpm to saturation (4 days). An antifungal drug was added to three cultures at a concentration equivalent to 0.5 times the MIC (0.25 μg/ml KTC, 0.5 μg/ml AMB) and incubated. Three control cultures were treated with an appropriate amount of dimethyl sulfoxide. Twelve hours after drug addition, three control cultures and three drug-treated cultures were harvested by filtration through Whatman no. 1 filter paper, washed thoroughly with sterile water, and quickly frozen in liquid nitrogen until RNA preparation.
RNA preparation.
The frozen T. rubrum cells were ground into a powder with a mortar and pestle in liquid nitrogen to facilitate cell disruption. Total RNA was isolated using the QIAGEN RNeasy plant mini kit (QIAGEN, Inc., Valencia, CA) according to the manufacturer's instructions. The RNA concentration and purity were determined spectrophotometrically by measuring absorbance at 230, 260, 280, and 320 nm. The purity and integrity of the RNA were confirmed by agarose gel electrophoresis. Three independent sets of RNA from control and three independent sets of RNA from drug-treated cells were used to prepare six independent cDNA sets for AMB and KTC, respectively. An aliquot of poly(A)+ mRNA was isolated with the Oligotex mRNA mini kit (QIAGEN).
Microarray hybridization.
First-strand cDNA was synthesized using Superscript II RT (Life Technologies/Invitrogen, Carlsbad, CA). An aliquot of poly(A)+ mRNA from 20 μg of total RNA sample was added to oligo(dT) (18- to 21-mer). The reaction mixture was heated to 65°C for 5 min and quickly chilled on ice. First-strand buffer (5×; 50 mM Tris-HCl, pH 8.3, at room temperature, 75 mM KCl, and 3 mM MgCl2); 2× 0.1 M dithiothreitol; 10 mM (each) dATP, dCTP, dGTP, and TTP; and RNase OUT (40 U) were added to the mixture and incubated at 42°C for 2 min. Four hundred units of SuperScript II RT were added to the mixture and incubated at 42°C for an hour. To inactivate the reaction, the mixture was heated at 70°C for 15 min. Second-strand cDNA was synthesized as follows: 5× second-strand buffer [20 mM Tris-HCl, pH 6.9; 90 mM KCl; 4.6 mM MgCl2; 0.15 mM β-NAD+; 10 mM (NH4)2SO4]; 0.2 mM deoxynucleoside triphosphate mix; Escherichia coli DNA ligase (10 U); E. coli DNA polymerase I (40 U); and E. coli RNase H (2 U) were added to a first-strand reaction tube and incubated for 2 h at 16°C. To stop the reaction, 10 μl 0.5 M EDTA was added. Double-stranded cDNA (dscDNA) was purified using QIAquick columns (QIAGEN, Valencia, CA) by following the manufacturer's instructions. dscDNA was then fluorescently labeled using BioPrime DNA labeling system (Life Technologies/Invitrogen, Carlsbad, CA). Those representing RNA from drug-treated cells were labeled using Cy5, and those representing RNA from control cells were labeled using Cy3. dscDNA was added to 2.5× random primer solution (50 mM Tris-HCl, pH 6.8, 5 mM MgCl2, 10 mM 2-mercaptoethanol, 300 μg/ml oligodeoxyribonucleotide primer), boiled at 100°C for 5 min, and then chilled on ice. deoxynucleoside triphosphate mix for DNA labeling (10×; 0.12 mM [each] dATP, dGTP, dTTP; 0.06 mM dCTP; 1 mM Tris-HCl, pH 8.0; 0.1 mM EDTA), Cy3 (or Cy5) dCTP (0.06 mM), and Klenow fragment (40 U) were added. The mixture was briefly centrifuged and was incubated at 37°C overnight away from light. Five microliters of 0.5 M EDTA (pH 8.0) was used to stop the reaction. Labeled cDNA was purified using QIAquick columns and mixed with 2 μg poly(A), 5× Denhardt's, 3× SSC, 24 μg yeast tRNA, 25 mM HEPES, pH 7.0, and 0.25% SDS. The mixture was heated at 100°C for 2 min, cooled to room temperature, and applied to the array slides under glass coverslips. Hybridization was performed at 65°C overnight in a Micro hybridization incubator (Robbins Scientific, Sunnyvale, CA). Slides were washed in 2× SSC (0.1% SDS), 1× SSC, and 0.2× SSC sequentially and then were scanned at 5-μm resolution on a GenePix 4000B scanner (Axon Instruments, Inc). The Cy5- and Cy3-labeled DNA samples were scanned at 635 and 532 nm, respectively.
Data analysis.
GenePix 6.0 software (Axon Instruments, Inc.) was used for image analysis and data visualization. Prior to data analysis, signals were normalized using a locally weighted scatter plot smoothing regression (LOWESS) algorithm (33) in the MIDAS software package (http://www.tigr.org/software/tm4) (25), with the smoothing parameter set to 0.33. Genes were considered differentially expressed if (i) average expression changed by at least threefold in three independent experiments performed with triplicate RNA samples or (ii) the change in gene expression was in the same direction (“increased” or “decreased”) in three experiments.
Quantitative real-time RT-PCR.
Quantitative real-time reverse transcription (RT)-PCR was used to verify the microarray result. Aliquots of the RNA preparations from untreated and treated samples used in the microarray experiments were saved for quantitative real-time RT-PCR follow-up studies. First-strand cDNAs were synthesized from 2 μg of total RNA in a 100-μl reaction volume using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA) in accordance with the manufacturer's instructions. Quantitative real-time PCR experiments were performed in triplicate using the 7000 sequence detection system (Applied Biosystems, Foster City, CA). Independent PCRs were performed using the same cDNA for both the gene of interest and the 18S rRNA, using the SYBR green PCR master mix (Applied Biosystems). Gene-specific primers were designed for the genes of interest and the 18S rRNA using Primer Express software (Applied Biosystems) and are shown in Table 1. The PCR cycle consisted of AmpliTaq Gold activation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. A dissociation curve was generated at the end of each PCR cycle to verify that a single product was amplified using software provided with the 7000 sequence detection system. The changes in fluorescence of SYBR green I dye in each cycle were monitored by the system software, and the threshold cycle (CT) above the background for each reaction was calculated. The CT value of 18S rRNA was subtracted from that of the gene of interest to obtain a ΔCT value. The ΔCT value of an arbitrary calibrator (e.g., untreated sample) was subtracted from the ΔCT value of each sample to obtain a ΔΔCT value. The gene expression level relative to the calibrator was expressed as 2−ΔΔCT.
TABLE 1.
Gene-specific primers used for real-time RT-PCRs
| Target | Sequence (5′-3′) (directiona) | Amplicon size (bp) |
|---|---|---|
| 18S | CGCTGGCTTCTTAGAGGGACTAT (F) | 51 |
| TGCCTCAAACTTCCATCGACTT (R) | ||
| ERG6 | TCATTTCTGCCGCTTTGGTAT (F) | 70 |
| CGCAAGATAGTGTTCGTGTCTTG (R) | ||
| ERG11 | CACTTCCTTGCCCTGTAGAGATC (F) | 78 |
| GGAGTTTTCAATGTCAGCAAGGTT (R) | ||
| ERG26 | TCCCGCTGTCTGAGCATTATTA (F) | 90 |
| TGAAACCGTCCGTGATGGC (R) | ||
| ABC1 | CCAGCCTCCTCAGCCTCTTT (F) | 91 |
| GGTGGGGCGATTTGTTGTC (R) | ||
| phiA | GCTGTCTGTGAAGTCAAGCCTGG (F) | 51 |
| GAAGATGGCGTCCTTGGGTC (R) | ||
| GIT1 | TCGGCTACATAAGCGACCACT (F) | 76 |
| GCAGCGAATACGATCAGGATAATA (R) | ||
| FKS1 | TGCTGAGTTCGAGGAAATGACC (F) | 58 |
| TGGAAGACCTGGAGTGTATGGAG (R) | ||
| GAS1 | CGCTGCTGATGTCCGTGATA (F) | 51 |
| CAGGTAACGTAGTTCCAGGTGTCC (R) | ||
| DW701717 | ATGCCCACGAAGCAGACAAC (F) | 75 |
| CTTCCAGCAGAGTCCGTTCCT (R) | ||
| DW708548 | GTAGGTCCATAGCGGCGTCTC (F) | 90 |
| GCCTGGCTGGGGTTGATG (R) |
F, forward; R, reverse.
RESULTS
MIC determinations.
The susceptibilities of T. rubrum to antifungal compounds were determined. The antifungal drugs used were KTC and AMB. Based on those experiments mentioned above, the MICs for the two drugs were determined as 0.5 μg/ml for KTC and 1 μg/ml for AMB.
Construction of cDNA libraries.
A total of 3,816 contigs and 7,269 singlets were found, which constitute the 11,085 T. rubrum assembled EST database. The ESTs were annotated using the databases mentioned above. It is noteworthy that some important genes involved in the growth, metabolism, signal transduction, pathogenesis, and drug resistance of T. rubrum were identified, which could be used to determine important metabolic pathways in T. rubrum.
Global gene expression results.
In total, 387 genes were differentially expressed upon exposure to KTC; 200 of which showed a significant increase in expression and 187 of which showed a significant decrease in expression. The data sets have been exported to the GEO (GEO accession number GSE 5014). The distribution of KTC-responsive genes and their biological roles are shown in Table 2. Of the genes that showed a response to KTC, most were classed as “unknown function” (48.6%), and the others were classified as involved in amino acid transport and metabolism (6.5%), lipid, fatty acid, and sterol metabolism (4.4%), posttranslational modification, protein turnover, chaperones (4.4%), and carbohydrate transport and metabolism (3.9%). Some genes among the 387 genes differentially expressed upon exposure to KTC were listed in Table 3.
TABLE 2.
KTC-responsive genes grouped by functional classification
| Functional classification | Gene(s) or accession no. |
|---|---|
| Translation, ribosomal structure, and biogenesis | DW700253, DW693737, DHH1, DW701896, DW406697, DW698669, DW700103, DW702114, DW688959, EB801718, DW683699 |
| Transcription | ARO8, DHH1, SPS1, DW696767, DW701896, DW692217, DW699444, DW705142, DW704069, HIP1 |
| Signal transduction mechanisms | DW710420, SPS1, DW707020, DW406202, DW701611, DW692217, RBG2, DW701993, DW705142, DW684792, HIP1, DW696808 |
| Secondary metabolites biosynthesis, transport, and catabolism | EB801566, DW703471, DW679333, DW692145, DW696127, DW702612, TMT1, DW695862 |
| RNA processing and modification | DW710174 |
| Posttranslational modification, protein turnover, chaperones | GST, DW699217, DW702672, DW693794, DW678523, DW679113, DW679068, DW697385, QRI7, CLP, HMT1, DW682878, DW680862, DW710922, DW680089, DW709783, DW701227 |
| Nucleotide transport and metabolism | DW698280, DW704384, DW690402 |
| Nuclear structure | DW679616, CRM1/MSN5 |
| Lipid metabolism | ERG6, CPO, ERG25, ERG26, ERG24, ERG4, CPR(P450R), DW406350, INO1, ATG15, ERG3, ERG11, DW696584, DW700960, IVD, DW678488, ECH |
| Intracellular trafficking, secretion, and vesicular transport | VPS33/slp1, ATG15, DW696952, DW702677, DW692357, CRM1/MSN5, YKT6, DW685279 |
| Inorganic ion transport and metabolism | DW706483, SIT1, DW684278, DW707140, SUL1, DW701993, DW683256, DW684457, DW683754, DW705131 |
| Extracellular structures | DW681163 |
| Energy production and conversion | DW698110, DW678636, DW698333, NDH, DW703655, DW691155, IDH, ACO2, DW406745, DW703259, DW703411, DW697175, COX4, DW678789 |
| DNA replication, recombination, and repair | DHH1, SPS1, DW406049, DW701896, DW692217, DW701869, HIP1 |
| Defense mechanisms | DW709093, DW405954 |
| Cytoskeleton | DW406457, SLA2, CAP2 |
| Coenzyme metabolism | DW701101, DW703971, DW684388, DW406323, DW406862, DW703639, DW697919, DW697729, DW710087, PDX1, DW705554 |
| Chromatin structure and dynamics | NCA2, DW689402 |
| Cell envelope biogenesis, outer membrane | DW680921, DW699411 |
| Cell division and chromosome partitioning | NUF2 |
| Carbohydrate transport and metabolism | DW704437, SKN1, DW702573, DW680921, DW698094, SIT1, DW699411, DW707140, DW683904, DW682260, FBP, DW684457, DW707827, DW709791, DW705131 |
| Amino acid transport and metabolism | ERG26, ARO8, DW708866, DW678965, SIT1, DW406862, DW703560, DW707140, DW697919, IDH, DW699207, DW697889, DW697729, ILV6, SAT, GGT, DW707501, DW683263, DW703967, DW684457, DW705554, GAP1, DW705131, DW678242, DW696663 |
| Other function | ABC1, TVP18, DW682938, DW683010, ANK, DW679066, DW691457, DW707096, RCI, DW678846, PhiA, DCW1, DW683510, DW694309, DW693546 |
| Unknown | Data not shown |
TABLE 3.
Some genes differentially expressed upon exposure to KTC
| Function and GenBank accession no. | Gene symbol | Annotated function(s)a | Fold induction/ repressionb |
|---|---|---|---|
| Ergosterol biosynthesis | |||
| DW693552 | ERG11 | Lanosterol 14-alpha-demethylase | +3.6 |
| DW705150 | ERG26 | C-3 sterol dehydrogenase | +11.7 |
| DW695095 | ERG3 | Sterol C-5 desaturase | +3.6 |
| EB801453 | ERG6 | Sterol C-24 methyltransferase | +37.5 |
| DW692574 | ERG4 | Sterol C-24 reductase | +6.5 |
| EB801509 | ERG24 | Sterol C-14 reductase | +8.6 |
| EB801697 | ERG25 | Sterol C-4 methyloxidase | +11.9 |
| Lipid metabolism | |||
| EB801711 | CPO | Chloride peroxidase | +25.0 |
| DW692676 | INO1 | Myoinositol-1-phosphate synthase | +3.8 |
| DW679389 | CPR | NADPH-cytochrome P450 reductase | +5.5 |
| DW685502 | ATG15 | Lipase required for intravacuolar lysis of autophagic bodies | +3.7 |
| DW696584 | Acyltransferase | +3.6 | |
| DW700960 | Fatty acid desaturase | +3.6 | |
| DW406350 | Cytochrome P450 involved in gamma-hexachlorocyclohexane degradation | +4.0 | |
| DW684219 | IVD | Isovaleryl-CoA dehydrogenase | −3.6 |
| DW678488 | 3-Oxoacid CoA transferase 1 | −4.4 | |
| EB801635 | ECH | Enoyl-CoA hydratase | −12.7 |
| Cell wall biosynthesis | |||
| DW680301 | PhiA | Putative cell wall protein | −5.3 |
| DW687782 | DCW1 | Homologous to Dfg5p | −5.8 |
| Multidrug resistance, DW680156 | ABC1 | Protein similar to yeast ABC transporters | +6.3 |
| Translation, ribosomal structure, and biogenesis | |||
| DW406697 | 60S ribosomal protein L35A/L37 | −3.1 | |
| DW697947 | 60S ribosomal protein L36 | −3.9 | |
| DW702114 | 60S ribosomal protein L6 | −3.4 | |
| EB801718 | 60S ribosomal protein L3 | −3.4 | |
| DW679066 | 60S ribosomal protein yl16a | −3.1 | |
| DW683699 | 60S ribosomal protein 15.5 kDa/SUN13 | −8.3 | |
| DW688959 | 40S ribosomal protein S18 | −3.4 | |
| DW698669 | 40S ribosomal protein S10 | −3.2 | |
| DW700253 | 40S ribosomal protein S15/S22 | +5.0 | |
| DW693737 | Mitochondrial/chloroplast ribosomal protein L15/L10 | +4.8 |
CoA, coenzyme A.
+, induction; −, repression.
In total, 686 genes were differentially expressed upon exposure to AMB; 244 of which showed a significant increase in expression and 442 of which showed a significant decrease in expression. The distribution of AMB-responsive genes and their biological roles are shown in Table 4. Of the genes that showed a response to AMB, most were of unknown function (44.2%), and the others were classified as involved in amino acid transport and metabolism (7.7%), translation, ribosomal structure, and biogenesis (4.8%), lipid, fatty acid, and sterol metabolism (3.5%), and inorganic ion transport and metabolism (2.5%). Some genes among the 686 genes differentially expressed upon exposure to AMB were listed in Table 5.
TABLE 4.
AMB-responsive genes grouped by functional classification
TABLE 5.
Some genes differentially expressed upon exposure to AMB
| Function and GenBank accession no. | Gene symbol | Annotated function(s)a | Fold induction/ repressionb |
|---|---|---|---|
| Membrane transport | |||
| DW680706 | Git1 | Putative glycerophosphoinositol permease | +7.3 |
| DW706156 | Glycine betaine/l-proline ABC transporter | +4.3 | |
| DW684040 | Sulfate ABC transporter ATP-binding protein | +4.0 | |
| DW708548 | DMT | Permeases of the drug/metabolite transporter | +3.4 |
| DW406136 | Synaptic vesicle transporter | +3.1 | |
| DW679720 | CCH1 | Voltage-gated Ca2+ channels (alpha 1 subunits) | +7.3 |
| DW710650 | Outer membrane receptor proteins | +3.9 | |
| DW704599 | PTR2 | Dipeptide/tripeptide permease | −5.0 |
| DW697886 | GAP1 | Amino acid permease | −15.5 |
| DW700599 | Sugar phosphate permease | −4.5 | |
| DW684458 | Arabinose efflux permease | −4.1 | |
| DW692468 | Atp1a1 | Na+/K+ transporting ATPase alpha 1 polypeptide | −3.4 |
| DW704314 | NMT1 | Periplasmic components of the ABC-type nitrate/sulfonate/bicarbonate transport systems | −4.0 |
| DW699236 | Permease of the major facilitator superfamily | −3.4 | |
| DW686710 | Arsenite-translocating ATPase | −3.4 | |
| DW696869 | Co/Zn/Cd efflux system component | −3.4 | |
| DW680730 | MEAA | Ammonium transporter | −4.0 |
| DW692685 | Ca2+-transporting ATPase type 2C member 1 | −5.8 | |
| DW707140 | Siderochrome-iron transporter | −9.0 | |
| DW689541 | Ctr1 | Copper uptake transporter | −11.3 |
| Ribosomal structure and biogenesis | |||
| DW679628 | 60S ribosomal protein L19 | −3.4 | |
| DW686754 | 60S ribosomal protein L28 | −3.3 | |
| DW692688 | 60S ribosomal protein L11 | −4.4 | |
| DW679066 | 60S ribosomal protein yl16a | −3.2 | |
| DW680751 | Ribosomal protein L13 | −3.2 | |
| DW692644 | Ribosomal protein S18 | −3.2 | |
| DW703685 | Ribosomal protein HS6 type (S12/L30/L7a) | −5.5 | |
| DW698567 | Mitochondrial large ribosomal subunit YmL35 | −4.9 | |
| EB801718 | 60S ribosomal protein L3 and related proteins | +24.5 | |
| EB801473 | 60s ribosomal protein L34 | +6.8 | |
| EB801470 | 40s ribosomal protein S27 | +11.5 | |
| Stress response | |||
| DW701186 | Probable chaperone protein DnaK | +3.2 | |
| DW692280 | Chaperone HSP104 | ||
| DW681149 | GSS | Glutathione synthetase | +4.2 |
| EB801466 | AOX | Alternative oxidase, mitochondrial precursor | +38.3 |
| EB801455 | Putative stress response RCI peptide | +56 | |
| EB801465 | TDH1 | Glyceraldehyde-3-phosphate dehydrogenase | +8.3 |
| DW688240 | UBI4 | Ubiquitin | +3.9 |
| DW689028 | HSP10 | Cochaperonin GroES (HSP10) | −4.3 |
| DW678423 | Predicted nucleotide kinase/nuclear protein involved oxidative stress response | −3.1 | |
| DW710246 | GPX | Glutathione peroxidase | −5.4 |
| DW694000 | HOR2 | dl-Glycerol-3-phosphatase | −4.0 |
| DW678364 | YAR1 | Ankyrin repeat protein | −3.5 |
| Lipid, fatty acid, and sterol metabolism | |||
| DW696584 | Glycerol 3-phosphate acyltransferase | +5.7 | |
| EB801458 | Carnitine O-acyltransferase | +5.3 | |
| DW682185 | Acyl carrier protein phosphodiesterase | +4.5 | |
| DW684219 | IVD | Isovaleryl-CoA dehydrogenase | +3.7 |
| DW694010 | Peroxisomal D3,D2-enoyl-CoA isomerase | +3.2 | |
| DW688343 | Carboxylesterase type B | +3.1 | |
| DW684992 | Allophanate hydrolase subunit 1 | +3.1 | |
| DW703105 | Related to short-chain alcohol dehydrogenases | −5.2 | |
| DW697546 | Cytochrome P450 CYP4/CYP19/CYP26 subfamilies | −4.7 | |
| DW692574 | ERG4 | Ergosterol biosynthesis ERG4/ERG24 family | −4.7 |
| DW699008 | 3-Methylcrotonyl-CoA carboxylase | −4.5 | |
| DW705750 | Phosphomannomutase 2 | −4.3 | |
| EB801635 | Enoyl-CoA hydratase | −3.9 | |
| DW680143 | ACS | Acyl-coenzyme A synthetases/AMP (fatty) acid ligases | −3.6 |
| DW703927 | Phosphoethanolamine cytidylyltransferase | −3.5 | |
| DW708678 | Lysosomal and prostatic acid phosphatases | −3.8 | |
| DW710649 | Short-chain acyl-CoA dehydrogenase | −3.6 | |
| DW678450 | Phosphatidic acid-preferring phospholipase A1, contains DDHD domain | −3.5 | |
| DW683501 | 3-Oxo-5a-steroid 4-dehydrogenase | −3.4 | |
| DW698659 | IDI1 | Isopentenyldiphosphate isomerase | −3.3 |
| DW707302 | Enoyl-[acyl carrier protein] reductase | −3.2 | |
| DW690970 | 3-Hydroxyacyl-CoA dehydrogenase | −3.1 | |
| DW691391 | ELO2 | Fatty acid elongase | −3.1 |
| DW686379 | MVD1 | Mevalonate pyrophosphate decarboxylase | −3.1 |
| Cell wall biosynthesis | |||
| DW687269 | FKS1 | Catalytic subunit of 1,3-beta-d-glucan synthase | −3.5 |
| DW678357 | SUN4 | Cell wall protein related to glucanases | −6.0 |
| DW699324 | 1,3-Beta-glucan synthase catalytic subunit | −3.8 | |
| DW703981 | GAS1 | Beta-1,3-glucanosyltransferase | −4.2 |
| DW681613 | Glycosylphosphatidylinositol-anchored protein | −3.1 | |
| DW679073 | Integral membrane protein, interacts with FtsH | +3.9 | |
| Signal transduction | |||
| DW703091 | Mitogen-activated protein kinase MpkA | −3.9 | |
| DW699167 | STE7 | Mitogen-activated protein kinase kinase | −3.1 |
| DW695308 | Two-component phosphorelay intermediate involved in mitogen-activated protein kinase cascade regulation | −4.4 | |
| DW694457 | SPS1 | Serine/threonine protein kinase | −3.2 |
| Multidrug resistance | |||
| DW690470 | RND family efflux system component | +3.9 | |
| DW708548 | Permeases of the drug/metabolite transporter superfamily | +3.4 | |
| DW698195 | ABC-type multidrug transport system, ATPase component | −4.8 | |
| DW701717 | Na+-driven multidrug efflux pump | −5.1 |
CoA, coenzyme A.
+, induction; −, repression.
Validation of microarray data by real-time RT-PCR.
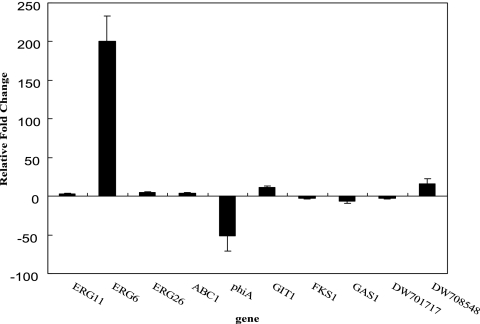
Real-time quantitative RT-PCR was conducted to validate microarray data using the same RNA from the original microarray experiment. Ten genes (5 per drug) were tested not only to confirm their roles in the response of T. rubrum to the respective drugs (e.g., ERG11, ERG6, and ERG26 for KTC; GIT1, FKS1, and GAS1 for AMB), but also to verify the novel responses identified in the present study (e.g., ABC1 and phiA for KTC; DW701717 and DW708548 for AMB.). The result of real-time PCR analysis was shown in Fig. 1. There was a strong positive correlation (r = 0.92) between the two techniques; 6 genes showed upregulation and 4 genes showed downregulation in response to drug treatment, which confirmed the reliability of the microarray data.
FIG. 1.
The relative change for 10 genes listed in Table 1 determined by quantitative real-time RT-PCR.
DISCUSSION
Gene expression responses to KTC.
The number and characteristics of the responsive genes are shown according to biological function in Table S1 in the supplemental material. In our large data pool, most responsive genes are classed as being of “unknown function” (188 genes), indicating that the EST has not previously been identified and there is no amino acid sequence homology with other proteins of known function.
In our experiment, some genes of the ergosterol biosynthesis pathway were upregulated in response to KTC, including genes encoding ERG11, ERG26, ERG3, ERG6, ERG4, ERG24, and ERG25. ERG11 encodes the azole target enzyme lanosterol demethylase, whereas ERG26, ERG3, ERG6, ERG24, ERG4, and ERG25 are functional downstream of ERG11, which indicates that their induction is in response to ergosterol depletion. Changes in the transcription levels of genes of the ergosterol pathway are in agreement with previous studies showing that this pathway is the target of azoles (1, 17). The most highly differentially expressed gene in this category was ERG6, which showed an ∼37-fold induction in response to KTC. ERG6 catalyzes a biosynthetic step not found in humans, and disruption of this gene in Saccharomyces cerevisiae has been shown to result in several compromised phenotypes, most markedly increased permeability. Inhibitors of the ERG6 gene product would make the cell increasingly susceptible to antifungal agents that would normally be excluded from the fungus and would allow for clinical treatment at lower dosages (14). We note that ERG6 may be a potential drug target of antifungal agents.
In addition to the ergosterol biosynthesis genes, transcription of seven additional genes involved in lipid metabolism was also increased, including CPO, INO1, CPR, ATG15, DW696584, DW700960, and DW406350. CPR (P450R) is required for microsomal eukaryotic cytochrome P-450 (CYP) monooxygenase activity, transferring both or sometimes just the first electron required for these reactions (38). The CYP enzymes are involved in the metabolism of foreign compounds such as lipophilic pollutants, pesticides, and drugs, as well as in many biosynthetic reactions, such as steroid, alkaloid, and terpenoid biosynthesis. Overexpression of CYP51 and FUS has been shown to produce different levels of KTC resistance in wild-type cells, indicating that the availability of CPR may limit the potential of overproduction of CYP51 as a mechanism of resistance to azole antifungal agents (37). Three genes involved in lipid metabolism were repressed in this study, including IVD, DW678488, and ECH.
In addition, KTC induced the transcription of the multidrug resistance gene ABC1. The observation that KTC induced transcription of ABC1 in T. rubrum is a key finding of this study. ABC1-encoded protein is similar to yeast ABC transporters (Pdr5 and Cdr1) believed to be involved in multidrug resistance, and ABC1 gene transcripts are inducible by toxic drugs. It has previously been shown that treatment of wild-type Magnaporthe grisea cultures with the antifungal compounds miconazole and metconazole markedly increase ABC1 transcript levels. An alternative hypothesis is that Abc1 provides a defense function during early stages of pathogenesis by acting as an efflux pump to provide resistance to antimicrobial compounds (32).
Our results showed that expression of PhiA was repressed by a factor of 5.3. It has been reported that cell wall protein encoded by the phiA gene of Aspergillus nidulans has a critical role in the development of normal phialides, with phiA-deficient mutants displaying abnormal phialides and marked reductions in conidiation. Melin et al. propose a more challenging hypothesis in which the fungus might sense an attack by toxic metabolites, and that synthesis of PhiA is initiated to produce conidia and ensure survival (19).
We found that transcription of the genes encoding eight ribosomal proteins (L35A/L37, L36, L6, L3, yl16a, 15.5kD/SNU13, S18, and S10) was downregulated significantly. Small reductions in the expression of ribosomal protein genes allow energy to be redistributed to allow increased expression of genes involved in protective responses, while maintaining a basal level of protein synthesis (34). It should be noted that not all ribosomal protein transcripts are repressed. Ribosomal protein S15/S22 and mitochondrial/chloroplast ribosomal protein L15/L10 were upregulated in our study.
We noted that DCW1, which encodes a putative mannosidase, was repressed significantly upon exposure to KTC. DCW1 is required for cell wall biosynthesis during bud formation and is homologous to Dfg5p. The results of both homozygote triplication tests and conditional expression strategies indicate that dfg5 and dcw1 mutations are synthetically lethal (28).
In total, our results showed that the azole antifungal inhibited cytochrome P450-dependent enzymes that were involved in the biosynthesis of cell membrane sterols.
Gene expression responses to AMB.
The number and characteristics of the responsive genes grouped according to biological function are shown in Table S2 in the supplemental material. As with KTC, most responsive genes (303 genes) were of “unknown function.”
Of the characterized genes, most were classified as involved in transport. Transcription of genes associated with membrane transport was upregulated in this study, including genes encoding GIT1, DW706156, DW684040, DMT, DW406136, CCH1, and DW710650. But some genes that encode proteins involved in membrane transport were downregulated in response to AMB, including genes encoding PTR2, GAP1, DW700599, DW684458, Atp1a1, NMT1 DW699236, DW686710, DW696869, MEAA, ATP2C1, DW707140, and CTR1. The polyenes are thought to intercalate into membranes, forming a channel through which cellular components, especially potassium ions, leak, and thereby destroy the proton gradient within the membrane (35). It is possible that damage to the cell wall would cause defects in the plasma membrane and affect transport of small molecules through the membrane.
In our experiments, it was found that AMB reduced the transcription of some genes that encode ribosomal proteins, such as L19, L28, L11, DW679066, L13, L4, S18, HS6-type (S12/L30/L7a), and YmL35. But AMB increased the expression level of the genes that encode L3, L34, and S27. Similar results have previously been obtained by Zhang et al. (40).
It was found that AMB induces genes that encode stress response-related proteins in T. rubrum, including HSP70, HSP104, EB801455, and UBI4; oxidative stress proteins such as GSS and AOX; and ethanol stress proteins such as TDH1. At the same time, AMB repressed the transcription of some stress-response-related genes, including HSP10, DW678423, and GPX, and osmotic stress protein HOR2.
In the present study, the transcription of several genes involved in lipid, fatty acid, and sterol metabolism was increased, such as DW696584, EB801458, DW682185, IVD, DW694010, DW688343, DW684992, and some genes were repressed, including DW703105, DW697546, ERG4, DW699008, DW705750, EB801635, ACS, DW703927, DW708678, DW710649, DW678450, DW683501, IDI1, DW707302, DW690970, ELO2, and MVD1. The polyenes are a class of antifungal drugs that target cell membranes containing ergosterol. The specificity of the drugs for ergosterol-containing membranes is thought to be due to an interaction between AMB and ergosterol in the membrane, although the mechanism of this interaction is unknown. Downregulation of certain ergosterol biosynthesis genes in response to AMB may be indicative of an attempt by the organism to use alternate sterols or sterol intermediates in the cell membrane (17).
The transcription of some genes involved in maintenance of cell wall integrity and signal transduction pathways was also repressed, including FKS1, SUN4, DW699324, GAS1, DW681613, SPS1, DW703091, STE7, DW703091, and DW695308. However, transcription of DW679073 was increased. In yeast, FKS1 encodes a subunit of the yeast 1,3-β-glucan synthase (18) and GAS1 is involved in remodeling of 1,3-β-glucan (23), loss of either of these two plasma-membrane-localized proteins results in significantly reduced levels of 1,3-β-glucan in the wall and in the formation of viable but swollen cells (23, 24).
Several multidrug resistance genes were affected in our experiments. Transcription of DW690470 and DW708548 was increased, whereas transcription of the gene DW698195 and DW701717 was reduced.
Responses in genes involved in regulation of cellular transport is in agreement with the mechanism of action of AMB, which binds membrane ergosterol to form pores that disrupt the membrane and cause leakage of ions and small molecules from the cell (12). The leakage of ions and nutrients owing to formation of pores in the membrane is partly compensated by increased expression of transmembrane transporters (40). The regulation of oxidative stress-response genes with AMB is consistent with the proposed oxidative damage caused by this antifungal agent (33, 34). The changes of transcription of some genes involved in maintenance of cell-wall integrity and signal transduction pathways are in agreement with previous observations that cell wall components can affect interactions between AMB and the cytoplasmic membrane (11). The data presented here indicate that membrane reconstruction, cell stress, and improvement of cell-wall integrity are the main responses of T. rubrum to AMB.
In conclusion, our knowledge of the molecular biology of T. rubrum is still in its infancy, and it is difficult to elucidate the entire biology of T. rubrum. The conclusions drawn in this study are only preliminary, and more genes will be identified and annotated. The T. rubrum microarray studies described here have revealed the gene expression changes with two classes of antifungal agents to be consistent with their known mechanisms of action. There were also some specific findings in T. rubrum that differ from previous reports, this may be the main interest of the T. rubrum microarray studies. The study lays the groundwork for antifungal drug development using microarray studies to identify gene expression profiles.
Supplementary Material
Acknowledgments
Financial support for this work came from The National High Technology Research and Development Program of China (accession number 2001AA223021) and National Key Technologies R&D Programme (accession number 2002BA711A14).
Footnotes
Published ahead of print on 23 October, 2006.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. S. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278:34998-35015. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bammert, G. F., and J. M. Fostel. 2000. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cervelatti, E. P., M. S. Ferreira-Nozawa, R. Aquino-Ferreira, A. L. Fachin, and N. M. Martinez-Rossi. 2004. Electrophoretic molecular karyotype of the dermatophyte Trichophyton rubrum. Genet. Mol. Biol. 27:99-102. [Google Scholar]
- 5.Cleveland, W. S. 1979. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 74:829-836. [Google Scholar]
- 6.De Hoog, G. S., J. Guarro, J. Gene, and M. J. Figueras. 2000. Atlas of clinical fungi, 2nd ed. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands, and University Rovirai Virgili, Reus, Spain.
- 7.Fernández-Torres, B., A. J. Carrillo, E. Martín, A. Del Palacio, M. K. Moore, A. Valverde, M. Serrano, and J. Guarro. 2001. In vitro activities of 10 antifungal drugs against 508 dermatophyte strains. Antimicrob. Agents Chemother. 45:2524-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández-Torres, B., F. J. Cabañes, A. J. Carrillo-Muñoz, A. Esteban, I. Inza, L. Abarca, and J. Guarro. 2002. Collaborative evaluation of optimal antifungal susceptibility testing conditions for dermatophytes. J. Clin. Microbiol. 40:3999-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernández-Torres, B., I. Inza, and J. Guarro. 2003. In vitro activities of the new antifungal drug eberconazole and three other topical agents against 200 strains of dermatophytes. J. Clin. Microbiol. 41:5209-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallis, H. A., R. H. Drew, and W. W. Pickard. 1990. Amphotericin B: 30 years of clinical experience. Rev. Infect. Dis. 12:308-329. [DOI] [PubMed] [Google Scholar]
- 11.Ghannoum, M. A., and L. B. Rice. 1999. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12:501-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartsel, S., and J. Bolard. 1996. Amphotericin B: new life for an old drug. Trends Pharmacol. Sci. 17:445-449. [DOI] [PubMed] [Google Scholar]
- 13.Jackson, C. J., R. C. Barton, and E. G. V. Evans. 1999. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions. J. Clin. Microbiol. 37:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen-Pergakes, K. L., M. A. Kennedy, N. D. Lees, R. Barbuch, C. Koegel, and M. Bard. 1998. Sequencing, disruption, and characterization of the Candida albicans sterol methyltransferase (ERG6) gene: drug susceptibility studies in erg6 mutants. Antimicrob. Agents Chemother. 42:1160-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemna, M. E., and B. E. Elewski. 1996. A U. S. epidemiologic survey of superficial fungal diseases. J. Am. Acad. Dermatol. 35:539-542. [DOI] [PubMed] [Google Scholar]
- 16.Larone, D. H. 2002. Medically important fungi: a guide to identification, 4th ed. American Society for Microbiology, Washington, DC.
- 17.Liu, T. T., R. E. B. Lee, K. S. Barker, R. E. Lee, L. Wei, R. Homayouni, and P. D. Rogers. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49:2226-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazur, P., N. Morin, W. Baginsky, M. El-Sherbeini, J. A. Clemas, J. B. Nielsen, and F. Foor. 1995. Differential expression and function of two homologous subunits of yeast 1,3-β-D-glucan synthase. Mol. Cell. Biol. 15:5671-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melin, P., J. Schnurer, and E. G. Wagner. 2003. Characterization of phiA, a gene essential for phialide development in Aspergillus nidulans. Fungal Genet. Biol. 40:234-241. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee, P. K., S. D. Leidich, N. Isham, I. Leitner, N. S. Ryder, and M. A. Ghannoum. 2003. Clinical Trichophyton rubrum strain exhibiting primary resistance to terbinafine. Antimicrob. Agents Chemother. 47:82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi. Approved standard M38-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 22.Niewerth, M., and H. C. Korting. 2000. The use of systemic antimycotics in dermatotherapy. Eur. J. Dermatol. 10:155-160. [PubMed] [Google Scholar]
- 23.Popolo, L., and M. Vai. 1999. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim. Biophys. Acta 1426:385-400. [DOI] [PubMed] [Google Scholar]
- 24.Ram, A. F. J., S. S. C. Brekelmans, L. J. W. M. Oehlen, and F. M. Klis. 1995. Identification of two cell cycle regulated genes affecting the β-1,3-glucan content of cell wall in Saccharomyces cerevisiae. FEBS Lett. 358:165-170. [DOI] [PubMed] [Google Scholar]
- 25.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 26.Sanglard, D. 2002. Resistance of human fungal pathogens to antifungal drugs. Curr. Opin. Microbiol. 5:379-385. [DOI] [PubMed] [Google Scholar]
- 27.Smith, K. J., M. Welsh, and H. Skelton. 2001. Trichophyton rubrum showing deep dermal invasion directly from the epidermis in immunosuppressed patients. Br. J. Dermatol. 145:344-348. [DOI] [PubMed] [Google Scholar]
- 28.Spreghini, E., D. A. Davis, R. Subaran, M. Kim, and A. P. Mitchell. 2003. Roles of Candida albicans Dfg5p and Dcw1p cell surface proteins in growth and hypha formation. Eukaryot. Cell 2:746-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Squeo, R. F., R. Beer, D. Silvers, I. Weitzman, and M. Grossman. 1998. Invasive Trichophyton rubrum resembling blastomycosis infection in the immunocompromised host. J. Am. Acad. Dermatol. 39:379-380. [DOI] [PubMed] [Google Scholar]
- 30.Suschka, S., B. Fladung, and H. F. Merk. 2002. Clinical comparison of the efficacy and tolerability of once daily canesten with twice daily nizoral (clotrimazole 1% cream vs. ketoconazole 2% cream) during a 28-day topical treatment of interdigital tinea pedis. Mycoses 45:91-96. [DOI] [PubMed] [Google Scholar]
- 31.Szekely, A., E. M. Johnson, and D. W. Warnock. 1999. Comparison of E-test and broth microdilution methods for antifungal drug susceptibility testing of molds. J. Clin. Microbiol. 37:1480-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urban, M., T. Bhargava, and J. E. Hamer. 1999. An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 18:512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanden Bossche, H., D. W. Warnock, B. Dupont, D. Kerridge, S. Sen Gupta, L. Improvisi, P. Marichal, F. C. Odds, F. Provost, and O. Ronin. 1994. Mechanisms and clinical impact of antifungal drug resistance. J. Med. Vet. Mycol. 32(Suppl. 1):189-202. [DOI] [PubMed] [Google Scholar]
- 34.Vanden Bossche, H., G. Willemsens, and P. Marichal. 1987. Anti-Candida drugs-the biochemical basis for their activity. Crit. Microbiol. Rev. 15:57-72. [DOI] [PubMed] [Google Scholar]
- 35.Vanden Bossche, H., P. Marichal, and F. C. Odds. 1994. Molecular mechanisms of drug resistance in fungi. Trends Microbiol. 2:393-400. [DOI] [PubMed] [Google Scholar]
- 36.Vander Straten, M. R., M. A. Hossain, and M. A. Ghannoum. 2003. Cutaneous infections dermatophytosis, onychomycosis, and tinea versicolor. Infect. Dis. Clin. N. Am. 17:87-112. [DOI] [PubMed] [Google Scholar]
- 37.Venkateswarlu, K., D. E. Kelly, N. J. Manning, and S. L. Kelly. 1998. NADPH cytochrome P-450 oxidoreductase and susceptibility to KTZ. Antimicrob. Agents Chemother. 42:1756-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vermilion, J. L., D. P. Ballou, V. Massey, and M. J. Coon. 1981. Separate roles for FMN and FAD in catalysis by liver microsomal NADPH-cytochrome P-450 reductase. J. Biol. Chem. 256:266-277. [PubMed] [Google Scholar]
- 39.Weitzman, I., and R. C. Summerbell. 1995. The dermatophytes. Clin. Microbiol. Rev. 8:240-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, L., Y. Zhang, Y. Zhou, S. An, Y. Zhou, and J. Cheng. 2002. Response of gene expression in Saccharomyces cerevisiae to amphotericin B and nystatin measured by microarrays. J. Antimicrob. Chemother. 49:905-915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.