Abstract
The K70E mutation in human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) has become more prevalent in clinical samples, particularly in isolates derived from patients for whom triple-nucleoside regimens that include tenofovir (TNV), abacavir, and lamivudine (3TC) failed. To elucidate the molecular mechanism by which this mutation confers resistance to these nucleoside RT inhibitors (NRTI), we conducted detailed biochemical analyses comparing wild-type (WT), K70E, and K65R HIV-1 RT. Pre-steady-state kinetic experiments demonstrate that the K70E mutation in HIV-1 RT allows the enzyme to discriminate between the natural deoxynucleoside triphosphate substrate and the NRTI triphosphate (NRTI-TP). Compared to the WT enzyme, K70E RT showed 2.1-, 2.3-, and 3.5-fold-higher levels of resistance toward TNV-diphosphate, carbovir-TP, and 3TC-TP, respectively. By comparison, K65R RT demonstrated 12.4-, 12.0-, and 13.1-fold-higher levels of resistance, respectively, toward the same analogs. NRTI-TP discrimination by the K70E (and K65R) mutation was primarily due to decreased rates of NRTI-TP incorporation and not to changes in analog binding affinity. The K65R and K70E mutations also profoundly impaired the ability of RT to excise 3′-azido-2′,3′-dideoxythymidine monophosphate (AZT-MP) and other NRTI-MP from the 3′ end of a chain-terminated primer. When introduced into an enzyme with the thymidine analog mutations (TAMs) M41L, L210W, and T215Y, the K70E mutation inhibited ATP-mediated excision of AZT-MP. Taken together, these findings indicate that the K70E mutation, like the K65R mutation, reduces susceptibility to NRTI by selectively decreasing NRTI-TP incorporation and is antagonistic to TAM-mediated nucleotide excision.
Nucleoside reverse transcriptase (RT) inhibitors (NRTI) are analogs of deoxyribonucleosides that lack the 3′-OH group of the ribose sugar. They were the first drugs used to treat human immunodeficiency virus type 1 (HIV-1) infection, and they remain integral components of essentially all antiretroviral regimens. Although combination therapies that contain one or more NRTI have profoundly reduced morbidity and mortality associated with AIDS, their long-term efficacies are limited by the selection of drug-resistant variants of HIV-1. Over the last twenty-five years, as antiretroviral therapies have evolved, the nature and pattern of drug resistance mutations identified in patients have also changed (32). In this regard, previously uncommon mutations have become more prevalent among patients experiencing treatment failure. For example, since the introduction of NRTI, such as tenofovir (TNV) and abacavir (ABC), which select for the K65R mutation in HIV-1 RT, the incidence of this mutation has steadily increased in clinical databases (17, 25, 31, 36).
Recently, the incidence of the K70E mutation in HIV-1 RT in clinical databases has also increased (16a). For example, Virco Laboratories reported that the prevalence of the K70E mutation increased in their database from 0.2% in 1999 to 0.5% in 2005. By comparison, the prevalence of the K65R mutation increased from 0.8% to 2.7% in the same time frame (32a). The K70E mutation was first identified following in vitro selection and analysis of HIV-1 resistant to the acyclic nucleoside phosphonate analog 9-[2-(phosphonomethoxy)ethyl]adenine (adefovir) (4). More recently, it was also selected in vitro by the d-enantiomer of beta-2′,3′-didehydro-2′,3′-dideoxy-5-fluorocytidine (12) and by the nucleotide analog phosphonomethoxy-2′-fluoro-2′,3′-dideoxydidehydroadenosine (4a). The K70E mutation was also detected in clinical trials of adefovir dipivoxil for HIV-1 infection (23, 24). However, after development of adefovir for treatment of HIV-1 infection was stopped in November 1999, K70E was no longer reported as a resistance mutation in HIV-1 genotype interpretations, and it is still not included in some of the most widely used mutation lists (16). Recently, several reports have documented the emergence of the K70E mutation in patients being treated with TNV in combination with other NRTI (5, 25a). For example, the K70E mutation was selected in 10% of antiretroviral-naïve subjects receiving TNV, ABC, and lamivudine (3TC) triple-NRTI therapy in the ESS 30009 study (25a).
In light of the reemergence of the K70E mutation in clinical samples, we were interested in elucidating the molecular mechanism by which this mutation confers resistance to TNV, ABC, and 3TC. This paper reports the results of detailed biochemical studies of the impact of the K70E mutation, compared to that of the K65R mutation, on nucleotide analog incorporation and excision by HIV-1 RT.
MATERIALS AND METHODS
Enzymes.
The M41L, K65R, K70E, L210W, and T215Y mutations were introduced into wild-type (WT) HIV-1LAI RT (28) by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). Full-length sequencing of mutant RTs was performed to confirm the presence of the desired mutations and to exclude adventitious mutations introduced during mutagenesis. WT and mutant recombinant HIV-1 RTs were overexpressed and purified to homogeneity as described previously (19, 20). RT concentrations were determined spectrophotometrically at 280 nm using an extinction coefficient (ɛ280) of 260 450 M−1 cm−1, and the active site concentration was calculated from pre-steady-state burst experiments (18). All experiments described below were performed using corrected active site concentrations.
DNA substrates.
DNA oligomers were synthesized by Integrated DNA Technologies (Coralville, IA). A 20-nucleotide DNA primer (P20; 5′-TCGGGCGCCACTGCTAGAGA-3′) and a 57-nucleotide DNA template (T57; 5′-CTCAGACCCTTTTAGTCAGAATGGAAANTCTCTAGCAGTGGCGCCCGAACAGGGACA-3′) were used in all experiments. Four DNA templates were synthesized, each of which had a different nucleotide at position 30 (N). The T57 template that was synthesized with guanine at position 30 also contained a thymidine at position 34. This strategy allowed us to evaluate the kinetics of single-nucleotide incorporation or excision for different NRTI-TP using the same primer (see below). P20 was 5′ radiolabeled with [γ-32P]ATP (GE Healthcare) and T4 polynucleotide kinase and then subjected to further purification by denaturing polyacrylamide gel electrophoresis using 7 M urea-16% polyacrylamide gels. 5′-32P-labeled P20 was annealed to T57 by adding a 1:1.5 molar ratio of primer to template at 90°C and allowing the mixture to slowly cool to ambient room temperature.
Nucleotides.
3′-azido-2′,3′-dideoxythymidine triphosphate (AZT-TP) and 3TC-TP were purchased from TriLink Biotechnologies (San Diego, CA) and Sierra Bioresearch (Tucson, AZ), respectively. Tenofovir diphosphate (TNV-DP) and carbovir triphosphate (CBV-TP; active metabolite of ABC) were kindly provided by Michael Miller (Gilead) and Randall Lanier (GlaxoSmithKline), respectively. All other nucleotides were purchased from GE Healthcare.
Pre-steady-state kinetic experiments.
A rapid-quench instrument (RQF-3 instrument; Kintek Corporation, Clarence, PA) was used for pre-steady-state experiments with reaction times ranging from 5 ms to 30 min. The typical experiment was performed at 37°C in 50 mM Tris-HCl (pH 8.0) containing 50 mM KCl, 10 mM MgCl2, and various concentrations of nucleotides. All concentrations reported refer to the final concentrations after mixing. Three hundred nanomolar active site HIV-1 RT was preincubated with 50 nM DNA substrate (P20-T57), prior to rapid mixing with nucleotide and divalent metal ions to initiate the reaction that was quenched with 0.5 M EDTA. The quenched samples were then mixed with an equal volume of gel loading buffer (98% deionized formamide, 10 mM EDTA, 1 mg/ml each of bromophenol blue and xylene cyanol), denatured at 85°C for 5 min, and the products were separated from the substrates on a 7 M urea-16% polyacrylamide gel. The disappearance of substrate (20-mer) and the formation of product (21-mer) were quantified using a GS525 Molecular Imager (Bio-Rad Laboratories, Inc., Hercules, CA).
Data analysis.
Data obtained from kinetic assays were fitted by nonlinear regression using Sigma Plot software (Jandel Scientific) using the appropriate equations (15). The apparent burst rate constant (kobs) for each particular concentration of deoxynucleoside triphosphate (dNTP) was determined by fitting the time courses for the formation of product (21-mer) using the following equation: [21-mer] = A[1 − exp(−kobst)], where A represents the burst amplitude and t is time (s). The turnover number (kpol) and apparent dissociation constant for dNTP (Kd) were then obtained by plotting the apparent catalytic rates (kobs) against dNTP concentrations and fitting the data with the following hyperbolic equation: kobs = (kpol[dNTP])/([dNTP] + Kd).
Assay of RT-catalyzed phosphorolysis of NRTI-MP terminated primers.
The P20 primer was 5′-end labeled with [γ-32P]ATP and annealed to T57 as described above. The primer was chain terminated by incubation with WT RT and 100 μM of the appropriate nucleotide (AZT-TP, 3TC-TP, TNV-DP, or ABC-TP) for 30 min at 37°C. 32P-labeled, chain-terminated 21-nucleotide primer was further purified by extraction of the appropriate band after 7 M urea-16% acrylamide denaturing gel electrophoresis. The purified chain-terminated primer was reannealed to T57 for use in phosphorolysis experiments. Phosphorolytic removal of NRTI-MP was achieved by incubating 300 nM active site RT with 50 nM of the chain-terminated template/primer (T/P) complex of interest in 50 mM Tris-HCl, pH 8.0, and 50 mM KCl. The reaction was initiated by the addition of 3.0 mM ATP or 150 μM pyrophosphate (PPi) and 10 mM MgCl2. For the AZT-MP excision assays, 1 μM TTP and 5 μM ddCTP were also included in the reaction mixture. For CBV-MP and TNV excision assays, 1 μM dGTP (CBV-MP) or 1 μM dATP (TNV), 1 μM TTP, and 5 μM ddCTP were included in the reaction mixture. For 3TC-MP excision assays, 1 μM dCTP, 1 μM TTP, and 5 μM ddATP were included in the reaction mixture. After defined incubation periods, aliquots were removed and processed as described previously (2).
Molecular modeling.
A molecular model of K70E HIV-1 RT was constructed using the X-ray crystallographic coordinates for the RT-T/P-TTP and RT-T/P-TNV-DP ternary complexes (13, 31; Brookhaven Protein Databank entries 1rtd and 1t05). Introduction of the K70E mutation into WT RT and energy minimization experiments were carried out using the Molecular Operating Environment program (Chemical Computing Group, Montreal, Quebec, Canada). Charges were calculated using the Gasteiger method, and iterative minimizations were carried out using the AMBER 99 force field until the energy difference between iterations was less than 0.0001 kcal/mol per Å.
RESULTS
Two kinetically distinct mechanisms of HIV-1 resistance to NRTI have been described previously (6, 30). One mechanism involves selective decreases in NRTI-TP versus normal dNTP incorporation during viral DNA synthesis. This resistance mechanism has been termed NRTI discrimination. The second mechanism involves the selective removal of the chain-terminating NRTI-MP from the prematurely terminated DNA chain (1, 21). This mechanism has been termed NRTI excision. To define the mechanism by which the K70E mutation in HIV-1 RT confers resistance to NRTI, we designed and performed independent experiments that assessed the abilities of WT and mutant RTs to incorporate NRTI-TP versus normal dNTP substrate or to excise the NRTI-MP from the 3′ end of the chain-terminated DNA primer.
Incorporation of dNTP and NRTI-TP.
Pre-steady-state analyses were carried out to elucidate the interactions of the natural dNTP substrates (dATP, dCTP, dGTP, and TTP) and nucleotide analogs (TNV-DP, 3TC-TP, CBV-TP, and AZT-TP) with the polymerase active sites of WT, K65R, and K70E HIV-1 RT (Table 1). These experiments defined the maximum rates of nucleotide incorporation (kpol), the nucleotide dissociation constants (Kd), and the catalytic efficiencies of incorporation (kpol/Kd). The kpol/Kd values for the incorporation of the natural substrates by WT or K70E HIV-1 RT were essentially identical, suggesting that the K70E mutation does not adversely affect the structure or function of the polymerase active site. By comparison, the catalytic efficiency values for K65R HIV-1 RT were decreased 1.4- to 2.8-fold, depending on the identity of the dNTP substrate. As noted by others (27), this decrease is primarily due to a lower maximum rate of dNTP incorporation (kpol) by K65R and not decreased dNTP binding affinity (Kd). The selectivity of RT, which is defined with the equation (kpol/Kd)dNTP/(kpol/Kd)analog, is an indication of the ability of the WT or mutant RT to discriminate between the natural dNTP substrate and the NRTI-TP. The WT enzyme favors incorporation of the natural dNTP over the nucleotide analogs (selectivity > 1), with the exception of AZT-TP. The K70E and K65R mutations independently increased the selectivity of RT for the natural substrate versus that for the nucleotide analogs (Table 1). The observed resistance (n-fold), which is the ratio of the selectivity of the mutant and WT enzymes, ranged from 1.7- to 3.5-fold for K70E RT and 3.2- to 13.1-fold for K65R RT (Table 1). For both mutant RTs, nucleotide analog resistance could be primarily attributed to decreased kpol rates, although small increases in Kd (∼1.5-fold) were noted for 3TC-TP (K65R and K70E RT) and TNV-DP (K65R RT).
TABLE 1.
Pre-steady-state kinetic constants for binding and incorporation of the natural dNTP substrates and NRTI-TP by WT, K65R, and K70E HIV-1 RTa
| dNTP/nucleotide analog substrate incorporated by WT or mutant HIV-1 RT | Kinetic value for incorporation of:
|
Selectivityb | Resistancec | |||||
|---|---|---|---|---|---|---|---|---|
| dNTP
|
Nucleotide analog
|
|||||||
| kpol (s−1) | Kd (μM) | kpol/Kd (μM−1 s−1) | kpol (s−1) | Kd (μM) | kpol/Kd (μM−1 s−1) | |||
| dATP/TNV-DP | ||||||||
| WT | 24.4 ± 3.9 | 3.22 ± 1.17 | 7.58 | 6.16 ± 1.0 | 4.29 ± 1.0 | 1.44 | 5.3 | |
| K65R | 12.6 ± 1.9 | 3.19 ± 0.68 | 3.95 | 0.45 ± 0.06 | 7.31 ± 2.7 | 0.06 | 65.8 | 12.4 |
| K70E | 18.0 ± 2.8 | 3.25 ± 0.94 | 5.50 | 2.09 ± 0.05 | 5.11 ± 1.5 | 0.41 | 13.3 | 2.5 |
| dCTP/3TC-TP | ||||||||
| WT | 1.22 ± 0.65 | 1.14 ± 0.37 | 1.07 | 0.023 ± 0.004 | 0.37 ± 0.20 | 0.06 | 17.8 | |
| K65R | 0.83 ± 0.20 | 1.08 ± 0.27 | 0.77 | 0.002 ± 0.001 | 0.61 ± 0.16 | 0.0033 | 233.3 | 13.1 |
| K70E | 1.10 ± 0.09 | 0.95 ± 0.32 | 0.86 | 0.010 ± 0.007 | 0.69 ± 0.13 | 0.014 | 61.4 | 3.5 |
| dGTP/CBV-TP | ||||||||
| WT | 7.53 ± 1.08 | 4.20 ± 1.36 | 1.79 | 0.63 ± 0.22 | 3.95 ± 1.83 | 0.16 | 11.2 | |
| K65R | 3.89 ± 0.72 | 4.00 ± 2.30 | 0.97 | 0.031 ± 0.009 | 4.33 ± 2.15 | 0.007 | 134.4 | 12.0 |
| K70E | 4.98 ± 1.30 | 4.42 ± 1.98 | 1.12 | 0.17 ± 0.08 | 4.64 ± 2.35 | 0.036 | 31.4 | 2.8 |
| TTP/AZT-TP | ||||||||
| WT | 8.08 ± 0.98 | 2.10 ± 0.71 | 3.84 | 8.78 ± 0.24 | 1.99 ± 0.99 | 4.41 | 0.87 | |
| K65R | 3.25 ± 0.20 | 2.38 ± 0.97 | 1.36 | 1.05 ± 0.31 | 2.18 ± 1.66 | 0.48 | 2.8 | 3.2 |
| K70E | 7.70 ± 1.25 | 2.05 ± 0.32 | 3.75 | 4.83 ± 0.97 | 2.31 ± 1.13 | 2.09 | 1.8 | 1.7 |
Data are the means ± standard deviations from at least three independent experiments.
Selectivity is defined as (kpol/Kd)dNTP/(kpol/Kd)NRTI-TP.
Resistance (n-fold) is calculated as selectivitymutant/selectivityWT.
Excision of NRTI-MP.
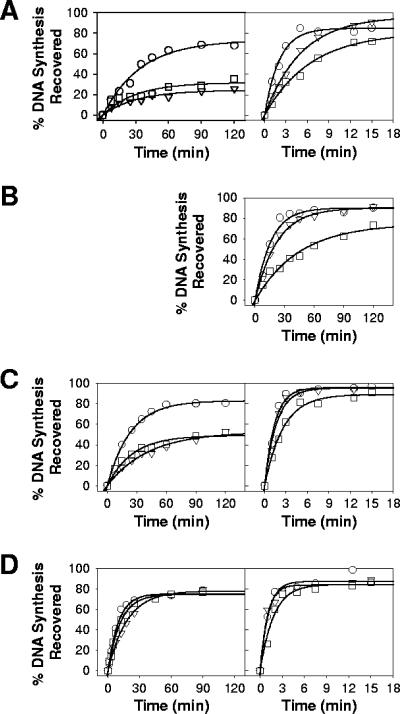
The NRTI-MP excision activities of WT, K65R, and K70E HIV-1 RT were investigated by measuring the abilities of WT or mutant RT to excise AZT-MP, 3TC-MP, CBV-MP, or TNV from the 3′ terminus of the primer and to rescue DNA synthesis, as described previously (2). Excision assays were carried out using either ATP or pyrophosphate as the phosphate donor (Fig. 1; Table 2). For ATP-mediated excision reactions (Fig. 1, left-hand panel), both the K65R and K70E enzymes showed decreased capacities to excise AZT-MP and CBV-MP compared with the WT enzyme (Table 2). The ability of K65R and K70E RT to excise TNV did not appear to be as severely attenuated. Neither mutant (K65R or K70E mutant) nor WT RT showed appreciable excision of 3TC-MP (data not shown). For PPi-mediated excision (Fig. 1, right-hand panel), K65R RT excised each of the NRTI-MP with efficiency lower than that of WT or K70E enzyme (Table 2). This result is consistent with a previous study that reported decreased pyrophosphorolytic activity for K65R HIV-1 RT (29). K70E RT also exhibited decreased rates of PPi-mediated excision for AZT-MP and 3TC-MP compared to the WT enzyme but not for CBV-MP or TNV.
FIG. 1.
Isotherms for ATP (left-hand panel)- or PPi (right-hand panel)-mediated phosphorolytic excision of AZT-MP (A), 3TC-MP (B), CBV-MP (C), and TNV (D) carried out by WT (ο), K65R (□), or K70E (▿) HIV-1 RT. None of the enzymes exhibited ATP-mediated excision of 3TC-MP (data not shown). Data are the means of three or four independent experiments.
TABLE 2.
Kinetic rates of ATP- and PPi-dependent excision of chain-terminating NRTI-MP by WT, K65R and K70E HIV-1 RTa
| RT excising indicated nucleotide analog |
kATPb
|
kPPic
|
||
|---|---|---|---|---|
| Mean ± SD (min−1) | Ratio (WT/mutant) | Mean ± SD (min−1) | Ratio (WT/mutant) | |
| AZT-MP | ||||
| WT | 0.029 ± 0.005 | 0.48 ± 0.03 | ||
| K65R | 0.008 ± 0.001 | 3.6 | 0.15 ± 0.02 | 3.2 |
| K70E | 0.007 ± 0.001 | 4.1 | 0.19 ± 0.02 | 2.5 |
| 3TC-MP | ||||
| WT | NDd | 0.064 ± 0.008 | ||
| K65R | NDd | 0.022 ± 0.006 | 2.9 | |
| K70E | NDd | 0.045 ± 0.007 | 1.4 | |
| CBV-MP | ||||
| WT | 0.042 ± 0.011 | 0.71 ± 0.09 | ||
| K65R | 0.019 ± 0.007 | 2.2 | 0.39 ± 0.07 | 1.8 |
| K70E | 0.024 ± 0.007 | 1.8 | 0.61 ± 0.06 | 1.2 |
| TNV | ||||
| WT | 0.100 ± 0.020 | 0.91 ± 0.12 | ||
| K65R | 0.084 ± 0.009 | 1.2 | 0.55 ± 0.08 | 1.7 |
| K70E | 0.055 ± 0.007 | 1.8 | 0.90 ± 0.10 | 1.0 |
Data are the means ± standard deviations from at least three independent experiments.
kATP, maximum rate of nucleotide incorporation with ATP-mediated excision.
kPPi, maximum rate of nucleotide incorporation with PPi-mediated excision.
ND, no detectable rate of excision.
Antagonism between K70E and TAMs.
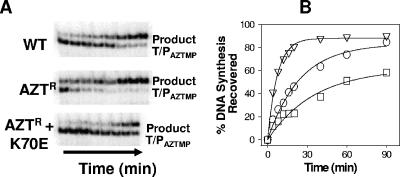
RT containing thymidine analog mutations (TAMs) shows an increased capacity to unblock AZT-MP-terminated primers in the presence of physiological concentrations of ATP (3, 21). Since the K70E mutation reduced the ability of RT to excise AZT-MP, we hypothesized that this mutation may antagonize excision by TAMs. To test this hypothesis, the K70E mutation was introduced into an AZT-resistant enzyme (Aztr RT) with M41L, L210W, and T215Y substitutions. ATP-mediated AZT-MP excision activities of WT, Aztr, and Aztr K70E RT were examined, as described above. As expected, the Aztr enzyme showed increased excision of AZT-MP compared with WT enzyme (Fig. 2). By contrast, the ability of Aztr K70E RT to carry out the same reaction was severely compromised and weaker than that of WT RT (Fig. 2).
FIG. 2.
ATP-mediated excision of AZT-MP and rescue of DNA synthesis carried out by WT, Aztr (M41L/L210W/T215Y), and Aztr K70E RT. (A) ATP-mediated unblocking reaction analyzed by denaturing gel electrophoresis. Excision products were analyzed at 4, 8, 12, 16, 20, 40, 60, and 90 min. (B) Isotherms for ATP-mediated phosphorolytic excision of AZT-MP by WT (ο), Aztr (▿), and Aztr K70E (□) HIV-1 RT. Data are the means of three independent experiments.
DISCUSSION
The K70E mutation in HIV-1 RT has been detected more frequently in clinical samples, particularly in patients for whom triple-nucleoside regimens that include TNV, ABC, and 3TC failed (5, 16a, 25a). In this study, we provide the first insights into the molecular mechanism by which this mutation confers resistance to NRTI. Furthermore, we provide a direct comparison of the K70E mutation with K65R, a mutation that has also become more prevalent following the widespread use of TNV and ABC (17, 25, 32, 37).
Our pre-steady-state kinetic data demonstrate that both the K65R and K70E mutations in HIV-1 RT increase the enzymes' abilities to discriminate between the natural dNTP substrate and the NRTI-TP. The levels of RT resistance conferred by the K70E mutation were 2.5-, 3.5-, and 2.8-fold for TNV-DP, 3TC-TP, and CBV-TP, respectively. These resistance values correlate well with data from viral drug susceptibility assays (4, 4a). By comparison, the K65R mutation conferred greater resistance (12- to 13.1-fold) to the same nucleotide analogs, which is consistent with prior reports (7, 9, 14, 27). The K65R mutation was selected in 53% of the patients receiving TNV/ABC/3TC in the ESS 30009 study, whereas the K70E mutation was selected in only 10% of the patients (25a). The K70E and K65R mutations were not present on the same viral genome when both mutations were detected in plasma, suggesting negative interaction between the mutations (20a). Therefore, the higher frequency of the K65R mutation in the ESS 30009 study is most likely explained by resistance conferred by the K65R mutation to ABC, 3TC and TNV being greater than that conferred by the K70E mutation. Differences in viral replication fitness between the mutants could contribute, but the K65R mutation (34, 36) has been reported to decrease viral replication fitness to a greater degree than the K70E mutation (4, 20a).
The catalytic efficiency of NRTI-TP (and dNTP) incorporation is driven by two kinetic parameters: (i) the affinity of the nucleotide for the RT polymerase active site (Kd) and (ii) the kpol. In general, NRTI-TP discrimination is achieved by the resistance mutation affecting only one of these kinetic parameters. For example, the M184V and V75T mutations in HIV-1 RT increase the NRTI-TP Kd without significantly impacting the kpol (9, 10, 26). Other mutations, such as Q151M and L74V, decrease the kpol without affecting the Kd (7, 8). Consistent with prior reports, our data show that K65R predominantly exerts its discrimination effects by selectively decreasing the kpol (9, 11, 27), although small changes in Kd were noted in this study and others (14, 27). Our results show that the K70E mutation has the same mechanism of discrimination as the K65R, L74V, and Q151M mutations, in that it predominantly exerts its effects by decreasing the kpol for the NRTI-TP. Residues K65 and K70 both reside in the β3-β4 loop in the “fingers” subdomain of the 66-kDa subunit of HIV-1 RT. In the crystal structure of the ternary HIV-1 RT-T/P-TTP and RT-T/P-TNV-DP complexes, the ζ-amino group of K65 interacts with the γ-phosphate of the bound nucleotide substrate (13, 31). Modeling studies suggest that the K65R mutation in HIV-1 RT prevents optimal positioning of the NRTI-TP in the active site, which results in decreased catalytic efficiency of incorporation (27, 29). In the ternary RT-T/P-dNTP complexes, the side chain of K70 does not interact with either the bound nucleotide or the nucleic acid substrate and exhibits minimal noncovalent interactions with other amino acid residue side chains. However, our preliminary modeling studies show that if the lysine at position 70 is replaced with glutamic acid, the δ-oxygen atom comes sufficiently close (<4 Å) to the ɛ-amino group of K65 to form a noncovalent bond (data not shown). We hypothesize that this putative interaction between K65 and E70 may alter the location of K65, thereby affecting NRTI-TP incorporation.
HIV-1 RT has the innate capacity to unblock NRTI-MP-terminated primers in the presence of physiological concentrations of ATP or PPi (1, 22). Biochemical studies show that TAMs confer resistance to AZT and other NRTI by accelerating the rate at which the terminal AZT-MP (or other NRTI-MP) is removed through phosphorolytic cleavage (3, 21). Both the K65R (this study; 25, 29, 35) and K70E (this study) mutant enzymes exhibit rates of ATP- or PPi-mediated phosphorolytic excision that are either significantly reduced or similar (in the case or PPi) to the WT enzyme. These data rule out the possibility that excision contributes to the NRTI resistance conferred by either of these mutations. Furthermore, when the K70E mutation was introduced into an AZT-resistant enzyme with M41L, L210W, and T215Y substitutions, the excision rates of AZT-MP were substantially reduced, indicating phenotypic antagonism between the K70E mutation and TAMs. Similarly, we have recently demonstrated antagonistic interactions between the K65R mutation and TAMs (24a, 25).
In conclusion, our mechanistic studies demonstrate that the K70E mutation, like the K65R mutation, confers resistance to ABC, 3TC, and TNV via a nucleotide discrimination phenotype. However, the level of resistance conferred by the K70E mutation to these nucleotides is less than that conferred by the K65R mutation. The K70E mutation also profoundly impairs the ability of RT to excise AZT-MP, suggesting that the K70E mutation is similar to the K65R mutation in antagonizing the phenotypic effects of TAMs.
Acknowledgments
This work was supported by a grant from the National Cancer Institute (SAIC contract 20XS190A).
Footnotes
Published ahead of print on 6 November 2006.
REFERENCES
- 1.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 2.Arion, D., N. Sluis-Cremer, and M. A. Parniak. 2000. Mechanism by which phosphonoformic acid resistance mutations restore 3′-azido-3′-deoxythymidine (AZT) sensitivity to AZT-resistant HIV-1 reverse transcriptase. J. Biol. Chem. 275:9251-9255. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherrington, J. M., A. S. Mulato, M. D. Fuller, and M. S. Chen. 1996. Novel mutation (K70E) in human immunodeficiency virus type 1 reverse transcriptase confers decreased susceptibility to 9-[2-(phosphonomethoxy)ethyl]adenine in vitro. Antimicrob. Agents Chemother. 40:2212-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Cihlar, T., A. Ray, D. Boojamra, L. Zhang, H. Hui, D. Grant, K. White, M. Desai, N. Parkin, and R. Mackman. 2006. GS 9148: a novel nucleotide active against HIV-1 variants with drug resistance mutations in reverse transcriptase, abstr. 45. Abstr. 13th Conf. Retrovir. Opportunistic Infect.
- 5.Delaugerre, C., L. Roudiere, G. Peytavin, C. Rouzioux, J. P. Viard, and M. L. Chaix. 2005. Selection of a rare resistance profile in an HIV-1-infected patient exhibiting a failure to an antiretroviral regimen including tenofovir DF. J. Clin. Virol. 32:241-244. [DOI] [PubMed] [Google Scholar]
- 6.Deval, J., J. Courcambeck, B. Selmi, J. Boretto, and B. Canard. 2004. Structural determinants and molecular mechanisms for the resistance of HIV-1 RT to nucleoside analogues. Curr. Drug Metab. 5:305-316. [DOI] [PubMed] [Google Scholar]
- 7.Deval, J., J. M. Navarro, B. Selmi, J. Courcambeck, J. Boretto, P. Halfon, S. Garrido-Urbani, J. Sire, and B. Canard. 2004. A loss of viral replicative capacity correlates with altered DNA polymerization kinetics by the human immunodeficiency virus reverse transcriptase bearing the K65R and L74V dideoxynucleoside resistance substitutions. J. Biol. Chem. 279:25489-25496. [DOI] [PubMed] [Google Scholar]
- 8.Deval, J., B. Selmi, J. Boretto, M. P. Egloff, C. Guerreiro, S. Sarfati, and B. Canard. 2002. The molecular mechanism of multidrug resistance by the Q151M human immunodeficiency virus type 1 reverse transcriptase and its suppression using alpha-boranophosphate nucleotide analogues. J. Biol. Chem. 277:42097-42104. [DOI] [PubMed] [Google Scholar]
- 9.Deval, J., K. L. White, M. D. Miller, N. T. Parkin, J. Courcambeck, P. Halfon, B. Selmi, J. Boretto, and B. Canard. 2004. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J. Biol. Chem. 279:509-516. [DOI] [PubMed] [Google Scholar]
- 10.Feng, J. Y., and K. S. Anderson. 1999. Mechanistic studies examining the efficiency and fidelity of DNA synthesis by the 3TC-resistant mutant (184V) of HIV-1 reverse transcriptase. Biochemistry 38:9440-9448. [DOI] [PubMed] [Google Scholar]
- 11.Feng, J. Y., F. T. Myrick, N. A. Margot, G. B. Mulamba, L. Rimsky, K. Borroto-Esoda, B. Selmi, and B. Canard. 2006. Virologic and enzymatic studies revealing the mechanism of K65R- and Q151m-associated HIV-1 drug resistance towards emtricitabine and lamivudine. Nucleosides Nucleotides Nucleic Acids 25:89-107. [DOI] [PubMed] [Google Scholar]
- 12.Hammond, J. L., U. M. Parikh, D. L. Koontz, S. Schlueter-Wirtz, C. K. Chu, H. Z. Bazmi, R. F. Schinazi, and J. W. Mellors. 2005. In vitro selection and analysis of human immunodeficiency virus type 1 resistant to derivatives of beta-2′,3′-didehydro-2′,3′-dideoxy-5-fluorocytidine. Antimicrob. Agents Chemother. 49:3930-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 14.Jeffrey, J. L., J. Y. Feng, C. C. Qi, K. S. Anderson, and P. A. Furman. 2003. Dioxolane guanosine 5′-triphosphate, an alternative substrate inhibitor of wild-type and mutant HIV-1 reverse transcriptase. Steady state and pre-steady state kinetic analyses. J. Biol. Chem. 278:18971-18979. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, K. A. 1995. Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Methods Enzymol. 249:38-61. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, V. A., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, A. Telenti, and D. D. Richman. 2005. Update of the drug resistance mutations in HIV-1: fall 2005. Top. HIV Med. 13:125-131. [PubMed] [Google Scholar]
- 16a.Kagan, R., L. Ross, M. Winters, T. Merigan, P. Heseltine, and M. Lewinski. 2005. Adefovir-associated HIV-1 RT mutation K70E in the age of tenofovir. Antivir. Ther. 10:S103. [Google Scholar]
- 17.Kagan, R. M., T. C. Merigan, M. A. Winters, and P. N. Heseltine. 2004. Increasing prevalence of HIV-1 reverse transcriptase mutation K65R correlates with tenofovir utilization. Antivir. Ther. 9:827-828. [PubMed] [Google Scholar]
- 18.Kati, W. M., K. A. Johnson, L. F. Jerva, and K. S. Anderson. 1992. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267:25988-25997. [PubMed] [Google Scholar]
- 19.Le Grice, S. F., C. E. Cameron, and S. J. Benkovic. 1995. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 262:130-144. [DOI] [PubMed] [Google Scholar]
- 20.Le Grice, S. F., and F. Gruninger-Leitch. 1990. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur. J. Biochem. 187:307-314. [DOI] [PubMed] [Google Scholar]
- 20a.Lloyd, R., J. Huong, E. Rouse, P. Gerondelis, M. Lim, M. Shaefer, A. Rodriguez, J. Gallant, R. Lanier, and L. L. Ross. 2005. HIV-1 RT mutations K70E and K65R are not present on the same viral genome when both mutations are detected in plasma, abstr. 1066. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother.
- 21.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 22.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, M. D., K. E. Anton, A. S. Mulato, P. D. Lamy, and J. M. Cherrington. 1999. Human immunodeficiency virus type 1 expressing the lamivudine-associated M184V mutation in reverse transcriptase shows increased susceptibility to adefovir and decreased replication capability in vitro. J. Infect. Dis. 179:92-100. [DOI] [PubMed] [Google Scholar]
- 24.Mulato, A. S., P. D. Lamy, M. D. Miller, W. X. Li, K. E. Anton, N. S. Hellmann, and J. M. Cherrington. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from AIDS patients after prolonged adefovir dipivoxil therapy. Antimicrob. Agents Chemother. 42:1620-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Parikh, U. M., N. Sluis-Cremer, and J. W. Mellors. 2005. Kinetic mechanism by which thymidine analog mutations antagonize K65R in HIV-1 reverse transcriptase. Antivir. Ther. 10:S95. [DOI] [PubMed] [Google Scholar]
- 25.Parikh, U. M., L. Bacheler, D. Koontz, and J. W. Mellors. 2006. The K65R mutation in human immunodeficiency virus type 1 reverse transcriptase exhibits bidirectional phenotypic antagonism with thymidine analog mutations. J. Virol. 80:4971-4977. [DOI] [PMC free article] [PubMed]
- 25a.Ross, L., P. Gerondelis, Q. Liao, B. Wine, M. Lim, M. Shaefer, A. Rodriguez, K. Limoli, W. Huang, N. T. Parkin, J. Gallant, and R. Lanier. 2005. Selection of the HIV-1 reverse transcriptase mutation K7OE in antiretroviral-naive subjects treated with tenofovir/abacavir/lamivudine therapy. Antivir. Ther. 10:S102. [Google Scholar]
- 26.Selmi, B., J. Boretto, J. M. Navarro, J. Sire, S. Longhi, C. Guerreiro, L. Mulard, S. Sarfati, and B. Canard. 2001. The valine-to-threonine 75 substitution in human immunodeficiency virus type 1 reverse transcriptase and its relation with stavudine resistance. J. Biol. Chem. 276:13965-13974. [DOI] [PubMed] [Google Scholar]
- 27.Selmi, B., J. Boretto, S. R. Sarfati, C. Guerreiro, and B. Canard. 2001. Mechanism-based suppression of dideoxynucleotide resistance by K65R human immunodeficiency virus reverse transcriptase using an alpha-boranophosphate nucleoside analogue. J. Biol. Chem. 276:48466-48472. [DOI] [PubMed] [Google Scholar]
- 28.Shi, C., and J. W. Mellors. 1997. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 41:2781-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sluis-Cremer, N., D. Arion, N. Kaushik, H. Lim, and M. A. Parniak. 2000. Mutational analysis of Lys65 of HIV-1 reverse transcriptase. Biochem. J. 348:77-82. [PMC free article] [PubMed] [Google Scholar]
- 30.Sluis-Cremer, N., D. Arion, and M. A. Parniak. 2000. Molecular mechanisms of HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs). Cell Mol. Life Sci. 57:1408-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuske, S., S. G. Sarafianos, A. D. Clark, Jr., J. Ding, L. K. Naeger, K. L. White, M. D. Miller, C. S. Gibbs, P. L. Boyer, P. Clark, G. Wang, B. L. Gaffney, R. A. Jones, D. M. Jerina, S. H. Hughes, and E. Arnold. 2004. Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nat. Struct. Mol. Biol. 11:469-474. [DOI] [PubMed] [Google Scholar]
- 32.Valer, L., L. Martin-Carbonero, C. de Mendoza, A. Corral, and V. Soriano. 2004. Predictors of selection of K65R: tenofovir use and lack of thymidine analogue mutations. AIDS 18:2094-2096. [DOI] [PubMed] [Google Scholar]
- 32a.Van Houtte, M., M. Staes, A. M. Geretti, T. Pattey, and L. Bacheler. 2006. Tenofovir drug resistance and mutational pathways associated with the RT mutation K70E in HIV-1, abstr. 94. Abstr. 4th Eur. HIV Drug Resist. Workshop.
- 33.Wainberg, M. A., B. G. Brenner, and D. Turner. 2005. Changing patterns in the selection of viral mutations among patients receiving nucleoside and nucleotide drug combinations directed against human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 49:1671-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber, J., B. Chakraborty, J. Weberova, M. D. Miller, and M. E. Quinones-Mateu. 2005. Diminished replicative fitness of primary human immunodeficiency virus type 1 isolates harboring the K65R mutation. J. Clin. Microbiol. 43:1395-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White, K. L., N. A. Margot, J. K. Ly, J. M. Chen, A. S. Ray, M. Pavelko, R. Wang, M. McDermott, S. Swaminathan, and M. D. Miller. 2005. A combination of decreased NRTI incorporation and decreased excision determines the resistance profile of HIV-1 K65R RT. AIDS 19:1751-1760. [DOI] [PubMed] [Google Scholar]
- 36.White, K. L., N. A. Margot, T. Wrin, C. J. Petropoulos, M. D. Miller, and L. K. Naeger. 2002. Molecular mechanisms of resistance to human immunodeficiency virus type 1 with reverse transcriptase mutations K65R and K65R+M184V and their effects on enzyme function and viral replication capacity. Antimicrob. Agents Chemother. 46:3437-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winston, A., S. Mandalia, D. Pillay, B. Gazzard, and A. Pozniak. 2002. The prevalence and determinants of the K65R mutation in HIV-1 reverse transcriptase in tenofovir-naive patients. AIDS 16:2087-2089. [DOI] [PubMed] [Google Scholar]