Abstract
Glycopeptides such as vancomycin are the treatment of choice for infections due to methicillin-resistant Staphylococcus aureus. This study describes the identification of high-level vancomycin-resistant S. aureus (VRSA) isolates in a polymicrobial biofilm within an indwelling nephrostomy tube in a patient in New York. S. aureus, Enterococcus faecalis, Enterococcus faecium, Micrococcus species, Morganella morganii, and Pseudomonas aeruginosa were isolated from the biofilm. For VRSA isolates, vancomycin MICs ranged from 32 to >128 μg/ml. VRSA isolates were also resistant to aminoglycosides, fluoroquinolones, macrolides, penicillin, and tetracycline but remained susceptible to chloramphenicol, linezolid, rifampin, and trimethoprim-sulfamethoxazole. The vanA gene was localized to a plasmid of ∼100 kb in VRSA and E. faecium isolates from the biofilm. Plasmid analysis revealed that the VRSA isolate acquired the 100-kb E. faecium plasmid, which was then maintained without integration into the MRSA plasmid. The tetracycline resistance genes tet(U) and tet(S), not previously detected in S. aureus isolates, were identified in the VRSA isolates. Additional resistance elements in the VRSA isolate included a multiresistance gene cluster, ermB-aadE-sat4-aphA-3, msrA (macrolide efflux), and the bifunctional aminoglycoside resistance gene aac(6′)-aph(2")-Ia. Multiple combinations of resistance genes among the various isolates of staphylococci and enterococci, including vanA, tet(S), and tet(U), illustrate the dynamic nature of gene acquisition and loss within and between bacterial species throughout the course of infection. The potential for interspecies transfer of antimicrobial resistance genes, including resistance to vancomycin, may be enhanced by the microenvironment of a biofilm.
Staphylococcus aureus, a major cause of potentially life-threatening infections acquired in health care and community settings, has developed resistance to most classes of antimicrobial agents. A dramatic increase in the number of health care-associated infections due to methicillin-resistant S. aureus (MRSA) in the 1990s (33) and the recent emergence of MRSA in community-associated infections (20, 23, 31) highlight the success of this species as a pathogen and its ability to adapt under pressure from antimicrobial agents. Glycopeptides such as vancomycin provide effective therapy against most multidrug-resistant strains of S. aureus.
Although vancomycin resistance was first reported for enterococci in 1988, the first clinical isolate of high-level vancomycin-resistant S. aureus (VRSA) was not isolated until June 2002 (in Michigan) (MIC = 1,024 μg/ml) (2, 41). This was closely followed by the identification of another VRSA isolate in Pennsylvania in September 2002 (MIC = 32 μg/ml) (3). These isolates were associated with chronic skin ulcers, and vancomycin resistance was mediated by Tn1546-like elements, most likely acquired from coinfecting strains of vancomycin-resistant enterococci (VRE).
The genetic exchange of antimicrobial resistance determinants among enterococci and staphylococci is well documented (16, 18, 29, 30). The resistance genes are typically found on conjugative plasmids or transposons. One requirement for the conjugative transfer of mobile genetic elements is cell-to-cell contact between donor and recipient. To facilitate this contact, enterococci have highly evolved conjugative systems that are responsible for the dissemination of antimicrobial resistance and virulence factors. These systems include the secretion of bacterial sex pheromones, small peptides that induce a mating response resulting in the aggregation or clumping of the cells (36).
Cell-to-cell contact occurs naturally in microbial biofilms. Microbial cells attached to a surface produce an extracellular polymeric substance that supports a highly structured microbial community (for reviews, see references 22 and 37). Cells within this matrix have increased tolerance to antimicrobial agents, making it difficult or impossible to eradicate the biofilm once it becomes established (13). Many species of microorganisms colonize and form biofilms on a variety of indwelling medical devices (12, 14).
This report describes the microbial community within a nephrostomy tube biofilm from which VRSA strains were isolated. VRSA, MRSA, and VRE isolates from various patient sites as well as the biofilm were analyzed by molecular techniques to identify potential donors of vancomycin resistance genes, possible recipient strains of S. aureus, and the mechanism of vancomycin resistance in VRSA.
MATERIALS AND METHODS
Isolation and identification of bacterial strains.
An isolate of vancomycin-resistant S. aureus was recovered from the urine of a 63-year-old female patient at a long-term care facility in New York (4). Over the next 4 weeks, additional isolates of staphylococci and enterococci were recovered from the patient's urine, gastrostomy tube site, nephrostomy tube biofilm, and rectal swabs. Species identification procedures using conventional biochemical tests included carbohydrate fermentation, enzyme production (alkaline phosphatase, β-glucosidase, β-galactosidase, β-glucuronidase, catalase, and coagulase), latex agglutination, and biochemicals (urea, nitrate, and Voges-Proskauer). In addition, DNA sequence analysis of the quinolone resistance-determining regions of the DNA gyrase genes gyrA and gyrB of the VRSA isolates were compared with analogous sequences from S. aureus ATCC 12600, the ATCC type strain.
Biofilm extraction.
The nephrostomy tube was in place for 63 days before it was removed from the patient, shipped on ice (overnight), and processed the following day at the Centers for Disease Control and Prevention (CDC). One-centimeter segments were cut from the proximal, medial, and distal portions of the tube, and the biofilm within the lumen of the tubing segments was extracted by washing with sterile phosphate-buffered saline. Biofilm suspensions from each segment were diluted in Butterfield buffer (Becton Dickinson Microbiology Systems, Sparks, MD [BD]) and inoculated onto four types of media: Columbia colistin nalidixic acid agar, MacConkey agar, mannitol salt agar, and tryptic soy agar with 5% sheep blood (all from BD). All plates were incubated at 35°C for 24 to 48 h. A total of 67 colonies with distinct morphologies were selected for identification and antimicrobial susceptibility testing.
Susceptibility testing.
MICs were determined by broth microdilution using cation-adjusted Mueller-Hinton broth (Difco) according to guidelines from the Clinical and Laboratory Standards Institute (CLSI) (formerly NCCLS) (28). Antimicrobial agents were obtained from the following manufacturers: chloramphenicol, doxycycline, gentamicin, minocycline, oxacillin, penicillin G, rifampin, tetracycline, and trimethoprim-sulfamethoxazole were obtained from Sigma Chemical Co. (St. Louis, MO); levofloxacin was obtained from Johnson & Johnson (Raritan, NJ); teicoplanin was obtained from Aventis Pharmaceuticals, Inc. (Somerset, NJ); erythromycin and vancomycin were obtained from Lilly Research Laboratories (Indianapolis, IN); and clindamycin was obtained from U.S. Pharmaceuticals (Rockville, MD).
Plasmids, PCR, and DNA sequence analysis.
Cultures of VRSA and VRE were grown in Mueller-Hinton broth containing 2 μg/ml vancomycin and 10 μg/ml gentamicin. MRSA plasmids were isolated from cultures grown in Mueller-Hinton broth without antibiotic selection. Plasmids were isolated using the QIAGEN Midi plasmid purification kit (QIAGEN Inc., Valencia, CA) according to the manufacturer's suggestion of prewarming the elution buffer to 50°C for elution of large plasmids. Whole-cell lysates were prepared as previously described (41). PCR mixtures (final volume, 50 μl) consisted of 50 pmol each primer, 100 μmol each deoxynucleoside triphosphate, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 1 U of AmpliTaq DNA polymerase LD (Applied Biosystems, Foster City, CA), and 5 μl cell lysate or 20 ng of plasmid. Amplification parameters included an initial denaturation step for 5 min at 95°C; 35 cycles of 95°C for 30 s, 20 s at the annealing temperature (Ta) (Table 1), and 30 s at 72°C; and a final extension step at 72°C for 7 min in a GeneAmp 9700 thermal cycler (Applied Biosystems). PCR products were purified with QIAquick spin columns (QIAGEN). DNA sequences of the PCR products were determined using dRhodamine dye terminator cycle sequencing (Applied Biosystems) and the same primers as those used for PCR amplification. DNA sequences of the forward and reverse strands were determined from independently amplified PCR fragments. DNA and derived amino acid sequences were analyzed with DNAsis for Windows (version 2.5; Hitachi Software Engineering Co., Ltd., South San Francisco, CA).
TABLE 1.
Oligonucleotide primers used in this study
| Genea | Sequence (5′-3′) | Ta (°C) | Reference or source |
|---|---|---|---|
| Aminoglycoside resistance | |||
| aac(6′)-aph(2") (F) | GAGCAATAAGGGCATACCAAAAATC | 56 | 24 |
| aac(6′)-aph(2") (R) | CCGTGCAATTGTCTTAAAAAACTGG | ||
| aac(6′)-Ii (F) | TGGCCGGAAGAATATGGAGA | 56 | 25 |
| aac(6′)-Ii (R) | GCATTTGGTAAGACACCTACG | ||
| ant(4′)-Ia (F) | GGAAGCAGAGTTCAGCCATG | 56 | 25 |
| ant(4′)-Ia (R) | TGCCTGCATATTCAACACGC | ||
| ant(6)-Ia (F) | CGGGAGAATGGGAGACTTTG | 56 | 25 |
| ant(6)-Ia (R) | CTGTGGCTCCACAATCTGAT | ||
| ant(9)-Ia (F) | GGTTCAGCAGTAAATGGTGGT | 60 | 25 |
| ant(9)-Ia (R) | TGCCACATTCGAGCTAGGGTT | ||
| aph(2")-Ib (F) | ATGGTTAACTTGGACGCTGAG | 60 | This study |
| aph(2")-Ib (R) | TTCCTGCTAAAATATAAACATCTCTGCT | ||
| aph(2")-Ic (F) | TGACTCAGTTCCCAGAT | 48 | 7 |
| aph(2")-Ic (R) | AGCACTGTTCGCACCAAA | ||
| aph(2")-Id (F) | GGTGGTTTTTACAGGAATGCCATC | 60 | 24 |
| aph(2")-Id (R) | CCCTCTTCATACCAATCCATATAACC | ||
| aph(3′)-IIIa (F) | CTGATCGAAAAATACCGCT | 60 | 25 |
| aph(3′)-IIIa (R) | ACAATCCGATATGTCGATGGAG | ||
| Macrolide resistance | |||
| erm(A)-43b (F) | TCTAAAAAGCATGTAAAAGAA | 50 | 38 |
| erm(A)-664 (R) | CTTCGATAGTTTATTAATATTAGT | ||
| erm(B)-42 (F) | GAAAGGGTACTCAACCAAATA | 50 | 38 |
| erm(B)-658 (R) | AGTAACGGTACTTAAATTGTTTAC | ||
| erm(C)-94 (F) | GATAATATCTTTGAAATCGGCTCA | 60 | This study |
| erm(C)-636 (R) | CCTGCATGTTTTAAGGAATTGTTA | ||
| msr(A)-994 (F) | GCAAATGGTGTAGGTAAGACAACT | 50 | 44 |
| msr(A)-1375 (R) | ATCATGTGATGTAAACAAAAT | ||
| Tetracycline resistance | |||
| tet(K)-473 (F) | TAGGGGGAATAATAGCACATT | 55 | This study |
| tet(K)-1060 (R) | AATCCGCCCATAACAAATA | ||
| tet(L)-403 (F) | AGGAAAATAGGGGTAAAGCAT | 55 | This study |
| tet(L)-917 (R) | CACCAATGTAGCCGAAAAT | ||
| tet(M)-595 (F) | GAACTCGAACAAGAGGAAAGC | 55 | 32 |
| tet(M)-1312 (R) | ATGGAAGCCCAGAAAGGAT | ||
| tet(O)-197 (F) | AACTTAGGCATTCTGGCTCAC | 55 | 32 |
| tet(O)-713 (R) | TCCCACTGTTCCATATCGTCA | ||
| tet(Q)-100 (F) | GGCTGTGTGGATAATGG | 50 | This study |
| tet(Q)-816 (R) | AGTCTCAGACTTCCGTCA | ||
| tet(U)-28 (F) | GATTGGCATGCGATGGTTC | 60 | This study |
| tet(U)-295 (R) | TCTCTGTCACATCCAACCC | ||
| tet(S)-796 (F) | GATGGTCAACGGCTTGTC | 50 | This study |
| tet(S)-1366 (R) | TGCCACTACCCAAAGGAA | ||
| tet(W)-117 (F) | GACAACGAGAACGGACACTATG | 55 | 1 |
| tet(W)-1341 (R) | AAGCGGGAGCGGCGTAACAGAC |
The ermB-aadE-sat4-aphA-3 gene cluster was amplified using primers LPP1 and LPP2, and the amplicon was restricted with EcoRV as described previously by Werner et al. (42). Primers designed for this study were based on gene sequences from GenBank. vanA, vanB, vanC, vanD were described previously by Clark et al. (9). Enterococcal ligase primers were described previously by Dutka-Malen et al. (15).
Numbers refer to nucleotide position in the gene sequence.
All isolates were screened for the following resistance determinants by PCR: vanA, vanB, vanC, vanD, tet(K), tet(L), tet(M), tet(O), tet(Q), tet(S), tet(U), tet(W), ermA, ermB, ermC, msrA, msrC, ant(4′)-Ia, ant(9)-Ia, ant(9)-Ib, ant(6)-Ia (previously designated aadE), aph(3′)-IIIa (previously designated aphA-3), aph(2")-Ib, aph(2")-Ic, aph(2")-Id, and the bifunctional enzyme aac(6′)-aph(2"). Oligonucleotide primer sequences, annealing temperatures, and references are listed in Table 1. Enterococcal isolates were also screened for aac(6′)-Ii, a chromosomally encoded enzyme proposed to be specific for, and produced by, all strains of E. faecium (11). Probes for vanA, tet(S), and tet(U) were generated with the PCR DIG probe synthesis kit (Roche, Indianapolis, IN) using relevant primers (Table 1). Plasmids were separated on 0.8% agarose gels and transferred to Zeta-Probe GT membranes (Bio-Rad). Southern blots were hybridized for 16 to 18 h at 65°C.
PFGE.
Genomic DNA was digested with SmaI restriction endonuclease, and the DNA fragments were separated by pulsed-field gel electrophoresis (PFGE) as described previously by McDougal et al. (27). Pulsed-field gel patterns were analyzed and compared using the BioNumerics software package (Applied Math, Kortrijk, Belgium) and interpreted using standard criteria (27).
RESULTS
Confirmation of VRSA.
The species of VRSA 595 was confirmed to be S. aureus by both conventional biochemical assays and DNA sequence analysis of gyrA and gyrB (data not shown). The gyrB sequence was identical to that of S. aureus type strain ATCC 12600. The gyrA gene sequence contained a single nucleotide change that resulted in a Ser-84-to-Leu amino acid substitution, consistent with the fluoroquinolone-resistant phenotype. Negative PCR results for enterococcal ligase genes (15) ruled out enterococcal contamination of S. aureus cultures.
Biofilm characterization.
A total of 67 colonies with distinct morphologies from the three segments of the nephrostomy tube were selected for identification and susceptibility testing. Enterococcus faecalis, Enterococcus faecium, Morganella morganii, Pseudomonas aeruginosa, and S. aureus were identified from each segment of the nephrostomy tube. In addition to these species, the proximal end of the tubing also contained Micrococcus spp. Based on susceptibility profiles, representative isolates of E. faecalis, E. faecium, and S. aureus from the biofilm were selected for analysis by PFGE, plasmid content, and mechanisms of antimicrobial resistance.
Typing by PFGE.
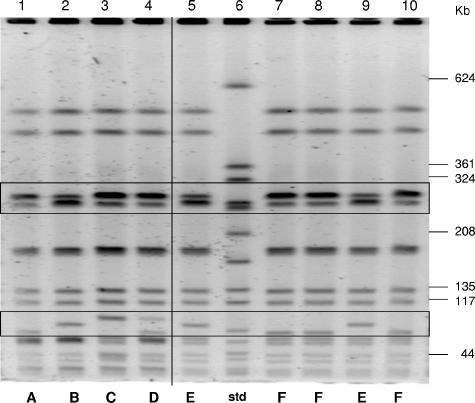
The SmaI restriction digests of genomic DNA from staphylococcal and enterococcal isolates recovered from the gastrostomy tube, nephrostomy tube, rectum, and urine of the patient were analyzed by PFGE. PFGE profiles of representative isolates of VRSA and MRSA from the nephrostomy tube are shown in Fig. 1. Four variants of VRSA (designated A, B, C, and D) and two variants of MRSA (designated E and F) were identified based on the genomic restriction profiles. All of the staphylococcal isolates were identified as pulsed-field type USA 800 (27). PFGE patterns of eight E. faecium isolates from the nephrostomy tube, urine, and rectum were also similar to each other, with three or fewer differences in SmaI restriction fragments (data not shown).
FIG. 1.
PFGE profiles of SmaI restriction fragments of VRSA and MRSA genomic DNA. A vertical line separates VRSA (lanes 1 to 4) from MRSA (lanes 5 and 7 to 10). The fragment size (in kilobases) is indicated for the SmaI digest of the reference strain, S. aureus NCTC 8325 (lane 6) (std). Boxed areas indicate regions with variations in the restriction profiles. VRSA variants are designated A to D, and MRSA variants are designated E or F.
Antimicrobial susceptibility testing.
Tables 2 and 3 present the antimicrobial susceptibility patterns of representative strains of S. aureus and enterococci, respectively, listed by the date of isolation and specimen source. All of the staphylococci isolated from patient samples and the nephrostomy tube were multidrug resistant (Table 2). Each isolate was resistant to fluoroquinolones, macrolides, and β-lactams. Susceptibility to clindamycin, gentamicin, and tetracycline varied among the isolates.
TABLE 2.
Representative S. aureus isolates: origins, antimicrobial MICs, and resistance genes
| Isolate | Site-datea | MIC (μg/ml)b
|
Resistance gene(s)c
|
ermB-aadE-sat4-aphA-3 gene cluster | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VAN | OXA | ERY | CLI | TET | DOX | LEV | GEN | van | tet | erm | Aminoglycosides | |||
| VRSA 595 | Urine-3/24 | 64 | >16 | >8 | >8 | >16 | 4 | 16 | 32 | vanA | tet(S), tet(U) | erm(B), msrA | aac(6′)-aph(2"), aadE, aph(3")-IIIa | + |
| MRSA 5737 | G. tube sited-4/2 | 1 | >16 | >8 | ≤0.12 | ≤0.5 | 0.5 | 16 | >64 | — | — | msrA | aac(6′)-aph(2"), ant(4′)-Ia, aph(3")-IIIa | − |
| MRSA 5739 | Rectum-4/2 | 1 | >16 | >8 | ≤0.12 | ≤0.5 | ≤0.25 | 16 | >64 | — | — | msrA | aac(6′)-aph(2"), ant(4′)-Ia, aph(3")-IIIa | − |
| MRSA 2513 | Biofilm-4/15 | 1 | >16 | >8 | >8 | ≤0.5 | ≤0.25 | 16 | ≤4 | — | — | erm(B), msrA | aadE, aph(3")-IIIa | + |
| MRSA 2530 | Biofilm-4/15 | 1 | >16 | >8 | ≤0.12 | ≤0.5 | ≤0.25 | 16 | ≤4 | — | — | msrA | aph(3")-IIIa | − |
| VRSA 2514 | Biofilm-4/15 | 32 | >16 | >8 | >8 | >16 | 4 | 16 | 64 | vanA | tet(S), tet(U) | erm(B), erm(C), msrA | aac(6′)-aph(2"), aadE, aph(3")-IIIa | + |
| MRSA 5735 | Urine-4/20 | 2f | >16 | >8 | >8 | 8 | 4 | 16 | 16 | vanA | tet(S), tet(U) | erm(B), msrA | aac(6′)-aph(2"), aadE, aph(3")-IIIa | + |
| MRSA 5736 | Urine-4/20 | 4f | >16 | >8 | >8 | 8 | 4 | 16 | >64 | vanA | tet(S), tet(U) | erm(B), msrA | aac(6′)-aph(2"), aadE, aph(3")-IIIa | + |
| MRSA 5733 | Urine-4/20 (cath.)e | 4f | >16 | >8 | >8 | 8 | 4 | 16 | >64 | vanA | tet(S), tet(U) | erm(B), msrA | aac(6′)-aph(2"), aadE, aph(3")-IIIa | + |
| VRSA 5734 | Urine-4/20 | >128 | >16 | >8 | >8 | 16 | 8 | 16 | >64 | vanA | tet(S), tet(U) | erm(B), msrA | aac(6′)-aph(2"), aadE, aph(3")-IIIa | + |
Date is shown as month/day.
CLI, clindamycin; DOX, doxycycline; ERY, erythromycin; GEN, gentamicin; LEV, levofloxacin; OXA, oxacillin; TET, tetracycline; VAN, vancomycin.
—, gene not detected by PCR.
G. tube site, gastrostomy tube site.
cath., catheterized urine specimen.
Indicates isolates with a vancomycin-susceptible or -intermediate phenotype but that were vanA+ by PCR.
TABLE 3.
Representative enterococcal isolates: origins, antimicrobial MICs, and resistance genes
| Isolate | Site-datea | MIC (μg/ml)b
|
Resistance gene(s)c
|
erm(B)-aadE-sat4-aphA-3 gene cluster | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VAN | PEN | ERY | TET | DOX | MIN | CHL | RIF | van | tet | erm | Aminoglycosides | |||
| E. faecalis 5744 | Urine-4/2 | 2 | 2 | >8 | >16 | 8 | 16 | 16 | 0.5 | — | tet(M) | erm(B) | aac(6′)-aph(2") | − |
| E. faecalis 5743 | Rectum-4/2 | 2 | 2 | >8 | >16 | 4 | 8 | 8 | 1 | — | tet(M) | erm(B) | aadE, aphA-3 | − |
| E. faecalis 5742 | Rectum-4/02 | 2 | 2 | >8 | >16 | 4 | 8 | 8 | 1 | vanA | tet(M) | erm(B) | aac(6′)-aph(2"), aadE, aphA-3 | + |
| E. faecium 2547 | Neph. tube-4/15 | 128 | 128 | >8 | >16 | 16 | 32 | 8 | >4 | vanA | tet(L), tet(M), tet(S), tet(U) | erm(B), msrC | aac(6′)-aph(2"), aac(6′)-Ii, aadE, aphA-3 | + |
| E. faecium 5749 | Urine-4/02 | 128 | 128 | >8 | >16 | 8 | 16 | 4 | >4 | — | tet(L), tet(M), tet(S), tet(U) | erm(B), msrC | aac(6′)-aph(2"), aac(6′)-Ii, aadE, aphA-3 | + |
| E. faecium 5750 | Urine (cath)-4/20 | 256 | 256 | >8 | >16 | 16 | 32 | 8 | 4 | vanA | tet(L), tet(M), tet(U) | erm(B), msrC | aac(6′)-aph(2"), aac(6′)-Ii, aadE | − |
| E. faecium 5751 | Urine (cath.)-4/20 | 256 | 256 | >8 | >16 | 16 | 16 | ≤2 | >4 | vanA | tet(L), tet(M), tet(S), tet(U) | erm(B), msrC | aac(6′)-Ii, aadE, aphA-3 | + |
| E. faecium 5752 | Urine (neph.)-4/20 | 128 | 128 | >8 | >16 | 16 | 32 | 8 | >4 | vanA | tet(L), tet(M), tet(S), tet(U) | erm(B), msrC | aac(6′)-aph(2"), aac(6′)-Ii, aadE, aphA-3 | + |
| E. faecium 5753 | Rectum-4/20 | 256 | 256 | >8 | >16 | 8 | 16 | 4 | >4 | vanA | tet(L), tet(M), tet(S), tet(U) | erm(B), msrC | aac(6′)-aph(2"), aac(6′)-Ii, aadE, aphA-3 | + |
| E. faecalis 5745 | Rectum-4/20 | 2 | 2 | >8 | >16 | 4 | 8 | 8 | 1 | vanA | tet(M) | erm(B) | aac(6′)-aph(2"), aadE, aphA-3 | + |
| E. faecalis 5746 | Rectum-4/20 | 2 | 2 | >8 | ≤0.5 | ≤0.25 | 0.12 | 16 | 2 | — | — | erm(B) | aac(6′)-aph(2"), aadE, aphA-3 | − |
Date is shown as month/day. Neph., nephrostomy; cath., catheter.
CHL, chloramphenicol; DOX, doxycycline; ERY, erythromycin; MIN, minocycline; PEN, penicillin; RIF, rifampin; TET, tetracycline; VAN, vancomycin.
—, gene not detected by PCR.
The first isolate of VRSA from the patient, designated VRSA 595, had an MIC of vancomycin of 64 μg/ml. This isolate was also resistant to aminoglycosides, macrolides, penicillin, tetracycline, and fluoroquinolones (Table 2) and intermediate to teicoplanin (MIC = 16 μg/ml) (not shown). VRSA 595 was susceptible to chloramphenicol, linezolid, rifampin, and trimethoprim-sulfamethoxazole (not shown). Another isolate, designated VRSA 5734, was isolated from the patient's urine 4 weeks after the isolation of VRSA 595. The MIC of vancomycin was consistently higher (>128 μg/ml) for this isolate. The stability of vancomycin resistance in these two isolates was investigated by sequential subcultures on nonselective media. VRSA 595 reverted to a vancomycin-susceptible phenotype after two subcultures. However, VRSA 5734 retained its vancomycin-resistant phenotype after 20 subcultures (data not shown). All of the VRSA isolates obtained from this patient retained the MRSA phenotype (MIC of oxacillin, >16 μg/ml).
The enterococcal isolates were also multidrug resistant (Table 3). In general, isolates of E. faecium had the same resistance profile as the VRSA isolates. E. faecium isolate 5749 was the only one that was susceptible to vancomycin. In contrast, among the isolates of E. faecalis, only isolates 5742 and 5745 were resistant to vancomycin. Unlike the E. faecium isolates, the E. faecalis isolates were susceptible to penicillin and rifampin, and one isolate, E. faecalis isolate 5746, was susceptible to most agents tested.
Molecular analyses of antimicrobial resistance mechanisms.
PCR primer sets for four vancomycin resistance determinants (vanA, vanB, vanC, and vanD) were used to investigate the mechanism of resistance. Only vanA amplicons were produced from the VRSA and VRE isolates (Tables 2 and 3). The vanA gene was also detected in three staphylococcal isolates that were susceptible to vancomycin (MIC ≤ 4 μg/ml) based on CLSI interpretation criteria available in 2004. Repeated PCR analysis by the New York Department of Health and CDC laboratories showed consistent amplification of vanA from whole-cell lysates and plasmid preparations from these three isolates, although the quantity of product was significantly less than that seen for the positive control (data not shown).
The VRSA isolates were tetracycline resistant. However, PCR amplifications for tet(K), tet(L), tet(M), and tet(O) were negative. Surprisingly, PCR was positive for tet(U), a gene previously described in a single isolate of E. faecium (35) (Table 3). The tet(S) gene was also amplified from each of the S. aureus isolates that possessed vanA and ermB, suggesting that these genes moved as a unit. Among the enterococcal isolates, only tet(M) was detected in isolates of E. faecalis. However, the E. faecium isolates harbored as many as four tetracycline resistance genes (Table 3). The tet(L), tet(M), tet(U), and tet(S) genes were detected in each E. faecium isolate except isolate 5750, which lacked the tet(S) gene. Because tet(S) is reported infrequently (26) and tet(U) has not previously been reported for S. aureus, the DNA sequences of these PCR products were determined and compared with the relevant gene sequences in GenBank [accession numbers AY534326 for tet(S) and EFU01917 for tet(U)]. The percentage of nucleotide identity was 99% for the tet(U) genes and 100% for tet(S) genes compared with the GenBank gene sequences (data not shown).
All S. aureus and enterococcal isolates in this study were resistant to macrolides (MIC of erythromycin, >8 μg/ml). msr(A) was detected in all of the staphylococcal isolates, and msr(C) was detected in each of the E. faecium isolates but in none of the E. faecalis isolates. ermB was identified in all of the enterococcal isolates and in each of the vanA-containing staphylococcal isolates (Tables 2 and 3). Two isolates of staphylococci contained both erm(B) and erm(C) (represented by VRSA 2514 in Table 2).
All of the staphylococci contained at least one aminoglycoside resistance determinant. The aph(3′)-IIIa determinant was present in all of the S. aureus isolates, while the gene for the bifunctional enzyme aac(6′)-aph(2") was present in all but one isolate (i.e., MRSA 2530). In addition, ant(6)-Ia (previously designated aadE) was present in all staphylococci isolated from urine or the nephrostomy tube biofilm but not in those isolated from the rectum and gastrostomy tube site. The number of aminoglycoside resistance genes varied among the enterococcal isolates. E. faecalis isolates had one to three aminoglycoside resistance determinants, and E. faecium isolates had three or four. As expected, aac(6′)-Ii was present only in the E. faecium isolates.
Based on the positive ermB and aphA-3 results, each isolate was examined for the ermB-aadE-sat4-aphA-3 gene cluster (42). A 5.5-kb fragment was amplified from staphylococci isolated from most of the urine and nephrostomy tube cultures but not from rectal or gastrostomy tube site cultures (Table 2). Among the enterococci, the gene cluster was amplified from two of four rectal isolates, three of four urine isolates, and the biofilm VRE isolate (Table 3). Restriction analysis of the 5.5-kb PCR products with EcoRV revealed a fragment pattern that corresponded with gene cluster type III (42) (data not shown).
Isolation and comparison of plasmids.
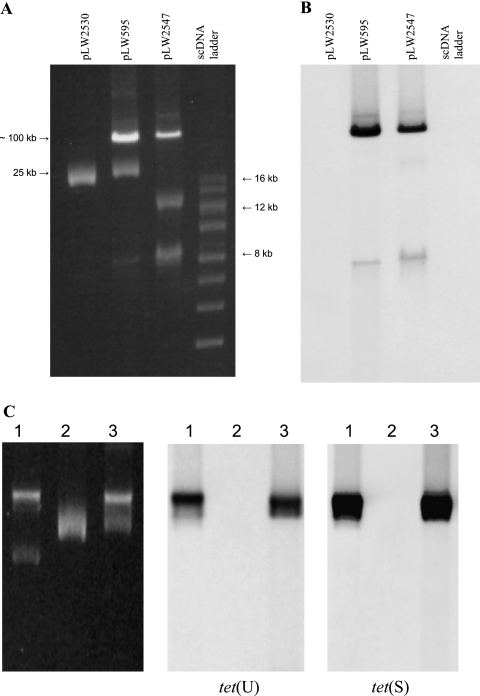
Plasmids were isolated from the VRSA isolate that was initially isolated from the patient (VRSA 595) and from potential donor and recipient strains from the biofilm, MRSA 2530 and VRE 2547 (Fig. 2A). The MRSA isolate harbored a single 25-kb plasmid. E. faecium isolate 2547 harbored at least two plasmids of 11 kb and ∼100 kb, and two plasmids (25 kb and ∼100 kb) were detected in VRSA 595. The 100-kb plasmid in the VRSA isolate was designated pLW595. The vanA, tet(S), and tet(U) genes were localized to the 100-kb plasmids from the VRE and VRSA isolates by Southern hybridization (Fig. 2B and C).
FIG. 2.
Plasmid content and Southern hybridization. (A) Uncut plasmids isolated from MRSA 2530, VRSA 595, and VRE (E. faecium 2547) were separated on a 0.8% agarose gel; the size standard shown is a supercoiled DNA (scDNA) ladder (size range, 2 to 16 kb; Invitrogen). The 25-kb size estimate is based on restriction fragment analysis of the MRSA 2530 plasmid; the 100-kb size is based on relative migration compared with the 120-kb plasmid from the Pennsylvania VRSA isolate and the 58-kb plasmid pLW1043 from the Michigan VRSA isolate (not shown). (B) Southern blot of A probed with a digoxigenin-labeled vanA gene fragment generated by PCR. (C) Southern blot to localize the tet(U) and tet(S) genes. Uncut plasmids from VRSA 595 (lane 1), MRSA 2530 (lane 2), and VRE 2547 (lane 3) were run in duplicate on the same gel and transferred to a Zeta-Probe membrane. The membrane was cut, and the two halves were hybridized with digoxigenin-labeled probes for either tet(U) or tet(S), as indicated.
Digestion with BglII restriction endonuclease linearized the MRSA plasmid, which allowed a direct comparison of the restriction fragments of the 100-kb plasmids from VRSA 595 and VRE 2547 (Fig. 3A). All of the BglII fragments below the linearized 25-kb plasmid in the VRSA digest were also seen in the VRE 2547 digest, suggesting that the 100-kb plasmid in the VRSA isolate was acquired from the VRE isolate.
FIG. 3.
Restriction analysis of VRSA, MRSA, and VRE plasmids from biofilm isolates. (A) BglII digests indicate a single restriction site in the MRSA plasmid. For VRSA 595, the additional fragments match those seen in the VRE plasmid. The “cured” VRSA is a vancomycin-susceptible derivative of VRSA 595. (B) EcoRI digests to compare MRSA 2530 plasmid fragments with those from the “cured” VRSA and VRSA 595.
Plasmid analysis of a vancomycin-susceptible derivative of VRSA 595, obtained after several subcultures on nonselective media, indicated that all of the enterococcal plasmid fragments were missing, and the remaining 25-kb plasmid had the same EcoRI restriction profile as the MRSA plasmid (Fig. 3B).
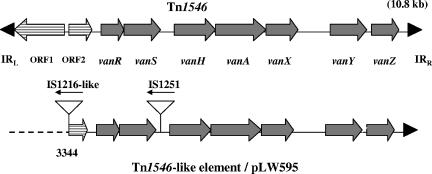
PCR and DNA sequence analysis identified a derivative of Tn1546, the 10.8-kb vanA prototype transposon, as part of the 100-kb plasmid of VRSA 595. Compared with the DNA sequence of Tn1546 (GenBank accession number M97297), the transposon within pLW595 was truncated by 3,343 nucleotides at the 5′ end, eliminating open reading frame 1 (ORF1) and part of ORF2 (Fig. 4). In place of the missing segment was an insertion sequence, IS1216V. Another insertion sequence, IS1251, was detected in the intergenic region between vanS and vanH.
FIG. 4.
Comparison of the vanA-containing genetic elements of pLW595 with the prototype Tn1546. ORF1 and ORF2 are represented by arrows filled with horizontal lines. Coding regions for van genes are shown as solid gray arrows. Triangles represent insertion sequences found in the Tn1546-like element from pLW595. Block arrows indicate the direction of transcription. Black arrowheads represent inverted repeat (IR) sequences.
DISCUSSION
Staphylococci, enterococci, and many other species of bacteria are known to attach to indwelling medical devices and form biofilms consisting of complex communities of single cells and microcolonies within a matrix of hydrated polysaccharides, proteins, and other macromolecules, including DNA (13, 43). Within this matrix, bacterial cells evade the host immune response and survive antimicrobial chemotherapy, resulting in persistent infections that are difficult to treat (36). Initially, biofilms may be composed of a single species, but the longer a medical device remains in place, the more likely that multiple species will be involved (12). The close contact of cells within a biofilm and the relative stability of the matrix have been demonstrated to facilitate gene transfer (8, 21, 22).
The microbial community of the biofilm associated with the emergence of VRSA in this patient included potential donors (VRE), recipients (MRSA), and VRSA transconjugants that harbored a variety of resistance determinants, including a Tn1546-like element (vanA). Both VRE and MRSA isolates were recovered from multiple specimens from a variety of body sites. One potential donor strain, E. faecium 5753, which harbored all of the resistance genes acquired by the MRSA isolates, was recovered from the patient's rectum, suggesting that the patient may have been colonized with the VRE donor. However, among the potential recipient strains of MRSA, those isolated from the rectum or gastrostomy tube site harbored a unique set of aminoglycoside resistance genes, suggesting that the MRSA isolates found in the nephrostomy tube biofilm may have originated from some other source.
Among the enterococcal isolates, the combination of vanA, tet(S), and tet(U) was detected only in isolates of E. faecium, suggesting that this species was the donor. Plasmid analyses from additional isolates and conjugation experiments will be needed to investigate this possibility. Although genetic transfer between E. faecium and S. aureus has been described previously (29), the transfer consisted of chromosomal DNA fragments and not plasmids. It is possible that among the many isolates associated with this infection, there may have been an E. faecalis strain with a plasmid identical or very similar to the E. faecium plasmid that harbored all of the genes transferred to the recipient MRSA isolate and that this strain was not among the colonies selected for characterization. However, we cannot rule out the possibility of plasmid transfer from E. faecium to MRSA facilitated by the close contact of cells within the biofilm.
Molecular characterization and DNA sequence analysis of the first VRSA isolate from Michigan in 2002 (41) revealed a composite plasmid that would have required two genetic events to occur, the transfer of the enterococcal plasmid to the recipient strain of S. aureus followed by a transposition event resulting in a composite plasmid composed of the staphylococcal plasmid and the enterococcal transposon. The requirement for two genetic events suggested that the number of unique VRSA isolates would be extremely limited. However, analyses of plasmids from VRSA 595 revealed that the enterococcal plasmid was transferred and maintained without integration into the staphylococcal chromosome or plasmid, a single genetic event. As a result, the probability of VRSA emerging from coinfections of VRE and MRSA has significantly increased.
The VRSA plasmid pLW595 also acquired tetracycline resistance determinants not previously recognized in isolates of S. aureus. The tet(U) gene has been described in a single report as a plasmid-encoded tetracycline resistance gene on a 1.9-kb plasmid, pKQ10, in E. faecium (35). In that report, in vitro transfer of tet(U) was sufficient to confer low-level resistance to tetracycline and minocycline. However, the mechanism of resistance remains unknown. The tet(S) gene has not previously been reported for either S. aureus or E. faecium. TetS, a ribosomal protection protein (5), was first described for a conjugative plasmid in Listeria and has been reported to be chromosomally encoded in E. faecalis (6) and as part of a Tn916-like element in Streptococcus intermedius (5). Conjugal transfer of tet(S) between strains of E. faecalis was reported to occur by the exchange of large chromosomal fragments (19). The appearance of tet(S) and tet(U) on a plasmid in E. faecium with the ability to transfer to S. aureus suggests that this combination of resistance genes may also be encountered with higher frequency in the future.
Among the vanA-containing staphylococci isolated from the patient, three isolates were susceptible to vancomycin by CLSI criteria available at the time these isolates were identified. Revised breakpoints from the CLSI now classify isolates with an MIC of 4 μg/ml vancomycin as intermediate (10). This genotype/phenotype combination may have been due to unstable plasmids or transposons that were lost from most of the cells. The high sensitivity of PCR amplification would detect vanA from a small subpopulation of cells, although the number of vanA-containing cells would be insufficient for accurate susceptibility testing. Perichon and Courvalin (34) previously reported that genetic instability with a high rate of spontaneous loss of the vancomycin resistance determinant was responsible for the low-level vancomycin-resistant phenotype of VRSA isolated in Pennsylvania in 2002. Alternatively, genetic rearrangement or deletion could have rendered the vanA operon nonfunctional or deficient in transcription.
Colonization with VRE and MRSA is not uncommon, especially among intensive care and long-term care patients (17, 40). These patients are also more likely to have indwelling medical devices that could harbor biofilms. Prior to this report, only two VRSA isolates have been described. The actual number of VRSA isolates may be greater than those reported to date because automated susceptibility testing methods used by many clinical laboratories failed to detect two of the first three VRSA isolates (39). The VRSA isolate from Pennsylvania in 2002 and VRSA 595 described in this report were reported to be vancomycin susceptible by automated methods (39). Subsequently, some manufacturers have modified their automated systems to improve VRSA detection. If the primary susceptibility testing method in a laboratory has not been validated for VRSA detection, supplementary testing of S. aureus, including MRSA, with vancomycin screen plates (brain heart infusion agar containing 6 μg/ml vancomycin) or other nonautomated broth- or agar-based MIC tests is recommended (http://www.cdc.gov/ncidod/dhqp/ar_visavrsa_algo.html).
In summary, VRSA isolates were recovered from a polymicrobial biofilm within an indwelling medical device that also contained vancomycin resistance donor strains (VRE) and recipient strains (MRSA). Unexpectedly, the apparent donor in this event appeared to be E. faecium and an unincorporated enterococcal plasmid was maintained coresident with a staphylococcal plasmid in the VRSA isolates. Resistance genes transferred from the enterococcal donor to the MRSA isolates included tet(S), tet(U), and a Tn1546-like element that was similar, but was not identical to, the Tn1546-like element identified in the Pennsylvania VRSA isolate. These data suggest the possibility that VRSA may emerge more frequently than previously expected, since a single genetic event can produce a stable VRSA isolate.
Acknowledgments
We thank Don Clewell for helpful discussions; Fred Tenover, Cliff McDonald, and Roberta Carey for manuscript review; Janice Carr for excellent technical assistance; and the generosity of Ron Skurray for providing pSK41 and Stephen Billington for providing pJGS259/tet(W).
Footnotes
Published ahead of print on 30 October 2006.
REFERENCES
- 1.Billington, S. J., J. G. Songer, and B. H. Jost. 2002. Widespread distribution of a Tet W determinant among tetracycline-resistant isolates of the animal pathogen Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 46:1281-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2002. Staphylococcus aureus resistant to vancomycin—United States, 2002. Morb. Mortal. Wkly. Rep. 51:565-567. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2002. Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. Morb. Mortal. Wkly. Rep. 51:902. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2004. Brief report: vancomycin-resistant Staphylococcus aureus—New York, 2004. Morb. Mortal. Wkly. Rep. 53:322-333. [PubMed] [Google Scholar]
- 5.Charpentier, E., G. Gerbaud, and P. Courvalin. 1993. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene 131:27-34. [DOI] [PubMed] [Google Scholar]
- 6.Charpentier, E., G. Gerbaud, and P. Courvalin. 1994. Presence of the Listeria tetracycline resistance gene tet(S) in Enterococcus faecalis. Antimicrob. Agents Chemother. 38:2330-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow, J. W., M. J. Zervos, S. A. Lerner, L. A. Thal, S. M. Donabedian, D. D. Jaworski, S. Tsai, K. J. Shaw, and D. B. Clewell. 1997. A novel gentamicin resistance gene in Enterococcus. Antimicrob. Agents Chemother. 41:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, B. B., C. Sternberg, J. B. Andersen, L. Eberl, S. Moller, M. Givskov, and S. Molin. 1998. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64:2247-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, N. C., R. C. Cooksey, B. C. Hill, J. M. Swenson, and F. C. Tenover. 1993. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob. Agents Chemother. 37:2311-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing, 16th informational supplement, M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10a.Clinical and Laboratory Standards Institute. 2004. Performance standards for antimicrobial susceptibility testing, M100-S14. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Costa, Y., M. Galimand, R. LeClercq, J. Duval, and P. Courvalin. 2006. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob. Agents Chemother. 37:1896-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donlan, R. M. 2001. Biofilms and device-associated infections. Emerg. Infect. Dis. 7:277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donlan, R. M., R. Murga, M. Bell, C. M. Toscano, J. H. Carr, T. J. Novicki, C. Zukerman, L. C. Corey, and J. M. Miller. 2001. Protocol for detection of biofilms on needleless connectors attached to central venous catheters. J. Clin. Microbiol. 39:750-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firth, N., and R. A. Skurray. 2000. Genetics: accessory elements and genetic exchange, p. 326-338. In V. A. Fishetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, DC.
- 17.Franchi, D., M. W. Climo, A. H. M. Wong, M. B. Edmond, and R. P. Wenzel. 1999. Seeking vancomycin-resistant Staphylococcus aureus among patients with vancomycin-resistant enterococci. Clin. Infect. Dis. 29:1556-1558. [DOI] [PubMed] [Google Scholar]
- 18.Francia, M. V., and D. B. Clewell. 2002. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1-like nic site, a specific relaxase and a possible TraG-like protein. Mol. Microbiol. 45:375-395. [DOI] [PubMed] [Google Scholar]
- 19.François, B., M. Charles, and P. Courvalin. 1997. Conjugative transfer of tet(S) between strains of Enterococcus faecalis is associated with the exchange of large fragments of chromosomal DNA. Microbiology 143:2145-2154. [DOI] [PubMed] [Google Scholar]
- 20.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, M. M. Farley, et al. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436-1444. [DOI] [PubMed] [Google Scholar]
- 21.Ghigo, J.-M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 22.Hausner, M., and S. Wuertz. 1999. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 654:3710-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiramatsu, K., K. Okuma, X. X. Ma, S. Hori, and M. Kapi. 2002. New trends in Staphylococcus aureus infections: glycopeptide resistance in hospital and methicillin resistance in the community. Curr. Opin. Infect. Dis. 15:407-413. [DOI] [PubMed] [Google Scholar]
- 24.Kao, S. J., I. You, D. B. Clewell, S. M. Donabedian, M. J. Zervos, J. Petrin, K. J. Shaw, and J. W. Chow. 2000. Detection of the high-level aminoglycoside resistance gene aph(2")-Ib in Enterococcus faecium. Antimicrob. Agents Chemother. 44:2876-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi, N., M. Alam, Y. Nishimoto, S. Urasawa, N. Uehara, and N. Watanabe. 2001. Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium. Epidemiol. Infect. 126:197-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lancaster, H., A. P. Roberts, R. Bedi, M. Wilson, and P. Mullany. 2004. Characterization of Tn916S, a Tn916-like element containing the tetracycline resistance determinant tet(S). J. Bacteriol. 186:4395-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.NCCLS/CLSI. 2005. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Document M7-A5. Clinical and Laboratory Standards Institute, Wayne, PA.
- 29.Noble, W. C., M. Rahman, T. Karade, and S. Schwarz. 1996. Gentamicin resistance gene transfer from Enterococcus faecalis and E. faecium to Staphylococcus aureus, S. intermedius and S. hyicus. Vet. Microbiol. 52:143-152. [DOI] [PubMed] [Google Scholar]
- 30.Noble, W. C., Z. Virani, and R. G. A. Cree. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 93:195-198. [DOI] [PubMed] [Google Scholar]
- 31.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbial. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsvik, B., I. Olsen, and F. C. Tenover. 1995. Detection of tet(M) and tet(O) using the polymerase chain reaction in bacteria isolated from patients with periodontal disease. Oral Microbiol. Immunol. 10:87-92. [DOI] [PubMed] [Google Scholar]
- 33.Panlilio, A. L., D. H. Culver, R. P. Gaynes, S. Bannerjee, T. S. Henderson, J. S. Tolson, and W. J. Martone. 1992. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975-1991. Infect. Control Hosp. Epidemiol. 13:582-586. [DOI] [PubMed] [Google Scholar]
- 34.Perichon, B., and P. Courvalin. 2004. Heterologous expression of the enterococcal vanA operon in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 48:4281-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ridenhour, M. B., H. M. Fletcher, J. E. Mortensen, and L. Daneo-Moore. 1996. A novel tetracycline-resistant determinant, tet(U), is encoded on the plasmid pKQ10 in Enterococcus faecium. Plasmid 35:71-80. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 37.Stoodley, P., D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 38.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDougal, J. M. Chaitram, S. K. McAllister, N. C. Clark, G. E. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warren, D. K., A. Nitin, C. Hill, V. J. Fraser, and M. H. Kollef. 2004. Occurrence of co-colonization or co-infection with vancomycin-resistant enterococci and methicillin-resistant Staphylococcus aureus in a medical intensive care unit. Infect. Control Hosp. Epidemiol. 25:99-104. [DOI] [PubMed] [Google Scholar]
- 41.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
- 42.Werner, G., B. Hildebrandt, and W. Witte. 2003. Linkage of erm(B) and aadE-sat4-aphA-3 in multiple-resistant Enterococcus faecium isolates of different ecological origins. Microb. Drug Resist. 9:S9-S16. [DOI] [PubMed] [Google Scholar]
- 43.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 44.Wondrack, L., N. Massa, B. V. Yang, and J. Sutcliffe. 1996. Clinical strain of Staphylococcus aureus inactivates and causes efflux of macrolides. Antimicrob. Agents Chemother. 40:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]