Abstract
Xanthomonas albilineans produces a family of polyketide-peptide compounds called albicidins which are highly potent antibiotics and phytotoxins as a result of their inhibition of prokaryotic DNA replication. Here we show that albicidin is a potent inhibitor of the supercoiling activity of bacterial and plant DNA gyrases, with 50% inhibitory concentrations (40 to 50 nM) less than those of most coumarins and quinolones. Albicidin blocks the religation of the cleaved DNA intermediate during the gyrase catalytic sequence and also inhibits the relaxation of supercoiled DNA by gyrase and topoisomerase IV. Unlike the coumarins, albicidin does not inhibit the ATPase activity of gyrase. In contrast to the quinolones, the albicidin concentration required to stabilize the gyrase cleavage complex increases 100-fold in the absence of ATP. The slow peptide poisons microcin B17 and CcdB also access ATP-dependent conformations of gyrase to block religation, but in contrast to albicidin, they do not inhibit supercoiling under routine assay conditions. Some mutations in gyrA, known to confer high-level resistance to quinolones or CcdB, confer low-level resistance or hypersensitivity to albicidin in Escherichia coli. Within the albicidin biosynthesis region in X. albilineans is a gene encoding a pentapeptide repeat protein designated AlbG that binds to E. coli DNA gyrase and that confers a sixfold increase in the level of resistance to albicidin in vitro and in vivo. These results demonstrate that DNA gyrase is the molecular target of albicidin and that X. albilineans encodes a gyrase-interacting protein for self-protection. The novel features of the gyrase-albicidin interaction indicate the potential for the development of new antibacterial drugs.
Albicidins are potent antibiotics and phytotoxins produced by the bacterial phytopathogen Xanthomonas albilineans. Albicidins selectively block prokaryote DNA replication (9, 10) and cause the characteristic chlorotic symptoms of sugarcane leaf scald disease by blocking chloroplast development (8, 11). They are pathogenesis factors with an important role in systemic invasion of host plants (48, 49).
Albicidins are bactericidal at nanomolar concentrations against a range of gram-positive and gram-negative bacteria, are not cytotoxic at 10 μM for cultured mammalian cells, and hold the potential to be novel clinical antibiotics. The major antimicrobial component produced in culture on rich media has a molecular mass of 842 Da and has a structure containing several aromatic rings (7). Biosynthesis involves a megasynthase with polyketide and nonribosomal peptide synthetase modules (26). Accessory proteins involved in synthesis and resistance functions are substantially clustered in the X. albilineans genome (24-26, 36).
Albicidin-resistant target mutants have not been obtained previously due to a high background frequency of mutations to resistance in Escherichia coli through changes to the Tsx nucleotide uptake channel (12). Other albicidin resistance mechanisms include antibiotic binding, enzymatic detoxification, and export (2, 14, 42, 48). Albicidins do not directly bind to or damage DNA, and no cross-resistance with other DNA replication inhibitors has previously been identified. The speed and specificity of inhibition of DNA replication by albicidin resemble those of DNA gyrase-inhibiting coumarin and quinolone antibiotics (10). In antibiotic checkerboard tests, there is slight synergy between albicidin and coumermycin and a slight interference of albicidin with nalidixic acid (C. Rathus and R. G. Birch, unpublished data).
Bacterial DNA gyrase is a topoisomerase with the unique ability to negatively supercoil DNA. It operates as an A2B2 complex of GyrA and GyrB subunits (17). The catalytic cycle for supercoiling involves the paired GyrA C-terminal domains in wrapping the DNA substrate around the enzyme and securing a DNA sequence to be reversibly cleaved (the G segment) by catalytic tyrosines in the GyrA N-terminal domains. The DNA to be transported through this break is directed through a series of three gates or clamps in the enzyme. In the resting conformation, the entry gate comprising the GyrB N termini is open, allowing entry of the transported DNA (T segment) into the cavity above the closed DNA gate, which comprises the GyrB C termini and the GyrA N termini with bound G-segment DNA. Binding of ATP closes the entry gate, clamping the T segment onto the DNA gate, which opens transiently to allow T-segment passage through the cleaved G segment. The energy stored in the protein-DNA (phosphotyrosine) covalent linkage is sufficient to allow the rejoining of the G segment, and ATP hydrolysis reopens the entry gate. The T segment is released from the cavity below the closed DNA gate through an exit gate, which is formed by the interface of amino acids in the central region of the paired GyrA peptides. ATP is essential for supercoiling, but strand passage can occur in the opposite direction for relaxation of supercoiled DNA without ATP.
Because its functions are essential in DNA replication, transcription, and gene regulation, DNA gyrase has become an important target for antibacterial antibiotics (22, 28). DNA gyrase has recently been shown to be essential also in plant organellar replication (15, 44). In this paper we show that DNA gyrase is the molecular target of albicidin, which has features of inhibition that differ from those of known antibiotic classes.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and antibiotics.
The bacterial strains and plasmids used for this study are listed in Table 1. Escherichia coli cultures were grown at 37°C in Luria-Bertani medium (37). Xanthomonas albilineans cultures were grown at 28°C in SMG3 medium (50). Albicidins were purified to single peaks by C18 high-pressure liquid chromatography, dissolved in methanol, and quantified by bioassay for their activities against E. coli K-12 strain MG1665, as described previously (12, 50). Under these growth conditions, earlier-eluting albicidin (α peak) predominates over the compound (β peak) described from complex media (7). They are closely related compounds, as indicated by their nuclear magnetic resonance spectra and by equivalent responses to all known albicidin resistance mechanisms, although the full structures have not yet been elucidated (M. J. Garson and R. G. Birch, unpublished data). The high-pressure liquid chromatography-purified α-peak albicidin was used in all experiments described here. Ciprofloxacin (Fluka) and novobiocin (Sigma) were dissolved in water.
TABLE 1.
Properties of strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| MG1665 | K-12 type strain (wild-type GyrA and GyrB) | |
| MLS83L | Isogenic to MG1665 except for GyrA S83L, quinolone resistant | W. M. Parks and A. Maxwell, unpublished data |
| B410 | Wild-type GyrA | 4 |
| B462 | B410 GyrA R462C, CcdB resistant | 4 |
| HB101 | Wild-type GyrB | 13 |
| CC6 | HB101 GyrB R136C, coumarin resistant | 16 |
| CC7 | HB101 GyrB R136S, coumarin resistant | 16 |
| Rosetta pLysS | Novagen | |
| X. albilineans | ||
| Xa13 | Wild-type albicidin producer from sugarcane (Queensland), Apr | 50 |
| LS155 | Wild-type albicidin producer from sugarcane (Queensland), Apr | 43 |
| Plasmids | ||
| pBluescript | Expression vector, native protein, bla (Apr) | Stratagene |
| pET-19b | Expression vector, His6 fusion protein, bla (Apr) | Invitrogen |
| pET19b-XaQnr | pET-19b containing albG, Apr | This study |
| pBR322 | Substrate DNA for DNA gyrase assays | 13 |
Gene cloning and characterization.
The albG gene from X. albilineans with flanking NdeI and BamHI sites was amplified by PCR with primers XaQnrF (5′-ATCAGGTTTCCACATATGCCGGCCAAGACCC-3′) and XaQnrR (5′-CGGGATCCAGGTTGCATCAGTCACACG-3′) (restriction sites are underlined). After digestion with NdeI and BamHI, the amplified fragment was cloned into pET-19b to give pET19b-XaQnr, which contained an N-terminal His6 tag to facilitate isolation of fusion proteins. Clones were tested for albicidin resistance by a microbial plate bioassay described previously (10).
DNA sequencing was performed by dideoxynucleotide chain termination (38) with a BigDye Terminator cycle sequencing kit (version 3.1) and a 373A DNA sequencer (PE Applied Biosystems).
Enzyme assays.
Unless specified, the DNA gyrase used was E. coli A2B2; the DNA substrate was pBR322 in the supercoiled, relaxed, or EcoRI-linearized form; and the assays were performed as described previously (20, 35, 39, 44). The methanol concentration in the reactions for testing of the albicidin dilution series was constant at 0.82% (vol/vol).
The gyrase supercoiling reaction mixtures (30 μl), which contained gyrase (3.4 nM), 1 mM ATP, and 5 nM (0.4 μg) relaxed pBR322 DNA, were incubated at 37°C for 30 to 60 min and then terminated by addition of 0.5 volume of STEB (20% [wt/vol] sucrose, 0.05 M Tris · HCl [pH 7.5], 0.05 M EDTA, 50 μg ml−1 bromophenol blue) and 2 volumes of chloroform-isoamyl alcohol (24:1), followed by electrophoretic analysis in 1% agarose gels. The 50% inhibitory concentrations (IC50s) for drug inhibition were determined as described previously (20).
Gyrase relaxation assays were performed similarly to the supercoiling assays, except that ATP and spermidine were omitted, the enzyme concentration was 10 nM, the substrate was supercoiled pBR322 DNA, and the samples were incubated at 37°C for 4 h. Relaxation assays catalyzed by topoisomerase IV were carried out by incubating enzyme (5 nM) and supercoiled pBR322 DNA (5 nM) with various concentrations of drug in 40 mM HEPES · KOH (pH 7.6), 10 mM magnesium acetate, 100 mM potassium glutamate, 10 mM dithiothreitol, 50 μg ml−1 bovine serum albumin, 1 mM ATP, 4 μg ml−1 tRNA, 1% (wt/vol) glycerol, 2% (vol/vol) dimethyl sulfoxide.
The DNA cleavage reaction mixtures contained gyrase (34 nM), 5 to 12 nM relaxed or supercoiled pBR322 DNA, 35 mM Tris · HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 2 mM dithiothreitol, 1.8 mM spermidine, 0.1 mg ml−1 bovine serum albumin, 6.5% (wt/vol) glycerol, and various concentrations of drug in the presence or the absence of 1 mM ATP or 5′-adenylyl β,γ-imidodiphosphate (ADPNP), a nonhydrolyzable ATP analog. Ciprofloxacin and CcdB were used at 5 μM as positive controls for stabilization of cleavage complexes. Linear product was released by the addition of sodium dodecyl sulfate (SDS) to 0.2% (wt/vol) and proteinase K to 0.1 mg ml−1, followed by incubation at 37°C for 30 min. Cleavage specificity reactions were performed as described previously (1).
The gyrase ATPase reaction mixtures contained 20 μM the N-terminal domain of GyrB (GyrB43) in 47 mM Tris · HCl (pH 7.5), 1 mM EDTA, 6% (wt/vol) glycerol, 26.6 mM KCl, 2 mM dithiothreitol, 5 mM MgCl2, 800 μM phosphoenolpyruvate, 400 μM NADH, 1% (vol/vol) pyruvate kinase-lactate dehydrogenase enzymes (Sigma), and 2 mM ATP; and the reactions were carried out at 25°C. For determination of the DNA-dependent ATPase activity, 70 nM gyrase was incubated with 2.8 nM linear pBR322 DNA under the conditions described above.
Qnr purification.
E. coli Rosetta pLysS cultures containing pET19b-XaQnr were grown at 37°C overnight. The starter cultures were inoculated at a 1:100 ratio into Luria-Bertani liquid medium containing ampicillin (100 μg ml−1) and chloramphenicol (30 μg ml−1). The cultures were grown at 30°C with shaking until they reached an optical density at 600 nm of 0.5. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the cultures were grown at 30°C for 4 h. Bacterial cells were harvested by centrifugation at 6,000 × g. Soluble proteins were released by incubation in 50 mM Tris · HCl (pH 7.5), 1 mM EDTA, 10% (wt/vol) glycerol, 0.02% (wt/vol) lysozyme, and 0.12% (wt/vol) Brij 35. The cell lysate was centrifuged at 100,000 × g for 1 h at 4°C; and the supernatant was dialyzed into 20 mM Tris · HCl (pH 7.5), 10% (wt/vol) glycerol, 150 mM NaCl, and 10 mM imidazole. The dialyzed lysate was loaded onto a 5 ml HiTrap chelating HP Ni column (Amersham), and the His-tagged protein was eluted with 600 mM imidazole. Fractions containing the 31-kDa protein were dialyzed into TGED buffer (44), flash frozen in liquid N2, and stored at −80°C.
RESULTS
Albicidin inhibits gyrase-mediated DNA supercoiling.
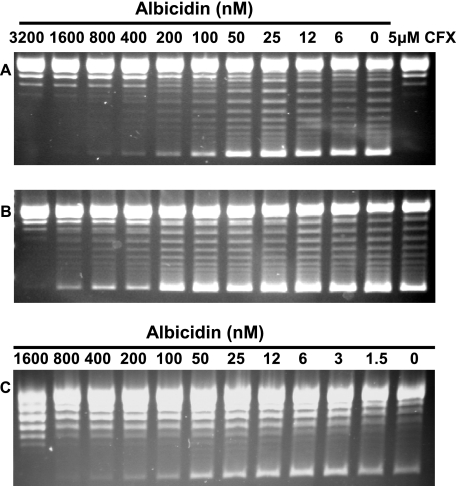
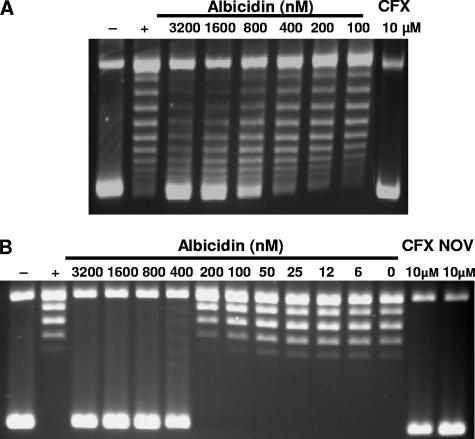
We found albicidin to be a potent inhibitor of DNA supercoiling catalyzed by E. coli DNA gyrase, with an IC50 value of ∼40 nM (Fig. 1A). It also inhibited DNA supercoiling mediated by Arabidopsis thaliana mitochondrial and chloroplast gyrases, with IC50s of ∼50 nM (Fig. 1C and data not shown). Albicidin inhibited the ATP-independent relaxation activity of E. coli DNA gyrase with an IC50 value of ∼600 nM (Fig. 2A) and the ATP-dependent relaxation reaction of topoisomerase IV with an IC50 value of ∼300 nM (Fig. 2B). It also inhibited decatenation by topoisomerase IV but did not affect the ATPase activity of DNA gyrase (data not shown).
FIG. 1.
Albicidin-mediated inhibition of DNA supercoiling reactions of DNA gyrases (3.4 nM) from E. coli MG1665 wild type (IC50 ∼40 nM) (A), E. coli with quinolone resistance-conferring gyrase mutation GyrA S83W (IC50, ∼200 nM) (B), and Arabidopsis thaliana mitochondria (IC50, ∼50 nM) (C). Lanes CFX, control treatment with the known DNA gyrase inhibitor ciprofloxacin.
FIG. 2.
Effect of albicidin on relaxation of supercoiled DNA by topoisomerases E. coli DNA gyrase (10 nM) (A) and E. coli topoisomerase IV (5 nM) (B). Controls were no enzyme (lanes −), no drug (lanes +), and the known gyrase inhibitors ciprofloxacin (lanes CFX) and novobiocin (lanes NOV).
Albicidin stabilizes the DNA gyrase cleavage complex in an ATP-dependent manner.
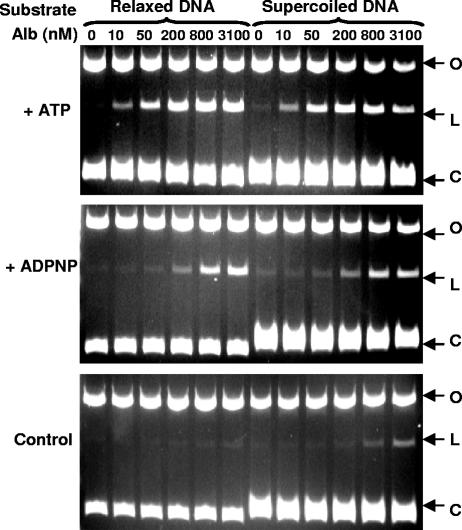
A number of agents, including quinolone drugs, CcdB, and microcin B17, can stabilize the covalent complex formed between gyrase and DNA. After SDS-proteinase K treatment, a linear DNA band in reaction mixtures containing antibiotic, gyrase, and DNA indicates stabilization of the gyrase-DNA cleavage complex (18, 28, 34). We found that the yield of linear DNA reached a plateau with an increasing albicidin concentration (Fig. 3). For convenience in this section, we use CC50 to mean the antibiotic concentration required to obtain half of the maximum yield of linear DNA observed in the reaction mixture containing albicidin, gyrase, ATP, and DNA. Albicidin stabilized the cleavage complex equally (CC50, ∼30 nM) with relaxed or supercoiled substrate in the presence of ATP and to a lesser extent (CC50, ∼300 nM) in the presence of ADPNP. In the absence of ATP or ADPNP, stabilization of the cleavage complex required a much higher albicidin concentration (CC50, ∼3,100 nM) when supercoiled DNA was used as the substrate and, apparently, an albicidin concentration at least an order of magnitude higher with relaxed DNA substrate (Fig. 3).
FIG. 3.
Ability of albicidin (Alb) to stabilize the gyrase-DNA cleavage complex in the presence or the absence of ATP or ADPNP and with relaxed or supercoiled DNA substrate. O, open circle DNA; L, linear DNA; C, closed circle DNA. The formation of linear DNA indicates stabilization of the cleavage complex. The DNA gyrase concentration was 34 nM. The reaction mixtures for the control panel did not include either ATP or ADPNP.
Albicidin and ciprofloxacin induced similar fragmentation patterns in linear pBR322 in the presence of DNA gyrase, indicating no major effect on the site specificity for cleavage (Fig. 4).
FIG. 4.
Cleavage of linear pBR3222 by DNA gyrase in the presence of ciprofloxacin (CFX) or albicidin (Alb). Lane M, molecular weight markers; lane SC, supercoiled pBR322; lanes for controls without antibiotic, linear pBR322 (lane 1), linear pBR322 extracted with phenol (lane 2), and linear pBR322 treated with SDS-proteinase K (lane 3). Reactions including antibiotic were 5 μM ciprofloxacin (lane 4), 10 μM ciprofloxacin (lane 5), 40 nM albicidin (lane 6), and 100 nM albicidin (lane 7).
Cross-resistance to known gyrase mutations.
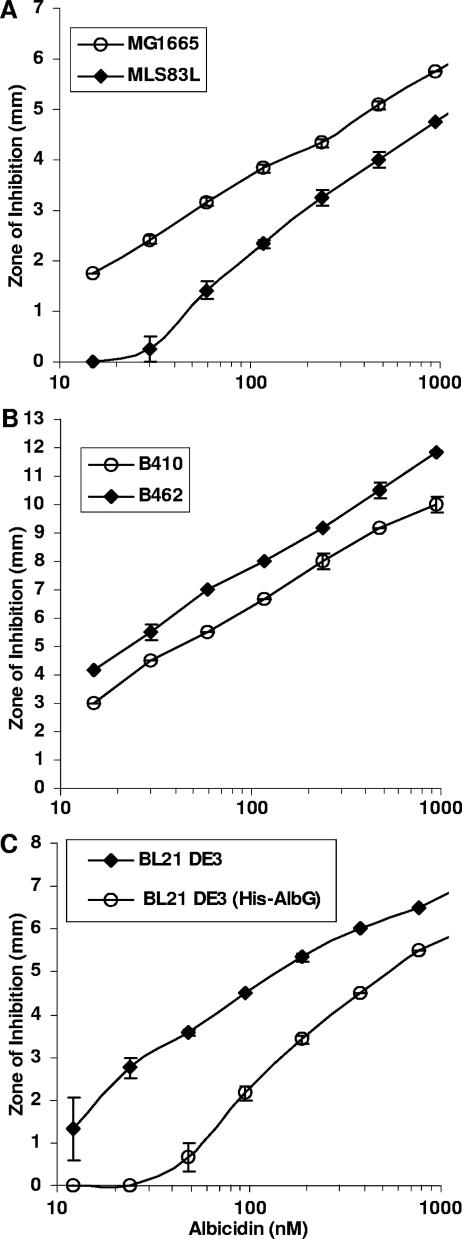
Spontaneous resistance to quinolone drugs commonly involves mutations that change Ser83 of GyrA to Leu or Trp (28, 47). We found that the E. coli quinolone-resistant strain MLS83L (GyrA S83L) showed a four- to sixfold increase in resistance to albicidin relative to that of its isogenic parent strain, MG1665, in the antimicrobial assay (Fig. 5A). Similarly, the IC50 for albicidin increased four- to sixfold with purified GyrA S83W enzyme relative to that with wild-type E. coli gyrase (Fig. 1B). Spontaneous resistance to coumarin drugs commonly involves mutations that alter Arg136 of GyrB (28). We found that coumermycin-resistant mutants CC6 (GyrB R136C) and CC7 (GyrB R136S) showed no significant difference in their sensitivities to albicidin relative to that of parent strain HB101 (data not shown).
FIG. 5.
(A) Albicidin cross-resistance by E. coli MG1665 and isogenic quinolone-resistant strain MLS83L (GyrA S83L); (B) increased sensitivity to albicidin resulting from the GyrA R462C (CcdB resistance) mutation in E. coli B462 relative to isogenic parent strain B410; (C) decreased sensitivity to albicidin in E. coli conferred by the albG gene from X. albilineans. The results are means with standard errors from three replicates.
Mutations that change Arg462 of GyrA to Cys confer resistance to CcdB (5). We found that E. coli B462 bearing this mutation showed a twofold increased sensitivity to albicidin compared with the sensitivities of its isogenic parent strain (strain B410) and wild-type strain MG1655 (Fig. 5B). Similarly, purified GyrA R462C protein showed twofold increased sensitivity to albicidin in assays testing for inhibition of supercoiling and for stabilization of the cleavage complex.
X. albilineans encodes a DNA gyrase-interacting protein (AlbG) that confers increased resistance to albicidin.
The albicidin biosynthetic gene cluster (36) contains a sequence that encodes a predicted product with 30 to 40% similarity to the quinolone resistance proteins Qnr and MfpA (32, 41). We cloned the X. albilineans homolog (here designated albG) into both pBluescript and pET19b vectors. This conferred a 30-fold increase in resistance to albicidin among E. coli cells grown without IPTG induction in the case of the pBluescript clone and a 4- to 6-fold increase in resistance in the case of the pET19b clone, which encodes a His6-AlbG fusion (Fig. 5C), with no change in sensitivity to ciprofloxacin (data not shown). Growth was inhibited in the presence of IPTG (50 μM). Purified His6-AlbG (0.65 μM) protected gyrase from the effects of albicidin in supercoiling assays, increasing the IC50 for albicidin by two- to fourfold, with little or no effect on the sensitivity to ciprofloxacin. In separate assays, His6-AlbG partially inhibited the supercoiling activity of DNA gyrase in the absence of albicidin (IC50, 6 μM) without evidently inhibiting the cleavage of supercoiled DNA by gyrase, which produces the linear DNA band in this assay (18). This is similar to the action of the MfpA protein, which mimics a B-form DNA and inhibits the supercoiling activity of DNA gyrase (23).
DISCUSSION
The results presented in this paper show that albicidin is a potent inhibitor of bacterial and plant organellar DNA gyrases and has a mechanism that likely differs at the molecular level from those of previously characterized gyrase-inhibiting antibiotics. For inhibition of supercoiling activity, the IC50 of albicidin (50 nM) is less than those of ciprofloxacin (700 nM) and novobiocin (250 nM) under the same conditions (20), and albicidin is as effective against the plant (A. thaliana) chloroplast and mitochondrial DNA gyrases as it is against the bacterial (E. coli) enzyme (Fig. 1).
Significance of a gyrase-inhibiting phytotoxin.
This is the first example of a bacterial phytotoxin with a known molecular target in plastid DNA replication. Other phytotoxins reported to target host nucleic acid biosynthesis are targetitoxin, which inhibits plastid RNA polymerase (40), and solanapyrone, which inhibits a nuclear DNA polymerase (30). Albicidin production probably serves a dual role for X. albilineans in its niche as a wound-infecting, systemic xylem-invading sugarcane pathogen. First, it is a potent antibiotic against secondary bacterial invaders. Interestingly, an albicidin detoxification gene confers resistance in Pantoea dispersa, which is widely associated with plants (48), and an albicidin exporter homolog exists in the genome of the sugarcane xylem-invading organism Liefsonia xyli (31). Second, albicidin is important for systemic invasion by X. albilineans, probably because interference with plastid function reduces the capacity of the plant to mount effective defense responses (6). Introduction of an organellar albicidin-resistant gyrase is a possible means of engineering plant resistance to sugarcane leaf scald disease.
Features of gyrase inhibition.
The effects of albicidin on reactions with purified enzymes are compared in Table 2. Unlike the coumarins, which interact primarily with the region around the GyrB binding site for ATP and function as competitive inhibitors of the ATPase activity needed for gyrase function (29), albicidin did not inhibit the ATPase activity of DNA gyrase. Albicidin did inhibit the ATP-independent relaxation reaction of gyrase, albeit with a higher IC50 than that for supercoiling, and it also inhibited ATP-dependent relaxation and decatenation by the related enzyme topoisomerase IV (Fig. 2B). Unlike quinolones, which apparently interact with the N-terminal domain of GyrA and cause rapid ATP-independent stabilization of the gyrase-cleaved DNA complex (21), albicidin required the presence of ATP for efficient stabilization of the complex (Fig. 3). In the absence of ATP, the CC50 values for cleavage increased from 30 nM by at least 100-fold for supercoiled substrate DNA and at least 1,000-fold for relaxed substrate DNA.
TABLE 2.
IC50 and CC50 values for albicidin in reactions with purified enzymes
| Enzyme reaction | Albicidin IC50 or CC50 (nM) |
|---|---|
| DNA supercoiling by DNA gyrase from | |
| wild-type E. coli | 40 |
| DNA supercoiling by DNA gyrase from | |
| Arabidopsis plastids or mitochondria | 50 |
| DNA supercoiling by DNA gyrase from | |
| quinolone-resistant (GyrA S83L) E. coli | 200 |
| ATP-dependent relaxation by topoisomerase | |
| IV from wild-type E. coli | 300 |
| ATP-independent relaxation by DNA gyrase | |
| from wild-type E. coli | 600 |
| ATPase activity of DNA gyrase from | |
| wild-type E. coli | Not inhibited |
| Stabilization of cleavage complex between | |
| DNA gyrase, relaxed or supercoiled DNA, | |
| and ATP (CC50) | 30 |
| Stabilization of cleavage complex between | |
| DNA gyrase, relaxed or supercoiled DNA, | |
| and ADPNP (CC50) | 300 |
| Stabilization of cleavage complex between | |
| DNA gyrase and supercoiled DNA without | |
| ATP or ADPNP (CC50) | 3,100 |
| Stabilization of cleavage complex between | |
| DNA gyrase and relaxed DNA without | |
| ATP or ADPNP (CC50) | >30,000 |
The ATP-dependent stabilization of the gyrase-DNA cleavage complex by albicidin is reminiscent of the effects of the plasmid-encoded peptides microcin B17 and CcdB. However, these peptides are slow poisons, apparently because their opportunity to enter and bind within the gyrase complex is transient during the catalytic cycle, while the DNA gate is open in the case of CcdB (19, 34, 39). As a result, they do not inhibit gyrase-mediated supercoiling under the usual assay conditions. For both microcin B17 and CcdB, dependence on ATP is greater for cleavage of relaxed DNA than for cleavage of supercoiled DNA (18, 33, 39). In contrast, albicidin proved to be an efficient inhibitor of gyrase-mediated supercoiling (Fig. 1), and dependence on ATP was similar for stabilization of the cleavage complex with relaxed and supercoiled substrate DNA (Fig. 3).
Mechanism of gyrase inhibition by albicidin.
Our results indicate that albicidin (like quinolones) must efficiently access and bind to its target sites within the gyrase complex without the need for multiple catalytic cycles. This is consistent with the small size of albicidins (molecular weight, ∼842) relative to those of the peptide poisons.
Interestingly, the nonhydrolyzed analogue ADPNP could partially substitute for ATP in facilitating albicidin-mediated stabilization of the gyrase-cleaved DNA complex, requiring an approximately 10-fold higher albicidin concentration for an effect equivalent to those required for reactions containing ATP with either supercoiled or relaxed DNA substrates. ADPNP apparently holds the entry gate of DNA gyrase in a closed conformation, supporting only a single cycle of supercoiling and slowing DNA relaxation (3, 46). The closing of the entry gate by ATP or ADPNP may be important to bring the gyrase-DNA complex into a conformation susceptible to action by albicidin, as envisaged for quinolones (21). The more stringent requirement for ATP by albicidin relative to that by quinolones implies that a different target conformation is formed during an ATP-mediated step in the supercoiling cycle. Albicidin may interfere with religation of the G segment once the DNA gate has opened. The absence of ATP in the relaxation assays may reduce the occurrence of the enzyme conformation susceptible to binding by albicidin. Analysis of albicidin binding to individual gyrase domains and analysis of the kinetics of the effects on gyrase reactions could help to elucidate the interaction.
Certain DNA gyrase mutants show albicidin resistance. Both quinolone and CcdB resistance-conferring mutations in E. coli altered the sensitivity to albicidin. Relative to the isogenic parent strains, the quinolone-resistant mutant (GyrA S83L) showed a fourfold increased albicidin resistance (Fig. 5A), whereas the CcdB-resistant mutant (GyrA R462C) showed a twofold increased albicidin sensitivity (Fig. 5B). S83L is close to the GyrA catalytic site for DNA cleavage and confers approximately 20-fold increased resistance to various quinolones, accompanied by decreased drug binding to the gyrase-DNA complex (21). R462 is in the central dimerization domain of GyrA, which also forms the exit gate in the A2B2 holoenzyme, and it stacks with W99 of CcdB to anchor this peptide poison as a wedge that holds the catalytic domain of gyrase in its open configuration (19). The R462C mutation confers high-level resistance to CcdB and blocks CcdB binding to gyrase (5, 18, 39). The conformational changes during the gyrase catalytic cycle have yet to be fully elucidated, and it is not apparent how a small molecule such as albicidin could simultaneously interact with the catalytic and the dimerization regions of GyrA. One or both of the mutations may affect the sensitivity to albicidin indirectly, for example, by altering the duration of the proposed ATP-mediated, albicidin-sensitive conformation during the catalytic cycle.
Self-protection against albicidin.
Antibiotics that stabilize the gyrase-cleaved DNA complex rapidly stop bacterial multiplication by blocking DNA replication (45), as previously observed for albicidin (10). Cell death probably involves DNA fragmentation at the unsealed DNA cleavage sites (27). It is not surprising that X. albilineans uses multiple self-protection mechanisms against this potent bactericide, including highly regulated biosynthesis (50) and efficient export (14). To these we can now add coordinated production with albicidin biosynthesis enzymes of a DNA-mimicking pentapeptide repeat protein that binds to DNA gyrase in competition with DNA (23) and protects DNA gyrase from inhibition by albicidin. This protein (AlbG) increases the resistance of E. coli DNA gyrase to albicidin both in vivo (Fig. 5C) and in vitro, and at higher concentrations it inhibits supercoiling by the E. coli gyrase in the absence of albicidin. In contrast to some other producers of gyrase-inhibiting antibiotics (20), there is no evidence of any resistant gyrase subunit in the antibiotic biosynthesis gene cluster (36). It will be interesting to examine the X. albilineans gyrase genes in the future and to determine the sensitivity of the X. albilineans gyrase both to albicidin and to AlbG.
Opportunity of a new class of gyrase-inhibiting antibiotics.
Given the evidence that albicidin is a potent gyrase inhibitor with action at the molecular level different from those of known antibiotic classes, further characterization of the structure and action of albicidin coupled with the growing insight into the albicidin biosynthetic machinery may open the possibility of a new family of antibacterial drugs.
Acknowledgments
This work was supported by a University of Queensland scholarship and travel funds for S.M.H.; work in A.M.'s laboratory was supported by BBRSC and the EU.
Footnotes
Published ahead of print on 30 October 2006.
REFERENCES
- 1.Barnard, F. M., and A. Maxwell. 2001. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob. Agents Chemother. 45:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basnayake, W. V. S., and R. G. Birch. 1995. A gene from Alcaligenes denitrificans that confers albicidin resistance by reversible antibiotic binding. Microbiology 141:551-560. [DOI] [PubMed] [Google Scholar]
- 3.Bates, A. D., M. H. Odea, and M. Gellert. 1996. Energy coupling in Escherichia coli DNA gyrase: the relationship between nucleotide binding, strand passage, and DNA supercoiling. Biochemistry 35:1408-1416. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, P. 1995. New ccdB positive-selection cloning vectors with kanamycin or chloramphenicol selectable markers. Gene 162:159-160. [DOI] [PubMed] [Google Scholar]
- 5.Bernard, P., and M. Couturier. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226:735-745. [DOI] [PubMed] [Google Scholar]
- 6.Birch, R. G. 2001. Xanthomonas albilineans and the antipathogenesis approach to disease control. Mol. Plant Pathol. 2:1-11. [DOI] [PubMed] [Google Scholar]
- 7.Birch, R. G., and S. S. Patil. 25. June 1985. Antibiotic and process for the production thereof. U.S. patent 4525354.
- 8.Birch, R. G., and S. S. Patil. 1987. Correlation between albicidin production and chlorosis induction by Xanthomonas albilineans, the sugarcane leaf scald pathogen. Physiol. Mol. Plant Pathol. 30:199-206. [Google Scholar]
- 9.Birch, R. G., and S. S. Patil. 1987. Evidence that an albicidin-like phytotoxin induces chlorosis in sugarcane leaf scald disease by blocking plastid DNA replication. Physiol. Mol. Plant Pathol. 30:207-214. [Google Scholar]
- 10.Birch, R. G., and S. S. Patil. 1985. Preliminary characterization of an antibiotic produced by Xanthomonas albilineans which inhibits DNA synthesis in Escherichia coli. J. Gen. Microbiol. 131:1069-1075. [DOI] [PubMed] [Google Scholar]
- 11.Birch, R. G., and S. S. Patil. 1983. The relation of blocked chloroplast differentiation to sugarcane leaf scald disease. Phytopathology 73:1368-1374. [Google Scholar]
- 12.Birch, R. G., J. M. Pemberton, and W. V. S. Basnayake. 1990. Stable albicidin resistance in Escherichia coli involves an altered outer membrane nucleoside uptake system. J. Gen. Microbiol. 136:51-58. [DOI] [PubMed] [Google Scholar]
- 13.Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 68:245-267. [DOI] [PubMed] [Google Scholar]
- 14.Bostock, J. M., G. Huang, S. M. Hashimi, L. H. Zhang, and R. G. Birch. 2006. A DHA14 drug efflux gene from Xanthomonas albilineans confers high-level albicidin antibiotic resistance in Escherichia coli. J. Appl. Microbiol. 101:151-160. [DOI] [PubMed] [Google Scholar]
- 15.Cho, H. S., S. S. Lee, K. D. Kim, I. Hwang, J. S. Lim, Y. I. Park, and H. S. Pai. 2004. DNA gyrase is involved in chloroplast nucleoid partitioning. Plant Cell 16:2665-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contreras, A., and A. Maxwell. 1992. GyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol. Microbiol. 6:1617-1624. [DOI] [PubMed] [Google Scholar]
- 17.Corbett, K. D., and J. M. Berger. 2004. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33:95-118. [DOI] [PubMed] [Google Scholar]
- 18.Critchlow, S. E., M. H. O'Dea, A. J. Howells, M. Couturier, M. Gellert, and A. Maxwell. 1997. The interaction of the F plasmid killer protein, CcdB, with DNA gyrase: induction of DNA cleavage and blocking of transcription. J. Mol. Biol. 273:826-839. [DOI] [PubMed] [Google Scholar]
- 19.Dao-Thi, M. H., L. Van Melderen, E. De Genst, H. Afif, L. Buts, L. Wyns, and R. Loris. 2005. Molecular basis of gyrase poisoning by the addiction toxin CcdB. J. Mol. Biol. 348:1091-1102. [DOI] [PubMed] [Google Scholar]
- 20.Flatman, R. H., A. J. Howells, L. Heide, H. P. Fiedler, and A. Maxwell. 2005. Simocyclinone D8, an inhibitor of DNA gyrase with a novel mode of action. Antimicrob. Agents Chemother. 49:1093-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heddle, J., and A. Maxwell. 2002. Quinolone-binding pocket of DNA gyrase: role of GyrB. Antimicrob. Agents Chemother. 46:1805-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heddle, J. G., F. M. Barnard, L. M. Wentzell, and A. Maxwell. 2000. The interaction of drugs with DNA gyrase: a model for the molecular basis of quinolone action. Nucleosides Nucleotides Nucleic Acids 19:1249-1264. [DOI] [PubMed] [Google Scholar]
- 23.Hegde, S. S., M. W. Vetting, S. L. Roderick, L. A. Mitchenall, A. Maxwell, H. E. Takiff, and J. S. Blanchard. 2005. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science 308:1480-1483. [DOI] [PubMed] [Google Scholar]
- 24.Huang, G., L. Zhang, and R. G. Birch. 2000. Albicidin antibiotic and phytotoxin biosynthesis in Xanthomonas albilineans requires a phosphopantetheinyl transferase gene. Gene 258:193-199. [DOI] [PubMed] [Google Scholar]
- 25.Huang, G., L. Zhang, and R. G. Birch. 2000. Analysis of the genes flanking xabB: a methyltransferase gene is involved in albicidin biosynthesis in Xanthomonas albilineans. Gene 255:327-333. [DOI] [PubMed] [Google Scholar]
- 26.Huang, G., L. Zhang, and R. G. Birch. 2001. A multifunctional polyketide-peptide synthase for albicidin biosynthesis in Xanthomonas albilineans. Microbiology 147:631-642. [DOI] [PubMed] [Google Scholar]
- 27.Malik, M., X. Zhao, and K. Drlica. 2006. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol. Microbiol. 61:810-825. [DOI] [PubMed] [Google Scholar]
- 28.Maxwell, A. 1997. DNA gyrase as a drug target. Trends Microbiol. 5:102-109. [DOI] [PubMed] [Google Scholar]
- 29.Maxwell, A., and D. M. Lawson. 2003. The ATP-binding site of type II topoisomerases as a target for antibacterial drugs. Curr. Top. Med. Chem. 3:283-303. [DOI] [PubMed] [Google Scholar]
- 30.Mizushina, Y., S. Kamisuki, N. Kasai, N. Shimazaki, M. Takemura, H. Asahara, S. Linn, S. Yoshida, A. Matsukage, O. Koiwai, F. Sugawara, H. Yoshida, and K. Sakaguchi. 2002. A plant phytotoxin, solanapyrone A, is an inhibitor of DNA polymerase beta and lambda. J. Biol. Chem. 277:630-638. [DOI] [PubMed] [Google Scholar]
- 31.Monteiro-Vitorello, C. B., L. E. A. Camargo, M. A. Van Sluys, J. P. Kitajima, D. Truffi, A. M. do Amaral, R. Harakava, J. C. F. de Oliveira, D. Wood, M. C. de Oliveira, C. Miyaki, M. A. Takita, A. C. R. da Silva, L. R. Furlan, D. M. Carraro, G. Camarotte, N. F. Almeida, H. Carrer, L. L. Coutinho, H. A. El-Dorry, M. I. T. Ferro, P. R. Gagliardi, E. Giglioti, M. H. S. Goldman, G. H. Goldman, E. T. Kimura, E. S. Ferro, E. E. Kuramae, E. G. M. Lemos, M. V. F. Lemos, S. M. Z. Mauro, M. A. Machado, C. L. Marino, C. F. Menck, L. R. Nunes, R. C. Oliveira, G. G. Pereira, W. Siqueira, A. A. de Souza, S. M. Tsai, A. S. Zanca, A. J. G. Simpson, S. M. Brumbley, and J. C. Setubal. 2004. The genome sequence of the gram-positive sugarcane pathogen Leifsonia xyli subsp. xyli. Mol. Plant-Microbe Interact. 17:827-836. [DOI] [PubMed] [Google Scholar]
- 32.Montero, C., G. Mateu, R. Rodriguez, and H. Takiff. 2001. Intrinsic resistance of Mycobacterium smegmatis to fluoroquinolones may be influenced by new pentapeptide protein MfpA. Antimicrob. Agents Chemother. 45:3387-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierrat, O. A., and A. Maxwell. 2003. The action of the bacterial toxin microcin B17—insight into the cleavage-religation reaction of DNA gyrase. J. Biol. Chem. 278:35016-35023. [DOI] [PubMed] [Google Scholar]
- 34.Pierrat, O. A., and A. Maxwell. 2005. Evidence for the role of DNA strand passage in the mechanism of action of microcin B17 on DNA gyrase. Biochemistry 44:4204-4215. [DOI] [PubMed] [Google Scholar]
- 35.Reece, R. J., and A. Maxwell. 1989. Tryptic fragments of the Escherichia coli DNA gyrase A protein. J. Biol. Chem. 264:19648-19653. [PubMed] [Google Scholar]
- 36.Royer, M., L. Costet, E. Vivien, M. Bes, A. Cousin, A. Damais, I. Pieretti, A. Savin, S. Megessier, M. Viard, R. Frutos, D. W. Gabriel, and P. C. Rott. 2004. Albicidin pathotoxin produced by Xanthomonas albilineans is encoded by three large PKS and NRPS genes present in a gene cluster also containing several putative modifying, regulatory, and resistance genes. Mol. Plant-Microbe Interact. 17:414-427. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain termination inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, A. B., and A. Maxwell. 2006. A strand-passage conformation of DNA gyrase is required to allow the bacterial toxin, CcdB, to access its binding site. Nucleic Acids Res. 34:4667-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinberg, T. H., and R. R. Burgess. 1992. Tagetitoxin inhibition of RNA polymerase III transcription results from enhanced pausing at discrete sites and is template dependent. J. Biol. Chem. 267:20204-20211. [PubMed] [Google Scholar]
- 41.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 99:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker, M. J., R. G. Birch, and J. M. Pemberton. 1988. Cloning and characterization of an albicidin resistance gene from Klebsiella oxytoca. Mol. Microbiol. 2:443-454. [DOI] [PubMed] [Google Scholar]
- 43.Wall, M. K., and R. G. Birch. 1997. Genes for albicidin biosynthesis and resistance span at least 69 kb in the genome of Xanthomonas albilineans. Lett. Appl. Microbiol. 24:256-260. [DOI] [PubMed] [Google Scholar]
- 44.Wall, M. K., L. A. Mitchenall, and A. Maxwell. 2004. Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc. Natl. Acad. Sci. USA 101:7821-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wentzell, L. M., and A. Maxwell. 2000. The complex of DNA gyrase and quinolone drugs on DNA forms a barrier to the T7 DNA polymerase replication complex. J. Mol. Biol. 304:779-791. [DOI] [PubMed] [Google Scholar]
- 46.Williams, N. L., A. J. Howells, and A. Maxwell. 2001. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 306:969-984. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyarse gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, L., and R. G. Birch. 1997. The gene for albicidin detoxification from Pantoea dispersa encodes an esterase and attenuates pathogenicity of Xanthomonas albilineans to sugarcane. Proc. Natl. Acad. Sci. USA 94:9984-9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, L., J. Xu, and R. G. Birch. 1999. Engineered detoxification confers resistance against a pathogenic bacterium. Nat. Biotechnol. 17:1021-1024. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, L., J. Xu, and R. G. Birch. 1998. Factors affecting biosynthesis by Xanthomonas albilineans of albicidin antibiotics and phytotoxins. J. Appl. Microbiol. 85:1023-1028. [DOI] [PubMed] [Google Scholar]