Abstract
The effect of paromomycin on the interaction of ribosomal subunits was studied. Paromomycin inhibited the antiassociation activity of initiation factor 3 (IF3). Furthermore, ribosomal subunits were associated to form 70S ribosomes by paromomycin even in the presence of 1 mM Mg2+. Paromomycin did not inhibit the binding of IF3 to the 30S ribosomal subunits. On the other hand, IF3 bound to the 30S subunits was expelled by paromomycin-induced subunit association (70S formation). These results indicate that the stabilization of 70S ribosomes by paromomycin may in part be responsible for its inhibitory effects on translocation and ribosome recycling.
Aminoglycosides cause miscoding (6, 18), stabilization of 70S ribosomes (4, 26), inhibition of translocation (2, 10, 16, 24), alanylation of transfer mRNA (37), and recycling of ribosomes (12, 13, 17). Although the mechanisms of the miscoding effect of paromomycin have been worked out previously (27-29), the exact mechanism of the effect of paromomycin on the translocation and recycling steps remains obscure. We investigated the effect of paromomycin on the interaction of subunits which plays an important role in both of these steps. We show that paromomycin inhibits the antiassociation activity of initiation factor 3 (IF3), which is an important component of the disassembly reaction of posttermination ribosomal complexes (14). This inhibitory effect comes from the fact that paromomycin strengthens the interaction between ribosomal subunits and even induces association of the subunits at low Mg2+ concentrations. Upon the association of subunits by paromomycin, the 30S-bound IF3 is displaced from the ribosomes. A possible mechanism of the inhibitory action of paromomycin on the translocation step is discussed.
MATERIALS AND METHODS
Buffers.
Buffer J consisted of 10 mM Tris-HCl, pH 7.4, 10 mM MgSO4, 50 mM KCl, and 1 mM dithiothreitol (DTT). Buffer R consisted of 10 mM Tris-HCl, pH 7.4, 8.2 mM MgSO4, 84 mM NH4Cl, and 0.5 mM DTT. Buffer S consisted of 10 mM Tris-HCl, pH 7.4, 1 mM MgSO4, 84 mM NH4Cl, and 0.5 mM DTT. Buffer S2 consisted of 10 mM Tris-HCl, pH 7.4, 4 mM MgSO4, 84 mM NH4Cl, and 0.5 mM DTT. Buffer S3 consisted of 10 mM Tris-HCl, pH 7.4, 6 mM MgSO4, 84 mM NH4Cl, and 0.5 mM DTT.
Ribosomes and factors.
Vacant ribosomes were prepared from Escherichia coli MRE600 (purchased from the University of Alabama Fermentation Facility, Birmingham) as described previously (21). Ribosome recycling factor (RRF) and elongation factor G (EF-G) were purified as described previously (11, 21) from E. coli DH5α harboring plasmid pRR2 (34) and E. coli JM83 harboring plasmid pECEG (15) (obtained from P. March), respectively. IF3 was purified from E. coli XL1-Blue harboring a plasmid expressing His-IF3 (35) (obtained from T. Ueda).
Sucrose density gradient ultracentrifugation (SDGC).
Unless otherwise mentioned, 0.6 A260 units (approximately 14 pmol) of 70S ribosomes (or the dissociated subunits) or 0.45 A260 units (approximately 31 pmol) of isolated 30S subunits in 275 μl of a reaction mixture (described in the figure legends) was applied on 4.5 ml of a 15% to 30% sucrose gradient prepared in the same buffer as the reaction mixture. Samples were centrifuged at 40,000 rpm (Beckman SW50.1 rotor) for 2.5 h at 4°C. For Fig. 1 and 3, the absorbance at 254 nm in the gradient was analyzed by use of an ISCO optical unit type 11 and a UA-6 detector (Teledyne Isco, Inc., Lincoln, NE) and areas corresponding to 30S, 50S subunits and 70S ribosomes were measured by ImageJ software (National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/). For Fig. 2 and 4, fractions were taken from the bottom of the tube (10 drops per fraction) and the absorbance at 260 nm was measured manually.
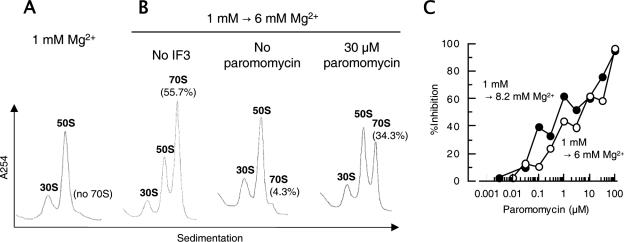
FIG. 1.
Paromomycin inhibits antiassociation activity of IF3. (A) 70S ribosomes (0.05 μM) were dissociated into subunits in buffer S (1 mM Mg2+ and other components) at 30°C for 7 min. (B) Subunits prepared as described for panel A were further incubated in the absence (left panel) or presence of IF3 (4.5 μM) (center panel) or IF3 (4.5 μM) and paromomycin (30 μM; Sigma, St. Louis, MO) (right panel) at 30°C for 5 min, and then magnesium acetate was added to 6 mM and the mixture was incubated at 30°C for an additional 10 min. The mixtures described for panels A and B were subjected to 15% to 30% SDGC (Beckman SW50.1 rotor at 40,000 rpm for 2.5 h). Sedimentation was from left to right. (C) Dose-response curves of paromomycin inhibition on the antiassociation activity of IF3 at 6 mM Mg2+ or 8.2 mM Mg2+. The following equation was used to calculate the inhibitory effect of paromomycin on the antiassociation activity of IF3: percent inhibition = [1 − (%70S1 − %70S2)/(%70S1 − %70S3)] × 100, where %70S1 is the percentage of 70S ribosomes in the absence of both IF3 and paromomycin at 6 mM (B, left panel) or at 8.2 mM (data not shown) Mg2+, %70S2 is the percentage of 70S ribosomes in the presence of both IF3 and paromomycin at 6 mM (B, right panel) or at 8.2 mM (data not shown) Mg2+, and %70S3 is the percentage of 70S ribosomes in the presence of only IF3 at 6 mM (B, center panel) or at 8.2 mM (data not shown) Mg2+.
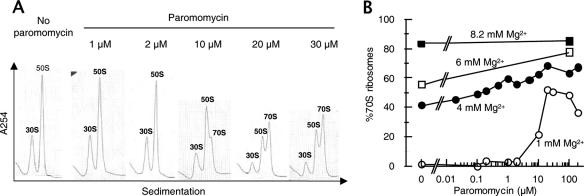
FIG. 3.
Paromomycin associates ribosomal subunits into 70S ribosomes. (A) Sedimentation pattern of ribosomes in the presence of paromomycin at 1 mM Mg2+. 70S ribosomes (0.05 μM) were dissociated into subunits in buffer S (1 mM Mg2+ and other components), various amounts of paromomycin (as indicated) were added, and the mixtures were incubated at 30°C for 10 min and analyzed as described in the legend for Fig. 1A. (B) Dose-response curves of subunit association by paromomycin at various Mg2+ concentrations. Ribosomal subunits were prepared in buffer S, various concentrations of paromomycin were added, the mixture was incubated at 30°C for 5 min, magnesium acetate was added to 4 mM (filled circles), 6 mM (open squares), or 8.2 mM (filled squares), and the mixture was incubated at 30°C for an additional 10 min and subjected to SDGC. Percentages of 70S ribosomes were calculated and plotted against paromomycin concentrations.
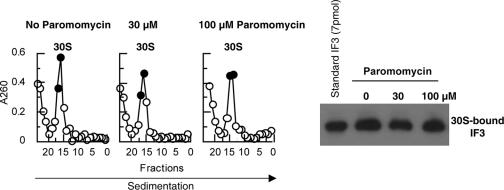
FIG. 2.
Paromomycin does not inhibit the binding of IF3 to 30S subunits. 30S subunits (0.45 A260 units) were incubated with IF3 (4.5 μM) in buffer R (8.2 mM Mg2+ and other components) followed by the addition of paromomycin or buffer. The mixture was subjected to SDGC as described in the legend for Fig. 1, except for the use of buffer R, and fractionated from the bottom of the tube (10 drops per tube). Absorbance at 260 nm was measured (left panel), and two fractions containing the peak of 30S subunits (filled circles) were collected and analyzed by Western blot analysis using anti-IF3 rabbit antiserum (right panel). Purified IF3 (7 pmol) was used as a standard.
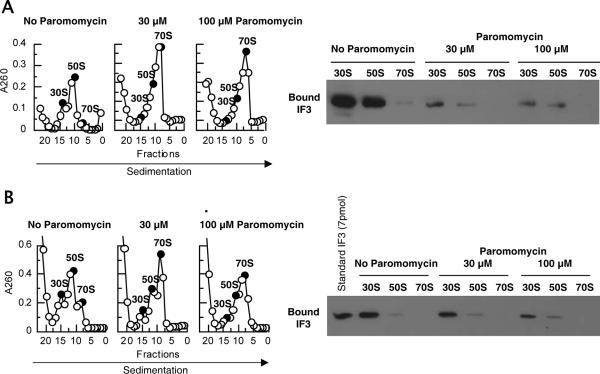
FIG. 4.
Bound IF3 is expelled from 30S subunits upon ribosomal association by paromomycin. (A) Binding of IF3 to ribosomes in the presence of paromomycin at 6 mM Mg2+. 70S ribosomes (0.05 μM) were dissociated into subunits in buffer S (1 mM Mg2+ and other components) at 30°C for 7 min, IF3 (4.5 μM) and paromomycin (0, 30 μM, or 100 μM as indicated) were added, the mixture was further incubated at 30°C for 5 min, magnesium acetate was then added to 6 mM, the mixture was incubated for 10 min and subjected to SDGC, and the presence of IF3 on 30S or 50S subunits and 70S ribosomes was examined as described in the legend for Fig. 2 except that the sucrose gradient was made in buffer S3 (6 mM Mg2+ and other components). The fractions (filled circles) were analyzed by Western blotting. (B) Binding of IF3 after the energy-dependent ribosomal splitting by RRF and EF-G in the presence of paromomycin. 70S ribosomes (0.05 μM) were incubated with RRF (1 μM), EF-G (1 μM), GTP (0.36 mM), and IF3 (4.5 μM) in buffer R (8.2 mM Mg2+ and other components) in the presence or absence of paromomycin (30 or 100 μM) at 30°C for 15 min. Samples were subjected to SDGC, and the presence of IF3 on 30S or 50S subunits and 70S ribosomes (filled circles) was examined as described for panel A except that the sucrose gradient was made in buffer R.
Binding of IF3.
Binding of IF3 to ribosomes and the subunits was analyzed essentially as described previously (14), with modifications as described below.
After the SDGC and fractionation were completed as described above, fractions containing 30S or 50S subunits or 70S ribosomes were collected. For Fig. 2, two fractions containing the peak of 30S subunits were collected, while for Fig. 4, only one fraction containing the peak of 30S or 50S subunits or 70S ribosomes was collected. Proteins in the fractions were precipitated by 10% trichloroacetic acid on ice for 1 h, washed in an ether-ethanol mixture (50% each), and then dried. Samples were suspended in 19 μl of Laemmli loading buffer with 1 μl of Tris base, boiled, and then subjected to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were blotted to a nylon membrane and analyzed by Western blotting. One-fourth (5 μl) (Fig. 2), one-half (10 μl) (Fig. 4B), or all (20 μl) (Fig. 4A) of the precipitated samples was analyzed. For Fig. 2 and 4B, 7 pmol of purified IF3 was also subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis as a standard. For the detection of IF3, rabbit antiserum against E. coli IF3, peroxidase-conjugated goat antibody against rabbit immunoglobulin G, and Amersham ECL detection reagents (GE Healthcare, Piscataway, NJ) were used. Rabbit antiserum against E. coli IF3 was raised by Rockland Immunochemicals, Inc. (Gilbertsville, PA), and used at a 1/5,000 dilution.
RESULTS
Paromomycin inhibits antiassociation activity of IF3.
We first examined if paromomycin inhibits the antiassociation activity of IF3. In the experiment depicted in Fig. 1, vacant ribosomes were dissociated into subunits at 1 mM Mg2+ (Fig. 1A) and exposed to 6 mM Mg2+ so that the subunits were reassociated into 70S ribosomes (Fig. 1B, left). If IF3 was present, the reassociation was inhibited and almost no 70S ribosome was observed (Fig. 1B, center). In the presence of paromomycin, however, certain subunits were reassociated into 70S ribosomes even in the presence of IF3 (Fig. 1B, right). The dose-dependent inhibitory effect of paromomycin is plotted in Fig. 1C. The inhibitory action of paromomycin on the antiassociation activity of IF3 could also be observed in the presence of 8.2 mM Mg2+ (Fig. 1C, filled circles). It is clear from this figure that paromomycin, even at a concentration as low as 0.1 μM, inhibits the antiassociation activity of IF3. It is noted that the inhibitory effect of paromomycin was slightly higher when association was induced at 8.2 mM Mg2+. Since this is the Mg2+ concentration at which paromomycin inhibited the disassembly of posttermination ribosomal complexes by RRF and EF-G (13), this could be the step which is influenced by paromomycin in the disassembly reaction.
Paromomycin does not inhibit binding of IF3 to the 30S ribosomal subunit.
The inhibition of the antiassociation activity of IF3 by paromomycin could be due to its inhibitory effect on the binding of IF3 to 30S subunits. To explore this possibility, the isolated 30S subunits were incubated with IF3 in the presence or absence of paromomycin at 8.2 mM (Fig. 2) or 1 mM (data not shown) Mg2+. The mixture was subjected to SDGC, and fractions were collected and examined for the presence of IF3 on 30S subunits by Western blot analysis. The result showed that even a high concentration of paromomycin (100 μM) does not inhibit the binding of IF3 to 30S subunits.
Ribosomal subunits are associated by paromomycin.
Having ruled out the possibility that paromomycin may inhibit the binding of IF3 to 30S subunits, the inhibitory effect of paromomycin on the antiassociation activity of IF3 may be due to the possibility that paromomycin associates the ribosomal subunits. To explore this possibility, the experiment depicted in Fig. 3A was performed. In this experiment, vacant ribosomes were incubated with increasing concentrations of paromomycin in the presence of 1 mM Mg2+ and the mixture was subjected to SDGC. It is noted that increasing amounts of 70S ribosomes were observed in the presence of paromomycin in a dose-dependent manner. The activity of paromomycin to associate subunits was plotted against the amount of paromomycin added (Fig. 3B, open circles). Fifty percent of subunits were associated into 70S ribosomes in 1 mM Mg2+ in the presence of 20 μM paromomycin. Increasing the concentration of paromomycin beyond 20 μM did not increase the association of subunits, indicating that some subunits are not sensitive to the paromomycin association activity. It is also noted that a small population of ribosomes (about 10%) remained as subunits in 8.2 mM Mg2+ even in the presence of a high concentration of paromomycin (Fig. 3B).
The association activity of paromomycin can be observed with all Mg2+ concentrations tested, except for 8.2 mM Mg2+, where almost all of the ribosomes are already associated. At 4 mM Mg2+, 70S ribosomes were 40%, and addition of 0.1 mM paromomycin increased this value to close to 70% (Fig. 3B), while addition of 2 mM Mg2+ (total, 6 mM Mg2+) increased 70S ribosomes to only 55% (Fig. 3B, open square at 0 μM paromomycin). This indicates that the association capacity of paromomycin is far greater than that of Mg2+ ions.
Bound IF3 is displaced upon ribosomal association by paromomycin.
The lack of the inhibitory effect of paromomycin on the binding of IF3 to the 30S subunit (Fig. 2) makes one wonder about what happens to the ribosome-bound IF3 when the antiassociation activity is inhibited by paromomycin, resulting in 70S ribosomes. To answer this question, the experiment depicted in Fig. 4A was performed. In this experiment, IF3 was incubated with 30S and 50S subunits in the presence or absence of paromomycin at a low (1 mM) Mg2+ concentration. The Mg2+ concentration was then raised to 6 mM, and the sedimentation pattern of formed 70S ribosomes (Fig. 4A, left panel) and the presence of IF3 on the ribosomes (Fig. 4A, right panel) were examined. It is clear that the 70S ribosomes which formed in the presence of paromomycin did not retain IF3, which was originally present on the 30S subunits. It appears that the 30S-bound IF3 was displaced by 50S ribosomes that associated with the 30S subunits because of paromomycin. The ribosomal association effect of paromomycin appears to be strong enough to expel the 30S-bound IF3.
We have also asked whether or not IF3 is bound to 70S ribosomes when the dissociation of 70S ribosomes by RRF, EF-G, and IF3 (14) is inhibited by paromomycin (Fig. 4B). In this experiment, 70S ribosomes were incubated with RRF, EF-G, GTP, and IF3 in 8.2 mM Mg2+ in the presence or absence of paromomycin. The splitting of 70S ribosomes by these three factors was inhibited by paromomycin (Fig. 4B, left panel), and the remaining 70S ribosome did not contain IF3 (Fig. 4B, right panel).
DISCUSSION
The best-known function of paromomycin is its miscoding effect. Paromomycin binds to the bulged neck of helix 44 (H44) around nucleotides 1406 to 1408 and 1492 to 1495 of 16S rRNA (3, 7, 25), and the mechanism of miscoding has been proposed through crystallographic (27, 28) and biochemical (20, 29) studies. Upon binding of paromomycin, nucleotides A1492 and A1493 of 16S rRNA flip out in a fashion similar to their flipping out in the presence of cognate tRNA and mRNA (3, 27). Nucleotides A1492 and A1493 interact with the second and the first codon-anticodon base pairs in the A site, respectively. Furthermore, binding of paromomycin to H44 rearranges the conformation of the 30S subunit and rotates the head and the spur towards the shoulder of the 30S subunit (closed form), which can also be observed upon binding of cognate tRNA (28). Thus, it appears that the presence of paromomycin together with near-cognate aminoacyl tRNA mimics the major conformational changes of ribosomes caused by cognate aminoacyl tRNA binding. Paromomycin also causes an error at the translation resuming point of transfer mRNA (37).
In this paper, we show a hitherto-unknown activity of paromomycin to cause association of ribosomal subunits. There are two other well-known agents, Mg2+ ions and polyamines, that cause the association of subunits. The ratio of bound Mg2+/rRNA-phosphate is about 0.1 with 30S and 0.08 with 50S subunits (39). With wheat germ ribosomes, the ratio becomes 0.5 under the Mg2+ ion saturation condition (36). Polacek and Barta (31) used the metal ion-induced rRNA cleavage method to determine the binding sites of Mg2+ on 16S and 23S rRNA. There are at least 5 and 17 specific binding sites of Mg2+ in 16S and 23S rRNA, respectively. Yet, the binding of the divalent ions to H44 of 16S rRNA was not detected in their study. In contrast, two Mg2+ ions were observed around A1492 and A1493 of 16S rRNA in the crystal structure of the 30S subunit (27). The paromomycin binding site appears to overlap with these Mg2+ binding sites.
Kakegawa et al. (19) examined the possible ribosomal proteins that bind polyamines (spermidine and spermine) and identified 25 proteins. Binding sites of spermine on 16S (1) and 23S (40) rRNA have also been reported previously. Importantly, the spermine-binding site on H44 of 16S rRNA (nucleotide 1411) (1) appears to be situated close to the paromomycin binding site. An antibiotic having spermidine-like moiety, edeine, also binds to the neck of H44 (around nucleotide 1498 of 16S rRNA) (30).
The fact that binding sites of these three “association reagents,” Mg2+, polyamines, and paromomycin, are shared suggests that they share the similar mechanism which involves H44. The H44 forms four intersubunit bridge regions, B2a, B3, B5, and B6, with the 50S subunit (9, 33, 41) and plays a role crucial for association of subunits (22, 32). Furthermore, neomycin, which belongs to the same aminoglycoside class as paromomycin, has been shown to bind to the 50S subunit (23). On the basis of these data, it is possible that paromomycin binds to both subunits and stabilizes one or more of the intersubunit bridges. We should hasten to add that, due to the large difference between the molecular sizes of paromomycin and Mg2+, the actual mechanisms of these two agents in the interaction of subunits must be different. Indeed, the association activity of paromomycin is much stronger than that of Mg2+ (Fig. 3) and it is strong enough to expel IF3 (Fig. 4), which may mimic the bridge region B2b (5) or may be located on the solvent side of 30S subunits (30). It is also possible that a large conformational change (closed conformation) of the 30S subunits upon binding of paromomycin (28) may play an important role for the association induced by paromomycin. However, these structural changes were observed at an Mg2+ concentration high enough to cause association of subunits, suggesting that the paromomycin-dependent structural change may not have a direct role in the association. It would be interesting to perform a cryoelectron microscopy study to see if EF-G binding causes ratcheting in the presence of paromomycin. In addition, extensive mutagenesis of the parts of H44 that are involved in the intersubunit interactions may help to substantiate our proposals.
Taken together, we propose that aminoglycoside binding to the ribosome makes it difficult for subunits to move relative to each other (8, 38), resulting in inhibition of translocation (2, 10, 16, 24). On the other hand, alternative possibilities of the mechanism of paromomycin inhibition on the translocation not related to the current observation are valid. For example, the reason could simply be because the affinity of A-site tRNA may be increased by paromomycin to the extent that it cannot be moved. However, another aminoglycoside, streptomycin, which also causes miscoding and inhibits translocation (16), did not alter the affinity of cognate aminoacyl tRNA, while it increased the affinity of near-cognate aminoacyl tRNA (18). Further studies are required to elucidate the mechanism of the translocation inhibition by paromomycin.
Acknowledgments
We thank Takuya Ueda (University of Tokyo, Tokyo, Japan) and Paul March (University of New South Wales, Sydney, Australia) for their kind gifts of plasmids coding for factors.
Footnotes
Published ahead of print on 6 November 2006.
REFERENCES
- 1.Amarantos, I., I. K. Zarkadis, and D. L. Kalpaxis. 2002. The identification of spermine binding sites in 16S rRNA allows interpretation of the spermine effect on ribosomal 30S subunit functions. Nucleic Acids Res. 30:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabanas, M. J., D. Vazquez, and J. Modolell. 1978. Inhibition of ribosomal translocation by aminoglycoside antibiotics. Biochem. Biophys. Res. Commun. 83:991-997. [DOI] [PubMed] [Google Scholar]
- 3.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 4.Cousin, M. A., D. Lando, T. Ojasoo, and J. P. Raynaud. 1977. Stability of 70S ribosomes in relation to misreading and antibacterial activity of aminoglycosides. Biochimie 59:59-63. [DOI] [PubMed] [Google Scholar]
- 5.Dallas, A., and H. F. Noller. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell 8:855-864. [DOI] [PubMed] [Google Scholar]
- 6.Davies, J., W. Gilbert, and L. Gorini. 1964. Streptomycin, suppression, and the code. Proc. Natl. Acad. Sci. USA 51:883-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fourmy, D., M. I. Recht, S. C. Blanchard, and J. D. Puglisi. 1996. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science 274:1367-1371. [DOI] [PubMed] [Google Scholar]
- 8.Frank, J., and R. K. Agrawal. 2000. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406:318-322. [DOI] [PubMed] [Google Scholar]
- 9.Gabashvili, I. S., R. K. Agrawal, C. M. Spahn, R. A. Grassucci, D. I. Svergun, J. Frank, and P. Penczek. 2000. Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell 100:537-549. [DOI] [PubMed] [Google Scholar]
- 10.Hausner, T. P., U. Geigenmuller, and K. H. Nierhaus. 1988. The allosteric three-site model for the ribosomal elongation cycle. New insights into the inhibition mechanisms of aminoglycosides, thiostrepton, and viomycin. J. Biol. Chem. 263:13103-13111. [PubMed] [Google Scholar]
- 11.Hirashima, A., and A. Kaji. 1972. Purification and properties of ribosome-releasing factor. Biochemistry 11:4037-4044. [DOI] [PubMed] [Google Scholar]
- 12.Hirashima, A., and A. Kaji. 1973. Role of elongation factor G and a protein factor on the release of ribosomes from messenger ribonucleic acid. J. Biol. Chem. 248:7580-7587. [PubMed] [Google Scholar]
- 13.Hirokawa, G., M. C. Kiel, A. Muto, M. Selmer, V. S. Raj, A. Liljas, K. Igarashi, H. Kaji, and A. Kaji. 2002. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 21:2272-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirokawa, G., R. M. Nijman, V. S. Raj, H. Kaji, K. Igarashi, and A. Kaji. 2005. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA 11:1317-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou, Y., E. S. Yaskowiak, and P. E. March. 1994. Carboxyl-terminal amino acid residues in elongation factor G essential for ribosome association and translocation. J. Bacteriol. 176:7038-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igarashi, K., H. Ishitsuka, and A. Kaji. 1969. Comparative studies on the mechanism of action of lincomycin, streptomycin and erythromycin. Biochem. Biophys. Res. Commun. 37:499-504. [DOI] [PubMed] [Google Scholar]
- 17.Ishino, T., K. Atarashi, S. Uchiyama, T. Yamami, Y. Saihara, T. Yoshida, H. Hara, K. Yokose, Y. Kobayashi, and Y. Nakamura. 2000. Interaction of ribosome recycling factor and elongation factor EF-G with E. coli ribosomes studied by the surface plasmon resonance technique. Genes Cells 5:953-963. [DOI] [PubMed] [Google Scholar]
- 18.Kaji, H., and A. Kaji. 1965. Specific binding of sRNA to ribosomes: effect of streptomycin. Proc. Natl. Acad. Sci. USA 54:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kakegawa, T., E. Sato, S. Hirose, and K. Igarashi. 1986. Polyamine binding sites on Escherichia coli ribosomes. Arch. Biochem. Biophys. 251:413-420. [DOI] [PubMed] [Google Scholar]
- 20.Karimi, R., and M. Ehrenberg. 1994. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur. J. Biochem. 226:355-360. [DOI] [PubMed] [Google Scholar]
- 21.Kiel, M. C., V. S. Raj, H. Kaji, and A. Kaji. 2003. Release of ribosome-bound ribosome recycling factor by elongation factor G. J. Biol. Chem. 278:48041-48050. [DOI] [PubMed] [Google Scholar]
- 22.Liiv, A., and M. O'Connor. 2006. Mutations in the intersubunit bridge regions of 23S rRNA. J. Biol. Chem. 281:29850-29862. [DOI] [PubMed] [Google Scholar]
- 23.Misumi, M., T. Nishimura, T. Komai, and N. Tanaka. 1978. Interaction of kanamycin and related antibiotics with the large subunit of ribosomes and the inhibition of translocation. Biochem. Biophys. Res. Commun. 84:358-365. [DOI] [PubMed] [Google Scholar]
- 24.Misumi, M., and N. Tanaka. 1980. Mechanism of inhibition of translocation by kanamycin and viomycin: a comparative study with fusidic acid. Biochem. Biophys. Res. Commun. 92:647-654. [DOI] [PubMed] [Google Scholar]
- 25.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 26.Moreau, N., C. Jaxel, and F. Le Goffic. 1984. Comparison of fortimicins with other aminoglycosides and effects on bacterial ribosome and protein synthesis. Antimicrob. Agents Chemother. 26:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogle, J. M., D. E. Brodersen, W. M. Clemons, Jr., M. J. Tarry, A. P. Carter, and V. Ramakrishnan. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897-902. [DOI] [PubMed] [Google Scholar]
- 28.Ogle, J. M., F. V. Murphy, M. J. Tarry, and V. Ramakrishnan. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111:721-732. [DOI] [PubMed] [Google Scholar]
- 29.Pape, T., W. Wintermeyer, and M. Rodnina. 2000. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 7:104-107. [DOI] [PubMed] [Google Scholar]
- 30.Pioletti, M., F. Schlunzen, J. Harms, R. Zarivach, M. Gluhmann, H. Avila, A. Bashan, H. Bartels, T. Auerbach, C. Jacobi, T. Hartsch, A. Yonath, and F. Franceschi. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 20:1829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polacek, N., and A. Barta. 1998. Metal ion probing of rRNAs: evidence for evolutionarily conserved divalent cation binding pockets. RNA 4:1282-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pulk, A., U. Maivali, and J. Remme. 2006. Identification of nucleotides in E. coli 16S rRNA essential for ribosome subunit association. RNA 12:790-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuwirth, B. S., M. A. Borovinskaya, C. W. Hau, W. Zhang, A. Vila-Sanjurjo, J. M. Holton, and J. H. Cate. 2005. Structures of the bacterial ribosome at 3.5 A resolution. Science 310:827-834. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu, I., and A. Kaji. 1991. Identification of the promoter region of the ribosome-releasing factor cistron (frr). J. Bacteriol. 173:5181-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimizu, Y., A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa, and T. Ueda. 2001. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19:751-755. [DOI] [PubMed] [Google Scholar]
- 36.Sperrazza, J. M., and L. L. Spremulli. 1983. Quantitation of cation binding to wheat germ ribosomes: influences on subunit association equilibria and ribosome activity. Nucleic Acids Res. 11:2665-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi, T., T. Konno, A. Muto, and H. Himeno. 2003. Various effects of paromomycin on tmRNA-directed trans-translation. J. Biol. Chem. 278:27672-27680. [DOI] [PubMed] [Google Scholar]
- 38.Valle, M., A. Zavialov, J. Sengupta, U. Rawat, M. Ehrenberg, and J. Frank. 2003. Locking and unlocking of ribosomal motions. Cell 114:123-134. [DOI] [PubMed] [Google Scholar]
- 39.Weiss, R. L., B. W. Kimes, and D. R. Morris. 1973. Cations and ribosome structure. 3. Effects on the 30S and 50S subunits of replacing bound Mg2+ by inorganic cations. Biochemistry 12:450-456. [DOI] [PubMed] [Google Scholar]
- 40.Xaplanteri, M. A., A. D. Petropoulos, G. P. Dinos, and D. L. Kalpaxis. 2005. Localization of spermine binding sites in 23S rRNA by photoaffinity labeling: parsing the spermine contribution to ribosomal 50S subunit functions. Nucleic Acids Res. 33:2792-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]