Abstract
Inoculum size and incubation time were varied during broth microdilution testing of the susceptibilities of 35 strains of lactic acid bacteria to six antibiotics. An increase in either parameter resulted in elevated MICs for all species. An inoculum of 3 × 105 CFU/ml is recommended to assess the antibiotic susceptibilities of these bacteria by using broth microdilution.
Lactic acid bacteria (LAB) are commonly used as food-processing aids and probiotics. Due to their genetic flexibility and widespread occurrence in the food chain and in the intestinal tract, LAB can act as potential reservoirs of antibiotic resistance genes that may be transferred to other bacteria, including human pathogens (4, 21). Thus, the presence of antibiotic resistance genes should be assessed before LAB are used in food applications. For this reason, standardized and reliable testing procedures are needed to define rational microbiological breakpoints for distinguishing between strains with and without acquired resistance genes (22).
There is currently no standard method for antibiotic susceptibility testing of LAB, although several broth microdilution methods have been used (5-7, 9, 16, 19, 20). At present, the Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) recommends broth microdilution for bacteria that grow aerobically (3) and for anaerobic bacteria belonging to the Bacteroides fragilis group (18). The inoculum size and incubation time are important parameters to evaluate during the development of broth microdilution methods (22) and have been extensively studied for several bacterial species (1, 8, 13, 14, 17) but not for nonenterococcal LAB. Other factors that may affect the susceptibility results are the incubation temperature and the composition of the atmosphere and the growth medium (12, 22). The poor growth of many LAB on established antibiotic susceptibility testing media such as Mueller-Hinton and Iso-Sensitest media has led to the recent development of LAB susceptibility test medium (LSM) (15). The present study was performed to evaluate the effects of the inoculum size and incubation time on antibiotic MICs for LAB using broth microdilution and LSM.
Twenty-nine LAB reference strains were tested: 27 Lactobacillus species, encompassing different phylogenetic groups and fermentation pathways, and 1 strain each of Lactococcus lactis subsp. lactis and Streptococcus thermophilus, both species widely used by the food industry (Table 1). Six clinical Lactobacillus isolates, recovered from cerebrospinal fluid, blood, dental caries, breast milk, intestines, and feces, respectively, were also tested. Enterococcus faecalis LMG 8222 was included as a quality control. Strains were obtained from the BCCM/LMG Bacteria Collection, Ghent University, Ghent, Belgium (n = 32), the German Collection of Microorganisms and Cell Cultures (DSMZ), Braunschweig, Germany (n = 3), and the American Type Culture Collection, Middlesex, United Kingdom (n = 1). All strains were grown anaerobically (AnaeroGen; Oxoid). The incubation temperatures and growth media used for each species are shown in Table 1.
TABLE 1.
LAB strains and growth conditions used for antibiotic susceptibility testing
| LAB strain (origin of clinical isolates) | Fermentation pathwaya | Phylogenetic groupb | Growth medium | Growth temp (°C) |
|---|---|---|---|---|
| Lactobacillus acidophilus LMG 9433T | A | L. delbrueckii | LSMc | 37 |
| Lactobacillus amylovorus LMG 9496T | A | L. delbrueckii | LSM | 37 |
| Lactobacillus delbrueckii subsp. bulgaricus LMG 6901T | A | L. delbrueckii | LSM | 37 |
| Lactobacillus gasseri LMG 9203T | A | L. delbrueckii | LSM | 37 |
| L. gasseri LMG 13047 (human feces) | A | L. delbrueckii | LSM | 37 |
| Lactobacillus helveticus LMG 6413T | A | L. delbrueckii | LSM | 37 |
| Lactobacillus johnsonii LMG 9436T | A | L. delbrueckii | LSM | 37 |
| L. johnsonii LMG 18175 (human intestine) | A | L. delbrueckii | LSM | 37 |
| Lactobacillus farciminis LMG 9200T | A | L. plantarum | LSM | 37 |
| Lactobacillus salivarius subsp. salivarius DSM 20555T | A | L. salivarius | LSM | 37 |
| Lactobacillus casei LMG 6904T | B | L. casei | LSM | 37 |
| Lactobacillus rhamnosus LMG 6400T | B | L. casei | LSM | 37 |
| L. rhamnosus LMG 10768 (blood) | B | L. casei | LSM | 37 |
| Lactobacillus paracasei subsp. paracasei LMG 13087T | B | L. casei | LSM | 37 |
| L. paracasei subsp. paracasei LMG 10774 (cerebrospinal fluid) | B | L. casei | LSM | 37 |
| Lactobacillus alimentarius LMG 9187T | B | L. plantarum | LSM | 37 |
| Lactobacillus pentosus LMG 10755T | B | L. plantarum | LSM | 28 |
| Lactobacillus plantarum LMG 6907T | B | L. plantarum | LSM | 28 |
| L. plantarum LMG 9206 (dental caries) | B | L. plantarum | LSM | 28 |
| Lactobacillus curvatus LMG 9198T | B | L. sakei | LSM | 28 |
| Lactobacillus sakei subsp. sakei LMG 9468T | B | L. sakei | LSM | 28 |
| Lactobacillus agilis LMG 9186T | B | L. salivarius | LSM | 37 |
| Lactobacillus bifermentans LMG 9845T | B | LSM | 37 | |
| Lactobacillus buchneri DSM 20057T | C | L. buchneri | LSM | 37 |
| Lactobacillus fructivorans LMG 9201T | C | L. buchneri | LSM | 37 |
| Lactobacillus hilgardii LMG 6895T | C | L. buchneri | LSM | 37 |
| Lactobacillus collinoides LMG 9194T | C | L. plantarum | LSM | 28 |
| Lactobacillus fermentum LMG 6902T | C | L. reuteri | LSM | 37 |
| Lactobacillus panis LMG 21658T | C | L. reuteri | LSM | 37 |
| Lactobacillus reuteri DSM 20016T | C | L. reuteri | LSM | 37 |
| L. reuteri ATCC 55730 (human breast milk) | C | L. reuteri | LSM | 37 |
| Lactobacillus suebicus LMG 11408T | C | L. reuteri | LSM | 37 |
| Lactobacillus brevis LMG 6906T | C | LSM | 37 | |
| Lactococcus lactis subsp. lactis LMG 6890T | ISTd | 32 | ||
| Streptococcus thermophilus LMG 6896T | IST + 1% lactose | 42 | ||
| Enterococcus faecalis LMG 8222 | LSM | 37 |
As summarized by Hammes and Vogel (11). Group A, obligately homofermentative lactobacilli; group B, facultatively heterofermentative lactobacilli; group C, obligately heterofermentative lactobacilli.
As defined by Hammes and Hertel (10).
Iso-Sensitest broth (90%; Oxoid) plus MRS broth (10%; Oxoid) adjusted to pH 6.7 (15).
Iso-Sensitest broth (Oxoid).
Broth microdilution was performed using ACE-ART VetMIC panels (National Veterinary Institute, Uppsala, Sweden). The 96-well microtiter plates contained six antibiotics, air dried in cabinets at approximately 30°C, in serial twofold dilution steps to determine MICs in the following ranges: oxytetracycline, 0.5 to 128 μg/ml; clindamycin, 0.12 to 8 μg/ml; streptomycin, 2 to 256 μg/ml; erythromycin, 0.12 to 16 μg/ml; gentamicin, 0.5 to 32 μg/ml; ampicillin, 0.12 to 8 μg/ml. Colonies from overnight cultures (20 to 24 h) were suspended in the corresponding growth medium (Table 1) to obtain four final densities ranging from 3 × 104 to 3 × 107 CFU/ml for each strain. Bacterial density was measured spectrophotometrically at 600 nm and verified by viable cell counts. Each well was filled with 100 μl of inoculum. The panels were covered with plastic lids and incubated anaerobically for 24 and 48 h. The MIC was defined, according to the manufacturer's recommendations, as the lowest antibiotic concentration for which there was no visible bacterial growth, i.e., the first well without a pellet.
The MICs of the quality control strain determined after 24 h at 3 × 105 CFU/ml were within the range reported by Klare et al. (15) for all antibiotics tested except erythromycin, which displayed a twofold increase in the MIC (data not shown). Interassay reproducibility was evaluated by assessing the susceptibilities of five reference strains (Lactobacillus acidophilus LMG 9433T, L. amylovorus LMG 9496T, L. plantarum LMG 6907T, L. rhamnosus LMG 6400T, and Lactobacillus sakei subsp. sakei LMG 9468T) to six antibiotics on five separate occasions. MICs determined at 3 × 105 CFU/ml after both 24 and 48 h incubation were within the accuracy limit of MIC standard tests (plus or minus one twofold dilution step) (2) for all strain-antibiotic combinations tested (data not shown).
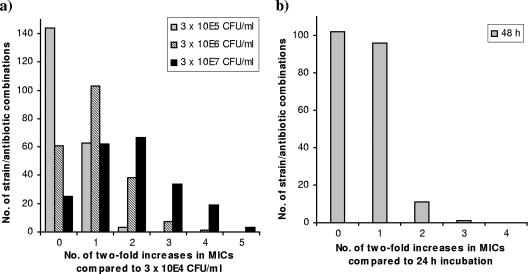
The MICs for all strains increased with inoculum size. Increasing the density from 3 × 104 to 3 × 105 CFU/ml resulted in identical MICs after 48 h of incubation for 144 (69%) of the 210 strain-antibiotic combinations, a twofold increase in MICs for 63 (30%) of the combinations, and a fourfold increase for the remaining 3 (1%) combinations (Fig. 1a). A similar stepwise increase in MICs was observed when the inoculum was further increased to 3 × 106 and 3 × 107 CFU/ml, respectively. The shift in the MIC due to increased inoculum size was independent of incubation time (data not shown).
FIG. 1.
Effects of increased inoculum size (a) and prolonged incubation time (b) on the MICs of six antibiotics for 35 LAB strains. (a) MICs obtained for the 210 strain-antibiotic combinations with different inoculum sizes, compared to MICs obtained with an inoculum size of 3 × 104 CFU/ml, after 48 h of incubation; (b) MICs obtained with an inoculum size of 3 × 105 CFU/ml after 48 h compared to 24 h of incubation. MICs above the test range are given as the concentration closest to the range, and MICs below the range are given as the lowest concentration.
All strains displayed similar increases in MICs when the incubation time was extended to 48 h. With an inoculum of 3 × 105 CFU/ml, 102 (49%) of the 210 strain-antibiotic combinations were unaffected by prolonged incubation, whereas MICs increased twofold for 96 (46%) of the combinations (Fig. 1b). For the remaining combinations (6%), fourfold MIC increases between 24 and 48 h of incubation were seen mainly with oxytetracycline and clindamycin, but these increases were all in the lower part of the MIC range (maximum, 2 to 8 μg/ml). The MIC increase with time was independent of inoculum size (data not shown). For the clinical isolates, the elevation in MICs due to increased inoculum size or incubation time was in accordance with the results for the LAB reference strains (data not shown).
The use of the two highest inoculum densities (3 × 106 and 3 × 107 CFU/ml) was generally associated with trailing growth, which prevented clear-cut end point readings. In addition, an inoculum of 3 × 104 CFU/ml resulted in poor growth for L. casei, L. collinoides, L. brevis, L. hilgardii, and L. buchneri after 24 h of incubation, making it difficult to determine MICs accurately. Thus, an inoculum size of 3 × 105 CFU/ml is recommended for broth microdilution susceptibility testing of LAB using LSM. This is in approximate agreement with the standardized inoculum size for aerobic bacteria (3) but is about three times lower than the density recommended for anaerobic bacteria (18). Sufficient growth for MIC determination was obtained at 3 × 105 CFU/ml after both 24 and 48 h of incubation for all strains tested. However, end points were more easily read after 48 h of incubation. In addition, LAB groups that need extended incubation for sufficient growth on LSM, such as most Bifidobacterium species (15), can be tested simultaneously when 48 h is used as the standard incubation time.
In conclusion, an increased inoculum size and an extended incubation time resulted in elevated antibiotic MICs for all LAB species, underlining the importance of controlled and standardized conditions for susceptibility testing of LAB. Hopefully, the results from this study will contribute to the development of a standard method for determining the antibiotic susceptibilities of LAB and to subsequent delineation of microbiological breakpoints for individual LAB species.
Acknowledgments
This work was supported by a grant from the European Commission, Sixth Framework Program (CT-2003-506214, “ACE-ART”). G.H. is a postdoctoral fellow of the Fund for Scientific Research, Flanders, Belgium (F.W.O.—Vlaanderen).
Footnotes
Published ahead of print on 23 October 2006.
REFERENCES
- 1.Boerner, J., K. Failing, and M. M. Wittenbrink. 1995. In vitro antimicrobial susceptibility testing of Borrelia burgdorferi: influence of test conditions on minimal inhibitory concentration (MIC) values. Zentbl. Bakteriol. 283:49-60. [DOI] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. Publication M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 3.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 4.Curragh, H. J., and M. A. Collins. 1992. High levels of spontaneous drug resistance in Lactobacillus. J. Appl. Bacteriol. 73:31-36. [Google Scholar]
- 5.Delgado, S., A. B. Florez, and B. Mayo. 2005. Antibiotic susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr. Microbiol. 50:202-207. [DOI] [PubMed] [Google Scholar]
- 6.Elliott, J. A., and R. R. Facklam. 1996. Antimicrobial susceptibilities of Lactococcus lactis and Lactococcus garvieae and a proposed method to discriminate between them. J. Clin. Microbiol. 34:1296-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Florez, A. B., S. Delgado, and B. Mayo. 2005. Antimicrobial susceptibility of lactic acid bacteria isolated from a cheese environment. Can. J. Microbiol. 51:51-58. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs, P. C., A. L. Barry, and S. D. Brown. 2001. Influence of variations in test methods on susceptibility of Haemophilus influenzae to ampicillin, azithromycin, clarithromycin, and telithromycin. J. Clin. Microbiol. 39:43-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, M., R. M. Wadowsky, and K. Barbadora. 1990. Recovery of vancomycin-resistant gram-positive cocci from children. J. Clin. Microbiol. 28:484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammes, W. P., and C. Hertel. 2003. The genera Lactobacillus and Carnobacterium. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, release 3.15, 3rd ed. Springer-Verlag, New York, NY. http://link.springer-ny.com/link/service/books/10125/. Accessed 25 October 2005.
- 11.Hammes, W. P., and R. F. Vogel. 1995. The genus Lactobacillus, p. 19-54. In B. J. B. Wood and W. H. Holzapfel (ed.), The genera of lactic acid bacteria, vol. 2. Blackie Academic & Professional, London, United Kingdom. [Google Scholar]
- 12.Huys, G., K. D'Haene, and J. Swings. 2002. Influence of the culture medium on antibiotic susceptibility testing of food-associated lactic acid bacteria with the agar overlay disc diffusion method. Lett. Appl. Microbiol. 34:402-406. [DOI] [PubMed] [Google Scholar]
- 13.Karlsson, M., C. Fellstrom, A. Gunnarsson, A. Landen, and A. Franklin. 2003. Antimicrobial susceptibility testing of porcine Brachyspira (Serpulina) species isolates. J. Clin. Microbiol. 41:2596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenny, G. E., and F. D. Cartwright. 1993. Effect of pH, inoculum size, and incubation time on the susceptibility of Ureaplasma urealyticum to erythromycin in vitro. Clin. Infect. Dis. 17(Suppl. 1):215-218. [DOI] [PubMed] [Google Scholar]
- 15.Klare, I., C. Konstabel, S. Müller-Bertling, R. Reissbrodt, G. Huys, M. Vancanneyt, J. Swings, H. Goossens, and W. Witte. 2005. Evaluation of new broth media for microdilution antibiotic susceptibility testing of lactobacilli, lactococci, pediococci, and bifidobacteria. Appl. Environ. Microbiol. 71:8982-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein, G., C. Hallmann, I. A. Casas, J. Abad, J. Louwers, and G. Reuter. 2000. Exclusion of vanA, vanB and vanC type glycopeptide resistance in strains of Lactobacillus reuteri and Lactobacillus rhamnosus used as probiotics by polymerase chain reaction and hybridization methods. J. Appl. Microbiol. 89:815-824. [DOI] [PubMed] [Google Scholar]
- 17.Liebers, D. M., A. L. Baltch, R. P. Smith, M. C. Hammer, and J. V. Conroy. 1989. Susceptibility of Legionella pneumophila to eight antimicrobial agents including four macrolides under different assay conditions. J. Antimicrob. Chemother. 23:37-41. [DOI] [PubMed] [Google Scholar]
- 18.NCCLS. 2004. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard M11-A6. NCCLS, Wayne, Pa.
- 19.Sidhu, M. S., S. Langsrud, and A. Holck. 2001. Disinfectant and antibiotic resistance of lactic acid bacteria isolated from the food industry. Microb. Drug Resist. 7:73-83. [DOI] [PubMed] [Google Scholar]
- 20.Swenson, J. M., R. R. Facklam, and C. Thornsberry. 1990. Antimicrobial susceptibility of vancomycin-resistant Leuconostoc, Pediococcus, and Lactobacillus species. Antimicrob. Agents Chemother. 34:543-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teuber, M., L. Meile, and F. Schwarz. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Leeuwenhoek 76:115-137. [PubMed] [Google Scholar]
- 22.White, D. G., J. Acar, F. Anthony, A. Franklin, R. Gupta, T. Nicholls, Y. Tamura, S. Thompson, E. J. Threlfall, D. Vose, M. van Vuuren, H. C. Wegener, M. L. Costarrica, et al. 2001. Antimicrobial resistance: standardisation and harmonisation of laboratory methodologies for the detection and quantification of antimicrobial resistance. Rev. Sci. Tech. Off. Int. Epizoot. 20:849-858. [DOI] [PubMed] [Google Scholar]