Abstract
β-Lactams are the drugs of choice for the treatment of infections caused by the important bacterial pathogen Streptococcus pneumoniae. The recent growth of resistance of this organism to penicillin observed worldwide is of the highest concern. In this study, using 887 surveillance pneumococcal isolates recovered in Poland from 1998 to 2002, we observed the increase in penicillin nonsusceptibility from 8.7% to 20.3%. All of the 109 penicillin-nonsusceptible S. pneumoniae (PNSP) isolates identified, together with 22 archival PNSP isolates from 1995 to 1997, were subsequently analyzed by susceptibility testing, serotyping, profiling of pbp genes, pulsed-field gel electrophoresis, and multilocus sequence typing (MLST). Four predominant serotypes, serotypes 6B, 9V, 14, and 23F, characterized 85.5% of the isolates. MLST revealed the presence of 34 sequence types, 15 of which were novel types. Representatives of seven multiresistant international clones (Spain23F-1, Spain6B-2, Spain9V-3, Taiwan23F-15, Poland23F-16, Poland6B-20, and Sweden15A-25) or their closely related variants comprised the majority of the study isolates. The spread of Spain9V-3 and its related clone of serotype 14/ST143 has remarkably contributed to the recent increase in penicillin resistance in pneumococci in the country.
Streptococcus pneumoniae (the pneumococcus) represents one of the leading bacterial infectious agents of respiratory tract infections (RTI) in humans, including acute otitis media, sinusitis, acute exacerbations of chronic bronchitis, and community-acquired pneumonia (5), and it is the main cause of mortality of patients affected by the latter disease (19). This pathogen is also frequently involved in life-threatening infections of the central nervous system; e.g., in Poland, it is responsible for 20.9% of the reported cases of bacterial meningitis (42). Recently, the situation has been aggravated worldwide by the appearance and spread of pneumococcal strains that have acquired resistance to several classes of antimicrobials, including β-lactams (2).
Resistance to penicillin and other β-lactam antibiotics in pneumococci is associated with modifications of genes encoding penicillin-binding proteins, enzymes involved in peptidoglycan synthesis that are molecular targets for β-lactams (4). In clinical isolates, resistance arises from the acquisition of foreign DNA from either viridans streptococci (11, 24) or other penicillin-nonsusceptible S. pneumoniae (PNSP) (7, 21) and, thus, from the formation of the so-called mosaic pbp genes. The acquisition of these resistance determinants by some clones further drives their spread under the selective pressure of extensive antibiotic consumption. Recently, 22 internationally disseminated pneumococcal clones that combine nonsusceptibility to penicillin with resistance to other classes of antimicrobials (multidrug resistance) have been recognized (28) (see www.sph.emory.edu/pmen). Two such clones were originally identified in Poland in the mid-1990s and were designated Poland23F-16 and Poland6B-20 (37).
The epidemiological situation concerning the penicillin nonsusceptibility of S. pneumoniae isolates in Poland has remained largely unstudied up to now, particularly at the molecular level. The aim of our study was therefore to characterize the population of PNSP strains isolated in the country from various infections with the use of molecular typing methods, such as pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST), as well as to investigate resistance determinants by PCR-restriction fragment length polymorphism (PCR-RFLP) of pbp genes and particularly clones in the context of β-lactam susceptibility phenotypes. (Parts of this work were presented at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, 14 to 17 September 2003.)
MATERIALS AND METHODS
Bacterial isolates, susceptibility testing, DNA isolation, and serotyping.
Eight hundred eighty-seven pneumococcal isolates were collected from 1998 to 2002 in 63 medical centers in 53 towns all over Poland as a part of a continuous surveillance system. These isolates were derived from 260 (29.3%) female and 496 (55.9%) male patients; otherwise, the gender was not reported. Twenty-four (2.7%) isolates were obtained from patients under 2 years of age. Additionally, 22 PNSP isolates obtained from 13 centers from 1995 to 1997 were included in the analysis. Altogether, the isolates were derived from the following clinical specimens: sputum (553 isolates), bronchoalveolar lavage (141), cerebrospinal fluid (126), and blood (54), with the remainder being obtained from other sources (pleural fluid, eye swab, sinus, and pus from middle ear). Isolates were reidentified on the basis of colony morphology, susceptibility to optochin (bioMérieux, Marcy l'Etoile, France), and bile solubility. Susceptibility to penicillin (Sigma Chemical Company, St. Louis, MO) was tested by the microdilution method and interpreted according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) (formerly NCCLS) with breakpoints of 0.12 μg/ml and 2 μg/ml for nonsusceptible and resistant phenotypes, respectively (30). Isolates with MICs within the range of 0.12 μg/ml to 1 μg/ml were considered intermediate to penicillin. All PNSP isolates identified were also tested against amoxicillin (SmithKline Beecham Pharmaceuticals, Worthing, United Kingdom), ceftriaxone (Sigma), cefepime (Bristol-Myers Squibb, New Brunswick, NJ), and meropenem (AstraZeneca, Macclesfield, United Kingdom) using CLSI guidelines. Total bacterial DNA was purified using the Genomic DNA Prep Plus kit (A&A Biotechnology, Gdynia, Poland). Serotyping was performed by PCR with primers specific for genes responsible for the biosynthesis of the 14, 19F, and 23F types of capsular polysaccharide (25). Isolates that were negative in those PCRs, all the isolates with a suspected serotype switch, and randomly selected representatives of the pneumococci serotyped by PCR were subjected to conventional immunological serotyping by capsule swelling reactions at the Statens Serum Institut, Copenhagen, Denmark.
The collection of the study isolates from 1998 to 2002 was used for two other analyses. The majority of the RTI isolates were screened for nonsusceptibility to ciprofloxacin, and susceptibilities and serotypes of five isolates that were both ciprofloxacin and penicillin nonsusceptible were reported previously (38). The isolates from cerebrospinal fluid and blood of patients with meningeal symptoms were subjected to a separate study in which 18 PNSP isolates were identified. Susceptibility profiles and serotypes of the meningitis-related isolates were described elsewhere previously (39).
PFGE, MLST, and PCR-RFLP of pbp genes.
DNA purification, its restriction digestion with the SmaI enzyme (Fermentas, Vilnius, Lithuania), and PFGE of the resulting DNA fragments were performed as described previously (26). Isolates sharing the same PFGE pattern were considered to be of the same PFGE type and subtype, whereas isolates with one to three band differences were classified into distinct subtypes of the same type. Isolates differing by more than three bands were reported as being separate types. The MLST analysis was performed according to a standard procedure involving the amplification and sequencing of seven housekeeping genes, as proposed previously by Enright and Spratt (12). The Internet-accessible database (www.mlst.net) was used for assigning allele numbers and, on the basis of the resulting allelic profiles, the sequence types (STs) of isolates. eBURST analysis (14) software (available at www.mlst.net) was used to estimate the relationships among the isolates and to construct a population snapshot, applying the definition according to which members of a clonal complex share six out of seven MLST loci (27, 41), i.e., which ones constitute single-locus variants (SLVs). The fingerprints of pbp1a, pbp2b, and pbp2x genes were determined by PCR-RFLP with gene-specific primers (7, 10, 29), followed by HinfI restriction enzyme (Fermentas) digestion and electrophoresis in 2% agarose gels (SeaKem; BMA, Rockland, ME). The resulting RFLP patterns were compared to their counterparts obtained for wild-type S. pneumoniae strain R6 (ATCC 27336). Each unique pattern was assigned a single Arabic number, and particular combinations of these patterns were assigned to the isolates in the following order: pbp1a-pbp2x-pbp2b.
In addition, the pneumococcal ATCC strains ATCC 700669 (Spain23F-1), ATCC 700670 (Spain6B-2), ATCC 700671 (Spain9V-3), ATCC 700906 (Taiwan23F-15), ATCC BAA-343 (Poland23F-16), ATCC BAA-612 (Poland6B-20), and ATCC BAA-661 (Sweden15A-25) were used for reference purposes in the PFGE analysis and PCR-RFLP of pbp genes. When a PCR-RFLP pattern of any of these strains differed from those found in the study group, it was designated “other” without additional numbering.
PFGE and MLST analyses of five ciprofloxacin- and penicillin-nonsusceptible isolates from RTI and of 18 penicillin-nonsusceptible isolates from patients with meningitis were reported previously (38, 39); however, PFGE was repeated and reinterpreted in this work because of the different context of the overall collection of the isolates.
Statistical analyses.
The diversity index was calculated as described previously by Grundmann et al. (18). The chi-square test with 95% confidence intervals was applied to assess the differences in nonsusceptibility frequencies in time and age groups; the two-tailed Spearman coefficient (r) was used in the analysis of correlation between antibiotic consumption and PNSP level, allowing a 1-year lag period between the consumption data and the PNSP prevalence.
RESULTS
Susceptibility to penicillin and other β-lactams.
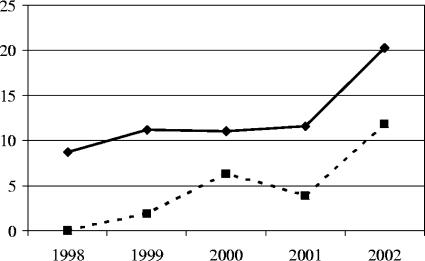
Altogether, 131 PNSP isolates from 39 centers in 32 towns were analyzed in this study, including 22 isolates from 1995 to 1997 and 109 isolates from 1998 to 2002 (12.3% of the isolates from this period). As shown in Fig. 1, the frequency of PNSP remained relatively stable over the period from 1998 to 2001 (8.1 to 11.7% of isolates) but then significantly increased in 2002 (20.3%; P = 0.03). The majority of PNSP isolates were derived from sputum samples (63 isolates; 48.1%), bronchoalveolar lavages (23 isolates; 17.6%), and cerebrospinal fluid (17 isolates; 13.0%). Fifteen isolates (11.5%) were obtained from children below 2 years of age; penicillin nonsusceptibility was therefore significantly associated with this age group (P < 0.001; chi-square test). The gender distribution did not differ from that for the entire collection; i.e., 32.1% and 55.7% of isolates were from female and male patients, respectively. The penicillin MIC50 and MIC90 values, calculated for all the isolates from the period from 1998 to 2002, were ≤0.015 μg/ml and 0.5 μg/ml, respectively. The majority of PNSP isolates (89 isolates; 67.9%) were resistant to penicillin (MIC range, 2 to 8 μg/ml); all these isolates, together with four isolates with intermediate penicillin resistance, were also resistant to cefuroxime. Resistance to amoxicillin, ceftriaxone, and cefepime was rare and occurred in 10, 8, and 2 isolates, respectively. As mentioned above, susceptibilities of ciprofloxacin-nonsusceptible isolates and meningitis-related isolates were described previously (38, 39).
FIG. 1.
Increase in frequency of PNSP in Poland from 1998 to 2002 (solid lines) and proportion of isolates belonging to the Spain9V-3 clone, ST413, and its SLVs among all the isolates in corresponding years (dashed lines).
Serotypes, PFGE types, and STs of PNSP.
Four serotypes, serotypes 6B, 9V, 14, and 23F, characterized the vast majority (85.5%) of all PNSP isolates (30, 20, 23, and 39 isolates, respectively). The other observed serotypes were serotypes 19A, 19F, 6A, 11A, 15A, and 24F. Four isolates were nontypeable (rough). The general coverages of the 7-valent and 23-valent vaccines were 87.8% and 88.6%, respectively, and in the group of isolates from children below 2 years of age, the 7-valent vaccine coverage was 93.3%.
PFGE analysis revealed the presence of 80 different DNA banding patterns, which were grouped into 27 types (Tables 1 and 2); 7 of these types were split further into 60 subtypes. The four predominant PFGE types (at least five isolates each) (9, 35), types 1, 4, 11, and 12, comprised 99 isolates (75.6%).
TABLE 1.
Clinical characteristics, serotypes, PFGE profiles, and STs of international PNSP clones
| Pneumococcal clone | STa | Allelic profilea,b | Serotype | PFGE type(s) | No. of isolates | Site(s) of isolationc,d | Yr of isolation | pbp profilee | MIC (μg/ml)f
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEN | AMX | CXM | CRO | FEP | MEM | |||||||||
| Spain23F-1 | 81 | 4-4-2-4-4-1-1 | 23F | 1a | 1-1-1 | |||||||||
| 81 | 4-4-2-4-4-1-1 | 23F | 1a, f, g, h, j, k, l, m, n, o, p, 2 | 20 | Sputum, BAL, CSF, blood | 1997-2002 | 1-1-1 | 2 | 1 | 4 | 1 | 1 | 0.5 | |
| 81 | 4-4-2-4-4-1-1 | 19A | 1d | 1 | BAL | 1997 | 1-1-1 | 2 | 2 | 8 | 2 | 1 | 1 | |
| 81 | 4-4-2-4-4-1-1 | 19F | 1b, c | 2 | Sputum | 1995, 2002 | 1-1-1 | 2 | 2 | 4-8 | 1 | 1 | 0.5 | |
| 1051 | 4-4-2-4-53-1-1 | 23F | 1i | 1 | Sputum | 1999 | 1-1-1 | 2 | 1 | 4 | 1 | 1 | 0.5 | |
| 1476 | 4-4-2-4-104-1-1 | 23F | 1f | 1 | Sputum | 1998 | 1-1-1 | 2 | 2 | 8 | 1 | 1 | 0.5 | |
| Spain6B-2 | 90 | 5-5-1-2-6-3-4 | 6B | 3a | 1-2-other | |||||||||
| 90 | 5-5-1-2-6-3-4 | 6B | 3a, b | 3 | CSF, sputum | 1999, 2001 | 1-2-2 | 2 | 1 | 4-8 | 1 | 1 | 0.5 | |
| Spain9V-3 | 156 | 7-11-10-1-6-8-1 | 9V | 4a | 1-1-1 | |||||||||
| 156 | 7-11-10-1-6-8-1 | 9V | 4a, b, e, f, g, h, i | 18 | Sputum, BAL, CSF, sinus | 1995, 1999, 2000-2002 | 1-1-1 | 2 | 0.5-2 | 2-8 | 0.5-1 | 0.5-1 | 0.25-0.5 | |
| 156 | 7-11-10-1-6-8-1 | 9V | 4h | 1 | CSF | 2002 | 2-1-1 | 1 | 1 | 4 | 0.5 | 0.5 | 0.5 | |
| 156 | 7-11-10-1-6-8-1 | 14 | 4b, c, d, e | 4 | Sputum, CSF | 2000, 2002 | 1-1-1 | 2 | 1 | 4-16 | 1-2 | 1 | 0.5 | |
| 156 | 7-11-10-1-6-8-1 | Rough | 5 | 1 | Sputum | 2002 | 1-1-1 | 2 | 1 | 8 | 1 | 1 | 0.25 | |
| 557 | 7-11-10-1-6-58-1 | 9V | 4e | 1 | CSF | 2000 | 1-1-1 | 2 | 1 | 2 | 0.5 | 0.5 | 0.5 | |
| Taiwan23F-15 | 242 | 15-29-4-21-30-1-14 | 23F | 7 | 3-1-3 | |||||||||
| 242 | 15-29-4-21-30-1-14 | 23F | 7 | 1 | Sputum | 2000 | 3-1-3 | 4 | 2 | 8 | 2 | 1 | 0.5 | |
| 242 | 15-29-4-21-30-1-14 | 23F | 8 | 1 | Blood | 2002 | 3-3-3 | 4 | 4 | 8 | 2 | 1 | 1 | |
| Poland23F-16 | 173 | 7-13-8-1-10-6-36 | 23F | 9 | 4-1-4 | |||||||||
| 173 | 7-13-8-1-10-6-36 | 23F | 9 | 1 | Blood | 1998 | 4-1-4 | 4 | 2 | 16 | 2 | 1 | 0.5 | |
| 173 | 7-13-8-1-10-6-36 | 23F | 10c | 1 | CSF | 1997 | 4-4-4 | 8 | 2 | 4 | 0.5 | 2 | 0.5 | |
| 173 | 7-13-8-1-10-6-36 | 23F | 10d | 1 | BAL | 1997 | 4-5-5 | 8 | 16 | >32 | 4 | 8 | 0.5 | |
| 272 | 7-13-8-1-10-6-37 | 23F | 11a | 3 | BAL, sputum | 2000 | 4-6-5 | 8 | 8-16 | ≥32 | 2-4 | 1-2 | 0.5 | |
| 272 | 7-13-8-1-10-6-37 | 23F | 11e, f | 3 | BAL, eye, sputum | 1998, 2000 | 4-7-5 | 8 | 8 | 4-16 | 1-2 | 1 | 0.25 | |
| 272 | 7-13-8-1-10-6-37 | 23F | 11b, c, d | 3 | CSF, blood | 2002 | 4-8-5 | 4-8 | 8 | 32 | 2-4 | 1 | 0.5 | |
| 1482 | 7-13-8-1-100-6-36 | 23F | 10a | 1 | Eye | 1997 | 5-4-4 | 0.5 | 0.25 | 1 | 0.25 | 0.25 | 0.06 | |
| 1506 | 7-13-8-1-10-6-162 | 23F | 10b | 1 | BAL | 1997 | 4-4-6 | 4 | 4 | 2 | 0.25 | 1 | 0.25 | |
| Poland6B-20 | 315 | 20-28-1-1-15-14-14 | 6B | 12a | WT-9-WT | |||||||||
| 315 | 20-28-1-1-15-14-14 | 6B | 12a, d, g | 14 | Sputum, ear, blood, BAL, CSF | 1995-1996, 1998, 2001-2002 | WT-9-WT | 0.12 | 0.06-0.12 | 0.12-0.25 | 0.06 | 0.06-0.12 | 0.06 | |
| 315 | 20-28-1-1-15-14-14 | 6B | 13 | 1 | Sputum | 1998 | 6-10-WT | 2 | 1 | 2 | 0.25 | 1 | 0.25 | |
| 315 | 20-28-1-1-15-14-14 | 6B | 12a | 2 | Sputum, sinus | 1999, 2001 | 7-11-WT | 0.5 | 0.25 | 0.25 | 0.25 | 0.25 | 0.12 | |
| 315 | 20-28-1-1-15-14-14 | 6B | 12c, e | 2 | Ear, sputum | 1995 | NT-7-3 | 2 | 1-2 | 2 | 1 | 1 | 0.25 | |
| 316 | 1-28-1-1-15-14-14 | 6B | 12b | 1 | Blood | 1998 | WT-9-WT | 0.12 | 0.12 | 0.12 | 0.06 | 0.06 | 0.06 | |
| 606 | 16-28-1-1-15-14-14 | 6B | 12d | 1 | CSF | 1998 | WT-9-WT | 0.12 | 0.12 | 0.06 | 0.06 | 0.06 | ||
| 1032 | 20-28-1-1-6-14-14 | 6B | 12h, i | 2 | Sputum | 2001 | WT-9-WT | 0.12 | 0.12 | 0.12 | 0.06 | 0.06 | 0.06 | |
| 1052 | 20-28-1-8-15-14-14 | 6B | 12f | 1 | Pleural fluid | 1996 | WT-9-WT | 0.12 | 0.12 | 0.06 | 0.06 | 0.06 | ||
| 1505 | 20-28-1-1-105-14-14 | 6B | 12b | 1 | Sputum | 2002 | WT-9-WT | 0.12 | 0.12 | 0.12 | 0.06 | 0.06 | 0.06 | |
| Sweden15A-25 | 63 | 2-5-36-12-17-21-14 | 15A | 14 | Other-12-4 | |||||||||
| 63 | 2-5-36-12-17-21-14 | 15A | 14 | 1 | Sputum | 1999 | 8-12-4 | 0.25 | ≤0.03 | 0.12 | 0.06 | 0.06 | ≤0.03 | |
New STs and alleles are underlined.
In the order aroE-gdh-gki-recP-spi-xpt-ddl.
BAL, bronchoalveolar lavage; CSF, cerebrospinal fluid.
Susceptibilities, serotypes, and STs of meningitis-related isolates were described in reference 39, and ciprofloxacin-nonsusceptible isolates (single isolates of ST81, ST276, and ST1477 and two isolates of ST156) were described in reference 38.
In the order pbp1a-pbp2x-pbp2b. NT, nontypeable; other, pattern unique to an ATCC clone.
PEN, penicillin; AMX, amoxicillin; CXM, cefuroxime; CRO, ceftriaxone; FEP, cefepime; MEM, meropenem.
TABLE 2.
Clinical characteristics, serotypes, PFGE profiles, and STs of other PNSP isolates
| STa | Allelic profilea,b | Serotype | Other countries of isolation | PFGE type | No. of isolates | Site(s) of isolationc,d | Yr of isolation | pbp profilee | MIC (μg/ml)f
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEN | AMX | CXM | CRO | FEP | MEM | |||||||||
| 490 | 2-13-9-1-6-19-14 | 6A | Bulgaria, Finland, Greece, Greenland, Sweden | 15 | 1 | Sputum | 2002 | WT-13-7 | 0.12 | 0.12 | 0.25 | 0.12 | 0.25 | 0.12 |
| 490 | 2-13-9-1-6-19-14 | 6B | 16 | 1 | BAL | 2002 | 5-1-3 | 1 | 0.5 | 2 | 0.5 | 0.5 | 0.25 | |
| 1049 | 2-25-9-1-6-19-14 | 6A | 15 | 1 | BAL | 2001 | WT-13-8 | 0.12 | 0.06 | 0.25 | 0.25 | 0.25 | 0.12 | |
| 1019 | 7-8-1-10-15-14-14 | 6A | 17 | 1 | Eye | 1997 | 5-14-WT | 0.5 | 0.5 | 2 | 0.12 | 0.5 | 0.06 | |
| 135 | 7-5-4-12-6-20-46 | 6B | Germany, Spain | 18, 19 | 2 | Eye, blood | 2002 | 5-15-WT | 0.25 | 0.12 | 0.25 | 0.25 | 0.25 | 0.06 |
| 1010 | 1-5-29-10-6-79-18 | 11A | 20 | 1 | Sputum | 1999 | NT-16-WT | 0.25 | 0.12 | 0.5 | 0.25 | 0.25 | 0.06 | |
| 143 | 7-5-10-18-6-8-1 | 14 | Hungary, Portugal | 4f, j, k, m, n, o, p, r, s, t, 6 | 14 | BAL, blood, CSF, pleural fluid, sputum | 1995-1996, 1999-2002 | 1-1-1 | 2 | 1-2 | 4-16 | 1-2 | 1-2 | 0.5 |
| 143 | 7-5-10-18-6-8-1 | 14 | 4l | 1 | BAL, sputum | 2001 | 1-6-1 | 2 | 1 | 8 | 1 | 1 | 0.5 | |
| 143 | 7-5-10-18-6-8-1 | 14 | 4f | 1 | Blood | 2002 | 1-11-1 | 2 | 0.25 | 4 | 1 | 0.5 | 0.5 | |
| 143 | 7-5-10-18-6-8-1 | 14 | 4j | 1 | CSF | 2002 | 1-17-1 | 2 | 2 | 16 | 2 | 1 | 0.5 | |
| 790 | 7-5-10-18-6-58-1 | 14 | Portugal | 4k | 1 | CSF | 1999 | 1-1-1 | 2 | 1 | 8 | 1 | 1 | 0.5 |
| 1477 | 7-5-10-18-6-145-1 | 14 | 4l | 1 | BAL | 2002 | 1-1-1 | 2 | 1 | 4 | 1 | 1 | 0.5 | |
| 1625 | 1-19-2-17-1-28-168 | 19A | 21 | 2 | BAL, sputum | 2001 | 8-WT-WT | 0.5 | 0.25 | 0.12 | 0.12 | 0.12 | 0.12 | |
| 1027 | 10-41-47-1-6-14-2 | 19F | 23 | 1 | Sputum | 1999 | 5-18-9 | 0.5 | 0.03 | 0.12 | 0.03 | 0.12 | 0.06 | |
| 277 | 7-13-8-6-6-12-8 | 23F | Iceland, The Netherlands | 24 | 1 | Sputum | 1999 | 9-18-11 | 0.5 | ≤0.03 | 0.25 | 0.06 | ≤0.03 | ≤0.03 |
| 230 | 12-19-2-17-6-22-14 | 24F | Italy, Portugal | 22d | 1 | Blood | 1998 | NT-19-WT | 0.5 | 0.06 | 0.5 | 0.12 | 0.12 | 0.06 |
| 276 | 2-19-2-17-6-22-14 | 19A | The Netherlands, Portugal | 22b | 1 | Sinus | 2001 | 8-1-WT | 1 | 0.5 | 4 | 1 | 0.5 | 0.25 |
| 276 | 2-19-2-17-6-22-14 | 19A | 22c | 1 | Sputum | 2002 | NT-1-WT | 1 | 0.5 | 4 | 1 | 1 | 0.25 | |
| 319 | 12-19-2-17-6-22-9 | 19A | 22a | 1 | Sinus | 1999 | NT-19-10 | 1 | 0.06 | 0.5 | 0.12 | 0.25 | 0.12 | |
| 317 | 2-10-50-29-6-19-59 | Rough | 25, 27 | 2 | Eye, BAL | 1995, 2001 | NT-20-WT | 0.5 | 0.5 | 0.5 | 0.12 | 0.5 | 0.06 | |
| 1473 | 2-10-50-29-6-19-160 | Rough | 26 | 1 | Sinus | 1996 | NT-21-WT | 0.5 | 0.06 | 0.5 | 0.12 | 0.25 | 0.06 | |
New STs and alleles are underlined.
In the order aroE-gdh-gki-recP-spi-xpt-ddl.
BAL, bronchoalveolar lavage; CSF, cerebrospinal fluid.
Susceptibilities, serotypes, and STs of meningitis-related isolates were described in reference 39, and ciprofloxacin-nonsusceptible isolates (single isolates of ST81, ST276, and ST1477 and two isolates of ST156) were described in reference 38.
In the order pbp1a-pbp2x-pbp2b. NT, nontypeable; other, pattern unique to an ATCC clone.
PEN, penicillin; AMX, amoxicillin; CXM, cefuroxime; CRO, ceftriaxone; FEP, cefepime; MEM, meropenem.
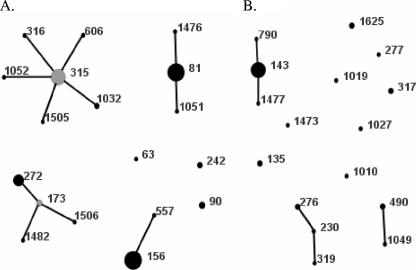
Among 34 STs found in the MLST analysis (Tables 1 and 2), 13 STs characteristic for 15 isolates were new. Of these STs, seven represented novel combinations of known alleles (STs 1010, 1019, 1027, 1032, 1049, 1051, and 1052), and six profiles contained new alleles (STs 1473, 1476, 1482, 1505, 1506, and 1625). Isolates of six main STs (STs 81, 156, 315, 143, and 272) with at least five isolates each constituted 70.2% of the PNSP group, and when their SLVs were also considered, the ratio increased to 81.7%. The diversity index was equal to 89.5% (confidence interval, 87.1% to 91.9%). The eBURST analysis showed the presence of 7 clonal complexes and 11 singletons (Fig. 2). The comparison of the isolates with the international multiresistant pneumococcal clones revealed that the isolates harboring allelic profiles that were identical to those of seven clones (Spain23F-1, Spain6B-2, Spain9V-3, Taiwan23F-15, Poland23F-16, Poland6B-20, Sweden15A-25) or their SLVs constituted the majority of the PNSP isolates (94 isolates; 71.8%) (Table 1 and Fig. 2A). Among the 37 remaining isolates (Table 2 and Fig. 2B), 19 isolates belonged to ST143 and serotype 14, and only these isolates showed penicillin resistance among the isolates that were not closely related to the international clones. As mentioned above, serotypes and STs of 5 ciprofloxacin-nonsusceptible PNSP isolates from patients with RTI and 18 penicillin-nonsusceptible isolates from patients with meningitis and their classification into the international clones have been reported previously (38, 39).
FIG. 2.
Population snapshot of 131 Polish PNSP isolates, as revealed by eBURST analysis. Lines indicate the presence of SLV links among particular STs symbolized by circles. The size of the circle corresponds to the number of isolates belonging to an ST. (A) International clones. (B) Other isolates.
Polymorphism of the pbp genes in PNSP.
Three pbp genes, pbp1a, pbp2x, and pbp2b, were studied by PCR-RFLP, yielding 10, 22, and 12 different restriction patterns, respectively (Tables 1 and 2). Some of the isolates produced wild-type (WT) RFLP patterns of the genes (22, 2, and 35 isolates, respectively), and in nine isolates, pbp1a was nontypeable (NT) due to the lack of an amplification product. For each of the three genes studied, a certain pattern (designated polymorph 1 in each case) was clearly predominant among the isolates (54.2% for pbp1a, 52.8% for pbp2b, and 54.2% for pbp2x), and some of the patterns were found in isolates unrelated by MLST (summarized in Table 3). Altogether, 29 different combinations of single-gene polymorphs (excluding those with nontypeable pbp1a) were discerned among the isolates, with the 1-1-1 profile being the most prevalent (65 isolates; 49.6%).
TABLE 3.
pbp patterns occurring in more than one unrelated ST
| Gene | Pattern | STsa |
|---|---|---|
| pbp1a | 1 | 81b (1051, 1476), 90,c 156d (557), 143 (790, 1477) |
| 5 | 135, 490, 1019, 1027, 1482e | |
| 8 | 63, 276, 1625 | |
| pbp2x | 1 | 81b (1051, 1476), 156d (557), 242,f 172,g 143 (790, 1477), 276, 490 |
| 6 | 143, 272e | |
| 7 | 315,h 272e | |
| pbp2b | 1 | 81b (1051, 1476), 156d (557), 143 (790, 1477) |
| 3 | 242,f 315,h 490 | |
| 4 | 63, 173,g 1482e |
SLVs of the main ST in the group are given in parentheses.
ST characteristic for the Spain23F-1 clone.
ST characteristic for the Spain6B-2 clone.
ST characteristic for the Spain9V-3 clone.
SLV of ST173, characteristic for the Poland23F-16 clone.
ST characteristic for the Taiwan23F-15 clone.
ST characteristic for the Poland23F-16 clone.
ST characteristic for the Poland6B-20 clone.
The Spain23F-1 group.
The Spain23F-1 group contained 25 isolates, most of which belonged to ST81 and its two SLVs that represented novel allelic profiles (ST1051 and ST1476) (Table 1). Serotype 23F was characteristic for the majority of the isolates; however, three isolates possessed serotype 19F or 19A. Of the two PFGE types discerned, type 1 was split into numerous subtypes (1a to 1p), with the subtype 1a pattern being identical to that of the ATCC Spain23F-1 strain. This group was uniform with respect to the pbp fingerprint (profile 1-1-1), which was the same as that for the reference ATCC strain. The isolates were resistant to penicillin (MIC, 2 μg/ml) and cefuroxime, intermediate to meropenem, susceptible or intermediate to ceftriaxone and cefepime, and susceptible to amoxicillin.
The Spain6B-2 group.
The Spain6B-2 group was represented by three isolates of ST90 and serotype 6B and a single PFGE type with two subtypes, one of which also characterized the ATCC Spain6B-2 strain (Table 1). The isolates shared their pbp1a and pbp2x RFLP patterns with the reference strain; however, they all produced another pbp2b pattern. The β-lactam susceptibility phenotypes of these isolates were the same as those of the Spain23F-1 representatives.
The Spain9V-3 group.
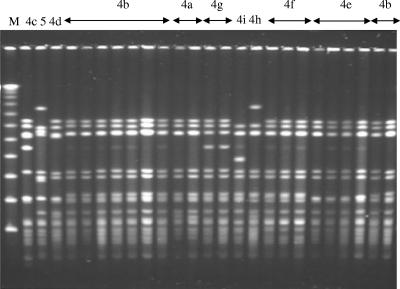
Out of 25 isolates that belonged to the Spain9V-3 group, most were of serotype 9V, four were of serotype 14, and a single isolate was rough (Table 1). All of the isolates belonged to ST156, except for a single ST557 isolate. Two PFGE types were observed, one of which was further differentiated into several subtypes (4a to 4i), with subtype 4a also being characteristic for the ATCC Spain9V-3 strain (Fig. 3). The majority of the isolates shared their pbp fingerprints with the Spain23F-1 clone, with the exception of a single isolate with the unique pbp1a pattern 2 and with a penicillin MIC of 1 μg/ml. In general, susceptibility to β-lactams in this group was the same as that among the Spain23F-1 isolates. Apart from a single isolate from 1995 and 2 isolates from 1999, 22 other representatives of this group were recovered in 2000 or later (P ≤ 0.001), with 9 isolates being recovered in 2002 (Fig. 1).
FIG. 3.
PFGE analysis of PNSP isolates belonging to the Spain9V-3 clone. M, molecular weight standard (Lambda Ladder; New England Biolabs, Beverly, MA). PFGE types and subtypes are indicated above the lanes.
The Taiwan23F-15 group.
Two isolates of ST242 and serotype 23F represented the Taiwan23F-15 group (Table 1). One of the isolates shared the PFGE pattern (PFGE type 7) and pbp fingerprint (profile 3-1-3) with the ATCC Taiwan23F-15 strain, whereas the other isolate varied in pbp2x (profile 3-3-3) and belonged to a different PFGE type. Both isolates were resistant to penicillin and cefuroxime, intermediate to ceftriaxone, and susceptible to cefepime but differed slightly in amoxicillin and meropenem susceptibilities.
The Poland23F-16 group.
All of the 14 isolates of the Poland23F-16 group had serotype 23F, and they belonged either to the main, presumably ancestral, ST173 or to one of its three SLVs (STs 272, 1482, and 1506) (Table 1). Three PFGE types with several subtypes (9, 10a to 10d, and 11a to 11f) were observed. Remarkably, seven different profiles of pbp genes were discerned among the isolates, and this divergence correlated with various susceptibility phenotypes. Except for a single ST1482 isolate with intermediate susceptibility to penicillin, the other isolates showed high-level resistance to penicillin (MICs, 4 to 8 μg/ml) and were resistant to cefuroxime. Moreover, 10 isolates were resistant to amoxicillin, 6 were resistant to ceftriaxone, and 2 were resistant to cefepime.
The Poland6B-20 group.
The Poland6B-20 group included 24 isolates of serotype 6B, classified into the major, probably ancestral, ST315 or one of its five SLVs (STs 316, 606, 1032, 1052, 1505) (Table 1). In the PFGE analysis, two types were present, one of which had several subtypes (12a to 12i). Similarly to the Poland23F-16 group, divergence of the pbp fingerprints (four profiles) correlating with various levels of resistance was observed. The majority of the isolates were intermediate to penicillin (MIC, 0.12 μg/ml) and showed modifications only in the pbp2x pattern; these isolates were uniformly susceptible to the other β-lactams tested. The group also included two isolates with increased penicillin MICs (0.5 μg/ml) and modified pbp1a (profile 7-11-WT) as well as three penicillin-resistant isolates with modifications of pbp2b and/or pbp1a patterns (6-10-WT and NT-7-3). These isolates showed cefuroxime resistance and increased MICs of other β-lactams, and they all differed from the remaining isolates by the type of the pbp2x pattern modification.
The Sweden15A-25 group.
A single isolate of ST63 and serotype 15A shared PFGE type 14 with the ATCC Sweden15A-25 strain but differed from it by the pbp1a PCR-RFLP pattern (Table 1). The isolate, similarly to the reference strain, was intermediate to penicillin and susceptible to other β-lactams.
The ST143 group.
All 19 isolates belonging to the ST143 group were of serotype 14 (Table 2). In the MLST analysis (Table 2), the major ST of these isolates, ST143, represented a double-locus variant of ST156 (in the gdh and recP loci), characteristic of the Spain9V-3 clone. In addition, this group contained two SLVs of ST143, ST790, and ST1477. Two PFGE types, types 6 and 4, the latter with 10 subtypes (4f and 4j to 4t) that belonged to the same PFGE type, type 4, as the representatives of Spain9V-3 were discerned. Although the majority of the isolates shared their pbp profile (1-1-1) with Spain23F-1 and Spain9V-3, three other variants of pbp2x were found in single isolates (profiles 1-6-1, 1-11-1, and 1-17-1). The group shared β-lactam susceptibility phenotypes with the three Spanish clones. Some of the isolates showed elevated MICs of cefuroxime (up to 16 μg/ml) but without obvious correlation to their pbp fingerprints. Although pneumococci of the ST143 group have been present in Poland since at least 1994 (37), they became more frequent from 2000 onwards (P = 0.025), following the same trend as that seen for Spain9V-3 (Fig. 1).
Other isolates.
The remaining 18 PNSP isolates (Table 2 and Fig. 1B) showed a significant degree of variability and included isolates of seven serotypes plus three rough isolates. Molecular analysis discerned 13 STs, 14 PFGE types, and nine combinations of pbp patterns among these isolates; for seven isolates, the complete pbp profiles were not determined due to the lack of pbp1a amplification. In the eBURST analysis, three clonal groups, ST490/1049, ST276/230/319, and ST317/1473, as well as six singletons, STs 1019, 135, 1010, 1625, 1027, and 277 (novel STs are underlined), were observed. All these isolates were intermediate to penicillin and susceptible to other β-lactams, except for two isolates of ST276 and single isolates of ST490 and ST1019, which were also cefuroxime resistant.
DISCUSSION
The first clinical isolates of pneumococci with reduced susceptibility to penicillin (MIC, 0.06 μg/ml) appeared in Australia in the 1960s (20) and were followed by resistant isolates, reported first in South Africa in 1977 (3). Since the 1980s, the prevalence of PNSP has been constantly increasing worldwide. In the recent Alexander Project involving 26 countries, the mean frequency of penicillin nonsusceptibility among isolates from 1998 to 2000 was estimated to be 31.7%, with 18.2% resistant and 13.5% intermediate isolates, and there were significant differences among particular countries (22). The overall rate of 12.3% PNSP and 8.9% resistant isolates from 1998 to 2002 placed Poland among countries with moderate resistance frequencies. However, a significant increase was observed in 2002, which clearly paralleled the increase in β-lactam consumption. As shown by the recent data from the European Surveillance of Antimicrobial Consumption Project, the consumption of penicillins and cephalosporins increased in Poland from 5.79 and 1.81 defined daily doses (DDDs) per 1,000 inhabitants per day, respectively, to 11.2 and 2.29 DDDs in the period from 1997 to 2001 and dropped slightly in 2002 to 9.86 and 2.04 DDDs, respectively (www.esac.ua.ac.be) (17). When our preliminary susceptibility data for 2003 (16.5% PNSP) were included, antibiotic consumption and PNSP prevalence correlated with each other (r = 0.83; P = 0.04) (A. Skoczyńska et al., unpublished). In Poland, penicillins and cephalosporins constitute the main types of antimicrobials used in outpatient treatment (56.3%), and the level of consumption of these antimicrobials in 2002 was very similar to that reported in Spain (17), which presumably promoted the spread of resistance (1).
The dissemination of representatives of two related clones, Spain9V-3 and ST143, appears to be responsible for the increase in resistance observed in 2002, while the frequencies of other clones remained relatively stable (Fig. 1). These two expanding clones differ by two loci in MLST and usually have a different serotype, i.e., serotypes 9V and 14, respectively. It was proposed that Spain9V-3 may represent the ancestral clone for ST143 as well as, independently, for the serotype 14 variants of Spain9V-3 (6) also found in this study. An isolate of ST144 and serotype 14 (45), which represents an SLV of both ST156 (Spain9V-3) and ST143, presumably constitutes a link between the two clones. The similarity of the two clones was also revealed by PFGE in which the majority of their representatives belonged to the same type, although they belonged to different subtypes.
Apart from the two clones, the specific clonal structure of Polish PNSP was dominated by a few other clones, all associated with the international clones. Such a high degree of clonality seems to be a characteristic feature of PNSP in general (9, 16, 23, 35, 41) and is associated, at least in part, with the complexity of the mechanism of acquisition of penicillin resistance determinants, which is why the acquisition occurs relatively rarely. The resulting PNSP clones, having gained the selective advantage, subsequently spread in bacterial populations and gradually diversify through mutations and recombination with DNA of other pneumococci (27). Accordingly, we have observed multiple SLVs (including new STs), PFGE types, and subtypes and very probable examples of serotype switching (8) among the isolates related to several major clones such as Spain23F-1, Spain6B-2, Spain9V-3, and Taiwan23F-15.
Variability in pbp profiles within clonal groups was also observed, which may be explained in two ways. It could be associated with clone diversification, e.g., pbp2x pattern 1 instead of pattern 3 in a single isolate related to Spain6B-2, which may have acquired the gene from one of the commonly occurring PNSP clones, Spain23F-1 or Spain9V-3. Such isolates, which differ from the main clone by one of the pbp fingerprints, are considered variant members of the clone (41, 45). Alternatively, the differences in the pbp patterns may have resulted from independent acquisitions of resistance determinants by a susceptible strain. Such an explanation seems to be especially likely in the case of the two clonal groups Poland23F-16 and Poland6B-20. Similar to the Hungary19A-6 clone (34), these two groups are remarkably divergent in their pbp profiles. The term “hitchhiking effect” (13) was proposed to describe the impact of the pbp2b transfer on the recombination frequency of the adjacent ddl gene, which was used in MLST. Alleles 36, 37, and 162 of this locus differ from each other by several nucleotides and so far have been found only in isolates related to Poland23F-16. A part of allele 36 that was present in the presumably ancestral ST173 was divergent from pneumococcal sequences (13), and alleles 37 (in ST272) and 162 (in ST1506) showed 95% and 99% homology, respectively, to the ddl gene from Streptococcus mitis (36). Isolates of both these STs also differed from ST173 in the pbp2b pattern, suggesting the acquisition of this gene together with the ddl allele. In general, the significant number of unrelated STs sharing the same pbp patterns within the analyzed group (Table 3) may indicate both independent DNA acquisition events from commensal streptococci and genetic exchange of these genes among pneumococci (7).
Similar to the findings of others (15, 31, 40), a relatively small number of serotypes was characteristic for the majority of Polish PNSP, which resulted in very good coverage by the 7-valent conjugated vaccine. Therefore, the introduction of childhood vaccination program should hopefully lead to a reduction in the frequency of resistance (44), especially along with more prudent antimicrobial use, as seen recently in Spain (32). However, the circulation of non-vaccine-type PNSP, such as 24F/ST230 (33, 43) and Sweden15A-25, as well as capsule switching may compromise the effect of the vaccine in the future and highlights the need for continuous surveillance of circulating PNSP isolates.
Acknowledgments
We thank Stephen Murchan for the critical reading of the manuscript and Anna Klarowicz for excellent technical assistance.
This study was partially financed by a grant from the Polish Committee for Scientific Research (3P0A 062 23). We acknowledge the use of the pneumococcal MLST database, which is located at the Imperial College, London, and is funded by the Wellcome Trust.
Footnotes
Published ahead of print on 16 October 2006.
REFERENCES
- 1.Albrich, W. C., D. L. Monnet, and S. Harbarth. 2004. Antibiotic selection pressure and resistance in Streptococcus pneumoniae and Streptococcus pyogenes. Emerg. Infect. Dis. 10:514-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appelbaum, P. C. 2002. Resistance among Streptococcus pneumoniae: implications for drug selection. Clin. Infect. Dis. 34:1613-1620. [DOI] [PubMed] [Google Scholar]
- 3.Appelbaum, P. C., A. Bhamjee, J. N. Scragg, A. F. Hallett, A. J. Bowen, and R. C. Cooper. 1977. Streptococcus pneumoniae resistant to penicillin and chloramphenicol. Lancet ii:995-997. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, C., F. Chi, S. Rahid, and R. Hakenbeck. 2004. Mechanisms for penicillin resistance in Streptococcus pneumoniae: penicillin-binding proteins, gene transfer, and cell wall metabolism, p. 339-349. In E. I. Tuomanen, T. J. Mitchell, D. A. Morrison, and B. G. Spratt (ed.), The pneumococcus. ASM Press, Washington, DC.
- 5.Carroll, K. C. 2002. Laboratory diagnosis of lower respiratory tract infections: controversy and conundrums. J. Clin. Microbiol. 40:3115-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffey, T. J., M. Daniels, M. C. Enright, and B. G. Spratt. 1999. Serotype 14 variants of the Spanish penicillin-resistant serotype 9V clone of Streptococcus pneumoniae arose by large recombinational replacements of the cpsA-pbp1a region. Microbiology 145:2023-2031. [DOI] [PubMed] [Google Scholar]
- 7.Coffey, T. J., C. G. Dowson, M. Daniels, J. Zhou, C. Martin, B. G. Spratt, and J. M. Musser. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255-2260. [DOI] [PubMed] [Google Scholar]
- 8.Coffey, T. J., M. C. Enright, M. Daniels, J. K. Morona, R. Morona, W. Hryniewicz, J. C. Paton, and B. G. Spratt. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73-83. [DOI] [PubMed] [Google Scholar]
- 9.Corso, A., E. P. Severina, V. F. Petruk, Y. R. Mauriz, and A. Tomasz. 1998. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb. Drug Resist. 4:325-337. [DOI] [PubMed] [Google Scholar]
- 10.Doern, G. V., K. P. Heilmann, H. K. Huynh, P. R. Rhomberg, S. L. Coffman, and A. B. Brueggemann. 2001. Antimicrobial resistance among clinical isolates of Streptococcus pneumoniae in the United States during 1999-2000, including a comparison of resistance rates since 1994-1995. Antimicrob. Agents Chemother. 45:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowson, C. G., T. J. Coffey, C. Kell, and R. A. Whiley. 1993. Evolution of penicillin resistance in Streptococcus pneumoniae: the role of Streptococcus mitis in the formation of a low affinity PBP2 in S. pneumoniae. Mol. Microbiol. 9:635-643. [DOI] [PubMed] [Google Scholar]
- 12.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 13.Enright, M. C., and B. G. Spratt. 1999. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol. Biol. Evol. 16:1687-1695. [DOI] [PubMed] [Google Scholar]
- 14.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenoll, A., G. Asensio, I. Jado, S. Berron, M. T. Camacho, M. Ortega, and J. Casal. 2002. Antimicrobial susceptibility and pneumococcal serotypes. J. Antimicrob. Chemother. 50(Suppl. S2):13-19. [DOI] [PubMed] [Google Scholar]
- 16.Gherardi, G., C. G. Whitney, R. R. Facklam, and B. Beall. 2000. Major related sets of antibiotic-resistant pneumococci in the United States as determined by pulsed-field gel electrophoresis and pbp1a-pbp2b-pbp2x-dhf restriction profiles. J. Infect. Dis. 181:216-229. [DOI] [PubMed] [Google Scholar]
- 17.Goosens, H., M. Ferech, R. V. Stichele, M. Elseviers, and the ESAC Project Group. 2005. Outpatient antibiotic use in Europe and association with resistance. Lancet 365:579-587. [DOI] [PubMed] [Google Scholar]
- 18.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez, F., M. Masia, J. C. Rodriguez, C. Mirete, B. Soldan, S. Padilla, I. Hernandez, F. De Ory, G. Royo, and A. M. Hidalgo. 2005. Epidemiology of community-acquired pneumonia in adult patients at the dawn of the 21st century: a prospective study on the Mediterranean coast of Spain. Clin. Microbiol. Infect. 11:788-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansman, D., and M. M. Bullen. 1967. A resistant pneumococcus. Lancet ii:264-265. [DOI] [PubMed] [Google Scholar]
- 21.Hauser, C., S. Aebi, and K. Mühlemann. 2004. An internationally spread clone of Streptococcus pneumoniae evolves from low-level to higher-level penicillin resistance by uptake of penicillin-binding protein gene fragments from nonencapsulated pneumococci. Antimicrob. Agents Chemother. 48:3563-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs, M. R., D. Felmingham, P. C. Appelbaum, R. N. Grunberg, and the Alexander Project. 2003. The Alexander Project 1998-2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52:229-246. [DOI] [PubMed] [Google Scholar]
- 23.Klugman, K. P. 2002. The successful clone: the vector of dissemination of resistance in Streptococcus pneumoniae. J. Antimicrob. Chemother. 50(Suppl. S2):1-5. [DOI] [PubMed] [Google Scholar]
- 24.Laible, G., B. G. Spratt, and R. Hakenbeck. 1991. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 5:1993-2002. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence, E. R., D. B. Griffiths, S. A. Martin, R. C. George, and L. M. C. Hall. 2003. Evaluation of semiautomated multiplex PCR assay for determination of Streptococcus pneumoniae serotypes and serogroups. J. Clin. Microbiol. 41:601-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefévre, J. C., G. Faucon, A. M. Sicard, and A. M. Gasc. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 31:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maynard Smith, J., E. J. Feil, and N. H. Smith. 2000. Population structure and evolution dynamics of pathogenic bacteria. BioEssays 22:1115-1122. [DOI] [PubMed] [Google Scholar]
- 28.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hackenbeck, W. Hryniewicz, J. C. Lefévre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munoz, R., T. R. Coffey, M. Daniels, C. G. Dowson, G. Laible, J. Casal, R. Hakenbeck, M. Jacobs, J. M. Musser, B. G. Spratt, and A. Tomasz. 1991. Intercontinental spread of multiresistant clone of serotype 23F Streptococcus pneumoniae. J. Infect. Dis. 164:302-306. [DOI] [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 6th ed. M7-A6, M100-S13. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 31.Nichol, K. A., G. G. Zhanel, and D. J. Hoban. 2003. Molecular epidemiology of penicillin-resistant and ciprofloxacin-resistant Streptococcus pneumoniae in Canada. Antimicrob. Agents Chemother. 47:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oteo, J., E. Lazaro, F. J. de Abajo, F. Baquero, J. Campos, and Spanish Members of the European Antimicrobial Resistance Surveillance System. 2004. Trends in antimicrobial resistance in 1,968 invasive Streptococcus pneumoniae strains isolated in Spanish hospitals (2001 to 2003): decreasing penicillin resistance in children's isolates. J. Clin. Microbiol. 42:5571-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pantosti, A., G. Gherardi, M. Conte, F. Faella, G. Dicuonzo, and B. Beall. 2002. A novel, multiple drug-resistant, serotype 24F strain of Streptococcus pneumoniae that caused meningitis in patients in Naples, Italy. Clin. Infect. Dis. 35:205-208. [DOI] [PubMed] [Google Scholar]
- 34.Reichmann, P., A. Konig, A. Marton, and R. Hakenbeck. 1996. Penicillin-binding proteins as resistance determinants in clinical isolates of Streptococcus pneumoniae. Microb. Drug Resist. 2:177-181. [DOI] [PubMed] [Google Scholar]
- 35.Richter, S. S., K. P. Heilmann, S. L. Coffman, H. K. Huynh, A. B. Brueggemann, M. A. Pfaller, and G. V. Doern. 2002. The molecular epidemiology of penicillin-resistant Streptococcus pneumoniae in the United States, 1994-2000. Clin. Infect. Dis. 34:330-339. [DOI] [PubMed] [Google Scholar]
- 36.Romeo, P., R. Lopez, and E. Garcia. 2004. Characterization of LytA-like N-acetylmuramoyl-l-alanine amidases from two new Streptococcus mitis bacteriophages provides insights into the properties of the major pneumococcal autolysin. J. Bacteriol. 186:8229-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadowy, E., J. Zhou, E. Meats, M. Gniadkowski, B. G. Spratt, and W. Hryniewicz. 2003. Identification of multidrug-resistant Streptococcus pneumoniae strains isolated in Poland by multilocus sequence typing. Microb. Drug Resist. 9:81-86. [DOI] [PubMed] [Google Scholar]
- 38.Sadowy, E., R. Izdebski, A. Skoczyńska, M. Gniadkowski, and W. Hryniewicz. 2005. High genetic diversity of ciprofloxacin-nonsusceptible isolates of Streptococcus pneumoniae in Poland. Antimicrob. Agents Chemother. 49:2126-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadowy, E., A. Skoczynska, J. Fiett, M. Gniadkowski, and W. Hryniewicz. 2006. Multilocus sequence types, serotypes, and variants of the surface antigen PspA in Streptococcus pneumoniae isolates from meningitis patients in Poland. Clin. Vaccine Immunol. 13:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Setchanova, L., and A. Tomasz. 1999. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates from Bulgaria. J. Clin. Microbiol. 37:638-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi, Z.-Y., M. C. Enright, P. Wilkinson, D. Griffiths, and B. G. Spratt. 1998. Identification of three major clones of multiply antibiotic-resistant Streptococcus pneumoniae in Taiwanese hospitals by multilocus sequence typing. J. Clin. Microbiol. 36:3514-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skoczyńska, A., P. Kriz, H. B. Konradsen, and W. Hryniewicz. 2000. Characteristics of the major etiologic agents of bacterial meningitis isolated in Poland in 1997-1998. Microb. Drug Resist. 6:147-153. [DOI] [PubMed] [Google Scholar]
- 43.Sousa, N. G., R. Sá-Leão, M. I. Crisóstomo, C. Simas, S. Nunes, N. Frazão, J. A. Carriço, R. Mato, I. Santos-Sanches, and H. de Lencastre. 2005. Properties of novel international drug-resistant pneumococcal clones identified in day-care centers of Lisbon, Portugal. J. Clin. Microbiol. 43:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talbot, T. R., K. A. Poehling, T. V. Hartert, P. G. Arbogast, N. B. Halasa, E. Mitchel, W. Schaffner, A. S. Craig, K. M. Edwards, and M. R. Griffin. 2004. Reduction in high rates of antibiotic-nonsusceptible invasive pneumococcal disease in Tennessee after introduction of the pneumococcal conjugate vaccine. Clin. Infect. Dis. 39:641-648. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, J., M. C. Enright, and B. G. Spratt. 2000. Identification of the major Spanish clones of penicillin-resistant pneumococci via the Internet using multilocus sequence typing. J. Clin. Microbiol. 38:977-986. [DOI] [PMC free article] [PubMed] [Google Scholar]