Abstract
A multiple-antibiotic-resistant Salmonella enterica serovar Kentucky strain was found to contain SGI1-K, a variant form of the Salmonella genomic island 1 (SGI1) with an In4-type class 1 integron that contains only one cassette array, aacCA5-aadA7, and an adjacent mercury resistance module. Part of the 3′-conserved segment (3′-CS) of the integron, together with the inverted short segment from the right-hand end of the integron transposition module normally found between the 3′-CS and IS6100 in In4 family integrons, has been removed by an IS6100-mediated deletion. IRt, the right-hand inverted repeat found at the outer end of the integron, abuts a mercury resistance region instead of the usual SGI1 backbone segment. The mer module is a hybrid of those found in Tn501 and Tn21. This mer region and a further uncharacterized segment of at least 10 kb appear to have been incorporated between IRt and the SGI1 backbone. These findings demonstrate that the multidrug resistance region in SGI1 can incorporate new DNA segments in the same way as multiple antibiotic resistance regions in plasmids.
In multiple-antibiotic-resistant strains of Salmonella enterica serovar Typhimurium DT104, the antibiotic resistance genes are frequently found clustered within a 43-kb integrated element known as the Salmonella genomic island 1 (SGI1) (4, 5). The original SGI1 isolated from Salmonella serovar Typhimurium DT104 strains contains five antibiotic resistance genes, all located within the boundaries of a complex class 1 integron (Fig. 1), recently designated In104 (18). Two of these genes, aadA2 and blaP1, are each within a gene cassette, but the cassettes are situated in two different attI1 sites that are separated by a region that includes the floR florfenicol resistance determinant and the tetRA(G) tetracycline resistance determinant (2, 4, 6). The fifth gene, sul1, is part of the 3′-conserved segment (3′-CS) of the integron. These five genes are responsible for the multiple antibiotic resistance phenotype conferred by SGI1, namely resistance to chloramphenicol and florfenicol (floR), ampicillin and carbenicillin (blaP1), streptomycin and spectinomycin (aadA2), sulfonamides (sul1), and tetracycline [tet(G) determinant]. SGI1 or variants of it have also been found in several other S. enterica serovars, consistent with horizontal gene transfer and mobilization of SGI1 by an IncC plasmid, as recently demonstrated (11). In all cases, SGI1 is found in the bacterial chromosome between the thdF and yidY genes (3, 9, 12-14, 16, 18, 22). A second integrated element (“retron phage”), located between one end of SGI1 and the yidY gene, is found only in Salmonella serovar Typhimurium strains (4, 5).
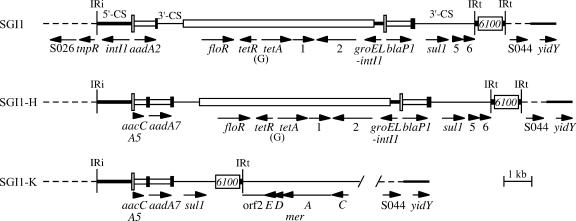
FIG. 1.
Integrons in SGI1 and variants. The map of In104, the integron in the original SGI1, was generated from the sequence in GenBank accession no. AF261825. For SGI1-H, the cassettes from AY458224 were substituted on the left. The complete sequence of the integron in SGI1-K and its immediate surroundings is from this study (AY463797.3). Dashed lines represent the surrounding SGI1 backbone, and the medium-width line on the right represents the adjacent chromosome. Narrow vertical bars represent the inverted repeats (IRi and IRt) of class 1 integrons. The 5′-CS, 3′-CS, and tni regions of class 1 integrons are indicated by lines of different thicknesses, with attI1 as a tall open box and gene cassettes as open boxes with a black bar at one end, indicating the 59-base element (59-be). IS6100 and the central, non-integron-derived region are open boxes. Arrows indicate the position and orientation of genes and open reading frames, which are named or numbered.
A number of variants of SGI1 containing different sets of resistance genes have also been identified (3, 9, 12-14, 18). These variants, SGI1-A to SGI1-J, appear to have gained, lost, or exchanged resistance genes by gaining and/or losing various segments of DNA, mostly as a result of events that could have occurred by homologous recombination (see references 3, 13, and 18). Some variants have lost the central region of In104 and include only one attI1 site containing gene cassettes. Others include gene cassettes that differ from those in SGI1, such as the dfrA1-orfC or the aacCA5-aadA7 pairs (13, 14, 18), or have a deletion encompassing an attI site (18). Further variants (3, 18) have gained an additional dfrA10 trimethoprim resistance gene associated with the potential insertion sequence known as CR1, which draws a variety of resistance genes into the complex class 1 integrons that contain it (28). Exchange of the cassette array and acquisition of CR1 (containing orf513) and the adjacent dfrA10 gene have both been shown experimentally to occur by homologous recombination (25, 27, 28). Recently, further variations that have lost other parts of SGI1 have been reported. One type appears to have been formed by the activity of CR1 and to have lost all of the sequence to the right of CR1 in the CR1-containing SGI1-A variant, with the deletion extending into the adjacent chromosomal genes (12). A variant in which the integrase-encoding end (IRi) of the class 1 integron abuts a different part of the SGI1 backbone has also been described (18). Variation in the structure of SGI1 thus serves as a paradigm for examining the evolution of multiple antibiotic resistance regions occurring within an integron that is found in a defined context and location, namely in the backbone of SGI1, which is integrated within the end of the thdF gene in the S. enterica chromosome.
We recently reported the sequence of an aacCA5-aadA7 gene cassette array in a class 1 integron from a multiple-antibiotic-resistant Salmonella enterica serovar Kentucky strain that was isolated in 2001 from spice imported into Australia from India (19). Here, this integron was localized to a variant SGI1 and both the complete structure of the integron and its context were investigated.
MATERIALS AND METHODS
Bacterial strains.
Salmonella serovar Kentucky strain SRC73 was isolated in 2001 from spice imported into Australia from India. It is resistant to ampicillin, gentamicin, streptomycin, spectinomycin, sulfathiazole, tetracycline, and nalidixic acid (19) and known to contain the cassette array aacCA5-aadA7 conferring resistance to gentamicin (aacCA5) and streptomycin and spectinomycin (aadA7), as well as the resistance genes blaTEM (ampicillin resistance), sul1 (sulfonamide resistance), strAB (streptomycin resistance), and tet(A) (tetracycline resistance), but not the aadA2 gene or tet(B), tet(G) determinants (19). The blaP1 (or blaP2) and floR genes were also absent (R. S. Levings, unpublished observations).
PCR mapping.
SRC73 cultures were grown overnight at 37°C on MacConkey agar (Becton Dickinson and Company). Whole-cell DNA was isolated using standard methods (31) and used as the template for PCR amplification reactions using various combinations of the primers listed in Table 1. Amplification was carried out in PCR buffer (Roche Molecular Biochemicals, Mannheim, Germany) containing each deoxynucleoside triphosphate (dNTP) at 160 μM, 20 pmol of each primer, approximately 10 to 50 ng of template, and 1 U of Taq DNA polymerase (Roche). Reaction conditions, described in detail elsewhere (18), were generally 94 to 96°C for 3 to 5 min, followed by 30 to 40 cycles of denaturation (94 to 96°C for 30 s), annealing (52 to 62°C for 30 to 60 s), and extension (72°C for 30 s for 2 min), and a final incubation at 72°C for 10 to 15 min. Products were separated on agarose gels, and sizes were estimated using 100-bp Plus (Fermentas, Vilnius, Lithuania) or Hyperladder I (Bioline, London, United Kingdom) and known amplification products from SGI1 as standards.
TABLE 1.
Characteristics of the primers used in this study
| Primer | Sequence (5′→3′) | Location | Nucleotide positions | Accession no. | Source or referencea |
|---|---|---|---|---|---|
| U7-L12 | ACACCTTGAGCAGGGCAAAG | thdF | 1-20 | AF261825.2 | 5 |
| LJ-R1 | AGTTCTAAAGGTTCGTAGTCG | int | 500-480 | AF261825.2 | 5 |
| RL-D3 | ATTGGTATGAGCCATGATGG | S024 | 20531-20550 | AF261825.2 | This study |
| S024-RV | GGTACGGTATCGCCTAAGTG | S024 | 21930-21911 | AF261825.2 | 9 |
| S026-FW | TCGGGTAATCTCAGCAGAGC | S026 | 25021-25040 | AF261825.2 | 9 |
| int-RV | GGGCATGGTGGCTGAAGGACC | intI1 | 27266-27246 | AF261825.2 | 9 |
| L2 | GACGATGCGTGGAGACC | intI1 | 27612-27628 | AF261825.2 | 30 |
| L3 | CTTGCTGCTTGGATGCC | 5′-CS | 27908-27892 | AF261825.2 | 21 |
| QS-1 | ATGAAAGGCTGGCTTTTTCTTG | qacEΔ1 | 28953-28974, 38413-38424 | AF261825.2 | 4 |
| QS-2 | TGAGTGCATAACCACCAGCC | sul1 | 29674-29655, 31934-31915 | AF261825.2 | 4 |
| sul1-F | GTGACGGTGTTCGGCATTCT | sul1 | 29297-29316, 35757-35776 | AF261825.2 | 17 |
| sul1-R | TTTACAGGAAGGCCAACGGT | sul1 | 39424-39405 | AF261825.2 | 17 |
| orf5-F | AGGTTGTGCGGCTGATGC | orf5 | 39776-39793 | AF261825.2 | 18 |
| orf5-R2 | CGAGTTCTAGGCGTTCTGC | orf5 | 40213-40195 | AF261825.2 | 18 |
| orf6-R | ACTATCTTCGGCCTTCACACG | orf6 | 40508-40488 | AF261825.2 | 18 |
| DB-T1 | TGCCACGCTCAATACCGAC | IS6100 | 41120-41138 | AF261825.2 | 3 |
| IS6100-Rv2 | AATGGTGGTTGAGCATGCC | IS6100 | 41475-41457 | AF261825.2 | 18 |
| MDR-Bb | GAATCCGACAGCCAACGTTCC | S044 | 41905-41884 | AF261825.2 | 3 |
| 104-RJ | TGACGAGCTGAAGCGAATTG | S044 | 42373-42392 | AF261825.2 | 5 |
| 104-D | ACCAGGGCAAAACTACACAG | yidY | 47130-47111 | AF261825.2 | 5 |
| RL-D1 | TTGTCCCTGATGAGACTGC | S044 | 41994-42012 | AF261825.2 | This study |
| RL-D2 | TCGGGATGATTGTGGCTCC | S044 | 42225-42207 | AF261825.2 | This study |
| 73QS-R1 | GTGGCAGCAACATCCTTTGG | aadA7 | 2843-2824 | AY463797.2 | This study |
| RH365 | CGATATGCACGCTCACC | Tn501mer | 3090-3074 | Z00027 | This study |
| RH383 | CTTCGTGAAATCAGCCCAG | Tn501mer | 384-366 | Z00027 | This study |
| RH389 | GTGCCGTCCAAGATCATG | Tn501mer | 1753-1770 | Z00027 | This study |
| RH444 | GACTCGCCGATGTTTCCACG | Tn501mer | 4303-4284 | Z00027 | This study |
| RH445 | GAACGCCTTGCCGTAATCG | Tn501mer | 2140-2158 | Z00027 | This study |
| RH446 | CTTACTGCGGTCAATCGTAGG | Tn501mer | 1860-1840 | Z00027 | This study |
| RH447 | CACACCAACTCAGACAGCACG | Tn501mer | 205-225 | Z00027 | This study |
| RH448 | CGGTGTTGGAACCCTATCG | Tn501mer | 753-771 | Z00027 | This study |
| CV-D1 | CAGCCGCAGTTCGTCTATG | Tn501mer | 2548-2566 | Z00027 | This studyc |
| CV-D2 | TCGTCAGGTAGGGGAACAAC | Tn501mer | 2941-2922 | Z00027 | This studyc |
Reference numbers are for the primer.
The sequence from AF261825.2 is GAATCCGACAGCCAACGCTTCC (the C in bold is not present in the primer).
Primer designed by C. Venturini.
DNA sequencing and sequence analysis.
PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Inc., Valencia, CA) following protocols supplied by the manufacturer. Automated sequencing was carried out as described previously (18, 28). The sequence of overlapping PCR fragments was determined on both strands for all regions except for parts of the 5′-CS and 3′-CS, where only one strand was sequenced. Assembly and analysis of sequences were carried out as described previously (28).
Southern hybridization.
Whole-cell DNA was digested with various restriction enzymes, and the fragments were resolved by electrophoresis through 0.7% agarose. The DNA was transferred to N+ nylon membrane (Amersham, Buckinghamshire, United Kingdom) by capillary action and was cross-linked to the membrane by baking at 80°C for 2 h. The membrane was exposed to denatured digoxigenin-labeled probes overnight at 42°C and then washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate for 15 min at room temperature and twice more stringently with 2× SSC-0.1% sodium dodecyl sulfate for 15 min at 68°C. Digoxigenin-labeled molecular mass markers (molecular marker set II; Roche Diagnostics Corporation) were included on all membranes, and bands were visualized fluorescently using standard protocols supplied by the manufacturer (Roche Diagnostics Corporation). Probes for intI1, merA, S024, and S044 were generated by PCR using primers listed in Table 1. The S024 probe (primers RL-D3 and S024-RV) is equivalent to p1-9 (4, 5) and was used to confirm that the XbaI restriction fragments carrying S012 to S026 were present.
Nucleotide sequence accession number.
The 4.36 kb of nucleotide sequence from the SGI1-K integron and flanking regions reported in this paper has been added to GenBank accession no. AY463797.1 to give AY463797.3. The sequences of the left- and right-hand boundaries of SGI1 (the thdF-int and S044-yidY junctions) are also included in GenBank accession no. AY463797.3.
RESULTS
Serovar Kentucky SRC73 contains a variant of SGI1.
Published primers (listed in Table 1) were used to detect the presence of SGI1 and the integron within it in the serovar Kentucky SRC73 strain. Products of the expected size (500 bp) were obtained with primers U7-L12 (in thdF) with LJ-R1 (in SGI1) and 104-RJ (in SGI1 and S044) with 104-D (in yidY), indicating that the standard junctions between an SGI1-like region and the thdF and yidY genes on the chromosome are present. The “retron phage” insertion found in SGI1-containing Salmonella serovar Typhimurium strains (4) was not present. To confirm this, the PCR products were sequenced. The sequences flanking SGI1 were, with the exception of a few base changes, identical to a continuous region extending from within the thdF gene to within the yidY gene in genome sequences of serovars Typhi, Paratyphi A, and Choleraesuis (GenBank accession no. AL627280, AE014613, CP000026, and AE017220). However, a 100-bp segment found adjacent to the right-hand end of the SGI is not present in the DT104 sequence or in the S. enterica serovar Typhimurium LT2 genome sequence (GenBank accession no. AF261825 and AE008879).
Using primers that detect the boundaries of the integron with the SGI1 backbone, namely S026-FW (in S026) with int-RV (in intI1) and DB-T1 (in IS6100) with MDR-B (in S044), only the S026-FW with int-RV product was obtained, indicating that the left end of an integron is at the normal position within the SGI1 backbone but that the right-hand end is not. The sequence of the S026-FW with int-RV product confirmed that the IRi end of the integron is in precisely the same position as in SGI1 (see GenBank accession no. AF261825).
The fact that a single 1.6-kb PCR product containing the aacCA5-aadA7 cassette array was obtained previously with primers L1 and R1 in the integron 5′-CS and 3′-CS (19) indicates that only one attI1 site is present in SRC73. Consistent with this conclusion, the tet(G) or floR genes that are found between the two attI1 sites in SGI1 were not detected using PCR primers within each gene and neither of the cassettes found in SGI1, aadA2 and blaP1, was present (see Table 1 in reference 18 for primer details). Hence, SRC73 contains a variant form of SGI1. Linkage of the aacCA5-aadA7 cassette array to the SGI1 backbone was demonstrated by PCR, using a primer in aadA7 (73QS-R1) with one in the SGI1 backbone (S026-FW). The variant therefore differs from its closest relative, SGI1-H (GenBank accession no. AY458224), found in serovar Newport (14), which contains two attI1 sites with the aacCA5-aadA7 cassette array replacing the aadA2 cassette of SGI1 (Fig. 1). Using the next available letter, the SGI1 variant in Salmonella serovar Kentucky was named SGI1-K.
Structure of the SGI1-K integron.
The complete sequence of the In104-K integron backbone (from IRi to IRt) was determined using overlapping PCR products as substrates (Fig. 1). The 5′-CS of In104-K contains the strong version of the Pc promoter [TTGACA(17)TAAACT], which drives transcription of the cassette genes (10), whereas both 5′-CS regions in the original SGI1 (GenBank accession no. AF261825) have Pc weak [TGGACA(17)TAAGCT], and a G residue replaces the more usual C at the 16th position of the 17-bp Pc spacer. The left-hand 5′-CS region in SGI1-H (AY458224), which has the same cassette array as SGI1-K, also has the strong version of Pc. This is consistent with a model in which, during formation of SGI1-H and SGI1-K, the new cassette array came into the integron within SGI1 by homologous recombination, bringing with it the Pc region and simultaneously removing the aadA2 cassette and associated Pc. However, SGI1-K has also lost the central region of the integron containing the floR and tet(G) resistance determinants together with the right-hand attI1 site and blaP1 cassette found in SGI1 and SGI1-H (Fig. 1).
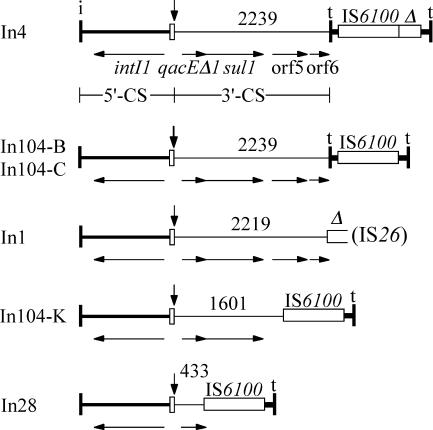
The primer 73L2-F1, situated in the aacCA5 gene, or sul1-F or orf5-F, in the 3′-CS, together with primer IS1600-Rv2 in IS6100 yielded PCR products, indicating that IS6100 is adjacent to the 3′-CS. However, the estimated sizes of the products (3.5 kb, 2.0 kb, and 1.0 kb, respectively) were all about 0.7 kb smaller than predicted for the standard In4-type integron configuration (26), which is present at the right-hand end of In104 in SGI1 (Fig. 1) and SGI1-A to SGI1-J. The sequence of the orf5-F/IS6100-Rv2 product revealed that only nucleotides 1 to 1601 of the standard 3′-CS, which includes the complete sul1 gene, immediately precede IS6100 (Fig. 2). Hence, 638 bp of the 3′-CS, which is 2,239 bp in In4 and In104, and the 123 bp of tni sequence (including IRt) found in inverse orientation between IS6100 and the 3′-CS of In4 and In104 have been lost. This indicates that an IS6100-mediated deletion event has occurred (Fig. 2), and similar deletions that remove the inverted tni end fragment as well as different lengths of the 3′-CS are found in other IS6100-containing class 1 integrons (29). There is also a single base difference (T218C) in the IS6100 of SGI1-K in SRC73 that is not seen in In104 from SGI1 or in any other example of IS6100 currently in GenBank.
FIG. 2.
Backbone structures of In4-type class 1 integrons and derivatives with IS6100-mediated deletions. Symbols are as in Fig. 1, and the vertical arrow marks the position at which the cassettes (not shown) are inserted. The extents of the 3′-CS are indicated above by the position of the last base present. Partial copies of IS6100 are shown as open boxes labeled Δ. The extents of genes are shown by horizontal arrows and are named under In4 only. The sources for the sequences used are as follows: In4, GenBank accession no. U12338; In104-B and In104-C, which were deduced from SGI1, AF261825; In1, AY046276; and In28, AF313472. In104-K is from this study (AY463797.3).
Though PCR products that were expected to include the IRt end of the integron (using primer DB-T1 with either MDR-B or 104-RJ) were not obtained, the sequence of the adjacent region was obtained fortuitously. In initial PCR mapping, the orf5-F primer paired with orf5-R2 gave a product, which was unexpected granted that the 3′-CS region in SGI1-K does not include the orf5-R2 primer site. However, the size of this product (1.6 kb) was larger than the expected product. Furthermore, a product of the same size was generated when the orf5-F primer was used with MDR-B. The sequence of the product obtained with orf5-F/MDR-B indicated that it was in fact generated by priming of orf5-F (AGGTTGTGCGGCTGATGC) at the correct position in the 3′-CS and again fortuitously in a sequence beyond the IRt end of the integron (see below). The 152-bp segment from the IRt end of class 1 integrons that is normally located to the right of IS6100 in In4 and In4-type integrons (26, 29) was present (Fig. 1 and 2), but it was not located adjacent to S044 as it is in SGI1 (Fig. 1) and most of the known SGI1 variants.
Context of the IRt end of the integron.
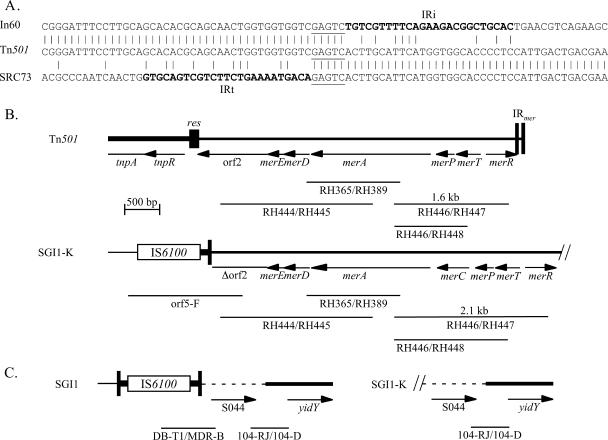
The sequence beyond IRt matched part of the Tn501 mercury resistance (mer) module, and the fortuitous priming site for orf5-F was identified as AGGgcGTGCtGgCTGATGC (mismatched bases in lowercase), which lies within the open reading frame orf2 in the mer module of Tn501 (7) and Tn5051 (23). IRt of the SRC73 integron abuts base 3925 of Tn501 (counting from the IRtnp end; 3925 corresponds to position 4430 in GenBank accession no. Z00027) (Fig. 3). This point is 191 bp from the resolution crossover position in the resI subsite of the res site. Three class 1 integrons for which sequence adjacent to the IRi end has recently been reported, AF355189 (15), AJ515707 (34), and AJ550807 and AJ628135 (24), appear to have been inserted at the same position in the Tn501 mer module. Though only the context of the IRi end of these integrons is available, IRi abuts position 3930 (from the IRtnp end) of Tn501 (or 196 bp from between A and T in the resI subsite), as expected when the 5-bp duplication formed during transposition is taken into account (Fig. 3A). However, the transposon within which they reside is more closely related to Tn5051 (GenBank accession no. Y177190) (23) which includes the same mer module as Tn501 with a different, but related, transposition (tnp) module.
FIG. 3.
Location of In104-K. (A) The sequence of the IRt boundary in SGI1-K is aligned with the sequences of Tn501 from GenBank accession no. Z00027 and the IRi boundary of integrons in AF355189 (In60), AJ515707, and AJ550807. The IRi and IRt regions are in boldface type, and the 5-bp duplication is underlined. (B) Structure of the adjacent mer segment. The map of the mer region of Tn501 (and Tn5051) is derived from Z00027, with the positions of specific PCR fragments indicated below. The SGI1-K map is derived from AY463797.3, the Tn501 mer sequence, and the Tn21 mer sequence. Features are as described in the legend to Fig. 1.
Extent of the mer region.
To examine if the complete Tn501 mer module is present in SGI1-K, further PCR mapping using pairs of primers either specific for Tn501 mer or that will detect Tn501 mer and other related mer modules (Table 1 and Fig. 3B) was carried out. The plasmid pVS1, which is the original source of Tn501 (33), was used as a positive control. With whole-cell DNA from SRC73 as a substrate, PCR products of the predicted size were obtained with RH444/RH445 and with RH365/RH389 (Fig. 3B), indicating that the mer region in SGI1-K extends to at least partway through the merA gene. However, a primer within this amplified region (RH446) paired with a primer in the merR gene (RH447) or a primer in the merT gene (RH448) yielded products 0.5 kb larger than the predicted size for Tn501 mer, suggesting that there is an insertion of approximately 0.5 kb within this segment. The sizes of these PCR fragments are consistent with the presence of a mer region containing the merC gene between merA and merP (20). The sequence of the merC region was identical to that of Tn21, and the mer module is a hybrid with the merD and -E genes and most of the merA gene derived from Tn501 and the merC, -T, and -R genes derived from Tn21 (GenBank accession no. AF071413). Consistent with the presence of a complete mer module, SRC73 was found to be resistant to mercuric chloride (20 μg/ml).
The right-hand end of SGI1-K.
One possible explanation for the configuration found in SGI1-K is that a plasmid (or nonreplicating circular molecule) containing a class 1 integron in the Tn501 or Tn5051 context has become incorporated into the SGI via homologous recombination between regions of identity (e.g., 5′-CS, 3′-CS, or IS6100) in the two integrons. However, if this were the case, a second integron should be present adjacent to S044. In SGI1 and most of its variants, the region between IRt of the integron and the right-hand end of SGI1 is 880 to 898 bp long and includes only a single gene, S044 (see Fig. 1). At least part of this region is present in SGI1-K, as the priming site for 104-RJ, which is located within S044, is present (Fig. 3C). However, a copy of IS6100 is not found near this end of SGI1 as the primer DB-T1 (in IS6100) paired either with MDR-B (in S044) or with 104-D (in yidY) did not yield products (Fig. 3C). This indicates that the final configuration of SGI1-K did not arise simply as the result of a single recombination event between identical regions (e.g., 5′-CS) in the integron in SGI and a plasmid containing a class 1 integron in the Tn5051 context, which would leave the IRt-S044 boundary intact. Hence, if the additional mer-containing segment entered in this way, further events must have occurred. Attempts to establish linkage between the mer module and S044 using long-range PCR were unsuccessful. Southern blots of SRC73 genomic DNA, digested with a variety of restriction enzymes, revealed an XbaI fragment of over 23 kb that hybridized to probes for intI1, merA, and S044. Assuming that the ends of this fragment are the XbaI sites in tnpR and S044 (see Fig. 1 for positions of these genes), a minimum size of 13 kb can be calculated for the region between the right-hand end of the mer region and S044.
DISCUSSION
SRC73, an S. enterica serovar Kentucky strain isolated in Australia in 2001 from a spice imported from India (19), represents the 13th Salmonella serovar that has been found to include an SGI1-type genomic island carrying multiple antibiotic resistance determinants (see reference 18). Though others have reported sequencing the boundaries between SGI1 and the chromosome for serovars Agona, Paratyphi B, and Albany (4, 11, 13), the sequences have not been released, nor were any differences between serovar Typhimurium and other serovars in the intergenic region between thdF and yidY mentioned. The sequence reported here for the chromosomal segments adjacent to the site of integration of SGI1-K matches those of serovars Typhi, Paratyphi A, and Choleraesuis in this region and SGI1-K is inserted within the end of the thdF gene. The first 100 bp adjacent to the right-hand end of SGI1-K is missing from the sequence of the S. enterica serovar Typhimurium genome (strain LT2) and from the region flanking SGI1 in the DT104 strain. It is replaced by the so-called “retron phage,” which lies adjacent to the end of thdF in LT2 or at the right-hand end of SGI1 in DT104 (5).
SGI1-K, the SGI1 variant found in SRC73, has undergone a number of changes relative to SGI1. It is related to the SGI1-H variant found in S. enterica serovar Newport (14) because both include the aacCA5-aadA7 cassette array together with a Pc promoter different from that found in most SGI1 variants. In both cases, the cassette array is likely to be derived from another DNA molecule—e.g., a plasmid, integrating element or other nonreplicating circle present in the same cell—with these cassettes having been gained by the SGI1 variants via homologous recombination within both CS (i.e., 5′-CS and 3′-CS) flanking cassettes in class 1 integrons (25, 27). An alternative is that for SGI1-K, a single crossover in the 5′-CS incorporated the complete extra molecule, which included a class 1 integron in the Tn501/Tn5051 context with subsequent events yielding the final configuration. The Salmonella serovar Newport strain containing the aacCA5-aadA7 cassette array in SGI1-H was isolated in France in 2001 from a patient recently returned from Egypt (14). The same cassette array has also been found in Vibrio fluvialis strains isolated in 2002 from patients with cholera in Calcutta, India (GenBank accession no. AB114632 and AY605484) (1, 32). It is also present in a Salmonella serovar Haifa strain isolated in Spain from a patient with traveler's diarrhea who had recently returned from Egypt (AY563051) (8) and serovar Kentucky isolates from Slovakia (AM039633). Thus, the aacCA5-aadA7 cassette array, which was only identified recently, has already been found in several countries and multiple species, demonstrating how readily cassette arrays containing new resistance genes can spread.
At least five further changes have clearly occurred in the course of the creation of SGI1-K from SGI1. First, the central region of the complex integron found in SGI1 including the right-hand attI1 site has been lost. This event is likely to have occurred by homologous recombination between the right- and left-hand copies of either the 5′-CS or the 3′-CS, but if SGI1-K is a derivative of SGI1-H, the recombination must have occurred in the 3′-CS. Second, an IS6100-mediated deletion has removed part of the 3′-CS. This event could have occurred either within the SGI or previously in the incoming Tn501 (Tn5051)-associated integron. Third, a mer module has become incorporated into the SGI. This change could have occurred simultaneously with the cassette exchange if only a single crossover were involved in that step. Fourth, the mer module, which is a hybrid between Tn501/Tn5051 mer and the merC-containing mer module from Tn21, has probably arisen by homologous recombination (23). The sequence switches from identity with Tn501 to identity with Tn21 in a short stretch of sequence identity near the beginning of the merA gene. Finally, the boundary between the IRt end of the integron and the SGI1 backbone has been disrupted. To explain the presence of only one integron and the absence of the standard boundary seen at the right-hand end of the integron in SGI1 and other SGI1 variants, it is necessary to invoke at least one further step, probably a deletion, that occurred after the postulated homologous recombination event incorporated a plasmid or nonreplication circular molecule into the SGI. However, further analysis of the large region (over 13 kb) between the mer module and S044 will be needed to resolve this. The locations of the remaining resistance genes known to be present in SRC73, namely blaTEM, strAB, and tet(A), also remain to be established, as plasmids were not detected using standard extraction procedures (19).
Acknowledgments
R.L. was supported in part by grants from the NSW Department of Primary Industries, the University of Wollongong, and the McGarvie Smith Trust and grant no. 402584 from the Australian National Health and Medical Research Council. S.R.P. was supported by grant no. 192108 from the NHMRC.
We thank Linda Falconer for technical assistance.
Footnotes
Published ahead of print on 6 November 2006.
REFERENCES
- 1.Ahmed, A. M., T. Nakagawa, E. Arakawa, T. Ramamurthy, S. Shinoda, and T. Shimamoto. 2004. New aminoglycoside acetyltransferase gene, aac(3)-Id, in a class 1 integron from a multiresistant strain of Vibrio fluvialis isolated from an infant aged 6 months. J. Antimicrob. Chemother. 53:947-951. [DOI] [PubMed] [Google Scholar]
- 2.Arcangioli, M. A., S. Leroy-Sétrin, J. L. Martel, and E. Chaslus-Dancla. 1999. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol. Lett. 174:327-332. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D. A., G. A. Peters, L. K. Ng, and M. R. Mulvey. 2000. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhymurium DT104. FEMS Microbiol. Lett. 189:285-291. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, N. L., T. K. Misra, J. N. Winnie, A. Schmidt, M. Seiff, and S. Silver. 1986. The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol. Gen. Genet. 202:143-151. [DOI] [PubMed] [Google Scholar]
- 8.Cabrera, R., J. Ruiz, F. Marco, I. Oliveira, M. Arroyo, A. Aladueña, M. A. Usera, M. T. Jiménez De Anta, J. Gascón, and J. Vila. 2004. Mechanism of resistance to several antimicrobial agents in Salmonella clinical isolates causing traveler's diarrhea. Antimicrob. Agents Chemother. 48:3934-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carattoli, A., E. Filetici, L. Villa, A. M. Dionisi, A. Ricci, and I. Luzzi. 2002. Antibiotic resistance genes and Salmonella genomic island 1 in Salmonella enterica serovar Typhimurium isolated in Italy. Antimicrob. Agents Chemother. 46:2821-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doublet, B., D. Boyd, M. R. Mulvey, and A. Cloeckaert. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55:1911-1924. [DOI] [PubMed] [Google Scholar]
- 12.Doublet, B., P. Butaye, H. Imberechts, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Salmonella genomic island 1 multidrug resistance gene clusters in Salmonella enterica serovar Agona isolated in Belgium in 1992 to 2002. Antimicrob. Agents Chemother. 48:2510-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doublet, B., R. Lailler, D. Meunier, A. Brisabois, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2003. Variant Salmonella genomic island 1 antibiotic resistance gene cluster in Salmonella enterica serovar Albany. Emerg. Infect. Dis. 9:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doublet, B., F.-X. Weill, L. Fabre, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 48:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubois, V., L. Poirel, C. Marie, C. Arpin, P. Nordmann, and C. Quentin. 2002. Molecular characterization of a novel class 1 integron containing blaGES-1 and a fused product of aac(3)-Ib/aac(6′)-Ib′ gene cassettes in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebner, P., K. Garner, and A. Mathew. 2004. Class 1 integrons in various Salmonella enterica serovars isolated from animals and identification of genomic island SGI1 in Salmonella enterica var. Meleagridis. J. Antimicrob. Chemother. 53:1004-1009. [DOI] [PubMed] [Google Scholar]
- 17.Leverstein-van Hall, M. A., A. Paauw, A.T.A. Box, H. E. M. Blok, J. Verhoef, and A. C. Fluit. 2002. Presence of integron-associated resistance in the community is widespread and contributes to multidrug resistance in the hospital. J. Clin. Microbiol. 40:3038-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levings, R. S., D. Lightfoot, S. R. Partridge, R. M. Hall, and S. P. Djordjevic. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levings, R. S., S. R. Partridge, D. Lightfoot, R. M. Hall, and S. P. Djordjevic. 2005. New integron-associated gene cassette encoding a 3-N-aminoglycoside acetyltransferase. Antimicrob. Agents Chemother. 49:1238-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maguire, A. J., D. F. Brown, J. J. Gray, and U. Desselberger. 2001. Rapid screening technique for class 1 integrons in Enterobacteriaceae and nonfermenting gram-negative bacteria and its use in molecular epidemiology. Antimicrob. Agents Chemother. 45:1022-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meunier, D., D. Boyd, M. R. Mulvey, S. Baucheron, C. Mammina, A. Nastasi, E. Chaslus-Dancla, and A. Cloeckaert. 2002. Salmonella enterica serotype Typhimurium DT 104 antibiotic resistance genomic island I in serotype Paratyphi B. Emerg. Infect. Dis. 8:430-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mindlin, S., G. Kholodii, Z. Gorlenko, S. Minakhina, L. Minakhin, E. Kalyaeva, A. Kopteva, M. Petrova, O. Yurieva, and V. Nikiforov. 2001. Mercury resistance transposons of Gram-negative environmental bacteria and their classification. Res. Microbiol. 152:811-822. [DOI] [PubMed] [Google Scholar]
- 24.Pagani, L., C. Colinon, R. Migliavacca, M. Labonia, J.-D. Docquier, E. Nucleo, M. Spalla, M. L. Bergoli, and G. M. Rossolini. 2006. Nosocomial outbreak caused by multidrug-resistant Pseudomonas aeruginosa producing IMP-13 metallo-β-lactamase. J. Clin. Microbiol. 43:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Partridge, S. R., H. J. Brown, and R. M. Hall. 2002. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrob. Agents Chemother. 46:1288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Partridge, S. R., C. M. Collis, and R. M. Hall. 2002. Class 1 integron containing a new gene cassette, aadA10, associated with Tn1404 from R151. Antimicrob. Agents Chemother. 46:2400-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sallen, B., A. Rajoharison, S. Desvarenne, and C. Mabilat. 1995. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of Enterobacteriaceae. Microb. Drug Resist. 1:195-202. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Srinivasan, V. B., R. K. Virk, A. Kaundal, R. Chakraborty, B. Datta, T. Ramamurthy, A. K. Mukhopadhyay, and A. Ghosh. 2006. Mechanism of drug resistance in clonally related clinical isolates of Vibrio fluvialis isolated in Kolkata, India. Antimicrob. Agents Chemother. 50:2428-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanisich, V. A., P. M. Bennett, and M. H. Richmond. 1977. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J. Bacteriol. 129:1227-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh, T. R., M. A. Toleman, W. Hryniewicz, P. M. Bennett, and R. N. Jones. 2003. Evolution of an integron carrying blaVIM-2 in Eastern Europe: report from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 52:116-119. [DOI] [PubMed] [Google Scholar]