Abstract
A screen for antifungal compounds from Lysobacter enzymogenes strain C3, a bacterial biological control agent of fungal diseases, has previously led to the isolation of heat-stable antifungal factor (HSAF). HSAF exhibits inhibitory activities against a wide range of fungal species and shows a novel mode of antifungal action by disrupting the biosynthesis of a distinct group of sphingolipids. We have now determined the chemical structure of HSAF, which is identical to that of dihydromaltophilin, an antifungal metabolite with a unique macrocyclic lactam system containing a tetramic acid moiety and a 5,5,6-tricyclic skeleton. We have also identified the genetic locus responsible for the biosynthesis of HSAF in strain C3. DNA sequencing of this locus revealed genes for a hybrid polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS), a sterol desaturase, a ferredoxin reductase, and an arginase. The disruption of the PKS-NRPS gene generated C3 mutants that lost the ability to produce HSAF and to inhibit fungal growth, demonstrating a hybrid PKS-NRPS that catalyzed the biosynthesis of the unique macrolactam system that is found in many biologically active natural products isolated from marine organisms. In addition, we have generated mutants with disrupted sterol desaturase, ferredoxin reductase, and arginase and examined the metabolites produced in these mutants. The work represents the first study of the genetic basis for the biosynthesis of the tetramic acid-containing macrolactams. The elucidation of the chemical structure of HSAF and the identification of the genetic locus for its biosynthesis establish the foundation for future exploitation of this group of compounds as new fungicides or antifungal drugs.
Heat-stable antifungal factor (HSAF) is a secondary metabolite produced by the bacterium Lysobacter enzymogenes strain C3 (originally called Stenotrophomonas maltophilia strain C3 [24]), a biological control agent originally isolated from grass foliage (7). Strain C3 was demonstrated in the field to reduce diseases caused by multiple fungal pathogens, including Bipolaris sorokiniana (28), Fusarium graminearum (26), Rhizoctonia solani (7), and Uromyces appendiculatus (27). It was also found to be effective in inhibiting the soilborne pathogens Pythium ultimum (12) and Magnaporthe poae (13) in greenhouse experiments. A search for antifungal factors in strain C3 led to the isolation of HSAF, which exhibits strong activity against a wide range of fungi (15).
The mechanism by which HSAF inhibits the growth of Aspergillus nidulans has been studied (16). HSAF disrupts the polarized growth of the fungus. Genetic analysis of A. nidulans mutants suggests that HSAF targets the biosynthesis of sphingolipids (16), which are ubiquitous components of eukaryotic cell membranes and signaling molecules involved in numerous cellular processes. Interestingly, HSAF appears to target a distinct group of sphingolipids that are required for polarized growth of filamentous fungi and appears to be absent from mammals and plants. Therefore, the antifungal mechanism of HSAF represents an unprecedented mode of action for an antifungal metabolite, and thus, the study of HSAF could lead to the development of novel fungicides or antifungal drugs.
The objectives of this study were to determine the chemical structure of HSAF and to identify the genetic locus in strain C3 that is required for the production of HSAF. The outcomes of the study will provide important knowledge for future application of HSAF and possible genetic engineering of the biosynthetic pathway to improve the production of HSAF or to produce new analogs.
MATERIALS AND METHODS
Bacterial and fungal strains, plasmids, and general DNA manipulations.
L. enzymogenes strain C3 and other bacterial strains were grown in Luria-Bertani (LB) broth medium or 1/10-strength tryptic soy broth (1/10 TSB; Sigma) at 28°C with shaking. Escherichia coli strain DH5α was used as the host for general DNA propagations. E. coli S17-1 was obtained from D. Y. Kobayashi (Rutgers University, New Brunswick, NJ). The plasmids used for cloning and sequencing included the pGEM-zf series from Promega (Madison, WI) and pANT841 (21). Plasmid preparation and DNA gel extraction were carried out with commercial kits (QIAGEN, Valencia, CA), and all other manipulations were performed by standard methods (22). The genomic DNA of L. enzymogenes strain C3 was prepared as described previously (12).
Cloning of genes for HSAF biosynthesis.
Genomic DNA was prepared from a polyketide synthase (PKS)-disrupted C3 mutant (see below). Approximately 10 μg of the DNA was digested with 50 units of PstI in a total volume of 50 μl. The PstI-digested DNA was precipitated by ethanol and resuspended in 20 μl H2O. The ligation reaction was performed by mixing 8 μl of the digested DNA with 1 μl of ligation buffer and 1 μl of T4 DNA ligase (5 units; Promega), and the mixture was incubated at 15°C for 24 h. An aliquot of 5 μl of the ligation mixture was used to transform E. coli DH5α, and the cells were spread onto LB plates containing 20 μg/ml gentamicin. Plasmid DNA was prepared from the developed colonies and screened by PCR assays with primers P1 and P2 (see below and Fig. 2). One of the rescued plasmids, pJQ200SK-5.3KB, which harbors a 5.3-kb DNA fragment, was sequenced. Similarly, the genomic DNA was digested with BamHI, SalI, or SacI; and individual plasmids were rescued upon self-ligation. The sequencing of these plasmids revealed an approximately 13.5-kb DNA region.
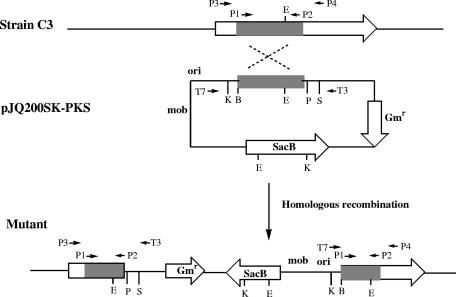
FIG. 2.
Scheme for PKS gene disruption in L. enzymogenes strain C3. A 407-bp fragment of the gene (shaded bar) was amplified from the genomic DNA and cloned into a gene disruption vector to produce pJQ200SK-PKS. PCR primers (primers P1, P2, P3, P4, T3, and T7) and their relative positions are shown as small arrows. B, BamHI; E, EcoRI; K, KpnI; P, PstI; S, SacI; Gmr, gentamicin resistance gene.
Construction of gene disruption vectors.
To construct the PKS disruption vector, two PCR primers, primers P1 (CCGCGGAAGTCGAAGCAGTA) and P2 (AAGGACTGCCTCACCGACAC), were designed on the basis of the preliminary data reported in Folman's Ph.D. dissertation (6). Folman obtained an 818-bp DNA fragment from a minitransposon (mini-Tn5 Cm) mutant of L. enzymogenes strain 3.1T8. The transposon insertion led to the loss of antifungal activity of strain 3.1T8. On the basis of this information, a 407-bp fragment was amplified by PCR from the genomic DNA of strain C3. The fragment was cloned into T vector (Promega) to confirm its identity by DNA sequencing and was then transferred to conjugation vector pJQ200SK (20) as an ApaI/SacI fragment to produce pJQ200SK-PKS (see Fig. 2). To construct the nonribosomal peptide synthetase (NRPS) module disruption vector, a 482-bp fragment was amplified from genomic DNA of strain C3 by PCR with primer NRPS1-F (GTTGAGCTGCTTGAGGAACC) and primer NRPS1-R (GATCTTCATGGCTTGGTACTCC). This fragment was cloned into the pGEM-T vector. The PCR product was released from the T vector by ApaI and SacI digestions and cloned into the same sites of pJQ200SK to produce pJQ200SK-NRPS. To disrupt the ferredoxin reductase (FNR) gene, a 373-bp internal fragment from the predicted gene (the Ferr fragment) was released by BamHI and SacI digestion from a subclone, pGEM3zf-3.0Kb. This fragment was cloned into the same sites of pJQ200SK to produce gene disruption construct pJQ200SK-Ferr. To disrupt the sterol desaturase gene, a 797-bp internal fragment from the putative gene (the SD fragment) was released by EagI digestion from a subclone, pGEM3zf-3.0Kb. This fragment was cloned into the NotI site of pANT841 to produce pANT-SD. The sterol desaturase fragment was released by SpeI and XhoI digestions and was cloned into the same sites of pJQ200SK to produce the sterol desaturase gene disruption construct pJQ200SK-SD. To disrupt the arginase gene, a 396-bp internal fragment from the predicted gene (the ARG fragment) was released by ApaI digestion from a subclone, pGEM3zf-2.3k. This fragment was cloned to the same site on pJQ200SK to produce the arginase disruption construct pJQ200SK-ARG.
Conjugal transfer of pJQ200SK constructs from E. coli S17-1 to L. enzymogenes strain C3.
pJQ200SK constructs were first introduced into E. coli S17-1 and then conjugally transferred from strain S17-1 to strain C3. To conjugally transfer the construct, a single colony of the S17-1 transformant was incubated in liquid LB medium supplemented with 20 μg/ml gentamicin at 37°C overnight. Strain C3 was also grown in liquid LB medium supplemented with 25 μg/ml kanamycin at 28°C for 48 h. Before conjugation, a fresh culture was initiated from each of the two seed cultures in liquid LB and incubated until the cell density reached an optical density at 595 nm of 0.7. A 1-ml aliquot of the cell suspension was taken from each culture, and the cells were collected by centrifugation at 3,000 × g for 3 min. The cells were suspended in 0.5 ml of 10 mM MgSO4, and the two cell suspensions were mixed. The mixture was spread onto five LB plates, and the plates were incubated at 28°C for 4 h. The cells were washed off the plates with 20 ml of 10 mM MgSO4, and 0.5 ml of the cells washed off of the plates was spread onto LB plates supplemented with 20 μg/ml kanamycin and gentamicin. The plates were incubated at 28°C for 2 days for selection. The colonies that developed on the selection medium were candidates of C3 gene disruption mutants.
Screening for gene disruption mutants.
To screen the gene-disrupted C3 mutants, genomic DNA was prepared from LB cultures of the individual candidates. For the PKS-disrupted mutants, primer P3 (GGTGTCGATCGACAGGCT) and primer P4 (AGCGACTACCGCGCGTTC), as well as universal primer T3 and primer T7 on vector pJQ200SK-PKS, were used for PCR. Primers P3 and P4 anneal to the flanking regions of the 407-bp fragment used in the homologous recombination (see Fig. 2). Thus, only the mutants that result from a homologous recombination produce a 453-bp PCR fragment when primers P3 and T3 are used and a 522-bp fragment when primers T7 and P4 are used. The wild type or mutants that resulted from random insertion did not produce any of the PCR fragments. To identify the NPRS-disrupted mutants, PCR was carried out with primer N1 (ACGCCAAGCTCGAAATTAAC) and primer N2 (ATCGAGCATTACCGCAACAG). The expected size of the PCR product is 695 bp, and this product produces two fragments of 225 bp and 470 bp upon SphI digestion. To identify the ferredoxin reductase gene-disrupted mutants, PCR was carried out with primer Ferr-1 (ACGACAGCCTTTTCAGCTTC) and primer T7. The expected size of the PCR product is 661 bp, and this product produces two fragments of 209 bp and 452 bp upon SacI digestion and two fragments of 272 bp and 389 bp upon SmaI digestion. To identify the sterol desaturase gene-disrupted mutants, PCR was performed with primer T7 and primer S5 (TGAACGGATTATCAGAGCAC). The expected product is a 957-bp fragment that produces three fragments of 670 bp, 171 bp, and 116 bp upon SphI digestion. To identify the arginase gene-disrupted mutants, PCR was conducted with primer T7 and primer Arg1 (GTTCGCATCCACCTCTTTTTC). The expected size is 606 bp, and this product produces two fragments of 294 bp and 312 bp upon SalI digestion.
Isolation and HPLC analysis of metabolites from strain C3 and mutants.
Strain C3 and individual mutant strains were cultured in 1/10 TSB (supplemented with 20 μg/ml gentamicin for mutant strains) at 28°C in a rotary shaker at 180 rpm for 4 days. The supernatant was collected from each culture by centrifugation at 10,000 × g at 4°C for 30 min. To analyze the metabolites excreted into the culture, the supernatant (10 to 20 μl) was directly injected into a high-pressure liquid chromatograph. A high-pressure liquid chromatography (HPLC) system (ProStar, model 210; Varian, Walnut Creek, CA) with an Alltima C18LL column (250 mm by 4.6 mm [inner diameter]; 5 μm; Alltech, Deerfield, IL). The mobile phases were water-trifluoroacetic acid (TFA; 100:0.025; vol/vol) (mobile phase A) and acetonitrile-TFA (100:0.025; vol/vol) (mobile phase B) with a gradient of 0 to 40% mobile phase B in mobile phase A in the first 10 min, 40 to 80% mobile phase B in mobile phase A from 10 to 15 min, 80% mobile phase B in mobile phase A from 15 to 20 min, 80 to 100% mobile phase B in mobile phase A from 20 to 21 min, 100% mobile phase B in mobile phase A from 21 to 23 min, and 100 to 0% mobile phase B in mobile phase A from 23 to 25 min. The flow rate was 1.0 ml/min. The peaks were detected at 220 nm on a UV-visible detector (ProStar, model 310; Varian). To prepare HSAF and its analogs for structural analysis, ammonium sulfate was added to the supernatant of the culture, with constant stirring until the final concentration reached 0.5 g/ml. The solution was incubated at 4°C for 12 h. The precipitate was collected by centrifugation at 10,000 × g at 4°C for 25 min and was dissolved in 100% methanol. The methanol fraction containing HSAF or its analogs was centrifuged at 10,000 × g at 25°C for 10 min, and the resultant supernatant was collected and dried with a Rotavapor. The residues were redissolved in 10% acetonitrile, and the solution was loaded on a Sep-Pak Vac 35-ml (10-g) C18 cartridge (Waters, Milford, MA) previously conditioned with 200 ml of 100% acetonitrile, followed by 200 ml of 10% acetonitrile. The cartridge was sequentially washed with 100 ml of 10% acetonitrile and 100 ml of 20% acetonitrile, and HSAF was eluted with 100 ml of 30% acetonitrile. HSAF was further purified by preparative HPLC (250 mm by 10 mm [inner diameter]; 5-μm) column with the same system described above. The fractions containing HSAF were combined, and the final pure HSAF was obtained by crystallization in methanol.
HR-MS and NMR analysis of the metabolites.
The melting point was measured with a Thomas-Hoover capillary melting point apparatus and was uncorrected. High-resolution electron-spray ionization mass spectrometry (MS) of the purified HSAF was performed on an API Qtrap 4000 instrument. The samples were directly infused in the mass spectrometer and were analyzed in the positive ion mode by using a Turbo ion spray source. The nuclear magnetic resonance (NMR) spectra (1H NMR and 1H-1H correlation spectroscopy, obtained by using a standard pulse program) were recorded on a Varian Mercury-400BB instrument in CD3OD and were calibrated by using the solvent peak (δH 3.31) as an internal reference.
Biological assays for antifungal activity.
One milliliter of the culture broth of the mutants or the wild type was collected and centrifuged on a bench-top centrifuge at 16,000 × g for 10 min. The supernatant was transferred to a new tube and was boiled for 10 min in a water bath. The sample was centrifuged at 16,000 × g for 10 min, and the supernatant was transferred to another tube and used for biological assays. The slide glass method was used for the assay (16). On a slide glass, 1 μl of spores (105/ml) of B. sorokiniana, Fusarium graminearum, or Fusarium verticillioides was mixed with 9 μl of culture broth. The slide glass was put inside a 15-mm petri dish with two pieces of wet Whatman paper at the bottom and was incubated at 25°C for 12 h. The inhibitory effect of the culture broth on spore germination was observed under a microscope. In control experiments, 1 μl of the spores was mixed with 9 μl of culture medium (also supplemented with 20 μg/ml of gentamicin in the cases of the controls for the mutants) and was incubated similarly. For the bioassays of purified HSAF and its analogs, the preparations were dried in a Speed-Vac and redissolved in 100% methanol. The methanol solution was first applied onto a slide glass and air dried. One microliter of B. sorokiniana spores and 9 μl of culture medium were then added directly to the spot. In control experiments, methanol was applied to the slide and similarly air dried.
RESULTS
Chemical structure of HSAF.
Strain C3 produced no detectable HSAF in rich media, such as LB or full-strength TSB. However, the strain produced HSAF in nutritionally limited media, such as 10% TSB. Following extraction with ammonium sulfate and methanol (16), we could obtain approximately 30 to 60 μg HSAF from 1 ml culture. Purified HSAF exhibited potent inhibitory activities against many fungal pathogens. At a concentration of 0.3 to 1.5 μM, HSAF arrested spore germination in B. sorokiniana and caused reduced hyphal extension or profuse branching of F. graminearum and F. verticillioides (data not shown).
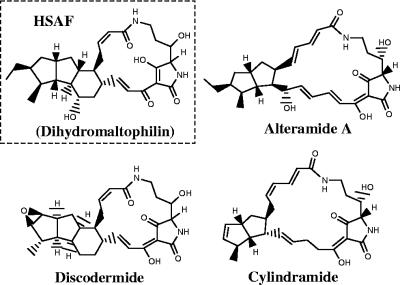
The purified HSAF was analyzed by mass spectrometry and one- and two-dimensional NMR. High-resolution electron-spray ionization mass spectrometry gave an m/z 513.2954 for [M(C29H40O6N2) + H]+ (calculated, 513.2959), which is 2 mass units heavier than the mass of the reference compound, maltophilin (purchased from EMC Microcollections GmbH, Tubingen, Germany) (Fig. 1). HSAF appeared as a colorless powder with a melting point of greater than 300°C when it was crystallized from methanol. 1H-NMR (in CD3OD) gave the following data: δ 8.02 (1H, br s, OH or NH, disappeared immediately after the addition of D2O or after the sample was kept standing for 2 days), 7.13 (1H, d, J = 13.2 Hz, H-10), 6.65 (1H, br dd, H-11), 6.05 (1H, br t, J = 10.0 Hz, H-25), 5.80 (1H, br d, J = 12.0 Hz, H-26), 3.97 (1H, br d, J = 6.0 Hz, H-4), 3.92 (1H, br s, H-5), 3.48 (2H, m, H-2a,24a), 3.36 (1H, m, H-14), 2.79 (1H, br t, J = 12.4 Hz, H-2b), 2.45 (1H, m, H-20), 2.08 to 2.19 (4H, m, H-12, 19a, 21a, 24b), 1.86 to 1.92 (2H, m, H-13a, 16), 1.72 (1H, m, H-22), 1.63 (2H, m, H-3a, 30a), 1.29 to 1.36 (5H, m, H-3b, 13b, 17, 18, 23), 1.23 (1H, q, J = 10.0 Hz H-15), 1.11 (3H, d, J = 6.4 Hz, H-29), 1.10 (1H, overlapping, H-30b), 0.96 (1H, m, H-21b), 0.90 (3H, t, J = 7.4 Hz, H-31), 0.86 (1H, overlapping, H-19b). These assignments, which were based on the results of the 1H-1H correlation spectroscopy experiment, are consistent with the reported values for dihydromaltophilin recorded in CD3OD-CDCl3 (3:1) (8). The coupling constants, which were well resolved for individual protons, are presented here, since the previous paper did not report these values. These data show that HSAF isolated from L. enzymogenes strain C3 has a chemical structure identical to that of dihydromaltophilin (Fig. 1), which was isolated from a Streptomyces species (8).
FIG. 1.
Structures of HSAF and several tetramic acid-containing macrolactams.
PKS-disrupted mutants lost the ability to produce HSAF and to inhibit fungal growth.
In order to identify the genetic locus required for the biosynthesis of HSAF in strain C3, we generated mutants through target-specific gene disruption by taking advantage of the partial sequence for a putative PKS gene isolated from L. enzymogenes strain 3.1T8 (6). Using two primers based on a region in the reported sequence, we amplified a 407-bp PCR product from the genomic DNA of strain C3. The DNA sequence of the fragment from strain C3 was 96.6% identical to that from strain 3.1T8. It also suggested that the 818-bp sequence reported in the earlier study likely contained sequence errors that resulted in frame shifts.
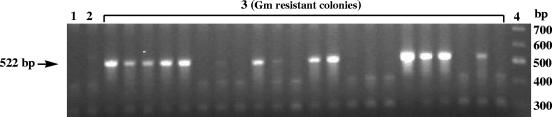
To determine if the putative PKS gene is involved in HSAF biosynthesis, we constructed a gene disruption vector, pJQ200SK-PKS, using the fragment amplified by PCR. Homologous recombination of the fragment with the corresponding region in the chromosome of C3 should generate PKS-disrupted mutants (Fig. 2). After mating of E. coli S17-1 carrying pJQ200SK-PKS with strain C3, 22 colonies developed on selection medium containing gentamicin and kanamycin. Among the colonies, 12 resulted from homologous recombination, as confirmed by PCR analysis. A 522-bp fragment was amplified from the genomic DNA prepared from the individual colonies by using primer T7 and primer P4 (Fig. 3), whereas a 453-bp fragment was obtained when primers P3 and T3 were used (data not shown). No PCR product was obtained from the wild type or the remaining 10 colonies, which presumably resulted from random insertion of the vector. The PCR products were sequenced, and the result confirmed that the colonies were PKS-disrupted mutants (Fig. 2).
FIG. 3.
Screening of the PKS gene-disrupted mutants by PCR assays. Genomic DNA was prepared from the gentamicin-resistant colonies and used as the template for PCR with primers P4 and T7 (Fig. 2). The expected size of the PCR product from homologous recombinants is 522 bp. Lane 1, control reaction without template; lane 2, wild type; middle unnumbered lanes, gentamicin (Gm)-resistant colonies; lane 4, 100-bp DNA ladder.
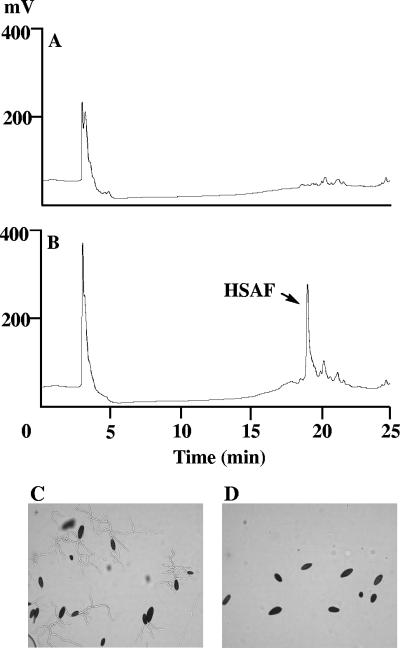
The PKS-disrupted mutants were examined for their antifungal activities and HSAF production. While the boiled culture broth of wild-type strain C3 completely inhibited spore germination in B. sorokiniana, the boiled culture broth of the PKS-disrupted mutants did not inhibit spore germination (Fig. 4). The supernatants from the mutant and the wild-type cultures were further analyzed by HPLC. The supernatant from the wild type exhibited a peak at 19.2 min that corresponded to HSAF, whereas the mutants did not produce HSAF (Fig. 4). The results demonstrate that the putative PKS gene is required for the biosynthesis of HSAF.
FIG. 4.
HPLC analysis and antifungal activity assay for the PKS-disrupted mutants. (A and B) HPLC traces of culture broths from mutant strain K19 (A) and wild type (B), with the peak of HSAF indicated; (C and D) activity assay for the culture broth from mutant strain K19 (C) and the wild type (D) by using Bipolaris sorokiniana as the test organism.
HSAF biosynthetic genes.
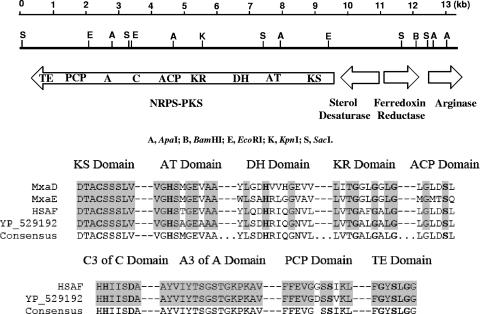
Following digestion of genomic DNA from the PKS-disrupted mutants with several restriction enzymes and rescue of the vector pJQ200SK by self-ligation, we sequenced the regions flanking the inserted vector. Sequences totaling about 13.5 kb were determined, and the analysis of the sequences indicated that this region contains four open reading frames: those for a hybrid polyketide synthase-nonribosomal peptide synthetase, a sterol desaturase, a ferredoxin reductase, and an arginase (Fig. 5). The hybrid PKS-NRPS contains nine domains: those for ketosynthase, acyltransferase, dehydratase, ketoreductase, acyl carrier protein, condensation, adenylation, peptidyl carrier protein, and thioesterase. All the highly conserved signature motifs within the domains were readily identifiable (Fig. 5). The PKS-NRPS contains 3,124 amino acid residues and is most similar (59% identity and 74% similarity from the start to the end) to the PKS-NRPS sequence from Saccharophagus degradans 2-40 (GenBank accession number YP_529192). The sequence also showed that the partial sequence of the putative PKS gene from strain 3.1T8 reported by Folman (6) is located at the ketosynthase domain of the hybrid PKS-NRPS.
FIG. 5.
Partial map of the 13.5-kb region of L. enzymogenes strain C3 hosting the HSAF biosynthetic genes. The predicted functions of the open reading frames are indicated. Restriction enzyme sites: A, ApaI; B, BamHI; E, EcoRI; K, KpnI; S, SacI. Domains of the hybrid PKS-NRPS: KS, ketosynthase; AT, acyltransferase; DH, dehydratase; KR, ketoreductase; ACP, acyl carrier protein; C, condensation; A, adenylation; PCP, peptidyl carrier protein; TE, thioesterase. The highly conserved signature motifs within the domains are shown, with the active-site residues highlighted in boldface letters. MxaD, PKS from Stigmatella aurantiaca (GenBank accession number AAK57188); MxaE, PKS from S. aurantiaca (GenBank accession number AAK57189); HSAF, the PKS-NRPS from L. enzymogenes C3; GenBank accession number YP_529192, PKS-NRPS sequence from Saccharophagus degradans 2-40 (GenBank accession number YP_529192).
Function of PKS-NRPS gene in HSAF synthesis.
To further confirm the role of the predicted hybrid PKS-NRPS in HSAF biosynthesis, we generated mutants with a disrupted NRPS module (at the C-domain region). A total of 32 putative NRPS-disrupted mutants were produced, 20 of which were confirmed by PCR assays with primer N1 and primer N2. A 695-bp fragment was amplified from the strains, and the fragment was cleaved by SphI to produce two fragments of 225 bp and 470 bp, as expected, from the mutants (data not shown). The PCR product was sequenced to further confirm the identities of the mutants. HPLC analysis of the culture broth of four NRPS-disrupted mutants showed that none of the mutants produced HSAF (Fig. 6B). Furthermore, none of the cultures had activity against B. sorokiniana in the spore germination assay (data not shown).
FIG. 6.
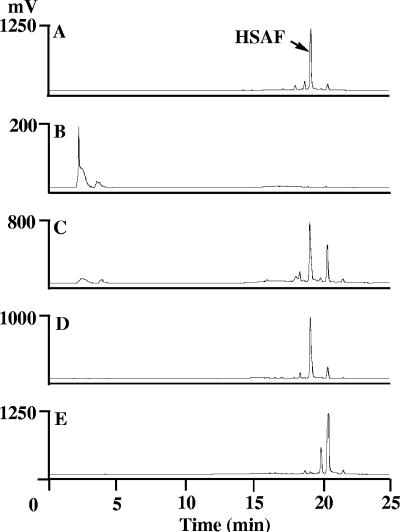
HPLC analysis of metabolites from mutants with disrupted gene for the NRPS module, ferredoxin reductase, arginase, and sterol desaturase. The results for crude extracts from wild-type strain C3 (A), NRPS-disrupted mutants (B), ferredoxin reductase-disrupted mutants (C), arginase-disrupted mutants (D) and sterol desaturase-disrupted mutants (E) are shown.
Functions of ferredoxin reductase, arginase, and sterol desaturase genes.
Three, 12, and 7 mutant strains with disruptions in the ferredoxin reductase, arginase, and sterol desaturase genes, respectively, were generated by mating of wild-type C3 with E. coli S17-1 carrying the respective pJQ200SK-gene construct. Insertion of the vector via homologous recombination into the target gene was confirmed in these strains by PCR analysis (data not shown). The mutants were evaluated for antifungal activities by the spore germination assay. Boiled culture broths from all of the mutant strains had inhibitory effects on the germination of B. sorokiniana spores similar to those of the broth from wild-type C3 strain and purified HSAF (data not shown). HPLC analysis of the broth from cultures of ferredoxin reductase- and arginase-disrupted mutants indicated that all of the mutants produced HSAF (retention time, 19.2 min), with the yields being similar to that of the wild type (Fig. 6C and D). This result was confirmed by liquid chromatography-MS, which showed that the compound produced by the mutants had the same molecular weight as HSAF (data not shown). Culture broth from the sterol desaturase-disrupted mutants did not contain HSAF but contained two dramatically increased metabolites (retention times, 19.9 and 20.4 min, respectively) (Fig. 6E). Activity tests showed that both metabolites were active in antifungal assays (data not shown). These two metabolites were also present in the mutants of ferredoxin reductase and arginase, as well as in the wild type, but at a much lower level. Interestingly, none of the PKS-NRPS-disrupted mutants produced these metabolites (Fig. 4B and Fig. 6B), indicating that these are probably precursors or shunt products of HSAF.
DISCUSSION
In this study, we identified the primary compound comprising the heat-stable antifungal activity in strain C3 to be dihydromaltophilin, an antibiotic first reported in a Streptomyces sp. (8) and an analog of xanthobaccins (17). Dihydromaltophilin belongs to a group of tetramic acid-containing macrolactam natural products, including the marine natural products cylindramide A (11), discodermide (9), and alteramide A (23) (Fig. 1). This group of metabolites has unique structural features and diverse biological activities, including anticancer, antibiotic, antiprotozoal, and antioxidant activities; and there has been extensive research interest in chemical total synthesis to prepare the scarce natural products (2-4, 10, 18). Our study represents the first attempt to investigate the molecular genetic basis for the biosynthesis of this group of natural products.
We were successful in identifying the genetic locus for HSAF biosynthesis in strain C3. The search was facilitated by the report of a putative PKS gene sequence in strain 3.1T8 (6). The high degree of sequence identity (96%) between the corresponding PKS gene regions in strains C3 and 3.1T8 and the fact that disruption of the PKS gene function in both strains resulted in the loss of antifungal activity suggest that strain 3.1T8 could also produce HSAF. Whether or not HSAF production is a common trait among strains of L. enzymogenes, however, is unknown. The absence of HSAF production in our PKS-NRPS-disrupted mutants demonstrates that the PKS-NRPS gene is required for the production of HSAF in strain C3. The result also implies that the biosynthesis of HSAF involves a hybrid polyketide-peptide mechanism.
Since the biosynthetic genes for secondary metabolites in microorganisms are generally found to cluster together at the same locus on a chromosome, we first used the PKS fragment as a probe to screen a genomic library based on the cosmid pLAFR3 (12) in our initial attempt to obtain the complete sequence for the PKS gene, as well as other genes required for HSAF biosynthesis. However, an extensive search of the library did not produce any positive clones, while the control experiments with a known ABC transporter as a probe did successfully identify three positive clones from the same library (data not shown). Subsequently, we constructed a new genomic library based on the cosmid SuperCos 1 (Stratagene). Again, using the PKS fragment as a probe, we isolated four positive clones from the new library, but none of the cosmid clones was stable in the host. The insert DNA in the cosmid vector was found to be either partially lost or rearranged during the regeneration and subcloning process. These results indicate that the HSAF biosynthetic genes might be toxic when they are expressed in the host. Since the direct screening of the genomic libraries did not lead to the identification of the biosynthetic genes, we took an alternative approach. We used the genomic DNA isolated from the gene-disrupted mutants to rescue plasmids that contained the homologous region as well as the flanking regions. The sequencing of the flanking regions eventually led to the identification of a gene cluster encoding a hybrid PKS-NRPS, a sterol desaturase, a ferredoxin reductase, and an arginase.
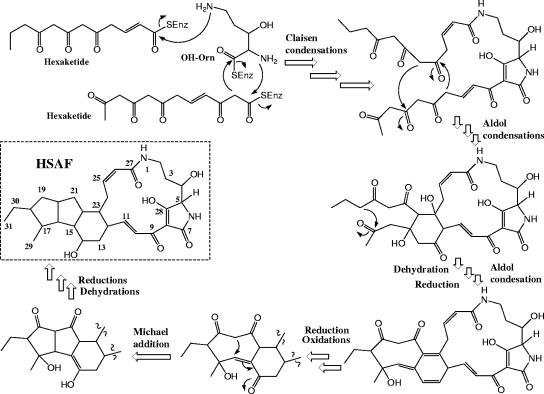
The chemical structure of HSAF suggests that an ornithine residue is incorporated into the lactam functionalities and that the biosynthesis of HSAF could involve both polyketide and nonribosomal peptide mechanisms, as seen in bleomycins and other natural products (5). This is supported by the identification of a hybrid PKS-NRPS gene. Indeed, a disruption of the NRPS module also led to the elimination of HSAF production in strain C3. This is direct evidence supporting the notion that the unique structure of the tetramic acid-containing macrolactam is derived from hybrid nonribosomal peptide and polyketide biosynthesis. A careful analysis of the adenylation domain of the NRPS module suggested that this domain is most similar to the ornithine-activating domain of gramicidin S-synthetase 2 from Brevibacillus brevis (14). Based on this information, we propose a possible biosynthetic pathway for HSAF (Fig. 7). Two PKS-synthesized hexaketide chains are condensed with the NRPS-activated ornithine via Claisen-type condensations to generate the basic skeleton of this group of natural products. Following a series of aldol condensations, reductions, and oxidations, the 5,5,6-membered tricyclic ring system is formed and fused to the tetramic acid-containing macrolactam ring system (Fig. 7). The different patterns of ring fusion within this group of natural products are intriguing (Fig. 1). For example, HSAF and discodermide have a three-ring (5,5,6-membered) system fused to the macrolactam, while ikarugamycin has a different pattern (5,6,5-membered system [structure not shown]) of the ring system fused to the macrolactam system. On the other hand, cylindramide and alteramide A have only a two-ring (5,5-membered) system fused to a larger lactam system (Fig. 1). It will be extremely interesting to exploit the properties of the enzymes involved in these post-PKS folding processes. As suggested by this biosynthetic pathway, multiple PKS modules are likely involved in the assembly of the two polyketide chains. The four-domain PKS module of the hybrid PKS-NRPS gene described in this study could catalyze the incorporation of the last two-carbon unit (C26-C27; Fig. 7) of the first polyketide chain. This is supported by the fact that this module lacks an enoylreductase domain, thus generating a carbon-carbon double bond at C25 and C26, and is physically linked to an NRPS, thus forming an amide bond between C27 and N1. Due to the instability of the cosmids carrying the gene cluster, we are taking the vector-rescuing approach to locate the remaining genes, including other PKS genes.
FIG. 7.
Proposed biosynthetic pathway for HSAF involving both polyketide and nonribosomal peptide mechanisms. SEnz, the thiol group of the phosphopantetheinyl cofactor of the carrier proteins (acyl carrier protein or peptidyl carrier protein) within the polyketide synthase or nonribosomal pepetide synthetase; OH-Orn, β-hydroxyornithine. Note that the sequence of the biosynthetic steps illustrated here is just one of several possibilities.
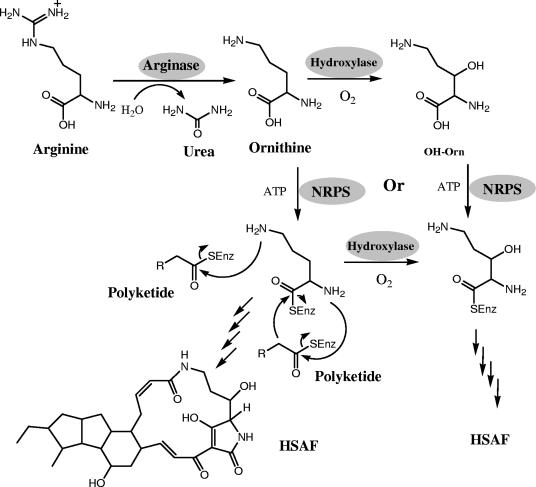
The presence of an arginase gene in the cluster also suggests that ornithine may be the direct substrate of NRPS. Arginase catalyzes the conversion of arginine to urea and ornithine and is one of the five enzymes of the urea cycle (1). This arginase is likely an enzyme dedicated to the biosynthesis of HSAF. It is not uncommon for “extra” copies of genes involved in primary metabolisms to be found in the gene clusters for secondary metabolisms (19), perhaps to provide better coordination of gene expression and enzymatic activities within the cluster. Thus, it is not surprising that the disruption of this gene did not affect the production of HSAF. This result suggests that the arginase for primary metabolism may be able to compensate for the arginase involved in HSAF production. Alternatively, the source of ornithine in the medium may be sufficient for HSAF production. The ornithine moiety in HSAF, as well as most of the tetramic acid-containing macrolactams (except ikarugamycin), carries a β-hydroxyl group (Fig. 1), which could be added prior to or after the incorporation of the amino acid by NRPS (Fig. 8). The determination of either of the pathways as illustrated in Fig. 8 needs further biochemical characterization of the NRPS genes.
FIG. 8.
Proposed mechanism for the formation of the two lactam moieties in HSAF. Note that the NRPS may use ornithine as a substrate and that the β-hydroxyl group may be added after the incorporation of the amino acid; alternatively, NRPS may use β-hydroxyornithine (OH-Orn) directly as substrate. SEnz, the thiol group of the phosphopantetheinyl cofactor of the carrier proteins (acyl carrier protein or peptidyl carrier protein) within the polyketide synthase or nonribosomal pepetide synthetase.
FNR is a FAD-containing protein and a member of the electron transfer chain in energy metabolism and photosynthesis. It uses the iron-sulfur-containing ferredoxin and NADP+ as substrates. The structure of HSAF suggests that several redox reactions are required during cyclization (Fig. 7). FNR is probably involved in this process. Apparently, the role of this FNR could be compensated for by other redox enzymes in the organism. This is supported by the fact that the disruption of the FNR gene in the HSAF gene cluster did not affect HSAF production. However, current data do not exclude the possibility that this FNR is not directly involved in HSAF biosynthesis.
Finally, sterol desaturase catalyzes the dehydrogenation reaction to introduce a carbon-carbon double bond into the sterol ring system, such as sterol C5-desaturase for a double bond at C-5-C-6 (25). Several oxidoreductions are implied in the biosynthesis of HSAF, and the sterol desaturase may play a role in the reactions. The disruption of this gene led to the elimination of HSAF production and a significant increase in the levels of two metabolites (Fig. 6). These two metabolites were also produced by the wild-type strain at a lower level, but their production, along with that of HSAF, was eliminated by the disruption of the PKS-NRPS gene. These results suggest that the metabolites may be precursors of HSAF and that the sterol desaturase is involved in the conversion of the metabolites to HSAF and its analogs.
Acknowledgments
We thank Sara Basiaga and Joseph Dumais, Department of Chemistry, University of Nebraska—Lincoln, for technical assistance with NMR analysis and Ashraf Raza, Department of Biochemistry, University of Nebraska—Lincoln, for technical assistance with MS analysis. We also thank Donald Kobayashi, Rutgers University, for donating E. coli strains and the C3 cosmid library.
This work was supported in part by an award from Nebraska Research Initiative and a pilot grant from Redox Biology Center, UNL, supported by NIH (grant P20RR17675). K.Z.-R. was supported by a CONACYT scholarship (no. 166245), Mexico; and X.-C.L. was supported by USDA/ARS Specific Cooperative Agreement 58-6408-2-0009. The research was performed in facilities renovated with support from NIH (grant RR015468-01).
Footnotes
Published ahead of print on 30 October 2006.
REFERENCES
- 1.Ash, D. E. 2004. Structure and function of arginases. J. Nutr. 134:2760S-2764S. [DOI] [PubMed] [Google Scholar]
- 2.Boeckman, R. K. J., C. H. Weidner, R. B. Perni, and J. J. Napier. 1989. An enantioselective and highly convergent synthesis of (+)-ikarugamycin. J. Am. Chem. Soc. 111:8036-8037. [Google Scholar]
- 3.Cramer, N., M. Buchweitz, S. Laschat, W. Frey, A. Baro, D. Mathieu, C. Richter, and H. Schwalbe. 2006. Total synthesis and NMR investigations of cylindramide. Chemistry 12:2488-2503. [DOI] [PubMed] [Google Scholar]
- 4.Cramer, N., S. Laschat, A. Baro, H. Schwalbe, and C. Richter. 2005. Enantioselective total synthesis of cylindramide. Angew. Chem. Int. Ed. Engl. 44:820-822. [DOI] [PubMed] [Google Scholar]
- 5.Du, L., Y. Cheng, G. Ingenhorst, G. Tang, Y. Huang, and B. Shen. 2003. Hybrid peptide-polyketide natural products: biosynthesis and prospects towards engineering novel molecules, p. 227-267. In J. K. Setlow (ed.), Genetic engineering—principles and methods, vol. 25. Kluwer Academic/Plenum Publishers, New York, NY. [DOI] [PubMed] [Google Scholar]
- 6.Folman, L. B. 2003. Biological control of Pythium aphanidermatum in soilless systems. Ph.D. thesis. University of Leiden, Leiden, The Netherlands.
- 7.Giesler, L. J., and G. Y. Yuen. 1998. Evaluation of Stenotrophomonas maltophilia strain C3 for biocontrol of brown patch disease. Crop Protect. 17:509-513. [Google Scholar]
- 8.Graupner, P. R., S. Thornburgh, J. T. Mathieson, E. L. Chapin, G. M. Kemmitt, J. M. Brown, and C. E. Snipes. 1997. Dihydromaltophilin; a novel fungicidal tetramic acid containing metabolite from Streptomyces sp. J. Antibiot. (Tokyo) 50:1014-1019. [DOI] [PubMed] [Google Scholar]
- 9.Gunasekera, S. P., M. Gunasekera, and P. McCarthy. 1991. Discodermide: a new bioactive macrocyclic lactam from the marine sponge Discodermia dissoluta. J. Org. Chem. 56:4830-4833. [Google Scholar]
- 10.Hart, A. C., and A. J. Phillips. 2006. Total synthesis of (+)-cylindramide A. J. Am. Chem. Soc. 128:1094-1095. [DOI] [PubMed] [Google Scholar]
- 11.Kanazawa, S., N. Fusetani, and S. Matsunaga. 1993. Bioactive marine metabolites. 45. Cylindramide: cytotoxic tetramic acid lactam from the marine sponge Halichondria cylindrata Tanita & Hoshino. Tetrahedron Lett. 34:1065-1068. [Google Scholar]
- 12.Kobayashi, D. Y., R. M. Reedy, J. D. Palumbo, J. M. Zhou, and G. Y. Yuen. 2005. A clp gene homologue belonging to the Crp gene family globally regulates lytic enzyme production, antimicrobial activity, and biological control activity expressed by Lysobacter enzymogenes strain C3. Appl. Environ. Microbiol. 71:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi, D. Y., and G. Y. Yuen. 2005. The role of clp-regulated factors in antagonism against Magnaporthe poae and biological control of summer patch disease of Kentucky bluegrass by Lysobacter enzymogenes C3. Can. J. Microbiol. 51:719-723. [DOI] [PubMed] [Google Scholar]
- 14.Krause, M., and M. A. Marahiel. 1988. Organization of the biosynthesis genes for the peptide antibiotic gramicidin S. J. Bacteriol. 170:4669-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, S. 2005. An antifungal secondary metabolite from Lysobacter enzymogenes strain C3: isolation, biological activity, mode of action and potential use in plant disease control. Ph.D. Thesis. University of Nebraska—Lincoln.
- 16.Li, S., L. Du, G. Yuen, and S. D. Harris. 2006. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 17:1218-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakayama, T., Y. Homma, Y. Hashidoko, J. Mizutani, and S. Tahara. 1999. Possible role of xanthobaccins produced by Stenotrophomonas sp. strain SB-K88 in suppression of sugar beet damping-off disease. Appl. Environ. Microbiol. 65:4334-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paquette, L. A., D. Macdonald, L. G. Anderson, and J. Wright. 1989. A triply convergent enantioselective total synthesis of (+)-ikarugamycin. J. Am. Chem. Soc. 111:8037-8039. [Google Scholar]
- 19.Proctor, R. H., D. W. Brown, R. D. Plattner, and A. E. Desjardins. 2003. Expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet. Biol. 38:237-249. [DOI] [PubMed] [Google Scholar]
- 20.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 21.Rajgarhia, V. B., and W. R. Strohl. 1997. Minimal Streptomyces sp. strain C5 daunorubicin polyketide biosynthesis genes required for aklanonic acid biosynthesis. J. Bacteriol. 179:2690-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Shigemori, H., M. A. Bae, K. Yazawa, T. Sasaki, and J. Kobayashi. 1992. Alteramide A, a new tetracyclic alkaloid from a bacterium Alteromonas sp. associated with the marine sponge Halichondria okadai. J. Org. Chem. 57:4317-4320. [Google Scholar]
- 24.Sullivan, R. F., M. A. Holtman, G. J. Zylstra, J. F. White, and D. Y. Kobayashi. 2003. Taxonomic positioning of two biological control agents for plant diseases as Lysobacter enzymogenes based on phylogenetic analysis of 16S rDNA, fatty acid composition and phenotypic characteristics. J. Appl. Microbiol. 94:1079-1086. [DOI] [PubMed] [Google Scholar]
- 25.Taton, M., T. Husselstein, P. Benveniste, and A. Rahier. 2000. Role of highly conserved residues in the reaction catalyzed by recombinant delta7-sterol-C5(6)-desaturase studied by site-directed mutagenesis. Biochem. 39:701-711. [DOI] [PubMed] [Google Scholar]
- 26.Yuen, G. Y., C. C. Jochum, L. E. Osborne, and Y. Jin. 2003. Biocontrol of fusarium head blight in wheat by Lysobacter enzymogenes C3. Phytopathology 93:S93. [Google Scholar]
- 27.Yuen, G. Y., J. R. Steadman, D. T. Lindgren, D. Schaff, and C. C. Jochum. 2001. Bean rust biological control using bacterial agents. Crop Protect. 20:395-402. [Google Scholar]
- 28.Zhang, Z., and G. Y. Yuen. 1999. Biological control of Bipolaris sorokiniana on tall fescue by Stenotrophomonas maltophilia C3. Phytopathology 89:817-822. [DOI] [PubMed] [Google Scholar]