Abstract
Molecular characterization of fluoroquinolone-resistant Streptococcus pneumoniae in Canada was conducted from 1997 to 2005. Over the course of the study, 205 ciprofloxacin-resistant isolates were evaluated for ParC and GyrA quinolone resistance-determining region (QRDR) substitutions, substitutions in the full genes of ParC, ParE, and GyrA, reserpine sensitivity, and serotype and by pulsed-field gel electrophoresis. Rates of ciprofloxacin resistance of S. pneumoniae increased significantly, from less than 1% in 1997 to 4.2% in 2005. Ciprofloxacin resistance was greatest in people >64 years of age and least in those <16 years of age. Significant increases were also noted in rates of resistance to gatifloxacin, gemifloxacin, levofloxacin, and moxifloxacin, to the current rates of 1.6%, 1.0%, 1.1%, and 1.0%, respectively. The most common genotype observed consisted of QRDR substitutions in GyrA (Ser81Phe) and ParC (Ser79Phe). Substitutions outside the QRDR of GyrA, ParC, and ParE were not associated with fluoroquinolone resistance in this study. Overall, 21% of isolates were reserpine-sensitive and were thus assumed to be efflux positive. The ciprofloxacin-resistant isolates belonged to 35 different serotypes, but 10 (19F, 11A, 23F, 6B, 22F, 12F, 6A, 14, 9V, and 19A) accounted for 72% of all isolates. The majority of the isolates were found to be genetically unrelated by pulsed-field gel electrophoresis. Within the observed clusters, there was considerable genetic heterogeneity with regard to fluoroquinolone resistance mechanisms and serotypes. Continued surveillance and molecular analysis of fluoroquinolone-resistant S. pneumoniae in Canada are essential for appropriate empirical treatment of infections and early detection of novel resistance mechanisms.
Streptococcus pneumoniae is a primary cause of respiratory tract infections, which result in substantial morbidity and mortality worldwide. Infections likely caused by S. pneumoniae frequently are empirically treated with penicillins and macrolides. Due to the increasing prevalence of resistance to the β-lactams and macrolides, the newer fluoroquinolones have been recommended for the empirical treatment of penicillin-resistant and multidrug-resistant S. pneumoniae (6). Fluoroquinolone resistance has remained low (<2%) in Canada for many years (8, 19). However, the percentage of S. pneumoniae organisms with reduced susceptibilities to the fluoroquinolones has increased recently (16). In order to monitor the emergence of fluoroquinolone resistance and investigate the mechanisms of resistance development, genotypic and phenotypic surveillance is essential.
Fluoroquinolones inhibit DNA replication by disrupting the activity of two essential enzymes, DNA gyrase and topoisomerase IV. These enzymes maintain the integrity of the genetic material during replication by altering the topological state of DNA. DNA gyrase and topoisomerase IV are A2B2 heterotetramers encoded by gyrA and gyrB (DNA gyrase) and parC and parE (topoisomerase IV). Resistance to fluoroquinolones in S. pneumoniae is mediated by two common mechanisms: chromosomal mutations in the quinolone resistance-determining regions (QRDR) of DNA gyrase and topoisomerase IV and by active efflux (17).
The purpose of this study was to conduct surveillance and molecular characterization of fluoroquinolone-resistant S. pneumoniae in order to monitor the resistance mechanisms as fluoroquinolone resistance increased in Canada. All Canadian ciprofloxacin-resistant S. pneumoniae isolates (MIC ≥4 μg/ml) collected between 1997 to 1998 and 2005 as part of an ongoing national surveillance program (Canadian Respiratory Organism Susceptibility Study [CROSS]) (18) were molecularly characterized by QRDR substitutions, reserpine-sensitive efflux, and serotype and by pulsed-field gel electrophoresis (PFGE) patterns. Additionally, the hypothesis that mutations outside the QRDR contribute to fluoroquinolone resistance was investigated.
MATERIALS AND METHODS
S. pneumoniae isolates.
Over 10,000 clinical isolates of S. pneumoniae were submitted to or isolated by the Department of Clinical Microbiology, Health Sciences Centre, Winnipeg, Manitoba, Canada, between August 1997 and December 2005 as part of CROSS (18). As part of CROSS, 25 medical centers from all regions of Canada collected 100 clinically significant S. pneumoniae respiratory tract isolates per year (one per patient) and shipped them to the reference laboratory (Health Sciences Centre, Winnipeg) on Amies charcoal swabs.
Antimicrobial susceptibilities.
MICs were determined as described by the Clinical and Laboratory Standards Institute guidelines (M7-A6 and M100-S15), using broth microdilution (9, 14). Test antibacterial agents were obtained as reference-grade powders from their various manufacturers.
Ciprofloxacin-resistant S. pneumoniae isolates.
All ciprofloxacin-resistant (MIC ≥ 4 μg/ml) isolates (n = 205) identified as part of CROSS were studied. A ciprofloxacin MIC of ≥4 μg/ml against S. pneumoniae is commonly considered to be the ciprofloxacin-resistant concentration. As the majority of S. pneumoniae isolates have ciprofloxacin MICs of ≤2 μg/ml, an MIC of ≥4 μg/ml is useful for the detection of upward shifts in MICs (8). Additionally, quinolone resistance-associated mutations begin to accumulate at a ciprofloxacin MIC of 4 μg/ml (8). Over the course of the study, 1,328 isolates with ciprofloxacin MICs of 2 μg/ml were collected and were not included in the molecular analysis as they had only a slight rise in MIC.
Full gene sequencing of gyrA, parC, and parE was conducted with 51 of the 205 aforementioned ciprofloxacin-resistant isolates. The 51 isolates were randomly selected such that isolates without QRDR substitutions in GyrA or ParC, isolates with GyrA QRDR substitutions, isolates with ParC QRDR substitutions, and isolates with QRDR substitutions in both GyrA and ParC were included in the study. Isolates were found to be representative of the study as they were collected from various years and from various provinces and represented 22 serotypes. Six randomly selected ciprofloxacin-susceptible S. pneumoniae isolates were included for comparison.
gyrA and parC QRDR and gyrA, parC, and parE amplification and sequencing.
QRDR amplification and sequencing were conducted using previously described primers and methodology (13, 19). S. pneumoniae ATCC 49619 was used as a control for the amplification procedure.
Amplification of the gyrA, parC, and parE genes was conducted using two previously described primers (13) and four primers designed throughout this study, as listed in Table S1 in the supplemental material. The amplification reaction mixtures consisted of 1.5 mM MgCl2-10× PCR buffer, 1 mM of each deoxynucleoside triphosphate, 10 μM of each primer, 1 mM MgCl2, 2.5 U of Taq DNA polymerase (GE Healthcare Bio-Sciences, Baie d'Urfe, Quebec, Canada), 10 ng of the DNA template, and sterile water to a final volume of 50 μl. Amplification conditions were 94°C for 5 min, 30 cycles of 94°C for 30 s, 50°C for 1 min (57°C for 45 s for gyrA), and 72°C for 3 min, with a final extension of 7 min at 72°C.
Sequencing reaction mixtures for the QRDR sequencing and the full gene sequencing were performed with 100 ng of purified PCR product using an ABI PRISM BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA). Sequencing analysis was conducted on an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA) in accordance with the manufacturer's instructions. The specimens were aligned and compared to the published sequence of S. pneumoniae R6 (ATCC BAA-255) using ABI PRISM SeqScape software (Applied Biosystems, Foster City, CA).
Reserpine efflux assay.
Reserpine-sensitive efflux was tested by using ciprofloxacin agar dilution in the presence and absence of a potential efflux inhibitor, reserpine. Two sets of Mueller-Hinton agar plates plus 5% sheep blood were made with doubling dilutions of ciprofloxacin. Reserpine (10 μg/ml) was added to one set of plates. Isolates with a fourfold or greater reduction in MIC in the presence of reserpine were considered positive for reserpine-sensitive efflux (5).
PFGE.
Genetic similarity of the isolates was tested by PFGE following previously described methods (15). Genomic DNA was prepared for PFGE as described previously by Nichol et al. (15). SmaI restriction fragments were resolved in a contour-clamped homogenous electric field apparatus (CHEF DRIII; Bio-Rad Laboratories, Hercules, CA) for 18 h (6 V/cm, 2- to 30-s switch times, included an angle of 120°). PFGE profiles were analyzed using BioNumerics version 3.5 (Applied Maths, Austin, Texas) software. A dendrogram was calculated by the unweighted pair group method with arithmetic averages (band tolerance, 1.5%, and dice coefficient, 1%). For the purpose of this study, isolates were defined as genetically indistinguishable, possibly related, or genetically unrelated if their PFGE profiles differed by 0, 1 to 3, or ≥4 bands, respectively (15). Isolates with more than 80% similarity on the dendrogram have been correlated with a ≤3-band difference, i.e., they are genetically related.
Serotyping.
Isolates were serotyped on the basis of capsular polysaccharide antigens by the quellung reaction following a standard methodology (2). Type-specific antisera were obtained from the Statens Seruminstitut (Copenhagen, Denmark). Global clones previously described by the Pneumococcal Molecular Epidemiology Network (www.sph.emory.edu/PMEN) were routinely serotyped as controls.
Statistical analysis.
The statistical significance between groups was determined by a two-tailed Fisher's exact test or chi-square analysis, as appropriate.
RESULTS
As described in Table 1, ciprofloxacin resistance of S. pneumoniae has increased significantly in Canada, from less than 1% in 1997 to 4.2% in 2005 (P, <0.0001). Smaller, but generally significant, increases in resistance rates with the various respiratory fluoroquinolones were observed throughout the study. The yearly resistance rates to ciprofloxacin, gatifloxacin, gemifloxacin, levofloxacin, and moxifloxacin are displayed in Table 1. Resistance rates increased from 0.6% to 1.6% for gatifloxacin between 1999 to 2000 and 2005, respectively (P = 0.04). Gemifloxacin resistance rates increased from 0.4% to 1.0% between 1999 to 2000 and 2005, respectively (P = 0.11). Resistance rates to levofloxacin increased from 0.2% in 1997 to 1998 to 1.1% in 2005 (P = 0.008). The moxifloxacin resistance rate was 0% in 1997 to 1998 and 1.0% in 2005 (P = 0.0005).
TABLE 1.
Summary of ciprofloxacin-resistant S. pneumoniae isolates collected as part of CROSS between 1997 and 2005 and the resistance mechanisms present in ciprofloxacin-resistant isolatesa
| Study yr (no. of isolates) | All isolates (n = 10,657)b
|
CIP-resistant isolates (n = 205)c
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rate (%) of bacterial resistance
|
n | % of QRDR genotype in
|
% of efflux | ||||||||||||||||||
| CIP
|
GAT | GEM | LVX | MOX | MDR | Isolates with no mutations | ParC | Isolates with ParC and GyrA | |||||||||||||
| Age distribution
|
Geographic distribution
|
||||||||||||||||||||
| Overall | <16 yr old (n = 2,057) | 16-64 yr old (n = 4,637) | >64 yr old (n = 3,963) | British Columbia (n = 1,019) | Alberta (n = 1,466) | Saskatchewan (n = 1,168) | Manitoba (n = 1,321) | Ontario (n = 2,330) | Quebec (n = 2,017) | Maritimes (n = 1,336) | |||||||||||
| 1997-1998 (1,152) | 0.6 | 0.0 | 0.2 | 1.4 | 0.7 | 1.1 | 0.0 | 0.8 | 0.0 | 0.9 | 0.4 | NT | NT | 0.2 | 0 | 0.0 | 4 | 25.0 | 25.0 | 50.0 | 50.0 |
| 1998-1999 (1,256) | 1.5 | 0.0 | 1.7 | 2.3 | 1.4 | 0.5 | 1.0 | 0.7 | 1.5 | 2.9 | 1.2 | NT | NT | 0.5 | 0.3 | 0.1 | 14 | 7.1 | 57.1 | 35.7 | 28.6 |
| 1999-2000 (1,511) | 1.5 | 0.3 | 0.7 | 3.3 | 3.4 | 0.7 | 2.2 | 1.7 | 0.9 | 1.5 | 1.0 | 0.6 | 0.4 | 0.9 | 0.2 | 0.1 | 22 | 9.1 | 27.3 | 35.7 | 22.7 |
| 2000-2001 (1,402) | 1.7 | 1.1 | 1.7 | 2.0 | 1.5 | 1.1 | 0.7 | 0.7 | 1.7 | 1.5 | 5.3 | 0.6 | 0.5 | 0.6 | 0.4 | 0.3 | 21 | 19.1 | 19.1 | 52.4 | 33.3 |
| 2001-2002 (1,546) | 1.7 | 0.0 | 1.5 | 2.5 | 1.4 | 3.1 | 0.7 | 2.2 | 1.6 | 1.5 | 1.4 | 1.0 | 0.1 | 1.2 | 0.1 | 0.1 | 21 | 14.3 | 14.3 | 71.4 | 14.3 |
| 2003 (1,279) | 2.3 | 0.5 | 1.5 | 3.8 | 2.0 | 1.1 | 0.0 | 5.5 | 2.6 | 1.9 | 2.7 | 0.8 | 0.3 | 1.4 | 0.3 | 0.5 | 31 | 3.2 | 19.4 | 77.4 | 16.1 |
| 2004 (1,290) | 3.7 | 0.0 | 2.9 | 6.3 | 1.1 | 2.5 | 2.7 | 5.2 | 3.7 | 6.3 | 1.6 | 1.6 | 0.7 | 1.9 | 0.5 | 0.2 | 43 | 9.3 | 27.9 | 60.5 | 18.6 |
| 2005 (1,221) | 4.2 | 1.0 | 2.9 | 7.6 | 4.3 | 5.1 | 0.6 | 3.6 | 4.9 | 4.2 | 6.5 | 1.6 | 1.0 | 1.1 | 1.0 | 0.3 | 49 | 24.5 | 24.5 | 49.0 | 18.8 |
| P value (1997-1998 vs 2005) | <0.0001 | 0.16 | 0.0011 | <0.0001 | d | d | d | d | d | d | d | 0.04e | 0.11e | 0.008 | 0.0005 | 0.13 | 1.0 | 1.0 | 1.0 | 0.19 | |
CIP, ciprofloxacin; GAT, gatifloxacin; GEM, gemifloxacin; LVX, levofloxacin; MOX, moxifloxacin; MDR, multidrug resistant (includes resistance to penicillin, ciprofloxacin, and a macrolide, tetracycline, or trimethoprim-sulfamethoxazole); NT, not tested.
Includes all S. pneumoniae isolates collected, where n indicates specific numbers of isolates for overall, age and geographic distribution, and fluoroquinolone resistance rates.
Ciprofloxacin-resistant S. pneumoniae isolates, excluding S. pneumoniae isolates that may have undergone interspecies recombination.
Statistical analysis not included due to large yearly fluctuations.
Statistical analysis between 1999 to 2000 and 2005.
Resistance rates were found to vary across the country, with the highest average levels found in Manitoba, followed by Quebec, Ontario, Nova Scotia, Alberta, New Brunswick, British Columbia, Saskatchewan, and Prince Edward Island. The yearly ciprofloxacin resistance rates for each province are listed in Table 1. Although all provinces appeared to have increasing resistance trends throughout the study, statistical analysis was not included as large yearly fluctuations occurred.
The majority of the ciprofloxacin-resistant S. pneumoniae isolates (62%) were isolated from elderly patients, >64 years of age, while 35% were from adults 16 to 64 years of age, and 3% were from children <16 years of age. The annual ciprofloxacin resistance rates in each age group are presented in Table 1. Statistically significant increases in ciprofloxacin resistance were observed between 1997 to 1998 and 2005 for the adult patients (P = 0.0011) and the elderly patients (P, <0.0001). Ciprofloxacin resistance was found to be greater for both the adult and the elderly groups than for the children (P, <0.0001 for both groups).
Throughout this study, 205 ciprofloxacin-resistant S. pneumoniae isolates were collected for molecular characterization. Additionally, 14 ciprofloxacin-resistant isolates that may have undergone interspecies recombination were identified throughout this study. As indicated in other recent studies, these isolates remain rare (4). These isolates had the Asn91Asp (ParC) and Ser114Gly (GyrA) substitutions previously reported for isolates that had undergone interspecies recombination with the viridans streptococci, likely S. oralis and S. mitis (3, 10). The isolates had variable bile (sodium deoxycholate) solubility and optochin sensitivity levels and may be better categorized as S. pseudopneumoniae (1). These isolates were excluded from the remainder of the molecular characterization, as it is unknown if these isolates are clinically like S. pneumoniae.
The majority of the ciprofloxacin-resistant S. pneumoniae isolates had substitutions in ParC and GyrA (Table 1). The most common genotype observed included substitutions Ser79Phe (ParC) and Ser81Phe (GyrA). All substitutions observed in the resistant isolates are presented in Table 2. As the ciprofloxacin MIC increased, the percentage of isolates with no QRDR substitutions or single substitutions in ParC decreased, and the percentage of isolates with QRDR substitutions in ParC and GyrA increased. Overall, 21% of the isolates were sensitive to reserpine and were presumed to be efflux positive. The majority of the isolates in this study that did not have QRDR substitutions but remained ciprofloxacin resistant were reserpine sensitive (68%). There were five isolates that had no QRDR substitutions and were reserpine insensitive. These isolates may be efflux positive but require a different efflux inhibitor for identification or have substitutions in gyrB or possess a currently unknown mechanism of resistance.
TABLE 2.
Serotype, reserpine sensitivity, and GyrA, ParC, and ParE mutational analysis of ciprofloxacin-resistant S. pneumoniae isolates by ciprofloxacin MICb
| Ciprofloxacin MIC (μg/ml) | Serotypes (n) | GyrA mutation
|
ParC mutation
|
ParE mutation (n)a | Reserpine sensitivity (n) | ||
|---|---|---|---|---|---|---|---|
| QRDR (n) | non-QRDR (n)a | QRDR (n) | non-QRDR (n)a | ||||
| 4 (n = 79) | 3 (1), 6A (4), 6B (5), 9N (3), 9V (3), 10A (1), 11A (13), 11F (1), 12F (5), 14 (2), 15A (1), 15B (4), 15C (1), 16F (1), 19A (2), 19F (14), 20 (1), 22F (5), 23A (1), 23F (5), 31 (1), 33F (1), 34 (1), 35F (1), NT (2) | S81F (14), E85A (1), E85K (1) | I711V (1), V486I (4), V489 (11), A653T (9) | A63T (1), D78A (1), S79A (2), S79C (1), S79F (31), S79Y (9), D83N (9), D83Y (2), S107Y (1), Y129S (1), K137N (16) | A189V (1), A282T (1), H373R (2), A394T (1), A450V (3), R443C (1), K473N (1), T493I (1), R569C (1), E589A (5), V608A (7), A724S (1), D822Y (2) | G34R (1), S132N (3), I162V (5), T216S (3), L290F (2), A326V (1), L374I (1), D435N (2), I460V (9) | Pos. (21), neg. (58) |
| 8 (n = 40) | 3 (4), 4 (2), 6A (4), 6B (3), 9V (3), 14 (3), 10A (3), 11A (4), 12F (2), 15A (1), 17F (1), 19A (2), 19F (2), 23F (5), 35B (1) | S81F (21), S81Y (1), E85L (4), E85G (1) | A191V (1), S237L (1), R295H (1), V486I (2), V489I (11), A653T (2), I711V (1), V768F (1) | Y59D (1), S79A (1), S79F (26), S79Y (3), D83Y (1), D83G (2), D83A (1), D83N (1), K137N (8) | A189V (1), D294L (1), H373R (1), A450V (3), I453Y (1), K473N (1), E589A (2), V608A (9), V608S (1), A724S (1), D822Y (2) | I162V (1), P166L (1), T212A (1), D217N (1), S399I (1), D435N (2), I460V (9), E474K (1) | Pos. (11), neg. (29) |
| 16 (n = 48) | 2 (1), 3 (1), 4 (2), 6A (2), 6B (6), 9A (1), 9N (3), 10A (1), 11A (1), 12F (7)14 (1), 15C (2), 18B (1), 18C (2), 19A (1), 19F (8), 22F (2), 23A (1), 23F (5) | S81F (34), S81Y (1), E85G (1), E85K (5) | S418T (1), V486I (4), V489I (4), E560D (1), A653T (1) I711V (2), L747F (1) S778L (1) | S79F (36), S79Y (6), D83N (3) D83Y (2), E120Q (1), K137N (16) | T257I (1), H373R (1), A394T (1), A394Y (1), A450V (1), K473N (1), R518H (1), E589A (2), V608A (3), M686I (1), A724S (1), D822Y (3) | I162V (1), P166L (1), D217N (1), G372R (1), D435N (1), I460V (8), E474K (1), A644T (1) | Pos. (6), neg. (42) |
| 32 (n = 38) | 6A (2), 6B (1), 9V (3), 11A (6), 11D (1), 12B (2), 14 (4), 19A (2), 19F (2), 20 (1), 22F (8), 23F (1), 33F (1), 35A (1), NT (3) | A17T (1), S81L (1), S81F (27), S81Y (3), E85K (7), S114G (1) | V489I (5), A653T (5) | S79F (24), S79Y (11), D83N (3), D83Y (1), K137N (15) | H373R (1), R443C (1), A450V (1), E589A (2), V608A (5) | I162V (2), T216S (3), I460V (2) | Pos. (5), neg. (32) |
Non-QRDR substitutions in GyrA and ParC and substitutions in ParE were determined for 17, 16, 13, and 5 isolates with ciprofloxacin MICs of 4 μg/ml, 8 μg/ml, 16 μg/ml, and 32 μg/ml, respectively.
n, number of isolates; Pos., positive; neg., negative.
To address the hypothesis that fluoroquinolone resistance may be due to resistance-associated mutations outside the QRDRs (7), full gene sequencing of gyrA, parC, and parE was conducted with 51 of the ciprofloxacin-resistant isolates with various fluoroquinolone resistance-related genotypes and phenotypes, as well as six ciprofloxacin-susceptible isolates for comparison purposes. Of the 51 ciprofloxacin-resistant isolates, 90% had non-QRDR substitutions in GyrA, 78% had non-QRDR substitutions in ParC, and 35% had non-QRDR substitutions in ParE. A large variety of non-QRDR mutations were observed. Overall, 46 different amino acid substitutions were observed: 12 in GyrA, 19 in ParC, and 15 in ParE, as displayed in Table 2. Substitutions occurring solely in ciprofloxacin-resistant isolates and appearing in 5% or more of the isolates included Val486Ile and Ile711Val in GyrA; His373Arg, Ala394Thr, Lys473Asn, and Ala724Ser in ParC; and Ser132Asn and Thr216Ser in ParE. None of the substitutions were associated with ciprofloxacin MIC or QRDR genotype. Non-QRDR substitutions were not associated with fluoroquinolone resistance in this study.
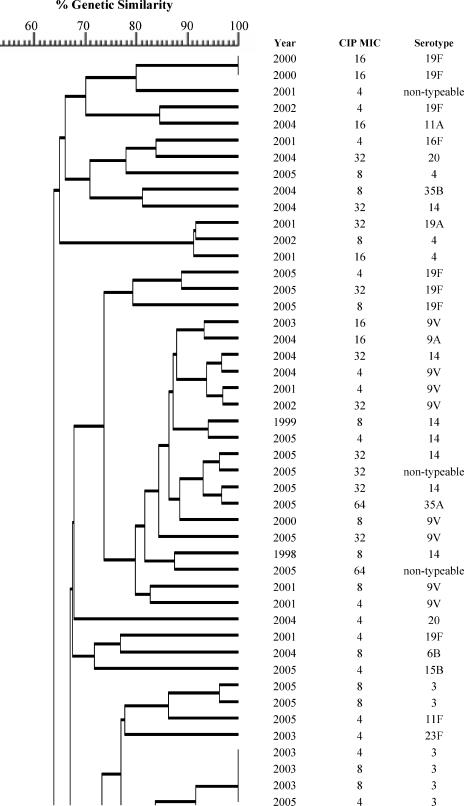
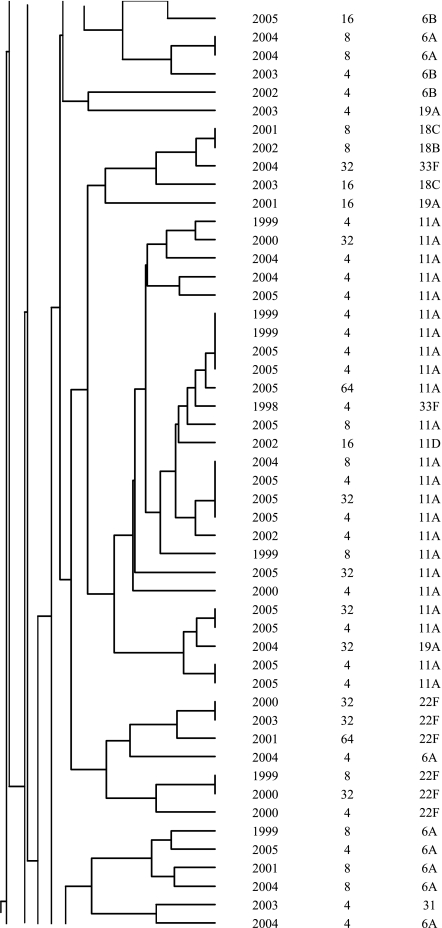
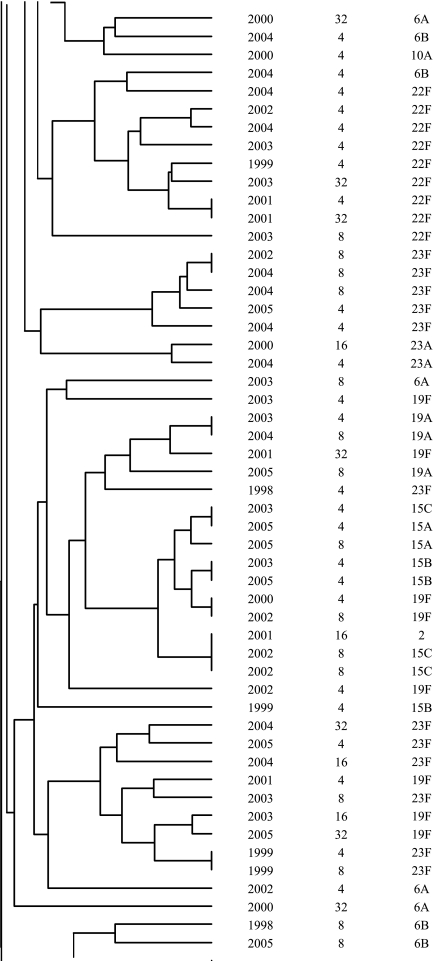
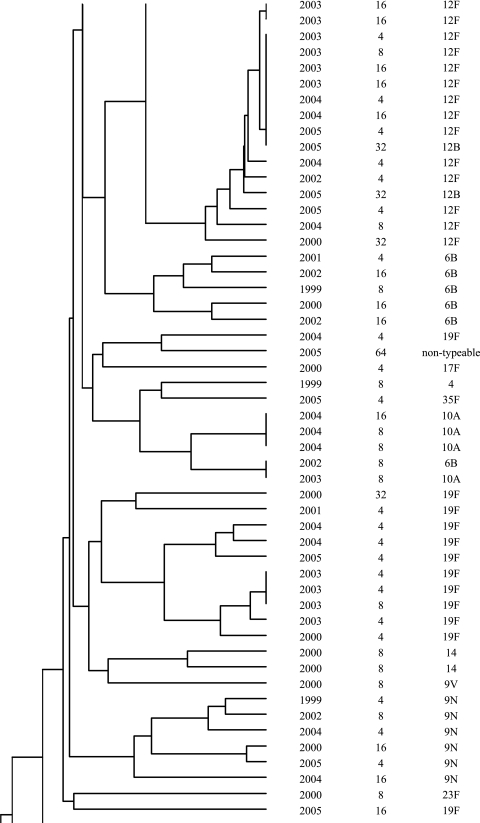
In addition to the identification of resistance-associated substitutions and reserpine-sensitive efflux in the ciprofloxacin-resistant isolates, characterization was performed by serotyping and PFGE to determine if fluoroquinolone resistance was clonal in Canada. The most common serotypes were, in order: 19F, 11A, 23F, 6B, 22F, 12F, 6A, 14, 9V, and 19A. The serotypes are listed in Fig. 1. Correspondingly to the serotyping, PFGE revealed significant heterogeneity among the ciprofloxacin-resistant isolates. As shown in Fig. 1, the majority of the isolates were genetically unrelated. Sixty-two percent of the ciprofloxacin-resistant isolates belonged to 14 different clusters of four or more organisms, and 33% belonged to 4 clusters of 10 or more isolates. Of the four large clusters, two consisted of isolates with a variety of serotypes and two were dominated by one particular serotype (11A or 12F). Within the clusters, there was considerable genetic heterogeneity with regard to fluoroquinolone resistance mechanisms.
FIG.1.
Dendrogram depicting genetic relatedness of 205 ciprofloxacin-resistant S. pneumoniae isolates on the basis of PFGE results.
DISCUSSION
Surveillance of fluoroquinolone resistance in S. pneumoniae in Canada was conducted between 1997 to 1998 and 2005. Over the course of the study, bacterial rates of resistance to the fluoroquinolones tested, ciprofloxacin, gatifloxacin, gemifloxacin, levofloxacin, and moxifloxacin, were found to have significant increases.
Ciprofloxacin resistance was found to be rare in children and highest in patients older than 64 years of age. Associations between fluoroquinolone resistance and advanced age have been previously reported and may be explained by the large amount of fluoroquinolone use in elderly patients (8).
Two hundred five ciprofloxacin-resistant isolates were molecularly characterized by QRDR substitutions, reserpine-sensitive efflux, and serotype and by PFGE. The most common genotype observed was that found in isolates with substitutions in both ParC and GyrA, followed by isolates with substitutions in ParC. A general trend observed was that as the ciprofloxacin MIC increased, the percentage of isolates with no QRDR substitutions or single substitutions in ParC decreased and the percentage of isolates with QRDR substitutions in ParC and GyrA increased.
Full gene sequencing of gyrA, parC, and parE was conducted to address the hypothesis that substitutions outside the QRDR may contribute to fluoroquinolone resistance. Throughout this analysis, three observations were made. First, no particular non-QRDR substitution appeared to correlate with ciprofloxacin MIC or QRDR genotype. Second, the greatest variety of non-QRDR substitutions was observed in ParC and the least in GyrA, but non-QRDR substitutions were most common in GyrA, followed by ParC and ParE in the ciprofloxacin-resistant isolates. This phenomenon was observed regardless of ciprofloxacin MIC. Third, non-QRDR substitutions may occur less frequently in isolates that already have a QRDR substitution in that particular gene. In conclusion, our study does not support the hypothesis that substitutions outside the QRDR are associated with fluoroquinolone resistance.
Overall, 21% of the isolates were sensitive to reserpine and presumed to be efflux positive. Recent studies have indicated that reserpine sensitivity may not identify fluoroquinolone efflux in S. pneumoniae (11, 12). Decreased accumulation in the presence of reserpine has not been observed in recent accumulation studies with potential efflux-positive isolates; thus, the utility of reserpine in the identification of efflux-positive isolates has been questioned (11, 12). Reserpine may identify certain phenotypes of S. pneumoniae that are not necessarily related to the organism's ability to efflux the fluoroquinolones. Thus, it may be that 21% of the isolates included in this study simply have a phenotype that responds differently to reserpine. However, until recently, it was a widely held assumption that reserpine could identify efflux-positive isolates and was accordingly used throughout this study.
The ciprofloxacin-resistant isolates belonged to 35 different serotypes, but 10 (19F, 11A, 23F, 6B, 22F, 12F, 6A, 14, 9V, and 19A) accounted for 72% of all isolates. The 23-valent polysaccharide pneumococcal vaccine recommended for adults over 64 years of age provides coverage against 79% (based on vaccine strains) and 91% (based on vaccine and vaccine-related strains) of the ciprofloxacin-resistant isolates identified in this study. Thus, improved utilization of the 23-valent vaccines has the potential to severely limit infections caused by fluoroquinolone-resistant isolates.
The majority of the ciprofloxacin-resistant isolates were found to be genetically unrelated by PFGE. Within the observed clusters, the isolates were largely heterogeneous in regard to QRDR substitutions and serotypes. The diversity of QRDR substitutions and serotypes observed in the clusters highlights the fact that although these isolates are genetically related, they have been subjected to different antimicrobial exposures and have undergone new mutational events as opposed to clonal dissemination. Similar heterogeneity of QRDR substitutions and serotypes within PFGE clusters has been reported among fluoroquinolone-resistant isolates from the United States (16). At present, clonal dissemination of fluoroquinolone-resistant S. pneumoniae is rare in Canada, but it has gradually increased throughout this ongoing surveillance program.
Continued surveillance and molecular analysis of fluoroquinolone-resistant S. pneumoniae in Canada are essential to identify the mechanisms of the increase in resistance and to permit an evaluation of methods aimed at limiting resistance development. As antimicrobial resistance patterns evolve worldwide, the combination of genotypic and phenotypic surveillance is essential for the early detection of resistance mechanisms.
Supplementary Material
Acknowledgments
Heather Adam is supported by the Canadian Institutes of Health Research, International Centre for Infectious Diseases Training program, and Kristen Schurek is supported by a Manitoba Research Health Council fellowship. This research was funded in part by the University of Manitoba and Abbott Laboratories Ltd., AstraZeneca Canada, Inc., sanofi aventis, Bayer Inc., Bristol-Myers Squibb Pharmaceutical Group, GlaxoSmithKline, Janssen-Ortho Inc., Merck Frosst Canada & Co., Pfizer/Pharmacia Canada Inc., and Wyeth-Ayerst Canada Inc.
Footnotes
Published ahead of print on 6 November 2006.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Arbique, J. C., C. Poyart, P. Trieu-Cuot, G. Quesne, M. da Gloria S. Carvalho, A. G. Steigerwalt, R. E. Morey, D. Jackson, R. J. Davidson, and R. R. Facklam. 2004. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J. Clin. Microbiol. 42:4686-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austrian, R. 1976. The quellung reaction, a neglected microbiologic technique. Mt. Sinai J. Med. 43:699-709. [PubMed] [Google Scholar]
- 3.Balsalobre, L., M. J. Ferrándiz, J. Liñares, F. Tubau, and A. G. de la Campa. 2003. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bast, D. J., J. C. S. de Azavedo, T. Y. Tam, L. Kilburn, C. Duncan, L. A. Mandell, R. J. Davidson, and D. E. Low. 2001. Interspecies recombination contributes minimally to fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:2631-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bast, D. J., D. E. Low, C. L. Duncan, L. Kilburn, L. A. Mandell, R. J. Davidson, and J. C. S. de Azavedo. 2000. Fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae: contributions of type II topoisomerase mutations and efflux to levels of resistance. Antimicrob. Agents Chemother. 44:3049-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush, K., and R. Goldschmidt. 2000. Effectiveness of fluoroquinolones against gram-positive bacteria. Curr. Opin. Investig. Drugs 1:22-30. [PubMed] [Google Scholar]
- 7.Canton, R., M. Morosini, M. C. Enright, and I. Morrissey. 2003. Worldwide incidence, molecular epidemiology and mutations implicated in fluoroquinolone-resistant Streptococcus pneumoniae: data from the global PROTEKT surveillance programme. J. Antimicrob. Chemother. 52:944-952. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D. K., A. McGeer, J. C. de Azavedo, and D. E. Low for the Canadian Bacterial Surveillance Network. 1999. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N. Engl. J. Med. 341:233-239. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. CLSI/NCCLS M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Ferrándiz, M. J., A. Fenoll, J. Liñares, and A. G. De La Campa. 2000. Horizontal transfer of parC and gyrA in fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marrer, E., A. T. Satoh, M. M. Johnson, L. J. V. Piddock, and M. G. P. Page. 2006. Global transcriptome analysis of the responses of a fluoroquinolone-resistant Streptococcus pneumoniae mutant and its parent to ciprofloxacin. Antimicrob. Agents Chemother. 50:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marrer, E., K. Schad, A. T. Satoh, M. G. P. Page, M. M. Johnson, and L. J. V. Piddock. 2006. Involvement of the putative ATP-dependent efflux proteins PatA and PatB in fluoroquinolone resistance of a multidrug-resistant mutant of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 50:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrissey, I., and J. George. 1999. Activities of fluoroquinolones against Streptococcus pneumoniae type II topoisomerases purified as recombinant proteins. Antimicrob. Agents Chemother. 43:2579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. M7-A6. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 15.Nichol, K. A., G. G. Zhanel, and D. J. Hoban. 2003. Molecular epidemiology of penicillin-resistant and ciprofloxacin-resistant Streptococcus pneumoniae in Canada. Antimicrob. Agents Chemother. 47:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter, S. S., K. P. Heilmann, S. E. Beekmann, N. J. Miller, C. L. Rice, and G. V. Doern. 2005. The molecular epidemiology of Streptococcus pneumoniae with quinolone resistance mutations. Clin. Infect. Dis. 40:225-235. [DOI] [PubMed] [Google Scholar]
- 17.Van Bambeke, F., J. M. Michot, J. Van Eldere, and P. M. Tulkens. 2005. Quinolones in 2005: an update. Clin. Microbiol. Infect. 11:256-280. [DOI] [PubMed] [Google Scholar]
- 18.Zhanel, G. G., L. Palatnick, K. A. Nichol, T. Bellyou, D. E. Low, and D. J. Hoban. 2003. Antimicrobial resistance in respiratory tract Streptococcus pneumoniae isolates: results of the Canadian Respiratory Organism Susceptibility Study, 1997 to 2002. Antimicrob. Agents Chemother. 47:1867-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhanel, G. G., A. Walkty, K. Nichol, H. Smith, A. Noreddin, and D. J. Hoban. 2003. Molecular characterization of fluoroquinolone resistant Streptococcus pneumoniae clinical isolates obtained from across Canada. Diagn. Microbiol. Infect. Dis. 45:63-67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.