Abstract
The anilinouracils (AUs) such as 6-(3-ethyl-4-methylanilino)uracil (EMAU) are a novel class of gram-positive, selective, bactericidal antibacterials which inhibit pol IIIC, the gram-positive-specific replicative DNA polymerase. We have linked various fluoroquinolones (FQs) to the N-3 position of EMAU to generate a variety of AU-FQ “hybrids” offering the potential for targeting two distinct steps in DNA replication. In this study, the properties of a hybrid, “251D,” were compared with those of representative AUs and FQs in a variety of in vitro assays, including pol IIIC and topoisomerase/gyrase enzyme assays, antibacterial, bactericidal, and mammalian cytotoxicity assays. Compound 251D potently inhibited pol IIIC and topoisomerase/gyrase, displayed gram-positive antibacterial potency at least 15 times that of the corresponding AU compound, and as expected, acted selectively on bacterial DNA synthesis. Compound 251D was active against a broad panel of antibiotic-resistant gram-positive pathogens as well as several gram-negative organisms and was also active against both AU- and FQ-resistant gram-positive organisms, demonstrating its capacity for attacking both of its potential targets in the bacterium. 251D also was bactericidal for gram-positive organisms and lacked toxicity in vitro. Although we obtained strains of Staphylococcus aureus resistant to the individual parent compounds, spontaneous resistance to 251D was not observed. We obtained 251D resistance in multiple-passage experiments, but resistance developed at a pace comparable to those for the parent compounds. This class of AU-FQ hybrids provides a promising new pharmacophore with an unusual dual mechanism of action and potent activity against antibiotic-sensitive and -resistant gram-positive pathogens.
The successful therapy of life-threatening disease caused by antibiotic-resistant bacteria has become a major challenge. Gram-positive bacteria with genomes containing a low proportion of guanine and cytosine (low-G+C gram-positive organisms), particularly Staphylococcus aureus, Enterococcus faecalis, Enterococcus faecium and Streptococcus pneumoniae (1, 17, 22), have been particularly problematic with respect to nosocomial infections and the development of resistance to traditional antibiotics. For example, Doern et al. (14) have recently reported an increase in penicillin-resistant S. pneumoniae in the clinic. Millichap et al. (29) have also reported a continuing rise in vancomycin-resistant enterococci. Even more troubling is the development of multidrug-resistant organisms. The recent isolation of vancomycin-resistant enterococci, which are also resistant to linezolid, leaves very few treatment options for these infections (34). Most alarmingly, six cases of vancomycin-resistant, methicillin-resistant S. aureus have now been confirmed by the CDC (13).
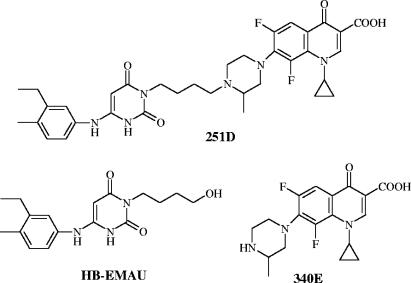
Clearly, new antimicrobial agents and targets are needed to combat an ever-increasing resistance problem among the low-GC gram-positive pathogens. A promising target is the bacterial replication-specific DNA polymerase III C (pol IIIC), the enzyme product of polC, a gene common to this class of organisms (20, 27). The promise of the target is based on three observations. First, its enzyme activity is absolutely essential for bacterial DNA replication (9). Second, inhibitors of bacterial DNA replication are bactericidal (15, 39). Third, the active site of pol IIIC has a unique structure that can be selectively attacked with inhibitory dGTP analogs of the so-called anilinouracil (AU) type (Fig. 1) (9). The pol IIIC inhibitory and antibacterial activities of the AU compounds can be enhanced by introduction of various 3 substituents, for example, the 3(4-hydroxybutyl) group of 3(4-hydroxybutyl)-6-(3-ethyl-4-methylanilino)uracil (HB-EMAU) (Fig. 1) (25, 38, 43). The most potent antibacterials have resulted from substitution of the 3 position of selected AUs with various fluoroquinolones (FQs) of the type shown in Fig. 1 (compound 340E) to generate a series of novel AU-FQ “hybrids” (Fig. 1) (43, 44). In this report, we describe in detail the properties of compound 251D, a representative hybrid composed of AU compound HB-EMAU linked to FQ compound 340E (Fig. 1), to define the mechanism of action, antibacterial properties, and therapeutic potential of this novel class of DNA replication inhibitors.
FIG. 1.
Structures of the hybrid compound, 251D, its anilinouracil component, 3-(4-hydroxybutyl)-6-(3-ethyl-4-methylanilino)uracil (HB-EMAU), and its fluoroquinolone component, {1-cyclopropyl-6,8-difluoro-7-[1-(3-methyl)piperazinyl]-4-oxo-1,4-dihydroquinoline}-3-carboxylic acid (340E).
MATERIALS AND METHODS
Bacterial strains.
A panel of 30 gram-positive organisms and 5 gram-negative organisms were used for determination of MICs. Bacillus cereus 4342 and 6464; Bacillus thuringiensis 10792 and 33679; Staphylococcus aureus 25923 and 13709 (Smith); Enterococcus faecalis 29212; vancomycin-resistant E. faecalis (VREF) 700802, 51575, and 51299; Enterococcus faecium 19434; Streptococcus pneumoniae 49619; penicillin-resistant S. pneumoniae (PRSP) 700670 (Spain), 700671 (France), 700673 (Hungary), 700674 (South Africa), and 700677 (Slovakia); Streptococcus pyogenes 12344; Stenotrophomonas maltophilia 13637; and Pseudomonas aeruginosa 27853 strains were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Bacillus subtilis (BD54) is a standard laboratory strain (27). Bacillus anthracis Sterne (attenuated strain) was provided by Colorado Serum Co. (Denver, CO). The sources of non-ATCC bacterial strains were as follows: S. aureus 1081, methicillin-resistant S. aureus (MRSA) 1094, 1123, 1132, and B42876, and vancomycin-resistant enterococcus (VRE) B42762 and 1644 are clinical isolates donated by Richard Ellison, University of Massachusetts Medical School; mupirocin-sensitive (2529) and mupirocin-resistant (777) S. aureus strains were from Lona Mody, University of Michigan Medical School; ciprofloxacin-resistant S. aureus was from David Hooper, Massachusetts General Hospital; linezolid-resistant VRE F118 was from Jason Prystowsky, Northwestern Memorial Hospital, Chicago, IL; Escherichia coli J53 and Klebsiella pneumoniae 5657 were donated by George Jacoby, Lahey Clinic. E. coli XL1-Blue was purchased from Stratagene (La Jolla, CA). HB-EMAU-resistant S. aureus was obtained as described previously (12). HB-EMAU-resistant and ciprofloxacin-resistant S. aureus was generated by isolating HB-EMAU-resistant colonies (12) from the ciprofloxacin-resistant strain provided by D. Hooper.
Enzymes.
The replicative DNA polymerases from Bacillus subtilis, pol IIIC and IIIE, were purified from recombinant strains as previously described in detail (5, 19). E. coli pol III core enzyme was purchased from Enzyco (Denver, CO). Calf thymus DNA polymerase α (pol α) and human DNA polymerase γ (pol γ) were purchased from Ulrich Hübscher, University of Zurich-Irchel, Switzerland.
The parC and parE subunits of B. subtilis topoisomerase IV and the gyrA and gyrB subunits of B. subtilis DNA gyrase were purified from recombinant strains and combined to form active heterodimers as described previously (4).
Chemicals.
HB-EMAU, 251D, and (1-cyclopropyl-6,8-difluoro-7-[1-(3-methyl)piperazinyl]-4-oxo-1,4-dihydroquinoline)-3-carboxylic acid (340E), the FQ component of 251D (Fig. 1) were synthesized as previously described (38, 42, 44). Ciprofloxacin was purchased from Mediatech, Herndon, VA. All other chemicals were reagent grade or better and obtained from commercial suppliers.
DNA polymerase activity assays.
Bacterial DNA polymerases were assayed in a 96-well plate format version of the standard DNA polymerase assay (2). Briefly, serial dilutions of compounds in dimethyl sulfoxide (DMSO) were added to the plates. A mixture containing 30 mM Tris, pH 7.5; 10 mM magnesium acetate; 4 mM dithiothreitol; 20% glycerol; 25 μM (each) dATP, dCTP, and dGTP; 10 μM dTTP (labeled at the 5-methyl group with 3H, 1.44 Ci/mmol); and 0.4 mg/ml of activated calf thymus DNA in a final volume of 25 μl was added to the drug dilutions. Assays were initiated by the addition of 0.025 to 0.06 units of enzyme (1 U is the amount required to incorporate 250 pmol of [3H]dTMP in a standard assay), incubated for 10 min at 30°C, and terminated by the addition of an ice-cold solution of 10% trichloroacetic acid containing 10 mM sodium pyrophosphate. Precipitated labeled DNA was collected on glass fiber filter plates (Millipore, Bedford, MA). The plates were washed with cold 1 M HCl and 100 mM sodium pyrophosphate, followed by cold 90% ethanol, dried, and counted in a MicroBeta Trilux liquid scintillation counter (Perkin Elmer, Wellesley, MA). Apparent inhibitor constants (Ki values) for compounds that are competitive with dGTP were determined directly by truncated assay in the absence of dGTP as described previously (40, 41).
The pol α and pol γ assays were performed as modifications of previously published assays (26, 32). Briefly, reactions were initiated by the addition of 0.025 to 0.06 units of pol α or pol γ (1 U is the amount required to incorporate 250 pmol of [3H]dTMP in a standard assay). Following incubation for 25 min at 37°C, the reaction was terminated by the addition of an ice-cold solution of 10% trichloroacetic acid containing 10 mM sodium pyrophosphate and worked up and analyzed as described above for the bacterial DNA polymerases. The bacterial versus mammalian enzymatic pol selectivity index for an inhibitor was determined as the ratio of its Ki value for pol IIIC and its Ki value for a given mammalian DNA polymerase.
DNA topoisomerase IV and gyrase activity assay.
DNA topoisomerase IV and gyrase activities were determined with a catenated DNA substrate prepared from the kinetoplast DNA of the insect trypanosome (Crithidia fasciculata) (8) supplied by Topogen (Port Orange, FL). Assays were performed as previously described in detail (4, 35). Reaction mixes (20 μl final volume) consisted of 40 mM Tris, pH 7.5, 6 mM magnesium chloride, 10 mM dithiothreitol, 100 mM potassium glutamate, 50 μg/ml bovine serum albumin, 1.5 mM ATP, 225 ng of DNA, and either a mixture of the two B. subtilis topoisomerase subunits, C and E, or the two B. subtilis gyrase subunits, A and B. Following incubation for 30 min at 30°C, the reaction was stopped by the addition of 5 μl of a mixture of 50% glycerol, 100 mM EDTA, pH 8.0, and 0.1% bromophenol blue. Products were separated by agarose gel electrophoresis in Tris-acetate-EDTA buffer by the method of Sambrook et al. (36). The appearance of linearized and decatenated supercoiled DNA was monitored by comparison with known linear and decatenated standards provided by Topogen.
Mechanism of action assays. (i) DNA polymerase.
The kinetics of inhibition of pol IIIC were measured as described above with the exception that the concentration of dGTP, the presumed competing substrate, was varied from 64 μM to 1 μM in twofold serial dilutions (including a 0 μM dGTP control). For each set of dGTP dilutions, inhibitor was added at an approximate concentration of 3×, 10×, or 30× the Ki value, obtained from the truncated assay. An inhibitor diluent control (DMSO) was included for each compound. Reaction velocities were determined at each dGTP concentration and used to create double reciprocal plots of velocity versus dGTP concentration. The plots were used to assess the mechanism of inhibition and calculate the apparent Ki value for the inhibitor (37).
(ii) DNA topoisomerase IV and gyrase.
The effects of a compound on the topoisomerase and gyrase activities were followed by adding DMSO stock solutions of the compound to achieve a series of concentrations (0 to 100 μg/ml) and a fixed concentration of DMSO (1%) as described previously (4). Individual samples from the inhibition assays were run on 1% agarose gels containing 0.5 μg/ml ethidium bromide and photographed at 302 nM on an UV light box using a Polaroid camera equipped with type 2B UV (Kodak) and deep yellow 15 (Tiffen) filters. Photographs were digitized by scanning and traced using NIH Image 1.62, which allows direct measurement of the area under each peak of the electropherogram tracing. The 50% inhibitory concentration (IC50) is defined as the concentration of compound that inhibits the decatenation of kinetoplast DNA by 50%.
(iii) Effect of compounds on macromolecule synthesis.
Compounds were examined at 5× MIC for their inhibitory effects on the synthesis of DNA, RNA, and protein in B. subtilis BD54 as the test organism. The assay exploited the incorporation of an appropriate radiolabeled precursor into a given macromolecule: [8-3H]adenine for DNA and RNA and l-[4,5-3H]leucine for protein (11, 39). Briefly, B. subtilis BD54 cultures were grown to log phase in C plus 50 media (10). At time zero, both the compound of interest in DMSO and the appropriate labeled synthetic precursor were added to the growing cells. At various time points, samples were collected from each culture and processed as described previously (11, 39). The radioactivity in the alkali-sensitive RNA was calculated as the difference between the total trichloroacetic acid (TCA)-insoluble radioactivity (i.e., DNA plus RNA) and the alkali-insensitive portion of the TCA-insoluble material (i.e., DNA). For protein synthesis, the incorporation of labeled leucine was assessed simply as TCA-precipitable radioactivity. The effect of compound on synthesis was based on the comparison to isotope incorporation in appropriate diluent controls.
Determination of MIC.
MICs were determined by the broth microdilution method outlined in the CLSI (formerly NCCLS) guidelines (31) with modifications to incubation conditions. Log-phase bacterial cultures were grown in cation-adjusted Mueller-Hinton broth (MHB) or MHB with 5% defibrinated horse blood (for Streptococcus species). Diluted bacterial cultures from log-phase growth were seeded into 96-well plates at a concentration of 1 × 105 CFU/ml, and plates were incubated at 37°C for 16 to 20 h with shaking. Streptococcus species were grown under the same conditions but in the presence of 5% humidified CO2. Test compound stock solutions in DMSO were diluted into media to achieve a final concentration of 1% DMSO. Cell growth was determined by measuring optical density (600 nm) in a microplate reader (Dynex Technologies, Chantilly, VA). MICs for antimicrobial compound-treated cultures were calculated as the lowest concentration of drug at which growth was not apparent, as measured by optical density at 600 nm. Values were determined in quadruplicate, and the highest MIC was listed as the MIC in data tables.
Determination of bactericidal activity.
The method to assess bactericidal potential was that used previously for AU compounds (39). Briefly, stock solutions of compounds in DMSO were diluted 100-fold to achieve 8×, 4×, or 2× the known MIC in MHB media containing the bacterial strain at a concentration of 1 × 106 CFU/ml. Inoculating cultures represented dilutions of mid log-phase growth. The cultures were incubated at 37°C for 24 h, and at various time points, 100-μl samples were removed and serially diluted in fresh MHB and then plated onto drug-free MHB-agar plates to determine the number of CFU/ml present in the sample (CFU/ml = number of colonies on the plate multiplied by the dilution factor and adjusted for a volume of 1 ml). The log10 value of CFU/ml was plotted versus time.
Determination of spontaneous mutation frequency.
The frequency of single-step spontaneous mutations to drug resistance of four bacterial strains was measured by a modification of the procedure of Barry et al. (7). Approximately 109 to 1010 CFU of log-phase bacteria were plated on 150-mm-diameter MHB agar plates containing compounds at 8× or 4× their respective MICs and incubated at 37°C for 48 h. In addition, several dilutions of each culture were also plated on drug-free media to provide accurate colony counts. Mutation frequencies were calculated by dividing the number of colonies growing on drug-containing plates by the total number of CFU that were plated (16). Each experiment was performed in triplicate on separate days, and the mutation frequencies represent an average of the three values obtained. To verify their stability, the resistant organisms were transferred several times in drug-free medium and then retested for resistance in MIC assays. All colonies were also tested with a panel of antibiotics (gentamicin, ciprofloxacin, vancomycin, methicillin, and rifampin) to verify the antibiotic resistance phenotype (i.e., target-based mutation) of each mutant (data not shown).
Multistep resistance selection.
Multistep resistance selection for 251D and its parent compounds, HB-EMAU and 340E, was performed as a modification of a previously described experiment (30). Ninety-six-well plates containing 150 μl of MHB and doubling dilutions of test compounds were inoculated with 5 × 105 CFU/ml of S. aureus (Smith) at compound concentrations ranging from 3 doubling dilutions above to 3 doubling dilutions below the MIC for each agent. The initial inoculum was prepared by diluting a mid-log-phase growth. Plates were incubated at 37°C for 24 h with shaking. Serial passages were performed by taking 50 μl from the well with growth at the highest compound concentration, growing overnight in compound-free media, and inoculating a fresh plate containing doubling dilutions of test compounds. Cultures from each passage were tested in an MIC assay against the selecting agent and the other two compounds.
Determination of mammalian cytotoxicity.
Cytotoxicity of the compounds was measured by plating MRC-5 cells, a human diploid fibroblast line (ATCC CLL-171), in 96-well plates (4 × 103 cells per well) in the presence or absence of compounds that had been added as a DMSO stock (final concentration of 1%). The latter culture and an identical control culture containing only DMSO were incubated at 37°C for 72 h in minimal essential medium (Gibco/Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum, and cell viability was tested with the vital stain MTS (28) according to the manufacturer's instructions (Promega, Madison, WI). Cytotoxicity was quantified as the CC50, the concentration of compound that inhibited 50% of conversion of MTS to formazan (28). The “therapeutic index” of a given agent is defined as the ratio of its mammalian cell cytotoxicity to its MIC against S. aureus Smith (i.e., CC50/MIC) in the presence of 10% fetal calf serum.
RESULTS
Target specificity and mechanism of hybrid action.
Compounds with the AU pharmacophore are generally highly selective inhibitors of pol IIIC (6), with no activity against the gram-positive pol IIIE or the related pol IIIE from a gram-negative organism. The results shown in Table 1 demonstrate that 251D, a representative of the AU-FQ hybrid pharmacophore, is a potent inhibitor of pol IIIC and, like HB-EMAU, is highly selective for pol IIIC, with little or no effect on gram-positive or gram-negative pol IIIE or the mammalian enzymes pol α and pol γ. The selectivity indices of 251D for the mammalian enzymes were >25,000 for pol α and 36,000 for pol γ, comparable to the corresponding indices of the “nonhybrid” compound HB-EMAU (>16,000 for both pol α and γ).
TABLE 1.
Inhibition of DNA polymerases
| Compound |
Ki (μM) fora:
|
||||
|---|---|---|---|---|---|
| B. subtilis pol IIIC | B. subtilis pol IIIE | E. coli pol IIIE | Calf thymus pol α | Human pol γ | |
| HB-EMAU | 0.063 | 117 | >1,008 | >1,008 | >1,008 |
| 251D | 0.019 | >482 | >482 | >482 | 684 |
Ki values were determined in a truncated assay (−dGTP) as described in Materials and Methods.
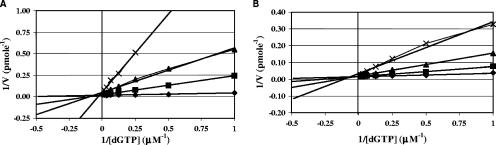
Compounds with the AU pharmacophore are specifically competitive with dGTP. To determine whether this mechanism also applies to the AU-FQ hybrid, we compared the effects of varying dGTP concentration on the anti-pol IIIC action of 251D and HB-EMAU. The results, which are summarized in the classical double reciprocal plots of Fig. 2, indicated that 251D, like the parent AU compound, HB-EMAU, is specifically competitive with dGTP, with a Ki calculated (37) from the plot of 0.025 μM, a value essentially identical to the Ki value obtained using the standard truncated assay, 0.019 μM (Table 1). The Ki for HB-EMAU calculated from the plot was 0.082 μM, a value essentially identical to the Ki value obtained using the standard truncated assay, 0.063 μM (Table 1).
FIG. 2.
Double reciprocal plots demonstrating the effect of varying dGTP concentrations on inhibition of pol IIIC. (A) 251D; (B) HB-EMAU. Inhibitor concentrations: ♦, no inhibitor; ▪, 3× Ki; ▴, 10× Ki; ×, 30× Ki.
We compared the effects of 251D and its parent FQ compound, 340E, as well as ciprofloxacin, nalidixic acid, and HB-EMAU on the classical FQ targets, topoisomerase and gyrase. The results are summarized in Table 2. Ciprofloxacin was a potent inhibitor of both enzymes but exhibited a fourfold-greater potency for the B. subtilis topoisomerase than for the gyrase as expected for gram-positive bacteria (4, 8, 35). In addition, nalidixic acid was a substantially less potent inhibitor of both enzymes, consistent with previously reported results (8, 35). Compound 340E was slightly less potent than ciprofloxacin against the DNA topoisomerase and equipotent with ciprofloxacin against the DNA gyrase. The AU-FQ hybrid, 251D, inhibited both the topoisomerase and gyrase with potencies lower than those of 340E and ciprofloxacin but greater than those of nalidixic acid. The parent AU, HB-EMAU, was essentially inactive against both enzymes.
TABLE 2.
Inhibition of B. subtilis topoisomerase IV and gyrase
| Compound | IC50 (μM) for:
|
|
|---|---|---|
| Topoisomerase IV | Gyrase | |
| 251D | 43.6 | 31 |
| 340E | 4.2 | 5.6 |
| Ciprofloxacin | 1.7 | 6.3 |
| Nalidixic acid | 1,090 | 149 |
| HB-EMAU | >500 | >500 |
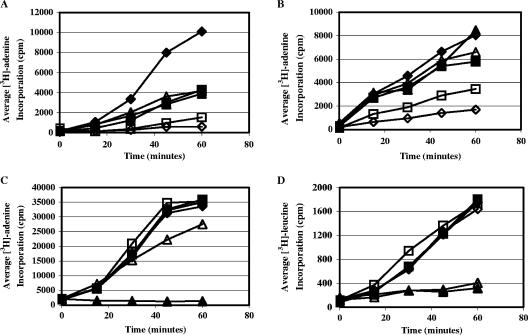
The AU pol IIIC inhibitors and the FQ topoisomerase/gyrase inhibitors are in general strongly selective for inhibition of replicative DNA synthesis in the whole bacterial cell compared with RNA or protein synthesis (11, 15). The effect of 251D on macromolecule synthesis in log-phase B. subtilis was compared to that of HB-EMAU and 340E by measuring DNA, RNA, and protein synthesis over a time course (60 min). All compounds were tested at 5× their respective MICs for B. subtilis. In addition, all compounds were tested at 1× their respective MICs in the DNA synthesis assay. We also included control compounds, rifampin, which is known to inhibit RNA synthesis, and chloramphenicol, which is known to inhibit protein synthesis. The results, displayed in Fig. 3, demonstrate that 251D, like HB-EMAU and 340E, potently inhibits DNA synthesis at a concentration of 5× MIC and, like HB-EMAU, potently inhibits DNA synthesis at 1× MIC with no effect on the synthesis of RNA or protein at 5× MIC. In addition, we demonstrated that rifampin and chloramphenicol, while not as specific for their targeted macromolecule synthesis at concentrations of 5× MIC, were unable to inhibit DNA synthesis at concentrations of 1× MIC. Unexpectedly, compound 340E was no more potent at inhibiting DNA synthesis than rifampin and chloramphenicol at 1× MIC, a finding that does not correspond with the results observed in a similar experiment with ciprofloxacin (M. M. Butler, unpublished results).
FIG. 3.
Effect of compounds on macromolecule synthesis in B. subtilis. Assays were performed using radiolabeled precursor ([3H]adenine for DNA and RNA and [3H]leucine for protein synthesis measurements) and B. subtilis strain BD54 as described in Materials and Methods using compounds at 5× or 1× their MICs as indicated. (A) DNA synthesis using compounds at 5× their respective MICs; (B) DNA synthesis using compounds at 1× their respective MICs; (C) RNA synthesis using compounds at 5× their respective MICs; (D) protein synthesis using compounds at 5× their respective MICs. DMSO control, filled diamonds; HB-EMAU, open diamonds; 340E, filled squares; 251D, open squares; rifampin, filled triangles; chloramphenicol, open triangles.
Antibacterial activity.
MICs of 251D and, for comparison, HB-EMAU and 340E were determined against a broad panel of gram-positive and gram-negative organisms. Among the gram-positive organisms, several were multidrug-resistant strains. We had a particular interest in exploring the potential of clinically derived, multiantibiotic-resistant gram-positive pathogens for displaying cross-resistance to the AU-FQ hybrid.
The results shown in Table 3 demonstrate that 251D has potent antibacterial activity against the entire panel of gram-positive organisms tested, being 5- to 15-fold more potent then the parent AU compound, HB-EMAU. Significantly, the hybrid potently inhibited growth of organisms that were resistant to 340E (i.e., some MRSA and VRE strains). In these organisms, MICs for 251D ranged from 0.625 to 5 μg/ml, as opposed to 10 to 80 μg/ml for 340E. The results shown in Table 3 indicate that 251D is active against mupirocin-sensitive and -resistant S. aureus strains, which are also resistant to both HB-EMAU and 340E. Compound 251D is also active against one strain of linezolid-resistant VREF, displaying MICs that were 8- to 64-fold lower than those of the parent compounds. Among the streptococcal strains, 251D was more potent than HB-EMAU and nearly equipotent with 340E in all cases.
TABLE 3.
Antibacterial activity in vitro
| Organism | MIC (μg/ml) of:
|
||
|---|---|---|---|
| 251D | HB-EMAU | 340E | |
| Bacilli | |||
| B. subtilis BD54 | 0.156 | 1.25 | 0.039 |
| B. cereus 4342 | 0.156 | 1.25 | 0.078 |
| B. cereus 6464 | 0.156 | 1.25 | 0.078 |
| B. thuringiensis 10792 | 0.156 | 1.25 | 0.156 |
| B. thuringiensis 33679 | 0.156 | 2.5 | 0.078 |
| B. anthracis Sterne | 0.156 | 2.5 | 0.039 |
| Staphylococci | |||
| S. aureus 25923 | 0.625 | 5 | 0.156 |
| S. aureus (Smith) 13709 | 0.313 | 5 | 0.078 |
| S. aureus 1081 | 0.625 | 5 | 10 |
| MRSA 1094 | 1.25 | 5 | 20 |
| MRSA B42876 | 2.5 | 5 | 80 |
| MRSA 1123 | 1.25 | 5 | 20 |
| MRSA 1132 | 0.625 | 5 | 0.156 |
| S. aureus mupirocin-sensitive 2529 | 5 | 20 | 80 |
| S. aureus mupirocin-resistant 777 | 2.5 | 10 | 40 |
| Enterococci | |||
| E. faecalis 29212 | 1.25 | 5 | 0.313 |
| VREF 700802 | 1.25 | 5 | 0.313 |
| VREF 51575 | 1.25 | 5 | 0.313 |
| VREF 51299 | 1.25 | 5 | 0.313 |
| E. faecium 19434 | 2.5 | 5 | 10 |
| Vancomycin-resistant E. faecium B42762 | 1.25 | 5 | 40 |
| Vancomycin-resistant E. faecium 1644 | 1.25 | 2.5 | 40 |
| VREF F118 (linezolid resistant) | 1.25 | 10 | 80 |
| Streptococci | |||
| S. pneumoniae 49619 | 0.625 | 2.5 | 0.313 |
| PRSP 700670 (Spain) | 0.625 | 5 | 0.313 |
| PRSP 700674 (S. Africa) | 0.625 | 5 | 0.625 |
| PRSP 700671 (France) | 0.313 | 2.5 | 0.313 |
| PRSP 700673 (Hungary) | 0.313 | 2.5 | 0.156 |
| PRSP 700677 (Slovakia) | 0.313 | 2.5 | 0.625 |
| S. pyogenes 12344 | 0.313 | 1.25 | 0.156 |
| Gram-negative strains | |||
| E. coli J53 | 2.5 | >80 | 0.01 |
| E. coli XL1-Blue (ciprofloxacin resistant) | 1.25 | >80 | 40 |
| K. pneumoniae 5657 | 20 | >80 | 0.156 |
| S. maltophilia 13637 | 10 | >80 | 0.078 |
| P. aeruginosa 27853 | >80 | >80 | 0.313 |
Although AU compounds do not significantly inhibit the growth of gram-negative bacteria (39), the presence of the FQ moiety in the hybrid compounds provides the possibility for endowing the AU-FQ hybrid with anti-gram-negative activity. As expected, HB-EMAU was inactive at a concentration of 80 μg/ml and 340E was active at concentrations of <1 μg/ml against several gram-negative bacterial strains (Table 3). The hybrid, 251D, did show moderate potency against standard gram-negative strains with MICs ranging from 1.25 to 20 μg/ml, with the exception of P. aeruginosa, where the compound was inactive at the highest concentration tested (Table 3). Interestingly, 251D displayed 32-fold-greater potency than 340E against E. coli XL1-Blue, a laboratory strain with a known gyrA mutation that renders it fluoroquinolone resistant (Stratagene).
To confirm the dual-target activity of 251D in intact bacteria, we assessed the potency of 251D against singly resistant (HB-EMAU or ciprofloxacin resistant) as well as doubly resistant (HB-EMAU and ciprofloxacin resistant) S. aureus. As shown in Table 4, 251D retains potent activity against S. aureus strains that are singly resistant to each of its component moieties. When tested against HB-EMAU-resistant S. aureus carrying a point-mutated form of pol IIIC (12), 251D was nearly as potent against this strain as it was against the wild-type parent strain (twofold increase in MIC). Compound 251D was also nearly as potent against a ciprofloxacin-resistant S. aureus carrying a topoisomerase mutation as it was against the isogenic strain carrying the wild-type topoisomerase (fourfold increase in MIC). Only when we tested a double pol IIIC/topoisomerase mutant that was both HB-EMAU- and ciprofloxacin-resistant did we show significant resistance to 251D (64-fold increase in MIC).
TABLE 4.
Effect of compounds on AU- and FQ-resistant S. aureus (Smith) strains
| Strain type | MIC (μg/ml) of:
|
||
|---|---|---|---|
| 251D | HB-EMAU | 340E | |
| Wild type | 0.313 | 5 | 0.078 |
| HB-EMAU resistant | 0.625 | >80 | 0.078 |
| Ciprofloxacin resistant | 1.25 | 5 | 5 |
| HB-EMAU resistant and ciprofloxacin resistant | 20 | >80 | 5 |
With the purpose of enumerating the advantages of the hybrid compound over the simple addition of two separate agents, we compared the antibacterial activity of 251D to an equimolar combination of the parent compounds. By virtue of its chemical linkage, the hybrid 251D contains equimolar concentrations of the parent compounds at any given concentration. As can be seen in Table 5, when the potencies of the parent compounds differ by 100-fold, as is the case for S. aureus (Smith), the MIC of the combination is approximately equal to the MIC of the most potent compound, and the MIC ratio between the hybrid and the combination is 1, that is, the hybrid and combination are equipotent. However, when the parent compounds are approximately equipotent but weak inhibitors, as is the case for MRSA, E. faecium, and VRE, the hybrid is much more potent than the combination, and the ratio ranges from 8 to 21, producing a meaningful advantage for the hybrid compound.
TABLE 5.
Effect on MICs of combining individual agents versus the linked hybrid
| Bacterial strain | MIC (μM) of:
|
Hybrid/combination ratio | |||
|---|---|---|---|---|---|
| 251D | HB-EMAU | 340E | HB-EMAU + 340E | ||
| S. aureus Smith ATCC 13709 | 0.88 | 28 | 0.26 | 0.88 | 1 |
| MRSA clinical isolate 1094 | 2.6 | 28 | 30 | 30 | 11.5 |
| E. faecium ATCC 19434 | 3.75 | 30 | 30 | 30 | 8.0 |
| VRE clinical isolate B42762 | 2.1 | 30 | >60 | 45 | 21.3 |
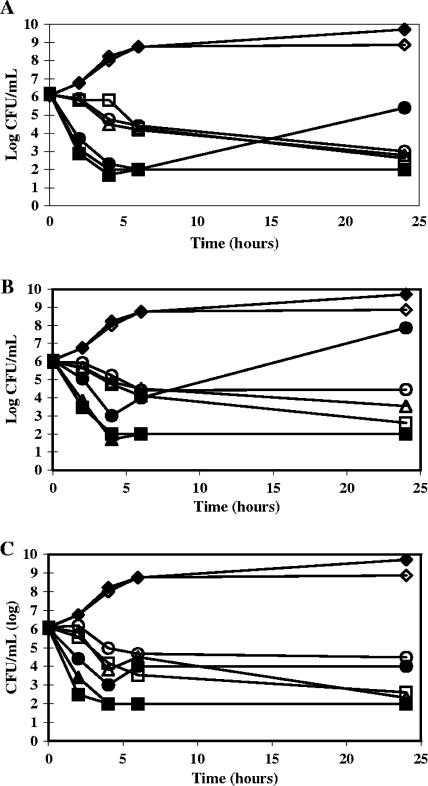
According to Pankey and Sabath (33), a compound is defined as bactericidal if it effects a 3-log or greater decrease in bacterial cell density after 24 h of incubation. The bactericidal potential of 251D was assessed in time kill assays with two representative gram-positive organisms, S. aureus and E. faecium. Each strain was grown to early log phase, diluted to 1 × 106 CFU/ml, and incubated for 24 h with 251D or its parent compounds at 8×, 4×, or 2× their corresponding MICs, and CFU/ml counts were measured at several time points, including 24 h. As shown in Fig. 4, 251D was clearly bactericidal for both test organisms at concentrations of 8× and 4× MIC and was bactericidal for E. faecium at 2× MIC (Fig. 4A). At 2× MIC, 251D displayed rapid bactericidality (within 2 h) for S. aureus followed by rebound growth at 24 h (Fig. 4A). Parent compound HB-EMAU was bactericidal for S. aureus at concentrations of 8× and 4× MIC and for E. faecium at a concentration of 8× MIC, with bacteriostatic activity observed with E. faecium at concentrations of 4× and 2× MIC. However, like the hybrid 251D, HB-EMAU at 2× MIC displayed rapid bactericidality (within 2 h) for S. aureus followed by rebound growth at 24 h (Fig. 4B). Finally, 340E was bactericidal for S. aureus and E. faecium at concentrations of 8× and 4× MIC and static at 2× MIC (Fig. 4C).
FIG. 4.
Time kill assays showing the bactericidal activities of 251D (A), HB-EMAU (B), and 340E (C). S. aureus (ATCC 13709), filled symbols; E. faecium (ATCC 19434), open symbols; control, diamonds; 8× MIC, squares; 4× MIC, triangles; 2× MIC, circles.
Resistance studies.
The frequency of development of spontaneous (single step) resistance to 251D was estimated using four representative gram-positive organisms. Table 6 summarizes the results and includes for comparison results for 340E and HB-EMAU (also reported previously [12]). At 8× MIC, mutation frequencies for HB-EMAU were in the range of 10−9 to 10−10, but we were unable to isolate 251D- or 340E-resistant strains using this method. Upon plating strains on agar containing compounds at 4× MIC, we were again unable to isolate 251D-resistant strains but obtained HB-EMAU resistance with all four strains and 340E-resistant S. aureus (Table 6). These results indicate that the frequency of development of resistance to 251D is lower than that of the parent AU compound and lower than or similar to the frequency of the parent FQ compound after a single passage.
TABLE 6.
Spontaneous resistance frequencies of compounds
| Strain | Spontaneous mutation frequency (1010) of strain at MIC multiplicity
|
|||||
|---|---|---|---|---|---|---|
| 251D
|
HB-EMAU
|
340E
|
||||
| 8× | 4× | 8× | 4× | 8× | 4× | |
| S. aureus 13709 | <1.1 | <5.6 | 7.3 | 160 | <1.1 | 5,100 |
| MRSA 1094 | <2.3 | <6.7 | 59 | 51,400 | <4.5 | <3.6 |
| E. faecium 19434 | <4.2 | <8.3 | 3.6 | 230 | <2.3 | <2.4 |
| VRE B42762 | <5.6 | <2.5 | 1,200 | 57 | <3.1 | <2.4 |
To ensure the isolation of resistant mutants and permit the examination of cross-resistance, we exposed S. aureus (Smith; 13709) to a range of twofold serial dilutions (3 dilutions above and 3 dilutions below the MIC) of 251D, HB-EMAU, and 340E for 17 passages in liquid medium. After each passage, isolates from the highest compound concentration that supported growth were subcultured in compound-free medium and then tested for susceptibility to 251D, HB-EMAU and 340E in an MIC assay. After 17 passages, S. aureus had developed a 16-fold increase in MIC for 251D (Table 7) as well as a >32-fold increase in MIC for HB-EMAU and a 16-fold increase in MIC for 340E. As expected, S. aureus selected to be resistant to 251D was also 16- to 32-fold resistant to its parent compounds, 340E and HB-EMAU. In addition, S. aureus selected to be resistant to either parent was also cross-resistant to 251D.
TABLE 7.
Development of resistance to compounds on liquid medium using the serial passage method with S. aureus
| Compound for passage | Compound for MIC test | Increase in MIC over wild-type S. aureus (fold) at passage:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Nonea | 3 | 6 | 9 | 12 | 15 | 17 | ||
| 251D | 251D | 1 | 4 | 8 | 16 | 16 | 16 | 16 |
| HB-EMAU | 1 | 1 | 2 | 4 | 8 | 16 | 32 | |
| 340E | 1 | 4 | 8 | 8 | 16 | 16 | 16 | |
| 251D | 1 | 2 | 4 | 4 | 2 | 4 | 4 | |
| HB-EMAU | HB-EMAU | 1 | 2 | 8 | >32b | >32 | >32 | >32 |
| 340E | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 251D | 1 | 2 | 4 | 4 | 2 | 4 | 4 | |
| HB-EMAU | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 340E | 340E | 1 | 4 | 8 | 16 | 8 | 8 | 16 |
Prior to passage.
Increase in MIC exceeded the solubility limit for this compound.
In vitro toxicity.
The cytotoxicity of 251D was compared with that of HB-EMAU and 340E using MRC-5 cells, a diploid human fibroblast cell line, in an MTS metabolism assay (28). The results, shown in Table 8, clearly indicate that the cytotoxicity of both 251D and 340E was low (CC50 = >80 μg/ml) and significantly lower than that of HB-EMAU (CC50 = 46 μg/ml). A predicted “therapeutic index,” the ratio between the CC50 and the antibacterial potency (MIC) for a typical sensitive organism, was calculated using MICs obtained in the presence of 10% fetal calf serum (FCS) to more closely mimic the potential protein binding conditions present in the cytotoxicity assay. The presence of 10% FCS caused a twofold increase in MIC for 251D, a fourfold increase for HB-EMAU, and no increase for 340E. Nevertheless, the predicted therapeutic index was higher for 251D than for HB-EMAU and was comparable to that of 340E (Table 8).
TABLE 8.
Mammalian cell (MRC-5) cytotoxicity
| Compound | CC50 (μg/ml) for MRC-5 | MICa for S. aureus (Smith) (μg/ml) | “Therapeutic index” (CC50/MIC) |
|---|---|---|---|
| 251D | >80 | 0.625 | >128 |
| HB-EMAU | 46 | 20 | 2.3 |
| 340E | >80 | 0.078 | >1,000 |
MICs were determined as described in Materials and Methods with the addition of 10% FCS to the growth medium to account for protein binding effects in the cytotoxicity assay.
DISCUSSION
Our goal for the development of the AU class of pol IIIC inhibitors is to maximize the antibacterial potency of this pharmacophore and enhance its potential for use in the treatment of gram-postive infections in vivo. By fusing the AU and FQ pharmacophores, we have generated not simply an improved pol IIIC inhibitor but a truly functional AU-FQ “hybrid” pharmacophore that retains the capacity to inhibit both of the targets of its parents. In sum, this novel compound class displays properties consistent with strong antibacterial potential. First, it has potent antibacterial and bactericidal activity against a broad range of low-GC gram-positive organisms. Second, it does not show cross-resistance to clinically derived strains that are resistant to commonly used antibiotics. Third, it demonstrates low in vitro toxicity in both enzyme selectivity and human cell cytotoxicity assays, a property that could possibly be translated to in vivo assays. Fourth, its capacity to hit two targets independently is expected to reduce the frequency of resistance development in the clinical setting. Finally, the hybrid pharmacophore exhibits striking synergistic antibacterial effects with respect to its AU and FQ components against several MRSA and VRE clinical strains.
The results confirm that 251D combines the in vitro target specificity of its AU and FQ components, inhibiting both topoisomerase/gyrase and pol IIIC, and maintains the specificity for inhibition of DNA synthesis and bactericidal mechanism of antibacterial activity of both parents. The anti-topoisomerase/gyrase activities of 251D are reduced by 10-fold relative to that of “free” 340E, which is most likely the result of steric hindrance caused by the moiety attached to the 7-piperazinyl substituent of the FQ. However, this reduced enzyme activity translates into only a fourfold decrease in antibacterial potency (in B. subtilis, for which enzyme and MIC data are presented). In contrast, the anti-pol IIIC potency of 251D is enhanced threefold by the presence of the FQ moiety. The reason for this enhancement is not clear, but it may derive from an FQ-induced increase in affinity of the AU moiety for the AU-specific binding pockets of pol IIIC. This increase in enzyme inhibitory potency can at least partially account for the eightfold increase in antibacterial potency of the hybrid versus the AU component. The limited structure activity relationship studies that have been performed with hybrid compounds so far (44) fail to resolve this discrepancy between target-related potency and antibacterial activity. We speculate below on three potential explanations for this phenomenon and are currently exploring these possibilities.
The antibacterial potency and the spectrum of activity of the hybrid compound are greater than those of the AU or FQ pharmacophores alone (Table 3). For example, unlike the corresponding AU, the hybrid was active against several gram-negative organisms. In addition, the hybrid provided an advantage over the FQ parent, 340E, against a variety of antibiotic-resistant gram-positive strains, including MRSA, mupirocin- and ciprofloxacin-resistant S. aureus, and linezolid-resistant and ciprofloxacin-resistant VRE. In several cases, 251D was more potent than either parent compound (by 4- to 64-fold). A dramatic example is E. coli XL1-Blue, a laboratory cloning strain that carries a gyrA mutation that renders the strain FQ resistant (Stratagene). In this case, the only targets of the hybrid molecule are the topo/gyrase enzymes, as the AU component is inactive against E. coli pol III (42). The MIC of 251D for this strain was 1.25, but the MIC for the active parent, 340E, was 40, representing a 32-fold difference. Typically, a single gyrA mutation confers two- to fourfold FQ resistance (23), and so other factors likely play a role in this strain's resistance to 340E and lack of resistance to 251D.
Perhaps the most surprising discovery of this study is that the hybrid 251D provides a striking advantage over both parent compounds administered together, exhibiting more potency than an equimolar combination of the parent compounds against strains for which the potencies of the parent compounds are equivalent (e.g., MRSA) (Table 5). We have found previously that combinations of AU and FQ compounds do not display synergy (W. LaMarr, unpublished result). Apparently, fusion of the AU and FQ components in a hybrid molecule creates a synergistic antibacterial effect, which is absent without the covalent linkage. This observation is not without precedent, as others have found that oxazolidinone-quinolone hybrids have greater potency than either parent when tested against antibiotic-resistant strains (18, 21). We postulate that the unexpected potency of 251D against strains that are less sensitive to both parent compounds is a result of (i) a greater ability to enter bacterial cells by diffusion (gram positive) or porin-mediated influx (gram negative) (ii) a reduced susceptibility to FQ-specific efflux pumps such as norA, norB, or norC, or (iii) increased potency against pol IIIC due to enhanced binding provided by the FQ moiety (as is likely the case with the quinolone moiety of the oxazolidinone-quinolone hybrid enhancing binding to the ribosomal target of linezolid) (21). Studies that address these possibilities are ongoing.
Measurements of cross-resistance among AU- and FQ-resistant mutants and double mutants strongly suggest that the hybrid acts in the intact cell by targeting both the polymerase and topoisomerase/gyrase activities (Table 4). For example, the hybrid compound is nearly as potent against either HB-EMAU or ciprofloxacin-resistant S. aureus strains as it is against the wild-type strain. In the case of a strain carrying a single mutation in the polC gene, in one of the four sites known to be capable of mutation to resistance (3), we saw a 16-fold increase in MIC for an AU compound, as was observed in a previously described experiment (11). Likewise, we observed a 64-fold increase in the FQ MIC in an FQ-resistant strain carrying dual gyrase and topoisomerase mutations as described previously (24). In contrast, marked resistance (64-fold) to 251D only occurred in the case of a double, i.e., HB-EMAUr/Cipr, resistant mutant (Table 4). This finding is consistent with the inability to generate single-step 251D-resistant mutants (Table 6) under conditions in which resistance was readily observed with the individual FQ and AU moieties. This low frequency of 251D-resistant mutations is consistent with the need to obtain mutations in each of two independent targets (i.e., polymerase and topoisomerase/gyrase) to yield high-level (>8-fold) AU-FQ resistance. Indeed, in serial passage experiments in which resistance to 251D was gradually selected, resistance to 340E developed by passage 3, while resistance to HB-EMAU appeared later in passage 9 (Table 7). Resistance to both parental compounds produced a cumulative effect, resulting in 16-fold resistance to 251D by passage 9.
In further support of the AU-FQ hybrid, we have recently demonstrated that members of this compound class are, in fact, efficacious when given intravenously in vivo in a murine staphylococcal infection model, confirming their potential as novel anti-infective agents (44). In sum, we have developed a potent antibacterial agent with a dual mechanism of action, which provides an advantage over single agents in the treatment of antibiotic-resistant gram-positive infections.
Acknowledgments
We thank Donald Moir, Microbiotix, Inc., for extensive editorial assistance. We also thank Richard Ellison, University of Massachusetts Medical School, Lona Mody, University of Michigan Medical School, David Hooper, Massachusetts General Hospital, Jason Prystowsky, Northwestern Memorial Hospital, and George Jacoby, Lahey Clinic, for donations of bacterial strains.
This work was supported in part by small business grant GM060828 from the National Institutes of Health (to GLSynthesis, Inc.) and by Shire BioChem Ltd., Laval, Canada.
Footnotes
Published ahead of print on 30 October 2006.
REFERENCES
- 1.Aeschlimann, J. R., E. Hershberger, and M. J. Rybak. 1999. Analysis of vancomycin population susceptibility profiles, killing activity, and postantibiotic effect against vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 43:1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes, M. H., and N. C. Brown. 1979. Antibodies to B. subtilis DNA polymerase III: use in purification and examination of homology among replication-specific DNA polymerases. Nucleic Acids Res. 6:1203-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, M. H., R. A. Hammond, C. C. Kennedy, S. L. Mack, and N. C. Brown. 1992. Localization of the exonuclease and polymerase domains of Bacillus subtilis DNA polymerase III. Gene 111:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, M. H., W. A. LaMarr, and K. A. Foster. 2003. DNA gyrase and DNA topoisomerase of Bacillus subtilis: expression and characterization of recombinant enzymes encoded by the gyrA, gyrB and parC, parE genes. Protein Expr. Purif. 29:259-264. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, M. H., S. D. Miller, and N. C. Brown. 2002. DNA polymerases of low-GC gram-positive eubacteria: identification of the replication-specific enzyme encoded by dnaE. J. Bacteriol. 184:3834-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes, M. H., P. M. Tarantino, Jr., P. Spacciapoli, N. C. Brown, H. Yu, and K. Dybvig. 1994. DNA polymerase III of Mycoplasma pulmonis: isolation and characterization of the enzyme and its structural gene, polC. Mol. Microbiol. 13:843-854. [DOI] [PubMed] [Google Scholar]
- 7.Barry, A. L., S. D. Brown, and P. C. Fuchs. 1996. In-vitro selection of quinolone-resistant staphylococcal mutants by a single exposure to ciprofloxacin or trovafloxacin. J. Antimicrob. Chemother. 38:324-327. [DOI] [PubMed] [Google Scholar]
- 8.Blanche, F., B. Cameron, F. X. Bernard, L. Maton, B. Manse, L. Ferrero, N. Ratet, C. Lecoq, A. Goniot, D. Bisch, and J. Crouzet. 1996. Differential behaviors of Staphylococcus aureus and Escherichia coli type II DNA topoisomerases. Antimicrob. Agents Chemother. 40:2714-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, N. C., L. W. Dudycz, and G. E. Wright. 1986. Rational design of substrate analogues targeted to selectively inhibit replication-specific DNA polymerases. Drugs Exp. Clin. Res. 12:555-564. [PubMed] [Google Scholar]
- 10.Brown, N. C., C. L. Weisseman III, and T. Matsushita. 1972. Inhibition of bacterial DNA replication by 6-p-hydroxyphenylazouracil. Nat. New Biol. 237:72-74. [DOI] [PubMed] [Google Scholar]
- 11.Butler, M. M., L. W. Dudycz, N. N. Khan, G. E. Wright, and N. C. Brown. 1990. Development of novel inhibitor probes of DNA polymerase III based on dGTP analogs of the HPUra type: base, nucleoside and nucleotide derivatives of N2-(3,4-dichlorobenzyl)guanine. Nucleic Acids Res. 18:7381-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler, M. M., D. J. Skow, R. O. Stephenson, P. T. Lyden, W. A. LaMarr, and K. A. Foster. 2002. Low frequencies of resistance among Staphylococcus and Enterococcus species to the bactericidal DNA polymerase inhibitor N(3)-hydroxybutyl 6-(3′-ethyl-4′-methylanilino) uracil. Antimicrob. Agents Chemother. 46:3770-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. April. 2006. Frequently asked questions about VISA/VRSA. Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/ncidod/dhqp/ar_visavrsa_FAQ.html.
- 14.Doern, G. V., K. P. Heilmann, H. K. Huynh, P. R. Rhomberg, S. L. Coffman, and A. B. Brueggemann. 2001. Antimicrobial resistance among clinical isolates of Streptococcus pneumoniae in the United States during 1999-2000, including a comparison of resistance rates since 1994-1995. Antimicrob. Agents Chemother. 45:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans, M. E., and W. B. Titlow. 1998. Selection of fluoroquinolone-resistant methicillin-resistant Staphylococcus aureus with ciprofloxacin and trovafloxacin. Antimicrob. Agents Chemother. 42:727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felmingham, D., R. R. Reinert, Y. Hirakata, and A. Rodloff. 2002. Increasing prevalence of antimicrobial resistance among isolates of Streptococcus pneumoniae from the PROTEKT surveillance study, and comparative in vitro activity of the ketolide, telithromycin. J. Antimicrob. Chemother. 50(Suppl. S1):25-37. [DOI] [PubMed] [Google Scholar]
- 18.Gordeev, M. F., C. Hackbarth, M. R. Barbachyn, L. S. Banitt, J. R. Gage, G. W. Luehr, M. Gomez, J. Trias, S. E. Morin, G. E. Zurenko, C. N. Parker, J. M. Evans, R. J. White, and D. V. Patel. 2003. Novel oxazolidinone-quinolone hybrid antimicrobials. Bioorg. Med. Chem. Lett. 13:4213-4216. [DOI] [PubMed] [Google Scholar]
- 19.Hammond, R., M. H. Barnes, S. Mack, J. Mitchener, and N. C. Brown. 1991. Bacillus subtilis DNA polymerase III: complete sequence, overexpression and characterization of the polC gene. Gene 98:29-36. [DOI] [PubMed] [Google Scholar]
- 20.Huang, Y.-P., and J. Ito. 1998. The hyperthermophilic bacterium Thermotoga maritima has two different classes of family C DNA polymerases: evolutionary implications. Nucleic Acids Res. 26:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubschwerlen, C., J. L. Specklin, D. K. Baeschlin, Y. Borer, S. Haefeli, C. Sigwalt, S. Schroeder, and H. H. Locher. 2003. Structure-activity relationship in the oxazolidinone-quinolone hybrid series: influence of the central spacer on the antibacterial activity and the mode of action. Bioorg. Med. Chem. Lett. 13:4229-4233. [DOI] [PubMed] [Google Scholar]
- 22.Huycke, M. M., D. F. Sahm, and M. S. Gilmore. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4:239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ince, D., and D. C. Hooper. 2001. Mechanisms and frequency of resistance to gatifloxacin in comparison to AM-1121 and ciprofloxacin in Staphylococcus aureus. Antimicrob Agents Chemother. 45:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ince, D., and D. C. Hooper. 2000. Mechanisms and frequency of resistance to premafloxacin in Staphylococcus aureus: novel mutations suggest novel drug-target interactions. Antimicrob. Agents Chemother. 44:3344-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhl, A., N. Svenstrup, C. Ladel, M. Otteneder, A. Binas, G. Schiffer, M. Brands, T. Lampe, K. Ziegelbauer, H. Rubsamen-Waigmann, D. Haebich, and K. Ehlert. 2005. Biological characterization of novel inhibitors of the gram-positive DNA polymerase IIIC enzyme. Antimicrob. Agents Chemother. 49:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longley, M. J., P. A. Ropp, S. E. Lim, and W. C. Copeland. 1998. Characterization of the native and recombinant catalytic subunit of human DNA polymerase γ: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry 37:10529-10539. [DOI] [PubMed] [Google Scholar]
- 27.Love, E., D. D'Ambrosio, and N. C. Brown. 1976. Mapping of the gene specifying DNA polymerase III of Bacillus subtilis. Mol. Gen. Genet. 144:313-321. [DOI] [PubMed] [Google Scholar]
- 28.Marshall, N. J., C. J. Goodwin, and S. J. Holt. 1995. A critical assessment of the use of microculture tetrazolium assays to measure cell growth and function. Growth Regul. 5:69-84. [PubMed] [Google Scholar]
- 29.Millichap, J., T. A. Ristow, G. A. Noskin, and L. R. Peterson. 1996. Selection of Enterococcus faecium strains with stable and unstable resistance to the streptogramin RP 59500 using stepwise in vitro exposure. Diagn. Microbiol. Infect. Dis. 25:15-20. [DOI] [PubMed] [Google Scholar]
- 30.Nagai, K., T. A. Davies, G. A. Pankuch, B. E. Dewasse, M. R. Jacobs, and P. C. Appelbaum. 2000. In vitro selection of resistance to clinafloxacin, ciprofloxacin, and trovafloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2740-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6. NCCLS, Wayne, PA.
- 32.Ottiger, H. P., and U. Hübscher. 1984. Mammalian DNA polymerase alpha holoenzymes with possible functions at the leading and lagging strand of the replication fork. Proc. Natl. Acad. Sci. USA 81:3993-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pankey, G. A., and L. D. Sabath. 2004. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of gram-positive bacterial infections. Clin. Infect. Dis. 38:864-870. [DOI] [PubMed] [Google Scholar]
- 34.Prystowsky, J., F. Siddiqui, J. Chosay, D. L. Shinabarger, J. Millichap, L. R. Peterson, and G. A. Noskin. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45:2154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saiki, A. Y., L. L. Shen, C. M. Chen, J. Baranowski, and C. G. Lerner. 1999. DNA cleavage activities of Staphylococcus aureus gyrase and topoisomerase IV stimulated by quinolones and 2-pyridones. Antimicrob. Agents Chemother. 43:1574-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Stryer, L. 1981. Biochemistry, 2nd ed. W. H. Freeman and Co., New York, NY.
- 38.Tarantino, P. M., Jr., C. Zhi, J. J. Gambino, G. E. Wright, and N. C. Brown. 1999. 6-Anilinouracil-based inhibitors of Bacillus subtilis DNA polymerase III: antipolymerase and antimicrobial structure-activity relationships based on substitution at uracil N3. J. Med. Chem. 42:2035-2040. [DOI] [PubMed] [Google Scholar]
- 39.Tarantino, P. M., Jr., C. Zhi, G. E. Wright, and N. C. Brown. 1999. Inhibitors of DNA polymerase III as novel antimicrobial agents against gram-positive eubacteria. Antimicrob. Agents Chemother. 43:1982-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright, G. E., and N. C. Brown. 1990. Deoxyribonucleotide analogs as inhibitors and substrates of DNA polymerases. Pharmacol. Ther. 47:447-497. [DOI] [PubMed] [Google Scholar]
- 41.Wright, G. E., and N. C. Brown. 1976. Inhibition of Bacillus subtilis DNA polymerase III by arylhydrazinopyrimidines. Novel properties of 2-thiouracil derivatives. Biochim. Biophys. Acta 432:37-48. [DOI] [PubMed] [Google Scholar]
- 42.Zhi, C., Z. Y. Long, J. Gambino, W. C. Xu, N. C. Brown, M. Barnes, M. Butler, W. LaMarr, and G. E. Wright. 2003. Synthesis of substituted 6-anilinouracils and their inhibition of DNA polymerase IIIC and gram-positive bacterial growth. J. Med. Chem. 46:2731-2739. [DOI] [PubMed] [Google Scholar]
- 43.Zhi, C., Z. Y. Long, A. Manikowski, N. C. Brown, P. M. Tarantino, Jr., K. Holm, E. J. Dix, G. E. Wright, K. A. Foster, M. M. Butler, W. A. LaMarr, D. J. Skow, I. Motorina, S. Lamothe, and R. Storer. 2005. Synthesis and antibacterial activity of 3-substituted-6-(3-ethyl-4-methylanilino)uracils. J. Med. Chem. 48:7063-7074. [DOI] [PubMed] [Google Scholar]
- 44.Zhi, C., Z. Y. Long, A. Manikowski, J. Comstock, W. C. Xu, N. C. Brown, P. M. Tarantino, Jr., K. A. Holm, E. J. Dix, G. E. Wright, M. H. Barnes, M. M. Butler, K. A. Foster, W. A. LaMarr, B. Bachand, R. Bethell, C. Cadilhac, S. Charron, S. Lamothe, I. Motorina, and R. Storer. 2006. Hybrid antibacterials. DNA polymerase-topoisomerase inhibitors. J. Med. Chem. 49:1455-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]