Abstract
Squalene epoxidase (SE) is the target of terbinafine, which specifically inhibits the fungal enzyme in a noncompetitive manner. On the basis of functional homologies to p-hydroxybenzoate hydroxylase (PHBH) from Pseudomonas fluorescens, the Erg1 protein contains two flavin adenine dinucleotide (FAD) domains and one nucleotide binding (NB) site. By in vitro mutagenesis of the ERG1 gene, which codes for the Saccharomyces cerevisiae SE, we isolated erg1 alleles that conferred increased terbinafine sensitivity or that showed a lethal phenotype when they were expressed in erg1-knockout strain KLN1. All but one of the amino acid substitutions affected conserved FAD/nucleotide binding sites. The G25S, D335X (W, F, P), and G210A substitutions in the FADI, FADII, and NB sites, respectively, rendered the SE variants nonfunctional. The G30S and L37P variants exhibited decreased enzymatic activity, accompanied by a sevenfold increase in erg1 mRNA levels and an altered sterol composition, and rendered KLN1 more sensitive not only to allylamines (10 to 25 times) but also to other ergosterol biosynthesis inhibitors. The R269G variant exhibited moderately reduced SE activity and a 5- to 10-fold increase in allylamine sensitivity but no cross-sensitivity to the other ergosterol biosynthesis inhibitors. To further elucidate the roles of specific amino acids in SE function and inhibitor interaction, a homology model of Erg1p was built on the basis of the crystal structure of PHBH. All experimental data obtained with the sensitive Erg1 variants support this model. In addition, the amino acids responsible for terbinafine resistance, although they are distributed along the sequence of Erg1p, cluster on the surface of the Erg1p model, giving rise to a putative binding site for allylamines.
Squalene epoxidase (SE; EC 1.14.99.7) is a mono-oxygenase that catalyzes the conversion of squalene to 2,3-oxidosqualene. For this reaction the enzyme requires molecular oxygen, flavin adenine dinucleotide (FAD), and, depending on the organism, either NADH or NADPH (for a recent review, see reference 25). Squalene epoxidases are essential for the synthesis of cholesterol in mammals and ergosterol in fungi and, thus, comprise useful and medically important targets that can be used to lower cholesterol levels (2) or to inhibit the growth of pathogenic fungi (25). Inhibitors of mammalian SE, such as NB598 and derivatives thereof (10, 30), have not reached the market. For the treatment of fungal infections, however, the allylamine derivatives naftifine and terbinafine have been applied successfully and were shown to specifically inhibit fungal squalene epoxidases (26, 27).
The first gene encoding SE was isolated from a terbinafine-resistant Saccharomyces cerevisiae mutant (11); and in recent years the SE-encoding genes of many other organisms were isolated and characterized, including those of pathogenic fungi, plants, mice, rats, and humans (24). Fungal squalene epoxidases are selectively inhibited by the allylamines terbinafine and naftifine in a noncompetitive manner with regard to the substrate squalene (27). In contrast, mammalian squalene epoxidases are competitively inhibited by NB598, which also belongs to the class of allylamines and contains the same side chain as terbinafine (10). This important difference in the mode of inhibition indicates that the structures of SEs of fungal and mammalian origins must differ at least with respect to the binding domains for the inhibitors (5, 6).
We have recently reported on terbinafine-resistant S. cerevisiae mutants, all of which carry unique amino acid substitutions in the Erg1 protein (13, 15). Interestingly, the amino acid residues affected are conserved in the squalene epoxidases of various origins and are distributed along the sequence of the Erg1 protein (25). The altered amino acids in the mutants were suggested to be part of the binding site for allylamines (15). Recently, terbinafine-resistant mutants of Trichophyton rubrum (21) and Aspergillus nidulans and Aspergillus fumigatus strains (23) were reported. These mutants carry mutations in SE genes that lead to a leucine replacement by phenylalanine in the position corresponding to F402L in yeast Erg1p, which has already been described to confer terbinafine resistance in S. cerevisiae (15). These new results emphasize the importance of particular amino acids in drug binding.
Although squalene epoxidases of various origins have been investigated with respect to substrate requirements, cofactors, and inhibitors, no structural model is available; and the domains responsible for enzymatic activity and inhibitor interactions are not well understood. p-Hydroxybenzoate hydroxylase (PHBH) of Pseudomonas fluorescens is the prototype of FAD-dependent hydroxylases and the only enzyme in this class of flavoproteins for which the three-dimensional structure is known (31). This enzyme contains two FAD fingerprint motifs (FADI and FADII) and a conserved sequence motif with a putative dual function in FAD/NAD(P)H binding (4). These domains are also found in squalene epoxidases. Recently, photoaffinity labeling of competitive squalene epoxidase inhibitors and site-directed mutagenesis of recombinant rat liver SE based on the PHBH model led to the identification of amino acids that are involved in substrate binding, the FAD interaction, catalytic activity, and protein stability (18, 19). These results provided the first evidence of the possible location of the substrate binding site in rat SE, which, however, is not a region whose sequence has a degree of homology to the sequences of other squalene epoxidases.
In order to elucidate the importance of specific amino acids for enzymatic activity and inhibitor effects on Erg1p, we isolated from S. cerevisiae erg1 alleles that encode functional squalene epoxidases and that confer a particular phenotype, such as terbinafine sensitivity. In a preliminary report we briefly described mutants which carry single amino acid substitutions in the conserved FADI and FADII domains, some of which conferred terbinafine sensitivity (24). Here we present a detailed analysis of these and other mutants and demonstrate that certain amino acid substitutions in yeast SEs lead to a reduction or a loss of enzymatic activity and to significantly increased sensitivity to allylamines. Although the overall homology between the SEs from yeasts and PHBH is only 15%, the enzymes share highly conserved regions, particularly in the FAD and nucleotide binding domains. We therefore built a homology model of the Erg1 protein of S. cerevisiae on the basis of the known structure of PHBH and can now assign the amino acids which cause a particular phenotype to certain domains in the model.
MATERIALS AND METHODS
Strains and growth conditions.
All strains and plasmids used in this study are summarized in Table 1. Escherichia coli XL1 carrying recombinant plasmids was grown in 2× tryptone yeast extract medium (29) in the presence of 100 μg/ml ampicillin at 37°C. S. cerevisiae KLN1 was grown under oxygen-limiting conditions (Anaerocult; Oxoid) in yeast extract-peptone-dextrose (YPD) medium (32) supplemented with ergosterol and Tween 80, as described previously (14). KLN1 with recombinant plasmids carrying wild-type ERG1 or erg1 alleles was grown in YPD medium under aerobic conditions. KLN1 with recombinant plasmids coding for nonfunctional SE was grown in yeast nitrogen base (YNB) minimal medium that contained all amino acids except leucine (32) and that was supplemented with ergosterol and Tween 80 under oxygen-limiting conditions. The components of the media were purchased from Difco, Merck, and Gibco BRL.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| E. coli XL1 | endA1 hsdR17 (hsdR hsdM+) supE44 thi-1 recA1 gyrA96 relA1 D(lac) [F′ proAB lacIqZDM15 Tn10 (Tetr)] | Stratagene |
| S. cerevisiae KLN1 | MATaERG1::URA3 leu2 ura3 trp1 | 14 |
| Plasmids | ||
| pRS315 | Centromere yeast/E. coli shuttle vector | K. Kuchler |
| pBIG1 | 2.3-kb PstI fragment with the wild-type ERG1 gene in pBluescript | 15 |
| pNS1 | 2.3-kb PstI fragment with the wild-type ERG1 gene in pRS315 | 15 |
| pG25S | pRS315 with 2.3-kb PstI fragment with allelic erg1G25S gene generated by site-directed mutagenesis | This study |
| pG27S | pRS315 with 2.3-kb PstI fragment with allelic erg1G27S gene generated by site-directed mutagenesis | This study |
| pG30S | pRS315 with 2.3-kb PstI fragment with allelic erg1G30S gene generated by random mutagenic PCR | This study |
| pL37P | pRS315 with 2.3-kb PstI fragment with allelic erg1L37P gene generated by random mutagenic PCR | This study |
| pR269G | pRS315 with 2.3-kb PstI fragment with allelic erg1R269G gene generated by random mutagenic PCR | This study |
| pD209A | pRS315 with 2.3-kb PstI fragment with allelic erg1D209A gene generated by site-directed mutagenesis | This study |
| pG210A | pRS315 with 2.3-kb PstI fragment with allelic erg1G210A gene generated by site-directed mutagenesis | This study |
| pG334A | pRS315 with 2.3-kb PstI fragment with allelic erg1G334A gene generated by site-directed mutagenesis | This study |
| pD335A | pRS315 with 2.3-kb PstI fragment with allelic erg1D335A gene generated by site-directed mutagenesis | This study |
| pD335W | pRS315 with 2.3-kb PstI fragment with allelic erg1D335W gene generated by site-directed mutagenesis | This study |
| pD335F | pRS315 with 2.3-kb PstI fragment with allelic erg1D335F gene generated by site-directed mutagenesis | This study |
| pD335P | pRS315 with 2.3-kb PstI fragment with allelic erg1D335P gene generated by site-directed mutagenesis | This study |
Mutagenesis of ERG1 gene and selection of mutants.
The wild-type ERG1 gene was randomly mutagenized in vitro by PCR amplification, as described previously (13). The PCR fragments were digested with PstI, cloned into the centromere vector pRS315, and transformed into E. coli XL1. Plasmid DNA was isolated from approximately 1,000 E. coli transformants by standard procedures (29), and the combined recombinant plasmids were transformed into S. cerevisiae KLN1 (14) by the protocol of Gietz et al. (9). Transformants expressing functional SE were selected by complementation of the aerobic growth defect of KLN1 on YPD agar plates (13). The transformants were tested for terbinafine sensitivity on YPD plates containing 10 μg/ml of terbinafine. Plasmid DNA was isolated from S. cerevisiae KLN1 transformants that were unable to grow on terbinafine-containing plates (34), amplified in E. coli XL1, and characterized by DNA sequence analysis. Recombinant plasmids pG30S, pL37P, and pR269G, which carry different erg1 alleles, were isolated by this method.
In a second approach, single point mutations were generated in the ERG1 gene by site-specific mutagenesis with modified oligonucleotides that carried the respective nucleotide exchanges. The amplified PCR fragments were used to replace the corresponding wild-type sequence in the ERG1 gene contained in recombinant plasmid pNS1 (13). The correct sequences were confirmed by DNA sequence analysis. By this method we constructed the erg1 alleles G25S, G27S, G334A, D335A, D335W, D335F, D335P, D209A, and G210A.
Drug susceptibility testing.
Drug susceptibility determination was performed in liquid YPD medium as described previously (15). Cultures were incubated for 24 h at 30°C with shaking in the presence of the inhibitors terbinafine (0.1 to 10 μg/ml), naftifine (2 to 50 μg/ml), ketoconazole (0.2 to 2 μg/ml), and cycloheximide (0.005 to 0.2 μg/ml). Control cultures of each strain were supplemented with the respective solvent. The growth of the yeast strains was measured by determining the optical density at 600 nm (OD600).
The susceptibilities of the yeast strains were also determined on YPD agar plates (15). The plates were incubated for 2 days at 30°C. The inhibitors tested were tolnaftate (2 to 20 μg/ml), itraconazole (2 to 20 μg/ml), and amorolfine (0.005 to 0.02 μg/ml).
DNA and RNA preparations and Northern blot analysis.
Plasmid DNA was isolated from the E. coli transformants by standard procedures (29) and from yeast cells by the method described by Strathern and Higgins (34). The DNA sequences were determined as described previously (15). The primers were purchased from VBC (Vienna, Austria). The Genetics Computer Group (GCG) program was applied to obtain sequence alignments (3).
Total RNA was isolated from mechanically disrupted yeast cells with the RNeasy Mini kit of QIAGEN, according to the manufacturer's protocol. RNA concentrations were determined spectrophotometrically. Northern blot analysis was performed as described previously (17), except that the ERG1- and ACT1-specific probes were labeled with digoxigenin (DIG) by using a DIG-High Prime DNA labeling kit (Roche Applied Science). The labeled probes were amplified by PCR, and the ERG1 and ACT1 genes in recombinant plasmids pNS1 and pYA301 (7) served as the templates. For hybridization and RNA detection, the reagents of the DIG-High Prime DNA II labeling and detection starter kit (Roche Applied Science) were used. DIG-labeled probes were detected with an α-DIG-11-dUTP-alkaline phosphatase conjugate, according to the manufacturer's instructions. Chemiluminescence signals were detected on AGFA Curix Ultra UV-G X-ray film and were scanned by using a Molecular Dynamics personal densitometer. The ERG1 mRNA signals were normalized to the ACT1 mRNA signals by using ImageQuant 5.1 software (Molecular Dynamics).
Protein stability and Western blot analysis.
The yeast strains were grown at 30°C in YPD medium to early log phase (OD600, between 0.5 and 0.6). To determine protein stability, cycloheximide was added to the cultures to give a final concentration of 100 μg/ml. At various time points, samples corresponding to 8 OD600 equivalents were removed, and the cells were pelleted by centrifugation at 3,500 × g for 1 min and frozen in liquid nitrogen. Whole-cell extracts were prepared by NaOH disruption, followed by trichloroacetic acid (TCA) precipitation, as described by Wendler et al. (36). The levels of the Erg1 protein expressed from various recombinant plasmids in KLN1 were determined in early-log-phase cultures grown aerobically in YPD medium or under conditions of oxygen limitation in YNB medium lacking leucine and supplemented with ergosterol and Tween 80. Four OD600 equivalents were removed, and whole-cell extracts were prepared as described above. TCA precipitates were dissolved in 100 μl final sample buffer, and 5-μl aliquots were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore). Western blot analysis was performed with anti-Erg1p antibodies (16) and anti-Sui2p antibodies, respectively. Erg1p migrates at 55 kDa; and Sui2p, which served as a loading control, migrates at 35 kDa.
Neutral lipid synthesis.
Cultures were grown in YPD medium with addition of 57,000 dpm/ml of [14C]acetate (specific activity, 37 mBq/mmol; UVVVR, Czech Republic) for lipid labeling. Cells were grown starting from a density of 1 × 106 cells/ml at 30°C with shaking. After 8 h (exponential phase) and 24 h (stationary phase), cells were collected by centrifugation, washed twice in distilled water, and diluted in water to a final concentration of 1 × 109 cells/ml; 1 ml of this suspension was taken for lipid extraction. Extraction and detection of lipids were performed as described previously (13). Thin-layer chromatography (TLC) plates were analyzed by using a personal molecular imager (FX; Bio-Rad) and were quantified by the use of QuantityOne software (Bio-Rad) or, alternatively, by determining the radioactivity in individual lipid spots by liquid scintillation counting.
Squalene epoxidase assay in vitro.
Yeast strains were grown aerobically in YPD medium or under oxygen-limiting conditions in YNB medium at 25°C to late log phase (OD600, between 1.5 and 2). The cells were harvested by centrifugation, washed with 10 mM Tris-HCl (pH 7.4), and disrupted in a glass bead mill. Whole-cell extracts were prepared by centrifugation at 12,000 × g for 20 min at 4°C. The protein concentration was determined by the method of Bradford by using bovine serum albumin as the standard. Three milligrams of total protein was incubated with [14C]mevalonic acid and the cofactor-substrate mix at 30°C for 2 h as described recently (8). The values given for each lipid species represent the percentage of the total intensity per lane averaged over at least three independent experiments.
Homology modeling.
PHBH from P. fluorescens (PDB code 1pbe) was identified by using the GenThreader program (12) as a suitable template for comparative modeling of Erg1p. This enzyme shares an overall sequence identity of only 15% with Erg1p but exhibits FAD-dependent mono-oxygenase activity similar to that of Erg1p. Homology models were generated by using the program Modeler 8v1 (28). The most significant step of the modeling process is to obtain the correct alignment of the target and the template sequences. The initial alignments of the PHBH and Erg1p sequences were obtained from the GenThreader results, which included (experimental or predicted) information on the secondary structures of both proteins. We adopted a recursive approach comprising manual adjustment of the sequence alignment and model building using the Modeler program (28). At each step, 10 models of Erg1p were generated. All these models were built in the presence of the FAD cofactor, and special restraints were applied for the secondary structure elements which were not present in the template but which were predicted to be present in Erg1p. Several cycles of manual realignment and model building were performed until no further improvement of the model could be achieved. The final Erg1p model was validated by using the program Prosa-II (33).
RESULTS
Isolation and characterization of erg1 alleles that confer terbinafine sensitivity.
In order to isolate erg1 alleles that confer altered terbinafine sensitivity in S. cerevisiae, the wild-type ERG1 gene was amplified by mutagenic PCR and the products were cloned into the centromere vector pRS315 and transformed into S. cerevisiae KLN1, a mutant that carries a disrupted ERG1 gene (14). As a result of screening for transformants which express a functional squalene epoxidase, we identified erg1 alleles that conferred either resistance or increased sensitivity to terbinafine. The resistance alleles have been described in a recent report (15). Three transformants that failed to grow on terbinafine-containing plates were isolated. Plasmid DNA was isolated from these yeast transformants and subjected to DNA sequence analysis. Each erg1 allele carried one unique nucleotide change that led to single amino acid substitutions in the Erg1 protein, two of which were localized to the highly conserved FADI binding domain in the N-terminal region of SE. The mutations in the isolated erg1 alleles lead to G30S (24) and L37P substitutions in the Erg1 protein (Table 1). The amino acids at both positions are highly conserved in squalene epoxidases of various origins (24). The mutation in the third erg1 allele causes an R269G substitution in the central part of the protein, again affecting a conserved amino acid in all squalene epoxidases (25).
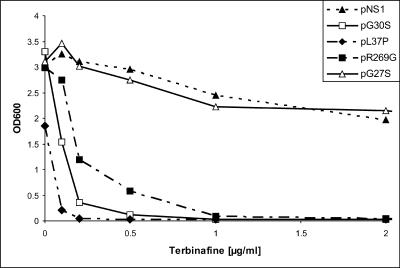
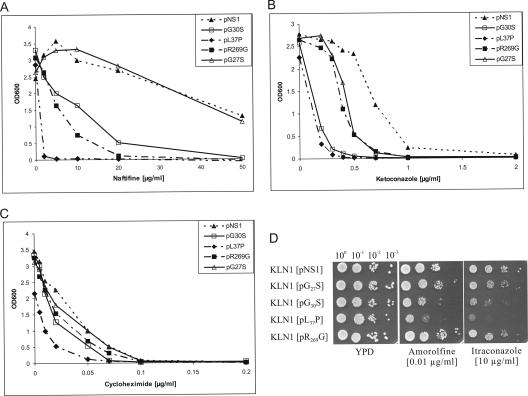
The terbinafine sensitivities of the KLN1 transformants carrying the mutated erg1 alleles G30S, L37P, and R269G were determined quantitatively by measuring the OD600 in liquid cultures after growth for 24 h at 30°C in the presence of the drug at the indicated concentrations (Fig. 1). The growth of KLN1(pNS1), which contained wild-type ERG1, was only slightly affected by 2 μg/ml of terbinafine, whereas growth of KLN1(pR269G) was completely inhibited at 1 μg/ml. The most sensitive strain was KLN1(pL37P), which could not grow in the presence of terbinafine at concentrations as low as 0.2 μg/ml. This strain also exhibited reduced growth in the absence of the inhibitor. The growth of KLN1(pG30S) was completely inhibited at 0.5 μg/ml, making this strain at least 10 times more sensitive than the wild type.
FIG. 1.
Growth of S. cerevisiae KLN1 strains carrying wild-type ERG1 (NS1) and various erg1 alleles on a centromere vector in the presence of terbinafine. Growth was monitored after 24 h of incubation at 30°C by measuring the OD600.
Since altered amino acid G30S is part of the conserved sequence 25GXGXXG30 of the FADI binding domain (24), we replaced the other two conserved glycine residues with serine by applying site-specific mutagenesis of the ERG1 gene, as described in Materials and Methods. The erg1 alleles were designated G25S and G27S, respectively (Table 1) (24). The Erg1 protein with the G27S exchange is functional, as shown by complementation of the aerobically lethal phenotype of S. cerevisiae KLN1, and does not alter the sensitivity to terbinafine (Fig. 1), whereas the Erg1 protein carrying the G25S amino acid substitution does not complement the KLN1 phenotype as a consequence of the expression of either an inactive or an unstable Erg1 protein variant (data not shown). Western blot analysis of cell extracts which were isolated from KLN1(pG25S) grown anaerobically showed that the steady-state levels of squalene epoxidase were comparable to those of the wild-type Erg1 protein in KLN1(pNS1) grown under the same conditions (data not shown). These data confirm that the G25S protein is expressed but is not functional.
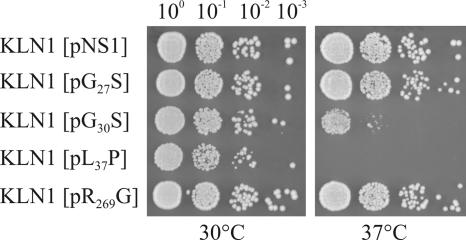
In addition to increased terbinafine sensitivity, the erg1 alleles L37P and, to a lesser degree, G30S also conferred temperature-sensitive growth phenotypes (Fig. 2). KLN1(pL37P) was unable to grow at 37°C, and KLN1(pG30S) showed reduced growth at the elevated temperature. In contrast, pR269G did not alter the growth behavior of KLN1 at the temperatures tested.
FIG. 2.
Temperature sensitivities of mutants expressing Erg1 protein variants. Overnight cultures of KLN1 carrying the wild-type ERG1 gene (pNS1) and four erg1 alleles on a centromere vector were prepared at 30°C in YPD medium. The OD600 of the overnight cultures was adjusted to 0.1 (100), and 5 μl of each of the 100 to 10−3 dilutions was spotted on YPD agar plates and grown for 2 days at 30°C and 37°C.
By alignment of the amino acid sequences of the squalene epoxidases, a highly conserved region termed FADII was assigned to amino acids 334GD335 in the Erg1 protein of S. cerevisiae (4, 25). D335 corresponds to D407 in rat SE, which has been reported to be part of the substrate binding site (19). The replacement of G334 or D335 by alanine (24) results in functional squalene epoxidases that confer either slightly increased or unaltered terbinafine sensitivity (data not shown). However, when D335 was replaced by tryptophan, phenylalanine, or proline (24), the Erg1 protein variants were expressed but had lost their functionality, as shown by the lack of complementation of the aerobic lethal phenotype of KLN1 (data not shown). This result suggests that the aspartate residue in the FADII fingerprint plays a crucial role in enzyme structure or function. A novel conserved motif in mono-oxygenases that is involved in nucleotide binding was recently described by Eppink et al. (4). This motif is also present in yeast SE and consists of 209DG210. Replacement of the aspartate residue by alanine does not extinguish enzymatic activity and confers wild-type sensitivity to terbinafine, whereas the Erg1p variant containing the G210A exchange, although it is expressed, is not functional (data not shown).
Squalene epoxidase variants exhibit reduced enzymatic activities.
To determine the enzymatic activities of the functional squalene epoxidase variants, crude cell extracts of KLN1 containing recombinant plasmids pNS1 (wild-type ERG1), pG30S, pG27S, pL37P, and pR269G were prepared from exponential cultures grown in YPD medium at 30°C. Since the G25S Erg1p variant did not complement the KLN1 phenotype, the strain expressing this protein was grown in YNB medium under oxygen-limiting conditions. Three milligrams of total cell extract protein was incubated with [14C]mevalonate, as described previously (8, 16). The nonsaponifiable lipids were extracted, separated by TLC, and quantified by phosphorimaging. The results are summarized in Table 2. In cell extracts of KLN1(pG30S), mevalonate was incorporated into sterol precursors and sterols, but squalene accumulated, while the amounts of radioactivity incorporated into squalene epoxide and lanosterol were reduced compared to the amounts incorporated in the wild-type extract of KLN1(pNS1). The effects were even more pronounced in cell extracts of KLN1(pL37P). These results prove that the Erg1 protein variants encoded by the L37P and G30S alleles exhibit reduced SE activities. The Erg1 protein expressed from the G25S allele did not show any enzymatic activity; 99% of the radioactivity was incorporated into squalene, and no reaction products were detected. This in vitro result explains the lack of complementation of the KLN1 phenotype by recombinant plasmid pG25S. SE variants G27S and R269G exhibited slightly reduced enzymatic activities, as shown by the modest accumulation of squalene, although the amount of squalene epoxide compares well to that of wild-type Erg1p.
TABLE 2.
Sterol intermediate composition in various ERG1 strainsa
| SIb | % of total radioactivity incorporated into sterol intermediates:
|
|||||
|---|---|---|---|---|---|---|
| pNS1 | pG27S | pG30S | pL37P | pR269G | pG25S | |
| S | 55.0 ± 9.0 | 73.1 ± 8.0 | 82.6 ± 9.0 | 93.4 ± 2.0 | 69.0 ± 8.5 | 99.4 ± 0.5 |
| SE | 6.7 ± 2.0 | 6.2 ± 2.0 | 3.0 ± 2.0 | 1.3 ± 0.7 | 5.8 ± 1.5 | NDc |
| L | 30.1 ± 9.0 | 16.1 ± 4.0 | 4.6 ± 2.0 | 1.1 ± 0.2 | 18.0 ± 5.5 | ND |
KLN1 strains carrying the indicated plasmids.
SI, sterol intermediate; S, squalene; SE, squalene epoxide; L, lanosterol.
ND, not detected.
Transcriptional up-regulation of erg1 expression in mutant strains.
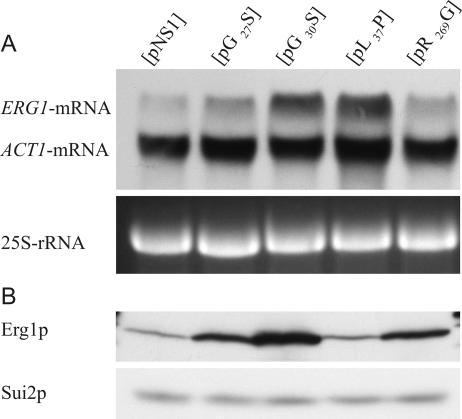
The reduced enzymatic activity of the Erg1 protein variants can lead to lower sterol levels in yeast cells, which, as described previously, induce the expression of the ERG1 gene at the transcriptional level (8, 17). We performed Northern blot analysis of KLN1 strains carrying the erg1 alleles G30S, L37P, G27S, and R269G and the wild-type ERG1 using ERG1- and ACT1-specific probes, essentially as described by Leber et al. (15). Wild-type ERG1 is expressed at a very low level, while the erg1-specific mRNA levels in mutant strains KLN1(pG30S) and KLN1(pL37P) are increased sevenfold over those in the wild type (Fig. 3A). The erg1-specific mRNA levels expressed from G27S and R269G alleles are only two times greater than those in the wild type. These results indicate that the limited enzymatic activity of the G30S and L37P SE variants shown in vitro (Table 2) leads to reduced (ergo)sterol levels and, thus, to feedback induction of erg1 expression. Corresponding to the slightly reduced enzymatic activity of the G27S and R269G Erg1 protein variants, the transcriptional up-regulation of erg1 expression is low in these strains.
FIG. 3.
Expression of erg1 alleles: mRNA and protein levels of terbinafine-sensitive mutants. Cultures of wild-type and terbinafine-sensitive KLN1 strains were grown in YPD medium to early log phase. (A) Northern blot analysis. Total RNA was extracted from mechanically disrupted cells and 10 μg of total RNA was separated by agarose gel electrophoresis. Hybridization was performed as described in Materials and Methods, using an ERG1-specific DIG-labeled DNA probe. The amounts of the ACT1 mRNA and 25S rRNA were used to normalize the amount of RNA loaded on the gel. (B) Western blot analysis. Whole-cell extracts were prepared from 4 OD600 equivalents by NaOH disruption, followed by TCA precipitation. Precipitates were dissolved in 100 μl final sample buffer, and 5-μl aliquots were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Western blot analysis was performed with anti-Erg1p antibodies and anti-Sui2p antibodies as the loading control.
When we analyzed steady-state protein levels by Western blot analysis, wild-type Erg1p corresponded well to the small amount of ERG1 mRNA, as shown in Fig. 3B. The squalene epoxidase level of the G30S variant was significantly increased, which reflects the elevated erg1 mRNA level. The levels of the G27S and R269G Erg1 proteins corresponded well to the slightly increased levels of their mRNAs. In contrast, the steady-state level of the L37P-encoded squalene epoxidase did not correspond to the amount of its mRNA (Fig. 3B). Although transcription was highly induced, the L37P protein level was very low and comparable to that of the wild type.
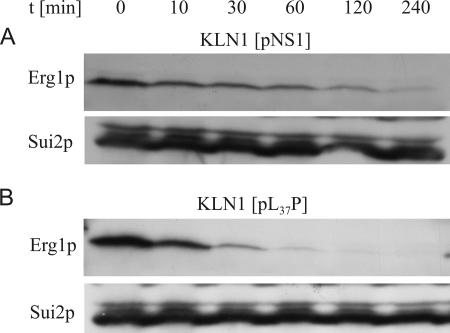
To elucidate the contradictory result for the L37P protein, we investigated the stability of this Erg1p variant with that of the wild-type protein as a control. Protein stability was measured over 4 h after cycloheximide addition, and Erg1p was detected by Western blotting. The results are shown in Fig. 4. The calculated half-life of the wild-type Erg1 protein is 60 min (Fig. 4A), whereas the L37P protein turned out to be very unstable, with a half-life of only 10 min (Fig. 4B). This result explains the low steady-state level of squalene epoxidase in KLN1(pL37P).
FIG. 4.
Stability of Erg1p from S. cerevisiae KLN1(pNS1) carrying wild-type ERG1 and terbinafine-sensitive mutant KLN1(pL37P). Strains were grown at 30°C in YPD medium to early log phase. After the inhibition of de novo protein biosynthesis by the addition of cycloheximide, samples of 8 OD600 equivalents were removed at the indicated time points (t). Whole-cell extracts were prepared by NaOH disruption, followed by TCA precipitation. Aliquots were subjected to Western blot analysis with anti-Erg1p antibodies and anti-Sui2p antibodies as the loading control.
In vivo sterol composition is altered in KLN1 carrying various erg1 alleles.
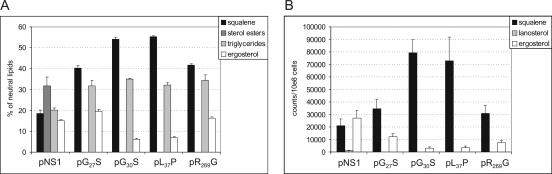
Elevated ERG1 mRNA levels are the result of transcriptional feedback induction by altered sterol composition. This mechanism was observed in S. cerevisiae when SE was inhibited by terbinafine or other ergosterol biosynthesis inhibitors (17), as well as in erg1 mutants whose SE activities were reduced (8). Since the SE variants encoded by erg1 alleles G30S and L37P and, to a much lesser degree, by erg1 alleles G27S and R269G exhibit reduced in vitro enzymatic activities, we determined neutral lipid levels by radiolabeling yeast cells with [14C]acetate in vivo. The strains were grown to exponential or stationary phase, the lipids were extracted, and their levels were determined as described previously (13). The results of the in vivo lipid analysis correlated very well with the SE activities in vitro. Early-log-phase cells of KLN1(pL37P) and KLN1(pG30S) showed the accumulation of squalene to high levels, a significant reduction of ergosterol, and a total absence of steryl esters (Fig. 5A). These changes indicate severe defects in ergosterol biosynthesis. The ergosterol contents in strains KLN1(pG27S) and KLN1(pR269G) were very similar to the ergosterol content of the wild-type strain, but also, these strains showed increased squalene levels and did not contain measurable amounts of steryl esters. The differences in sterol profiles were even more striking when nonsaponifiable lipids were analyzed by TLC (Fig. 5B). When cells were grown to stationary phase, the neutral lipid composition showed a certain adaptation of lipid profiles, indicating that the cells, especially those with moderate defects in ergosterol biosynthesis, such as KLN1(pG27S) and KLN1(pR269G), could balance their lipid contents after prolonged incubation (data not shown). The results for the sterol levels support our in vitro data on the limited enzymatic activity of the Erg1p variants and prove that the induction of erg1 mRNA expression in the sensitive strains (Fig. 3A) can be linked to decreased (ergo)sterol levels.
FIG. 5.
Sterol compositions of S. cerevisiae KLN1 strains carrying the wild-type ERG1 gene (NS1) and various erg1 alleles. Cultures were grown for 8 h in the presence of [14C]acetate in YPD medium to log phase. (A) Cellular lipids were isolated and separated by TLC as described in Materials and Methods. Relative radioactivity was expressed as the percentage of the label in neutral lipids. (B) Analysis of nonsaponifiable lipids by two-dimensional TLC. Radioactivity was determined by liquid scintillation counting of the spots corresponding to ergosterol, lanosterol, and squalene.
Only alleles L37P and G30S confer cross-sensitivity to sterol biosynthesis inhibitors.
The terbinafine hypersensitivity conferred by the mutant alleles may also be related to the observed decrease in sterol content in the plasma membrane, which can lead to increased general permeability for a variety of antifungal compounds, including terbinafine. Alternatively, low ergosterol levels may render mutant cells more susceptible to any additional interference with ergosterol biosynthesis. These possibilities were tested by estimating the sensitivities of mutant strains to several other inhibitors, such as the squalene epoxidase inhibitor naftifine; ketoconazole and itraconazole as inhibitors of lanosterol-14α-demethylase (Erg11p); the morpholine derivative amorolfine, which inhibits C-14 reductase (Erg24p) and C-8 isomerase (Erg2p) further downstream in the ergosterol biosynthesis pathway; and cycloheximide, a protein synthesis inhibitor. The results are shown in Fig. 6. The Erg1 protein encoded by allele L37P conferred high sensitivity not only to terbinafine (>10-fold greater sensitivity than the wild type, as shown in Fig. 1) but also to naftifine (>25-fold greater sensitivity than the wild type) and ketoconazole (approximately 5-fold greater sensitivity than the wild type). The sensitivities to itraconazole and amorolfine were also higher than those of any other mutant (Fig. 6D). The G30S allele also conferred increased sensitivity to ergosterol biosynthesis inhibitors, although to a lesser degree than L37P did (Fig. 6A, B, and D), but did not alter the sensitivity to cycloheximide (Fig. 6C). The multiple sensitivities of these mutant strains to ergosterol synthesis inhibitors suggest that reduced squalene epoxidase activity and, consequently, low ergosterol levels lead to increased sensitivity to any additional compound challenging ergosterol biosynthesis. KLN1(pL37P) was the most sensitive of all strains; and it also showed increased sensitivity to cycloheximide, which indicates that other factors are also possibly involved in the hypersensitive phenotype of this particular strain. KLN1(pG27S) was as sensitive as wild-type strain KLN1(pNS1) not only to terbinafine (Fig. 1) but also to naftifine, itraconazole, and amorolfine (Fig. 6), while only the sensitivity to ketoconazole was increased (approximately twofold) (Fig. 6B). The Erg1 protein encoded by allele R269G behaved somewhat differently. It conferred increased sensitivity to terbinafine (>5-fold; Fig. 1) and naftifine (>10-fold; Fig. 6A), whereas the susceptibility to non-squalene epoxidase inhibitors, such as amorolfine, itraconazole, and cycloheximide, remained unchanged and the sensitivity to ketoconazole was increased only 2-fold, which was in the same range as that conferred by G27S (Fig. 6). The results suggest that the sensitivity conferred by the R269G variant is restricted to squalene epoxidase inhibitors.
FIG. 6.
Cross sensitivity of S. cerevisiae KLN1 strains carrying various erg1 alleles. (A to C) In the presence of the indicated inhibitors, growth in liquid cultures was monitored after 24 h at 30°C by measuring the OD600; (D) overnight cultures of the strains were prepared at 30°C in YPD medium. The OD600 of the overnight cultures was adjusted to 0.1 (100), and 5 μl of each of the 100 to 10−3 dilutions was spotted on YPD agar plates, with or without indicated inhibitor, and grown for 2 days at 30°C.
Homology model of Erg1p.
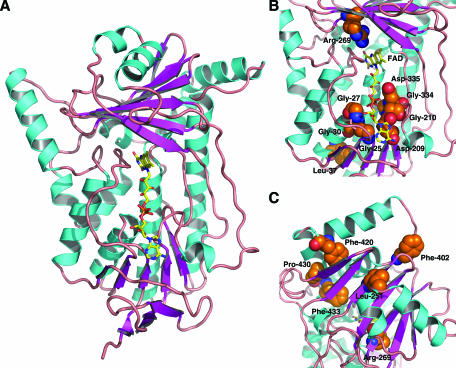
A homology model of Erg1p was built on the basis of the crystal structure of PHBH from P. fluorescens. According to a Prosa-II analysis (33), the overall structure of the model is correct (Fig. 7A). However, due to the low identity of only 15% between the target and the template sequences, fine details of the structure must be interpreted with caution.
FIG. 7.
Homology model of Erg1p from S. cerevisiae, based on the crystal structure of PHBH. (A) Schematic representation of the three-dimensional model. The FAD cofactor is shown as a yellow sticks model. C-terminal residues 440 to 496, which are predicted to form two α helices but which have no corresponding part in PHBH, are not shown. (B) Close-up view of the core cleft of Erg1p with the FAD cofactor. Amino acid side chains, for which mutations are described in the text, are displayed as CPK models (C, orange; N, blue; O, red). (C) Close-up view of a cluster of residues, alterations of which lead to terbinafine-resistant variants of Erg1p. This figure was prepared by using PyMol (http://www.pymol.org).
G30, together with G25 and G27, is part of the consensus motif for nucleotide binding in the FADI domain (24). In the homology model built in the presence of the FAD cofactor, these three residues are located close to the biphosphate and the adenosyl portion of the cofactor (Fig. 7B). Leucine 37 is part of an α helix, and the N-terminal end of this helix points toward the biphosphate group of the flavin cofactor. Amino acid residues D209 and G210 of the nucleotide binding domain and G334 and D335, which comprise the FADII motif, are also close to the biphosphate and ribitol moieties of the flavin cofactor but are located on the opposite side with respect to G25, G27, and G30. In the case of D209 and G210, both amino acids are located close to the cofactor in the region of its adenosyl moiety (Fig. 7B). The arginine residue at position 269 is located close to the isoalloxazine moiety of the flavin cofactor (Fig. 7B).
DISCUSSION
Allylamines have been widely used for the treatment of fungal infections caused by dermatophytes and other pathogens. The target enzyme in fungi, SE, has been known for a long time (26, 27); yet neither the domains that are involved in substrate, cofactor, or inhibitor binding nor the structure of the enzyme is known in detail. We have chosen the Erg1 protein of S. cerevisiae to investigate the roles of certain conserved domains and amino acids in enzymatic activity and drug susceptibility. To achieve this goal, the ERG1 gene was mutagenized by PCR and erg1 alleles that encode modified squalene epoxidases were isolated by functional complementation of the aerobic lethal phenotype of squalene epoxidase-deficient mutant KLN1. Squalene epoxidase variants that confer terbinafine resistance due to single amino acid substitutions were described recently (13, 15). In this report we present data on Erg1p variations that lead to increased sensitivity to terbinafine and Erg1p variants that exhibit reduced enzymatic activity. In addition, Erg1 variants with altered amino acids in conserved regions that failed to complement the KLN1 phenotype due to a lack of enzymatic activity are described.
Inhibition of S. cerevisiae by allylamines is accompanied by an accumulation of squalene and a lack of ergosterol, as well as by feedback induction of ERG1 expression (16). A similar effect has been observed in mutants that express Erg1 protein variants with reduced enzymatic activities (8). In such mutants expression of Erg1p is also induced at the transcription level (8); and the larger amounts of Erg1p variants still allow the synthesis of sufficient squalene epoxide, which is finally converted to ergosterol at levels that permit growth. When, however, such strains are treated with ergosterol biosynthesis inhibitors, very low concentrations of these compounds are now sufficient to further reduce ergosterol to levels at which the strains cannot grow any more (8). Such strains should exhibit increased sensitivities to ergosterol biosynthesis inhibitors in general. This phenotype is indeed observed in KLN1 carrying the erg1 allele G30S and is even more pronounced in KLN1 carrying the erg1 allele L37P. These strains are not only more sensitive to terbinafine (24) or the chemically related naftifine; the sensitivity was also increased to all other sterol biosynthesis inhibitors tested, such as ketoconazole and itraconazole, which represent members of the azole group and which inhibit lanosterol C-14 demethylase (Erg11p) downstream of SE in the ergosterol biosynthesis pathway, and amorolfine, which affects C-14 reductase (Erg24p) and C-8 isomerase (Erg2p). When SE activity was measured in vitro by incorporation of the early precursor mevalonate into squalene, squalene epoxide, and lanosterol, we could demonstrate that crude extracts containing G30S or L37P exhibited reduced SE activities. Squalene accumulated, and the levels of squalene epoxide and lanosterol were significantly reduced. The direct comparison of the enzymatic activities of these two Erg1p variants, however, is difficult, since the amount of Erg1p in the L37P extract was much lower than that in the G30S extract due to the high degree of instability of the former protein variant. In vivo sterol composition measurements of the mutant strains verify our in vitro data on reduced SE activity. Low ergosterol levels due to reduced Erg1p activity is obviously linked to terbinafine sensitivity. These results compare well to those obtained for the recently described mutant carrying the erg1-1 allele, which encodes an SE variant with reduced enzymatic activity and which confers terbinafine sensitivity (8). In addition, the decrease in ergosterol content caused by the interruption of the Candida glabrata ERG1 gene also increased the sensitivity of this organism to terbinafine and azoles (35). Similar effects were observed in Candida albicans when ergosterol levels were reduced by the limited expression of SE (22), but this mutant also became more susceptible to other inhibitors not involved in sterol synthesis due to reduced drug efflux activity.
According to the amino acid sequence alignment of the mono-oxygenases, the G30S substitution in the terbinafine-sensitive Erg1p variant affects an amino acid in the FADI domain (24). This domain represents a typical βαβ fold, in which G25, G27, and G30 are part of the consensus sequence for FAD binding (37). Since no structural data for squalene epoxidases are available, we built a homology model of yeast SE in the presence of the cofactor FAD (Fig. 7A), based on the crystal structure of PHBH from P. fluorescens (31). According to this model, the three conserved glycine residues are located close to the biphosphate and the adenosyl portion of the cofactor (Fig. 7B). At positions 25 and 30, the in silico replacement of glycine even with the rather small amino acid serine leads to severe steric clashes with surrounding residues and the cofactor, which may prohibit correct cofactor binding and which may influence enzymatic activity. SE variant G30S does indeed exhibit reduced enzymatic activity, and variant G25S is not functional, as shown by the lack of in vitro enzymatic activity. Qualitatively, position 27 appears to be able to accept at least smaller side chains, consistent with the G27S variant being similar to wild-type SE regarding sensitivity to allylamines. To our knowledge these are the first mutations in the FADI site in squalene epoxidases to confer a phenotype.
L37 is part of an α helix whose N-terminal end points toward the biphosphate group of the flavin cofactor (Fig. 7B) and interacts with the negative charge of this group through its dipole moment. Replacement of this residue by proline would definitely distort the α helix, possibly leading to structural rearrangements in this part of the protein structure. Both effects very likely affect the binding and positioning of the cofactor. This assumption is supported by the decreased stability and the weak enzymatic activity of the Erg1 protein variant.
The amino acid residues D209 and G210 of the nucleotide binding domain, as well as G334 and D335, which comprise the FADII motif (4), are also close to the biphosphate and ribitol moieties of the flavin cofactor but are located on the opposite side with respect to the FADI domain (Fig. 7B). The substitutions G334A and D335A lead to functional squalene epoxidases with wild-type properties, whereas mutations D335W and D335F result in nonfunctional proteins (24). Since the D335A substitution gives a functional protein, the negative charge at this position is not essential. The incorporation of large side chains at these positions, however, will most likely interfere with cofactor binding due to steric effects. A replacement by proline (D335P) also yields a nonfunctional SE, which may be rationalized by local structural changes in the polypeptide close to the active site of the enzyme. Interestingly, three amino acid residues were shown to be involved in substrate binding in rat SE. These residues are K399, R400, and D407, of which only D407 is conserved in squalene epoxidases; D407 corresponds to D335 in the S. cerevisiae SE (24, 25). In contrast to the Erg1 protein of yeasts, only triple mutants in which all three residues were replaced by amino acids with bulky groups showed decreased enzymatic activities (19).
D209 and G210 are located close to the cofactor in the region of its adenosyl moiety (Fig. 7B). The G210A mutation yields a nonfunctional enzyme, whereas replacement of D209 by alanine does not extinguish enzymatic activity and results in a protein with wild-type terbinafine sensitivity properties. In the model (Fig. 7B), the side chain of D209 is pointing away from the cofactor, and replacement of this residue by alanine does not create any nonfavorable interactions with the flavin and, thus, has minor effects on the function of the protein. In rat SE, replacement of the corresponding D284 residue by alanine was reported to lead to decreased enzymatic activity (18), but no information on the phenotype conferred by the reduced enzymatic activity was given. On the other hand, any side chain introduced at position 210 (G210X) would lead to unfavorable interactions with the biphosphate of the cofactor and might therefore explain the loss of enzymatic activity of the G210A-encoded SE, which we determined in vitro and by complementation analysis.
The Erg1 protein with the R269G variation behaves differently from the proteins of the terbinafine-sensitive variants with amino acid exchanges in the nucleotide binding domains. It shares nearly all properties with the G27S protein. The mutants express the same steady-state Erg1 protein and erg1 mRNA levels, they show comparable in vivo sterol compositions, and the enzymes exhibit identical in vitro activities. However, they confer different susceptibilities to allylamines. R269G leads to a 5 to 10 times higher sensitivity to the squalene epoxidase inhibitors terbinafine and naftifine, whereas G27S behaves like the wild type. The susceptibilities to other sterol biosynthesis inhibitors remained unchanged. This selective sensitivity to allylamines cannot be explained by additive or synergistic effects between the reduced Erg1 enzymatic activity and inhibitor actions that seem to play a major role in sensitivity to ergosterol biosynthesis inhibitors in G30S and L37P strains. The result for R269G leads to the speculation of whether direct interaction with allylamines or conformational changes in the protein are affected by the R269G substitution.
The arginine residue at position 269 is located close to the isoalloxazine moiety of the flavin cofactor (Fig. 7B). It is conserved among squalene epoxidases and is also present in PHBH (R220), where its supposed role is to stabilize the active conformations of the flavin cofactor and to maintain a positive electrostatic potential in the active site (20). Whether or not this is also true for Erg1p is a matter of discussion, because the observed effects of the R269G variant are not as dramatic as those seen for the more conservative substitution R220K in PHBH (20). Regarding our results on allylamine sensitivity, it seems rather unlikely that R269 is involved in a direct inhibitor interaction, due to its location within the molecule (Fig. 7B) and the knowledge that allylamines represent noncompetitive inhibitors of fungal squalene epoxidases. It is therefore conceivable that this amino acid alteration facilitates a conformational change of Erg1p upon terbinafine binding which leads to inactivation of the enzyme.
From our results on Erg1p-derived terbinafine resistance in yeast strains (13, 15), we can draw the conclusion that a small number of amino acids participate in drug binding. One of the amino acid substitutions in S. cerevisiae SE, F402L, has recently been reported to also confer resistance in pathogenic fungal species, such as T. rubrum and A. fumigatus, when the corresponding leucine was changed to phenylalanine (21, 23). Although the amino acids that confer terbinafine resistance in yeasts (L251F, F402L, F420L, P430S, and F433S) are distributed along the sequence of the Erg1 protein, in the three-dimensional model they cluster on the surface of Erg1p, which comprises a possible terbinafine binding site (Fig. 7C). Regarding the position of R269, it can also be concluded that this amino acid is not part of the potential drug binding domain. It should be stressed that no functional data for mutant variants were implemented in the building of the model, so that these results not only reveal more details on the terbinafine interaction on Erg1p but also show the quality and the value of the model presented.
With the structural model at hand, we can now alter specific positions in the Erg1 protein that might interfere with the enzymatic activity or terbinafine interaction in order to learn more about this enzyme. The use of hypersensitive yeast strains to determine the modes of action of antifungal compounds has recently been suggested by Buurman et al. (1). The novel mutant strains described in this report which express Erg1p variants that confer either general sensitivity to ergosterol biosynthesis inhibitors or, in the case of the R269G variant, selective sensitivity to squalene epoxidase inhibitors might be used to screen for novel inhibitors of ergosterol biosynthesis or, more specifically, of fungal squalene epoxidases.
Acknowledgments
We thank A. Stütz, Novartis Forschungsinstitut Vienna, Vienna, Austria, for terbinafine and naftifine and T. Kuchta, Food Research Institute, Bratislava, Slovak Republic, for itraconazole and amorolfine. The construction of strains by T. Kowatz, M. Beichel, M. Steindl, and N. Hojas is gratefully acknowledged. We thank V. Klobučníková for spot assays.
This work was financially supported in part by Fonds zur Förderung der wissenschaftlichen Forschung projects P14415 (to F.T.) and W901 (DK Molecular Enzymology to K.G.) and APVV 51-024904 and VEGA 2/5127/25 (to I.H.).
Footnotes
Published ahead of print on 16 October 2006.
REFERENCES
- 1.Buurman, E. T., B. Andrews, A. E. Blodgett, J. S. Chavda, and N. F. Schnell. 2005. Utilization of target-specific, hypersensitive strains of Saccharomyces cerevisiae to determine the mode of action of antifungal compounds. Antimicrob. Agents Chemother. 49:2558-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chugh, A., A. Ray, and J. B. Gupta. 2003. Squalene epoxidase as hypocholesterolemic drug target revisited. Prog. Lipid Res. 42:37-50. [DOI] [PubMed] [Google Scholar]
- 3.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eppink, M. H., H. A. Schreuder, and W. J. Van Berkel. 1997. Identification of a novel conserved sequence motif in flavoprotein hydroxylases with a putative dual function in FAD/NAD(P)H binding. Protein Sci. 6:2454-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favre, B., and N. S. Ryder. 1996. Characterization of squalene epoxidase activity from the dermatophyte Trichophyton rubrum and its inhibition by terbinafine and other antimycotic agents. Antimicrob. Agents Chemother. 40:443-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favre, B., and N. S. Ryder. 1997. Differential inhibition of fungal amd mammalian squalene epoxidases by the benzylamine SDZ SBA 586 in comparison with the allylamine terbinafine. Arch. Biochem. Biophys. 340:265-269. [DOI] [PubMed] [Google Scholar]
- 7.Gallwitz, D., and I. Sures. 1980. Structure of a split yeast gene: complete nucleotide sequence of the actin gene in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 77:2546-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Germann, M., C. Gallo, T. Donahue, R. Shirzadi, J. Stukey, S. Lang, C. Ruckenstuhl, S. Oliaro-Bosso, V. McDonough, F. Turnowsky, G. Balliano, and J. T. Nickels, Jr. 2005. Characterizing sterol defect suppressors uncovers a novel transcriptional signaling pathway regulating zymosterol biosynthesis. J. Biol. Chem. 280:35904-35913. [DOI] [PubMed] [Google Scholar]
- 9.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 10.Horie, M., Y. Tsuchiya, M. Hayashi, Y. Iida, Y. Iwasawa, Y. Nagata, Y. Sawasaki, H. Fukuzumi, K. Kitani, and T. Kamei. 1990. NB-598: a potent competitive inhibitor of squalene epoxidase. J. Biol. Chem. 265:18075-18078. [PubMed] [Google Scholar]
- 11.Jandrositz, A., F. Turnowsky, and G. Hogenauer. 1991. The gene encoding squalene epoxidase from Saccharomyces cerevisiae: cloning and characterization. Gene 107:155-160. [DOI] [PubMed] [Google Scholar]
- 12.Jones, D. T. 1999. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 287:797-815. [DOI] [PubMed] [Google Scholar]
- 13.Klobucnikova, V., P. Kohut, R. Leber, S. Fuchsbichler, N. Schweighofer, F. Turnowsky, and I. Hapala. 2003. Terbinafine resistance in a pleiotropic yeast mutant is caused by a single point mutation in the ERG1 gene. Biochem. Biophys. Res. Commun. 309:666-671. [DOI] [PubMed] [Google Scholar]
- 14.Landl, K. M., B. Klosch, and F. Turnowsky. 1996. ERG1, encoding squalene epoxidase, is located on the right arm of chromosome VII of Saccharomyces cerevisiae. Yeast 12:609-613. [DOI] [PubMed] [Google Scholar]
- 15.Leber, R., S. Fuchsbichler, V. Klobucnikova, N. Schweighofer, E. Pitters, K. Wohlfarter, M. Lederer, K. Landl, C. Ruckenstuhl, I. Hapala, and F. Turnowsky. 2003. Molecular mechanism of terbinafine resistance in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 47:3890-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leber, R., K. Landl, E. Zinser, H. Ahorn, A. Spok, S. D. Kohlwein, F. Turnowsky, and G. Daum. 1998. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol. Biol. Cell 9:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leber, R., R. Zenz, K. Schrottner, S. Fuchsbichler, B. Puhringer, and F. Turnowsky. 2001. A novel sequence element is involved in the transcriptional regulation of expression of the ERG1 (squalene epoxidase) gene in Saccharomyces cerevisiae. Eur. J. Biochem. 268:914-924. [DOI] [PubMed] [Google Scholar]
- 18.Lee, H. K., P. Denner-Ancona, J. Sakakibara, T. Ono, and G. D. Prestwich. 2000. Photoaffinity labeling and site-directed mutagenesis of rat squalene epoxidase. Arch. Biochem. Biophys. 381:43-52. [DOI] [PubMed] [Google Scholar]
- 19.Lee, H. K., Y. F. Zheng, X. Y. Xiao, M. Bai, J. Sakakibara, T. Ono, and G. D. Prestwich. 2004. Photoaffinity labeling identifies the substrate-binding site of mammalian squalene epoxidase. Biochem. Biophys. Res. Commun. 315:1-9. [DOI] [PubMed] [Google Scholar]
- 20.Moran, G. R., B. Entsch, B. A. Palfey, and D. P. Ballou. 1996. Evidence for flavin movement in the function of p-hydroxybenzoate hydroxylase from studies of the mutant Arg220Lys. Biochemistry 35:9278-9285. [DOI] [PubMed] [Google Scholar]
- 21.Osborne, C. S., I. Leitner, B. Hofbauer, C. A. Fielding, B. Favre, and N. S. Ryder. 2006. Biological, biochemical, and molecular characterization of a new clinical Trichophyton rubrum isolate resistant to terbinafine. Antimicrob. Agents Chemother. 50:2234-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasrija, R., T. Prasad, and R. Prasad. 2005. Membrane raft lipid constituents affect drug susceptibilities of Candida albicans. Biochem. Soc. Trans. 33:1219-1223. [DOI] [PubMed] [Google Scholar]
- 23.Rocha, E. M., R. E. Gardiner, S. Park, N. M. Martinez-Rossi, and D. S. Perlin. 2006. A Phe389Leu substitution in ergA confers terbinafine resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 50:2533-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruckenstuhl, C., A. Eidenberger, S. Lang, and F. Turnowsky. 2005. Single amino acid exchanges in FAD-binding domains of squalene epoxidase of Saccharomyces cerevisiae lead to either loss of functionality or terbinafine sensitivity. Biochem. Soc. Trans. 33:1197-1201. [DOI] [PubMed] [Google Scholar]
- 25.Ruckenstuhl, C., R. Leber, and F. Turnowsky. 2005. Squalene epoxidase as drug target. Res. Adv. Antimicrob. Agents Chemother. 5:35-51. [Google Scholar]
- 26.Ryder, N. S. 1991. Squalene epoxidase as a target for the allylamines. Biochem. Soc. Trans. 19:774-777. [DOI] [PubMed] [Google Scholar]
- 27.Ryder, N. S., and M. C. Dupont. 1985. Inhibition of squalene epoxidase by allylamine antimycotic compounds. A comparative study of the fungal and mammalian enzymes. Biochem. J. 230:765-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Sawada, M., K. Washizuka, and H. Okumura. 2004. Synthesis and biological activity of a novel squalene epoxidase inhibitor, FR194738. Bioorg. Med. Chem. Lett. 14:633-637. [DOI] [PubMed] [Google Scholar]
- 31.Schreuder, H. A., P. A. Prick, R. K. Wierenga, G. Vriend, K. S. Wilson, W. G. Hol, and J. Drenth. 1989. Crystal structure of the p-hydroxybenzoate hydroxylase-substrate complex refined at 1.9 Å resolution. Analysis of the enzyme-substrate and enzyme-product complexes. J. Mol. Biol. 208:679-696. [DOI] [PubMed] [Google Scholar]
- 32.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 33.Sippl, M. J. 1993. Recognition of errors in three-dimensional structures of proteins. Proteins 17:355-362. [DOI] [PubMed] [Google Scholar]
- 34.Strathern, J. N., and D. R. Higgins. 1991. Recovery of plasmids from yeast into Escherichia coli: shuttle vectors. Methods Enzymol. 194:319-329. [DOI] [PubMed] [Google Scholar]
- 35.Tsai, H. F., M. Bard, K. Izumikawa, A. A. Krol, A. M. Sturm, N. T. Culbertson, C. A. Pierson, and J. E. Bennett. 2004. Candida glabrata erg1 mutant with increased sensitivity to azoles and to low oxygen tension. Antimicrob. Agents Chemother. 48:2483-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wendler, F., H. Bergler, K. Prutej, H. Jungwirth, G. Zisser, K. Kuchler, and G. Hogenauer. 1997. Diazaborine resistance in the yeast Saccharomyces cerevisiae reveals a link between YAP1 and the pleiotropic drug resistance genes PDR1 and PDR3. J. Biol. Chem. 272:27091-27098. [DOI] [PubMed] [Google Scholar]
- 37.Wierenga, R. K., P. Terpstra, and W. G. Hol. 1986. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 187:101-107. [DOI] [PubMed] [Google Scholar]