Abstract
The emergence of antiviral-resistant cytomegalovirus (CMV) strains is a continuing clinical problem, with increased numbers of immunocompromised patients given longer-duration antiviral prophylaxis. Two previously unrecognized CMV DNA polymerase mutations (N408K and A834P) identified separately and together in at-risk lung and kidney transplant recipients and a third mutation (L737M) identified in a liver transplant recipient were characterized by marker transfer to antiviral-sensitive laboratory strains AD169 and Towne. Subsequent phenotypic analyses of recombinant strains demonstrated the ability of mutation N408K to confer ganciclovir (GCV) and cidofovir (CDV) resistance and of mutation A834P to confer GCV, foscarnet, and CDV resistance. Mutation L737M did not confer resistance to any of the antiviral agents tested. A recombinant strain containing both N408K and A834P demonstrated increased GCV and CDV resistance compared to the levels of resistance of the virus containing only the A834P mutation. The addition of mutation N408K in combination with A834P also partially reconstituted the replication impairment of recombinant virus containing only A834P. This suggests that perturbation of both DNA polymerization (A834P) and exonuclease (N408K) activities contributes to antiviral resistance and altered replication kinetics in these mutant strains. The identification of these multidrug-resistant CMV strains in at-risk seronegative recipients of organs from seropositive donors suggests that improved prophylactic and treatment strategies are required. The additive effect of multiple mutations on antiviral susceptibility suggests that increasing antiviral-resistant phenotypes can result from different virus-antiviral interactions.
Immunocompromised patients with life-threatening illnesses caused by human cytomegalovirus (CMV) can be treated with the antiviral agents ganciclovir (GCV), valganciclovir (valGCV), foscarnet (FOS), or cidofovir (CDV) (36, 41). The use of oral valaciclovir, GCV, or valGCV has also made the delivery of prophylactic anti-CMV therapy easier and has improved the outcomes for highly immunosuppressed patients (24, 34, 37). CMV strains resistant to antiviral agents arise under the pressure of antiviral selectivity, with very low levels of resistant genotypes detected in patients prior to antiviral treatment (20, 33). Therefore, the lengthy periods of antiviral administration required for effective inhibition of this latent virus provide the selective pressure for the emergence of antiviral-resistant CMV strains, with resistant CMV strains detected in 5 to 30% of immunocompromised patients treated for more than 2 months, depending on the patient group studied (3, 28, 31). In some patients, particularly severely immunocompromised children, antiviral-resistant CMV strains can emerge rapidly (within 6 weeks) after the start of antiviral treatment (18, 42). Other risk factors for resistant CMV infections include transplantation from a seropositive donor to a seronegative recipient (D+/R−), the use of prolonged low-dose oral antiviral prophylaxis or intermittent intravenous treatment, and the administration of highly potent immunosuppression (19, 27, 31).
CMV antiviral resistance develops via mutations in the UL97 protein kinase (responsible for GCV phosphorylation) or the CMV UL54 DNA polymerase (for reviews, see references 5 and 23). Mutations of the CMV UL97 protein kinase that confer GCV resistance have been well characterized and are localized at codons 460 (protein kinase functional domain VIb) and 520; in addition, single base changes and deletions within codons 590 to 607 have been found to confer GCV resistance (1, 6, 10, 13, 25, 43). This simplifies identification of the CMV UL97 protein kinase mutations associated with antiviral resistance by targeted PCR sequencing or restriction fragment length polymorphism analysis (6, 35, 39). Conversely, CMV DNA polymerase mutations associated with antiviral resistance occur throughout the functional domain region spanning codons 301 (DNA polymerase exonuclease I region) to 989 (domain V) (4, 7-9, 11, 14, 15, 42, 44, 46). Furthermore, previously unrecognized mutations of the CMV DNA polymerase continue to be identified in CMV strains isolated from patients receiving antivirals (17, 39, 46).
In this study, we characterized three novel mutations identified in CMV strains isolated from patients with clinically resistant CMV disease. Two of these, N408K (which occurs in DNA polymerase domain IV) and A834P (which occurs in DNA polymerase domain III), were identified in separate D+/R− lung transplant recipients from Australia and in a kidney transplant recipient from the United States. None of the patients responded to GCV, FOS, or CDV therapy (30, 39). The third mutation, L737M (DNA polymerase domain II), was identified in a liver transplant recipient receiving GCV (39). The phenotypic antiviral susceptibilities of recombinant CMV strains containing these mutations were assessed in vitro by two different assays, and the potential impacts of these mutations on DNA polymerase structure and function are discussed. These findings have implications for the management of D+/R− immunocompromised transplant recipients at high risk of developing multidrug-resistant CMV infections.
MATERIALS AND METHODS
Specimens, plasmid vectors, and viruses.
Previously reported DNA polymerase mutations (N408K, L737M, and A834P) were identified in EDTA-anticoagulated blood specimens from patients with clinical evidence of infection with antiviral-resistant CMV (30, 39). These specimens were obtained with patient consent, and testing was carried out following the ethical guidelines of the South Eastern Sydney and Illawarra Area Health Service. CMV laboratory strains AD169 and Towne were obtained from the American Type Culture Collection (ATCC; strains ATCC VR538 and ATCC VR977, respectively). Previously described pBluescript plasmid vectors containing either the AD169- or Towne-derived CMV UL54 DNA polymerase genes with engineered PmeI restriction sites were used to transfer the mutations to recombinant virus (7, 12). The construction of antiviral-sensitive AD169-derived and Towne-derived recombinant viruses used for marker transfer has been reported previously (7, 12). This includes the AD169-derived recombinant viruses T2211, which contains the secreted alkaline phosphatase (SEAP) reporter gene, a SwaI restriction site within the UL97 gene, and unique PmeI restriction sites within the UL54 DNA polymerase gene; T2233, which is a derivative of T2211 that contains the wild-type UL97 amino acid sequence; T2241, which is a derivative of T2211 that contains the wild-type UL54 DNA polymerase amino acid sequence; and the Towne-derived recombinant virus T1472, which contains UL54 DNA polymerase with unique PmeI restriction sites (Table 1) (7, 12).
TABLE 1.
Antiviral susceptibilities of AD169-derived mutants assessed by SEAP-based assaya
| Virus | DNA pol genotype | GCV
|
FOS
|
CDV
|
|||
|---|---|---|---|---|---|---|---|
| IC50 (μM) | Fold increaseb | IC50 (μM) | Fold increaseb | IC50 (μM) | Fold increaseb | ||
| T2241 | Wild type (AD169) | 1.4 ± 0.5 | 42 ± 14 | 0.3 ± 0.1 | |||
| T2211 | Wild type + PmeI | 1.3 ± 0.4 | 41 ± 12 | 0.2 ± 0.1 | |||
| T2233 | Wild type + PmeI | 1.4 ± 0.4 | 45 ± 8 | 0.3 ± 0.1 | |||
| Avg for wild types | 1.33 | 42.7 | 0.25 | ||||
| T2293 | N408K | 5.6 ± 1.6 | 4.2 | 32 ± 6 | 0.7 | 5.2 ± 1.5 | 21.0 |
| T2291 | A834P | 7.1 ± 3.6 | 5.4 | 274 ± 87 | 6.4 | 0.7 ± 0.4 | 3.0 |
| T2311 | N408K + A834P | 30.2 ± 8.8 | 22.7 | 308 ± 78 | 7.2 | 4.7 ± 2.3 | 18.8 |
Mean values and standard deviations were calculated from 7 to 23 replicate experiments.
Fold increase in IC50 compared to the average IC50 for wild-type sensitive strains.
Construction of transfer vectors containing DNA polymerase mutations.
DNA polymerase PCR products containing the N408K or A834P mutation or both the N408K and the A834P mutations were subcloned into the pBluescript plasmid vector containing AD169-derived CMV DNA polymerase, as described previously (7). Mutations L737M and A834P were also inserted into the CMV DNA polymerase of the Towne-derived pBluescript plasmid vector (7) by site-directed mutagenesis with a Quickchange site-directed mutagenesis kit (Stratagene) and 30- to 41-mer high-pressure liquid chromatography-purified oligonucleotides containing the mutation of interest (Proligo Australia). The clones were screened for the presence of the desired mutation by PCR sequencing (39), and the entire DNA polymerase gene from purified plasmid DNA was sequenced by using overlapping primers to confirm that no undesirable mutations had been introduced.
Marker transfer.
Mutations N408K, L737M, and A834P were transferred from the pBluescript plasmid vectors to AD169 SEAP (T2211) or Towne-derived recombinant virus (T1472) as described previously (7, 12). Briefly, this consisted of cotransfection of human foreskin fibroblast (HFF) cultures with XbaI-EcoRI-digested AD169- and Towne-derived pBluescript plasmid vectors containing the mutation of interest with PmeI-digested T2211 (AD169 SEAP) and T1472 Towne recombinant virus, respectively. The resultant virus cultures were sampled and tested for the presence of the N408K, L737M, or A834P mutation by PCR sequencing (7). Recombinant viruses were plaque purified at least once beyond the stage where the wild-type sequence was not detected by sequencing or restriction digestion analysis. This process resulted in AD169-derived recombinant viruses T2293 (containing N408K), T2291 (containing A834P), and T2311 (containing N408K and A834P) and Towne-derived recombinant viruses T2296 (containing L737M) and T2287 (containing A834P).
Antiviral susceptibility analysis of the AD169-derived mutants.
The GCV, FOS, and CDV susceptibilities of strain AD169-derived antiviral-sensitive strains T2211, T2233, and T2241 and recombinants T2293 (containing N408K), T2291 (containing A834P), and T2311 (containing N408K and A834P) were determined by the SEAP-based assay (12). Cell-free virus stock was inoculated at a multiplicity of infection of 0.01 to 0.03 into HFF cultures in 24-well plates and grown in the absence or presence of twofold increasing concentrations of antiviral. After 7 days of culture, aliquots of the supernatant from each well were analyzed for SEAP activity as described previously (12), and 50% inhibitory concentrations (IC50s) were calculated by linear regression analysis (12). Each assay was carried out multiple times (7 to 23 times), and the averages and standard deviations were calculated. The GCV, FOS, and CDV susceptibilities of AD169 wild-type virus (ATCC), T2293 (containing N408K), T2291 (containing A834P), and T2311 (containing N408K and A834P) were also determined by a plaque reduction assay (PRA) with human embryonic lung fibroblasts (29). Each of these assays was carried out in triplicate, and the averages and standard deviations were calculated. Recombinant viruses with a greater than twofold increase in IC50 compared to those for the wild-type sensitive control strains were considered antiviral resistant, as this has been shown to be reproducible in previous studies of recombinant viruses (7, 13).
Antiviral susceptibility analysis of the Towne-derived mutants.
The GCV, FOS, and CDV susceptibilities of wild-type Towne (ATCC) and Towne-derived strains T2296 (containing L737M) and T2287 (containing A834P) were determined by PRA (40). A cell-free virus stock (50 PFU) was inoculated onto 80 to 100% confluent human lung (MRC-5) fibroblasts in 48-well plates (two assays per plate), and the plates were centrifuged at 600 × g for 30 min. After 1 h of incubation at 37°C with 5% CO2, the supernatant was removed and virus was grown in duplicate in the presence of 0.625 to 160 μM GCV, 6.25 to 1,600 μM FOS, or 0.0625 to 16 μM CDV in medium containing 0.8% carboxymethyl cellulose as well as with medium without an antiviral. The plaque numbers were counted, and the IC50 values were calculated for each assay after 7 days of culture. Each assay was carried out multiple times (8 to 11 times), and the averages and standard deviations were calculated. Recombinant viruses with a greater than twofold increase in IC50 compared to those for the wild-type sensitive control strains were considered antiviral resistant (7).
In vitro replication assays.
The growth properties of mutant recombinant viruses over multiple cycles of replication were assessed by inoculating strains T2211 and T2241 (wild-type controls), T2291 (containing A834P), and T2311 (containing N408K and A834P) into 24-well HFF cultures at a multiplicity of infection of 0.01. These experiments were carried out on three separate dates with independent viral dilutions. Four wells were inoculated per strain and after a 90-min absorption period, the supernatant was removed and replaced with 1 ml minimum essential medium-3% fetal bovine serum. On days 1, 4, 5, 6, 7, and 8, 80-μl aliquots of the medium supernatant were removed and frozen for subsequent SEAP assay. At the end of the experiment, SEAP assays were performed with chemiluminescence, as described previously (12). The SEAP assay values observed over the time period were graphed, with points and error bars representing the means and standard deviations, respectively, of the four wells sampled per strain.
RESULTS
Antiviral susceptibilities of AD169-derived recombinant viruses.
SEAP-based analysis and PRA consistently demonstrated the ability of mutation N408K to confer GCV and CDV resistance but not FOS resistance to AD169-derived recombinant virus T2293 (containing N408K) (Tables 1 and 2). SEAP-based analysis indicated that N408K conferred high-level resistance to CDV, with a 21.0-fold increase in the CDV IC50 of recombinant T2293 compared to the IC50s for the wild-type sensitive strains. The fold increase in CDV resistance (5.4-fold compared to those for the wild-type strains) was less pronounced when the increase was determined by PRA, which is probably related to the higher IC50s for the wild-type strains obtained by PRA compared to those obtained by SEAP-based analysis. The A834P mutation conferred GCV, FOS, and CDV resistance to recombinant virus T2291, as determined by SEAP-based analysis and PRA. Like N408K, the ability of mutation A834P to confer GCV resistance to T2291 was less pronounced when resistance was measured by PRA (2.2-fold increase in the GCV IC50 compared to that at the baseline) but was more clearly evident by SEAP-based analysis (5.4-fold increase in the GCV IC50 compared to that at the baseline).
TABLE 2.
Antiviral susceptibilities of AD169-derived mutants assessed by PRAa
| Virus | DNA pol genotype | GCV
|
FOS
|
CDV
|
|||
|---|---|---|---|---|---|---|---|
| IC50 (μM) | Fold increaseb | IC50 (μM) | Fold increaseb | IC50 (μM) | Fold increaseb | ||
| AD169 | Wild type | 1.9 ± 0.5 | 78 ± 13 | 0.9 ± 0.3 | |||
| T2293 | N408K | 10 ± 3.0 | 5.3 | 122 ± 48 | 1.6 | 4.9 ± 1.3 | 5.4 |
| T2291 | A834P | 4.1 ± 2.0 | 2.2 | 431 ± 184 | 5.5 | 3.3 ± 1.3 | 3.7 |
| T2311 | N408K + A834P | 12.4 ± 1.8 | 6.5 | 732 ± 418 | 9.4 | 8.9 ± 4.4 | 9.9 |
Mean values and standard deviations were calculated from three replicate experiments.
Fold increase in IC50 compared to the average IC50 for wild-type sensitive strains.
The addition of N408K with A834P in recombinant T2311 produced 2.7- to 6.2-fold increases in GCV and CDV resistance compared to the levels of resistance of T2291 containing the single A834P mutation by both the SEAP assay and PRA (Tables 1 and 2). As expected, addition of N408K with A834P produced a negligible (1.1- to 1.7-fold) increase in FOS resistance, given the lack of FOS resistance produced by the N408K mutation in T2293 with a single mutation. The combined effect of the N408K and A834P mutations on antiviral resistance was less consistent when the resistance of the mutant with the single N408K mutation was compared to that of the mutant with dual mutations. These data suggest that N408K enhanced the GCV and CDV resistance of mutants containing A834P, but the effect was not consistently reciprocal.
Antiviral susceptibility of the Towne-derived recombinant viruses.
Mutation L737M did not confer GCV or FOS resistance to Towne-derived recombinant strain T2296 (Table 3). A twofold increase in CDV susceptibility compared to that of the sensitive wild-type Towne strain was demonstrated for T2296, but this virus was not considered CDV resistant, given that resistant viruses typically demonstrate a greater than twofold increase in antiviral sensitivity (7, 12). Towne-derived recombinant T2287, which contained the A834P mutation, showed resistance to all three drugs (GCV, FOS, and CDV) by PRA, similar to the findings presented above for the corresponding AD169-derived recombinant T2291.
TABLE 3.
Antiviral susceptibilities of Towne-derived mutants assessed by PRA
| Virus | DNA pol genotype | GCV
|
FOS
|
CDV
|
|||
|---|---|---|---|---|---|---|---|
| IC50 (μM) | Fold increaseb | IC50 (μM) | Fold increaseb | IC50 (μM) | Fold increaseb | ||
| Towne | Wild type | 4.9 ± 0.5 | 62.8 ± 13.6 | 0.3 ± 0.1 | |||
| T2296 | L737M | 5.4 ± 1.9 | 1.1 | 63.4 ± 12.9 | 1.0 | 0.6 ± 0.1 | 2.0 |
| T2287 | A834P | 30.1 ± 10.2 | 6.1 | 486.3 ± 7.9 | 7.7 | 1.8 ± 0.5 | 6.0 |
Mean values and standard deviations were calculated from 8 to 11 replicate experiments.
Fold increase in IC50 compared to the IC50 for the wild-type sensitive strain.
Replication fitness of recombinant viruses.
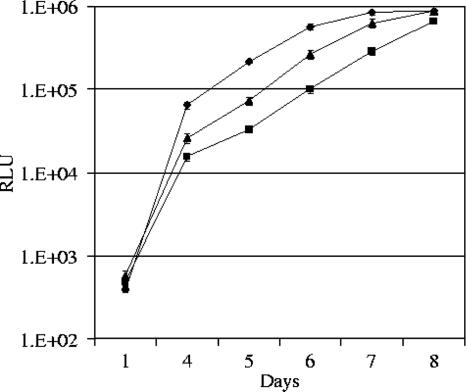
The amount of virus T2291 (containing A834P) measured by SEAP-based chemiluminescence after 4, 5, and 6 days in HFF culture was 4.2- to 6.5-fold less than that observed for the AD169-like wild-type strain (T2211) (Fig. 1). A similar decrease was observed when T2291 was compared to the second wild-type control strain, T2241 (results not shown). To ensure that the smaller amount of growth was not due to relative underinoculation, strain T2291 was adjusted on two of the setup dates so that its SEAP levels at day 1 were slightly greater than those for the wild-type strains. These initial reductions in virus growth became less pronounced by days 7 and 8, at which point the amount of wild-type virus had reached a maximum plateau, with 2.9- and 1.3-fold differences between the amounts for wild-type virus T2211 and strain T2291, respectively. Early attenuation was also observed for dual recombinant virus T2311, with a 2.1- to 2.9-fold reduction in viral output after 4 to 6 days of culture compared to that of the wild-type strain. By day 8 the viral output of strain T2311, which contained dual mutations, was equivalent to the maximum plateau for wild-type strain T2211. Similar results were obtained for each of the three experiments carried out on different dates. The SEAP activity at day 5 for the wild-type controls (T2211 or T2241) was consistently greater than that for T2311, which was in turn greater than that for T2291, with a >2-standard-deviation difference in the SEAP activity of T2291 and those of the other strains. T2311 consistently demonstrated higher SEAP activity levels than T2291 whether the SEAP levels for T2311 on day 1 were slightly higher or lower than those for T2291 (results not shown). The addition of mutation N408K in dual recombinant virus T2311 therefore improved the replication fitness of virus T2291, which contained only A834P.
FIG. 1.
Growth curve analysis of DNA polymerase mutant strains T2291 (▪; containing A834P) and T2311 (▴; containing N408K and A834P) versus wild-type antiviral-sensitive virus (⧫; T2211). Virus output (mean of four values) was quantified after 1, 4, 5, 6, 7, and 8 days of culture by a SEAP-based chemiluminescence assay (standard deviation error bars are indicated). RLU, relative light units. Similar results were obtained for each of the three experiments carried out on different dates.
DISCUSSION
The identification of multidrug resistance-conferring DNA polymerase mutations N408K (GCV and CDV resistance) and A834P (GCV, FOS, and CDV resistance) separately and together in three CMV strains infecting immunosuppressed CMV-seronegative transplant recipients emphasizes the increased risk of antiviral-resistant CMV acquisition in these patients (21, 27, 30, 31, 39). Consistent with the known risk factors for the emergence of these antiviral-resistant CMV strains, these transplant recipients had received long-term GCV therapy, potentially at subclinical doses (3, 16, 20, 27). The subsequent persistence of these strains may be attributed to the ongoing but intermittent antiviral (GCV, FOS, or CDV) treatment that these patients then received (30, 39). Consistent extended high-dose antiviral prophylaxis or observational preemptive therapy based on improved diagnostic assays may reduce the frequency of antiviral-resistant CMV strains in high-risk D+/R− transplant recipients (21). Unfortunately, all current CMV drugs may result in dose-limiting toxicity, and less toxic but effective anti-CMV therapy needs to be developed.
Consistent with our findings for mutation N408K (Tables 1 and 2), mutations of DNA polymerase domain IV-exonuclease II (ExoII) domain typically confer various levels of resistance to GCV and CDV but not resistance to FOS (14). The levels of GCV and CDV resistance among strains with the N408K mutation were similar to the published levels of GCV and CDV resistance among strains with the N408D mutation (14). This asparagine residue is highly conserved among α-like DNA polymerases and is located within the ExoII domain, which binds to single-stranded DNA and the metal ions required for excision of mismatched bases (2). It is interesting that substitution of a neutral polar residue (asparagine) to a basic residue (lysine) or acidic residue (aspartic acid) at this important codon has similarly contributed to GCV and CDV resistance.
The domain III amino terminus of herpesviruses consists of a DNA template binding region (47), immediately followed by residues important for pyrophosphate binding and nucleotide incorporation (9, 15, 44). Mutation A834P is situated within a putative α-helical region (α helix Q) at the less well defined carboxy end of DNA polymerase domain III (32, 45), where two other CMV mutations (T838A and G841A) and a herpes simplex virus (HSV) mutation (R842S) associated with antiviral resistance have been identified (22, 26, 42). The HSV R842S mutation has little effect on the interaction of the HSV DNA polymerase with dGTP (26), even though the neurovirulence of HSV is attenuated by this mutation in mice (38). This evidence and the reduced level of CMV replication in vitro produced by the A834P mutation shown here suggest that mutations toward the domain III carboxy terminus of DNA polymerase alter DNA template and substrate interactions through conformational changes in the catalytic core (32, 45). The close interactions of domain III and domain II residues in HSV- and RB69-derived DNA polymerase models support this hypothesis (26, 32, 45).
Qualitative antiviral resistance was consistently demonstrated for strains with the N408K and A834P mutations by two validated assays: the SEAP-based assay (which measures cell-free virus output to the medium) and PRA (which measures cell-associated virus replication) (12, 29). The different viral parameters measured by the two assays are likely to account for the quantitative differences in antiviral inhibitory concentrations indicated by the two assays, as suggested by the lower IC50 values demonstrated for even wild-type strain AD169 as measured by the SEAP-based assay compared with that measured by PRA (Tables 1 and 2). The greater dynamic range and increased number of replicates possible with the SEAP-based assay allow greater confidence in the mean values obtained. However, variations in results from laboratory to laboratory for standardized assays are well documented (29), requiring “in-house” comparisons to the relevant wild-type virus when qualitative antiviral resistance is determined by either method.
Further characterization of emerging DNA polymerase mutations that confer resistance to antiviral agents (such as N408K and A834P) and even those that result in antiviral-sensitive phenotypes (L737M) is necessary for the accurate and definitive detection of antiviral-resistant strains by rapid genotypic assays (35, 39). This and other studies are elucidating the total numbers of DNA polymerase mutations that confer antiviral resistance, but more investigations are required before this list is complete. As demonstrated here, investigation of novel antiviral resistance mutations increases our understanding of the functional regions of CMV DNA polymerase and will assist with the development of novel antiviral agents that inhibit CMV replication.
Acknowledgments
This work was supported by project grant 300532 from the National Health and Medical Research Council of Australia and U.S. National Institutes of Health grant AI39938.
We thank Gail Marousek, Laura Van Wechel, Heather Lichy, and Monique Nicolle for technical assistance.
Footnotes
Published ahead of print on 16 October 2006.
REFERENCES
- 1.Baldanti, F., D. Michel, L. Simoncini, M. Heuschmid, A. Zimmermann, R. Minisini, P. Schaarschmidt, T. Schmid, G. Gerna, and T. Mertens. 2002. Mutations in the UL97 ORF of ganciclovir-resistant clinical cytomegalovirus isolates differentially affect GCV phosphorylation as determined in a recombinant vaccinia virus system. Antivir. Res. 54:59-67. [DOI] [PubMed] [Google Scholar]
- 2.Bernad, A., L. Blanco, J. M. Lazaro, G. Martin, and M. Salas. 1989. A conserved 3′—5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59:219-228. [DOI] [PubMed] [Google Scholar]
- 3.Boivin, G., C. Gilbert, A. Gaudreau, I. Greenfield, R. Sudlow, and N. A. Roberts. 2001. Rate of emergence of cytomegalovirus (CMV) mutations in leukocytes of patients with acquired immunodeficiency syndrome who are receiving valganciclovir as induction and maintenance therapy for CMV retinitis. J. Infect. Dis. 184:1598-1602. [DOI] [PubMed] [Google Scholar]
- 4.Cannon, J. S., F. Hamzeh, S. Moore, J. Nicholas, and R. F. Ambinder. 1999. Human herpesvirus 8-encoded thymidine kinase and phosphotransferase homologues confer sensitivity to ganciclovir. J. Virol. 73:4786-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou, S. 1999. Antiviral drug resistance in human cytomegalovirus. Transplant. Infect. Dis. 1:105-114. [DOI] [PubMed] [Google Scholar]
- 6.Chou, S., A. Erice, M. C. Jordan, G. M. Vercellotti, K. R. Michels, C. L. Talarico, S. C. Stanat, and K. K. Biron. 1995. Analysis of the UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance. J. Infect. Dis. 171:576-583. [DOI] [PubMed] [Google Scholar]
- 7.Chou, S., N. S. Lurain, K. D. Thompson, R. C. Miner, and W. L. Drew. 2003. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32-39. [DOI] [PubMed] [Google Scholar]
- 8.Chou, S., G. Marousek, S. Guentzel, S. E. Follansbee, M. E. Poscher, J. P. Lalezari, R. C. Miner, and W. L. Drew. 1997. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J. Infect. Dis. 176:786-789. [DOI] [PubMed] [Google Scholar]
- 9.Chou, S., G. Marousek, D. M. Parenti, S. M. Gordon, A. G. LaVoy, J. G. Ross, R. C. Miner, and W. L. Drew. 1998. Mutation in region III of the DNA polymerase gene conferring foscarnet resistance in cytomegalovirus isolates from 3 subjects receiving prolonged antiviral therapy. J. Infect. Dis. 178:526-530. [DOI] [PubMed] [Google Scholar]
- 10.Chou, S., and C. L. Meichsner. 2000. A nine-codon deletion mutation in the cytomegalovirus UL97 phosphotransferase gene confers resistance to ganciclovir. Antimicrob. Agents Chemother. 44:183-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou, S., R. C. Miner, and W. L. Drew. 2000. A deletion mutation in region V of the cytomegalovirus DNA polymerase sequence confers multidrug resistance. J. Infect. Dis. 182:1765-1768. [DOI] [PubMed] [Google Scholar]
- 12.Chou, S., L. C. Van Wechel, H. M. Lichy, and G. I. Marousek. 2005. Phenotyping of cytomegalovirus drug resistance mutations by using recombinant viruses incorporating a reporter gene. Antimicrob. Agents Chemother. 49:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou, S., R. H. Waldemer, A. E. Senters, K. S. Michels, G. W. Kemble, R. C. Miner, and W. L. Drew. 2002. Cytomegalovirus UL97 phosphotransferase mutations that affect susceptibility to ganciclovir. J. Infect. Dis. 185:162-169. [DOI] [PubMed] [Google Scholar]
- 14.Cihlar, T., M. D. Fuller, and J. M. Cherrington. 1998. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J. Virol. 72:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cihlar, T., M. D. Fuller, A. S. Mulato, and J. M. Cherrington. 1998. A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture. Virology 248:382-393. [DOI] [PubMed] [Google Scholar]
- 16.Drew, W. L. 2000. Ganciclovir resistance: a matter of time and titre. Lancet 356:609-610. [DOI] [PubMed] [Google Scholar]
- 17.Ducancelle, A., J. Gravisse, S. Alain, A. M. Fillet, F. Petit, M. J. S. L. Pors, and M. C. Mazeron. 2005. Phenotypic characterisation of cytomegalovirus DNA polymerase: a method to study cytomegalovirus isolates resistant to foscarnet. J. Virol. Methods 125:145-151. [DOI] [PubMed] [Google Scholar]
- 18.Eckle, T., P. Lang, L. Prix, G. Jahn, T. Klingebiel, R. Handgretinger, B. Selle, D. Niethammer, and K. Hamprecht. 2002. Rapid development of ganciclovir-resistant cytomegalovirus infection in children after allogeneic stem cell transplantation in the early phase of immune cell recovery. Bone Marrow Transplant. 30:433-439. [DOI] [PubMed] [Google Scholar]
- 19.Emery, V. C. 2001. Investigation of CMV disease in immunocompromised patients. J. Clin. Pathol. 54:84-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emery, V. C., and P. D. Griffiths. 2000. Prediction of cytomegalovirus load and resistance patterns after antiviral chemotherapy. Proc. Natl. Acad. Sci. USA 97:8039-8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emery, V. C., A. F. Hassan-Walker, A. K. Burroughs, and P. D. Griffiths. 2002. Human cytomegalovirus (HCMV) replication dynamics in HCMV-naive and -experienced immunocompromised hosts. J. Infect. Dis. 185:1723-1728. [DOI] [PubMed] [Google Scholar]
- 22.Erice, A., C. Gil-Roda, J. L. Perez, H. H. Balfour, Jr., K. J. Sannerud, M. N. Hanson, G. Boivin, and S. Chou. 1997. Antiviral susceptibilities and analysis of UL97 and DNA polymerase sequences of clinical cytomegalovirus isolates from immunocompromised patients. J. Infect. Dis. 175:1087-1092. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert, C., and G. Boivin. 2005. Human cytomegalovirus resistance to antiviral drugs. Antimicrob. Agents Chemother. 49:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruber, S. A., J. Garnick, K. Morawski, D. H. Sillix, M. S. West, D. K. Granger, J. M. El-Amm, G. J. Alangaden, P. Chandrasekar, and A. Haririan. 2005. Cytomegalovirus prophylaxis with valganciclovir in African-American renal allograft recipients based on donor/recipient serostatus. Clin. Transplant. 19:273-278. [DOI] [PubMed] [Google Scholar]
- 25.Hanson, M. N., L. C. Preheim, S. Chou, C. L. Talarico, K. K. Biron, and A. Erice. 1995. Novel mutation in the UL97 gene of a clinical cytomegalovirus strain conferring resistance to ganciclovir. Antimicrob. Agents Chemother. 39:1204-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, L., K. K. Ishii, H. Zuccola, A. M. Gehring, C. B. Hwang, J. Hogle, and D. M. Coen. 1999. The enzymological basis for resistance of herpesvirus DNA polymerase mutants to acyclovir: relationship to the structure of alpha-like DNA polymerases. Proc. Natl. Acad. Sci. USA 96:447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isada, C. M., B. Yen-Lieberman, N. S. Lurain, R. Schilz, D. Kohn, D. L. Longworth, A. J. Taege, S. B. Mossad, J. Maurer, S. M. Flechner, S. D. Mawhorter, W. Braun, S. M. Gordon, S. K. Schmitt, M. Goldman, J. Long, M. Haug, and R. K. Avery. 2002. Clinical characteristics of 13 solid organ transplant recipients with ganciclovir-resistant cytomegalovirus infection. Transplant. Infect. Dis. 4:189-194. [DOI] [PubMed] [Google Scholar]
- 28.Jabs, D. A., B. K. Martin, M. S. Forman, J. P. Dunn, J. L. Davis, D. V. Weinberg, K. K. Biron, F. Baldanti, and H. Hu. 2001. Longitudinal observations on mutations conferring ganciclovir resistance in patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis: the Cytomegalovirus and Viral Resistance Study Group report number 8. Am. J. Ophthalmol. 132:700-710. [DOI] [PubMed] [Google Scholar]
- 29.Landry, M. L., S. Stanat, K. Biron, D. Brambilla, W. Britt, J. Jokela, S. Chou, W. L. Drew, A. Erice, B. Gilliam, N. Lurain, J. Manischewitz, R. Miner, M. Nokta, P. Reichelderfer, S. Spector, A. Weinberg, B. Yen-Lieberman, and C. Crumpacker. 2000. A standardized plaque reduction assay for determination of drug susceptibilities of cytomegalovirus clinical isolates. Antimicrob. Agents Chemother. 44:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levi, M. E., N. Mandava, L. K. Chan, A. Weinberg, and J. L. Olson. 2006. Treatment of multi-drug resistant cytomegalovirus retinitis with systematically administered leflunomide. Transplant. Infect. Dis. 8:38-43. [DOI] [PubMed] [Google Scholar]
- 31.Limaye, A. P., L. Corey, D. M. Koelle, C. L. Davis, and M. Boeckh. 2000. Emergence of ganciclovir-resistant cytomegalovirus disease among recipients of solid-organ transplants. Lancet 356:645-649. [DOI] [PubMed] [Google Scholar]
- 32.Liu, S., J. D. Knafels, J. S. Chang, G. A. Waszak, E. T. Baldwin, M. R. Deibel, D. R. Thomsen, F. L. Homa, P. A. Wells, M. C. Tory, R. G. Poorman, H. Gao, X. Qiu, and A. P. Seddon. 2006. Crystal structure of the herpes simplex 1 virus DNA polymerase. J. Biol. Chem. 281:18193-18200. [DOI] [PubMed] [Google Scholar]
- 33.Liu, W., C. Shum, D. F. Martin, B. D. Kuppermann, A. J. Hall, and T. P. Margolis. 2000. Prevalence of antiviral drug resistance in untreated patients with cytomegalovirus retinitis. J. Infect. Dis. 182:1234-1238. [DOI] [PubMed] [Google Scholar]
- 34.Lowance, D., H. H. Neumayer, C. M. Legendre, J. P. Squifflet, J. Kovarik, P. J. Brennan, D. Norman, R. Mendez, M. R. Keating, G. L. Coggon, A. Crisp, I. C. Lee, et al. 1999. Valacyclovir for the prevention of cytomegalovirus disease after renal transplantation. N. Engl. J. Med. 340:1462-1470. [DOI] [PubMed] [Google Scholar]
- 35.Lurain, N. S., A. Weinberg, C. S. Crumpacker, S. Chou, and the Adult AIDS Clinical Trials Group-CMV Laboratories. 2001. Sequencing of cytomegalovirus UL97 gene for genotypic antiviral resistance testing. Antimicrob. Agents Chemother. 45:2775-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattes, F. M., E. G. Hainsworth, A. F. Hassan-Walker, A. K. Burroughs, P. Sweny, P. D. Griffiths, and V. C. Emery. 2005. Kinetics of cytomegalovirus load decrease in solid-organ transplant recipients after preemptive therapy with valganciclovir. J. Infect. Dis. 191:89-92. [DOI] [PubMed] [Google Scholar]
- 37.Paya, C., A. Humar, E. Dominguez, K. Washburn, E. Blumberg, B. Alexander, R. Freeman, N. Heaton, M. D. Pescovitz, and Valganciclovir Solid Organ Transplant Study Group. 2004. Efficacy and safety of valganciclovir vs. oral ganciclovir for prevention of cytomegalovirus disease in solid organ transplant recipients. Am. J. Transplant. 4:611-620. [DOI] [PubMed] [Google Scholar]
- 38.Pelosi, E., F. Rozenberg, D. M. Coen, and K. L. Tyler. 1998. A herpes simplex virus DNA polymerase mutation that specifically attenuates neurovirulence in mice. Virology 252:364-372. [DOI] [PubMed] [Google Scholar]
- 39.Scott, G. M., M. A. Isaacs, F. Zeng, A. M. Kesson, and W. D. Rawlinson. 2004. Cytomegalovirus antiviral resistance associated with treatment induced UL97 (protein kinase) and UL54 (DNA polymerase) mutations. J. Med. Virol. 74:85-93. [DOI] [PubMed] [Google Scholar]
- 40.Scott, G. M., H. L. Ng, C. J. Morton, M. W. Parker, and W. D. Rawlinson. 2005. Murine cytomegalovirus resistant to antivirals has genetic correlates with human cytomegalovirus. J. Gen. Virol. 86:2141-2151. [DOI] [PubMed] [Google Scholar]
- 41.Soderberg-Naucler, C., and V. C. Emery. 2001. Viral infections and their impact on chronic renal allograft dysfunction. Transplantation 71:SS24-SS30. [PubMed] [Google Scholar]
- 42.Springer, K. L., S. Chou, S. Li, R. H. Giller, R. Quinones, J. E. Shira, and A. Weinberg. 2005. How evolution of mutations conferring drug resistance affects viral dynamics and clinical outcomes of cytomegalovirus-infected hematopoietic cell transplant recipients. J. Clin. Microbiol. 43:208-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan, V., C. L. Talarico, S. C. Stanat, M. Davis, D. M. Coen, and K. K. Biron. 1992. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature 358:162-164. [DOI] [PubMed] [Google Scholar]
- 44.Tchesnokov, E. P., C. Gilbert, G. Boivin, and M. Gotte. 2006. Role of helix P of the human cytomegalovirus DNA polymerase in resistance and hyper-susceptibility to the antiviral drug foscarnet. J. Virol. 80:1440-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, J., A. K. Sattar, C. C. Wang, J. D. Karam, W. H. Konigsberg, and T. A. Steitz. 1997. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell 89:1087-1099. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg, A., D. A. Jabs, S. Chou, B. K. Martin, N. S. Lurain, M. S. Forman, and C. Crumpacker. 2003. Mutations conferring foscarnet resistance in a cohort of patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis. J. Infect. Dis. 187:777-784. [DOI] [PubMed] [Google Scholar]
- 47.Ye, L. B., and E. S. Huang. 1993. In vitro expression of the human cytomegalovirus DNA polymerase gene: effects of sequence alterations on enzyme activity. J. Virol. 67:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]