Abstract
Rotavirus serotype G9 is recognized as the most widespread of the emerging serotypes, emerging since 1996 as a frequent cause of severe acute gastroenteritis in children from many countries covering all continents of the world. This study characterized serotype G9 strains collected in three widely separated Australian centers from 1997 to 2001. All G9 strains possessed the VP4 P[8] and VP6 subgroup II genes. The overall prevalence of the G9 strains increased in Australia, from 0.6% of the strains found in 1997 to 29% of the strains found in 2001. The prevalence of G9 relative to all other serotypes varied from year to year and with geographic location. In Melbourne (representing east coast urban centers), G9 made up 11 to 26% of all of the strains found from 1999 to 2001. In Perth (representing west coast urban centers), G9 made up less than 2% of the strains found in 1997 to 2000 but increased to 18.6% of the strains found in 2001. In Alice Springs (representing widely dispersed settlements in northern arid regions), G9 made up 0 to 5% of the strains found from 1997 to 2000 and was the dominant strain in 2001, making up 68.9% of all of the strains found. Three distinct antigenic groups based on reaction with neutralizing monoclonal antibodies (N-MAbs) were identified, including a dominant group (63%) that cross-reacted with the serotype G4 N-MAb. Phylogenetic analysis of the VP7-encoding gene from Australian strains, compared with a worldwide collection of G9 strains, showed that the Australian G9 strains made up a genetic group distinct from other serotype G9 strains identified in the United States and Africa. Future epidemiological studies of the occurrence of G9 strains should combine reverse transcription-PCR and typing with G1 to G4 and G9 N-MAbs to determine the extent of G9 and G4 cross-reactions among rotavirus strains, in order to assess the need to incorporate G9 strains into new candidate vaccines.
Rotaviruses are the major cause of severe gastroenteritis in young children worldwide. Vaccines are being developed to reduce the huge impact of the disease caused by rotavirus infection. The first vaccines were developed to provide specific protection against the four predominant serotypes of rotavirus, G1 to G4 (29), as these have been the most common serotypes causing severe disease in children globally since 1973 (40). Recent epidemiological studies in Bangladesh (49), Brazil (23, 32, 44), India (1), the United States (24, 42), and Malawi (13) show that other G types (G5, G6, G8, G9, and G10) can be identified as causes of severe disease and are of emerging importance in some communities. Serotype G9 is recognized as the most widespread of the “emerging” serotypes and has been identified since 1996 as a frequent cause of severe disease in hospitalized children in the United States, Japan, India, Bangladesh, France, Italy, Malawi, Nigeria, Australia, China, Thailand, and the United Kingdom (3, 7, 8, 12, 19, 26, 34, 37, 39, 41, 42, 47, 49, 50).
The rotavirus genome is composed of 11 segments of double-stranded RNA located inside the core of a triple-layered structure. The outer capsid proteins VP4 and VP7 elicit neutralizing antibody immune responses, creating both serotype-specific and cross-reactive immunity (18). Antigenic differences in VP4 and VP7 are the basis of the G (VP7 glycoprotein) and P (protease-activated VP4 protein) serotypes. To date, 9 P and 10 G serotypes have been identified in humans by cross-neutralization tests (18, 46, 48). Unlike rotavirus G typing, there are two designations of rotavirus type P because of incomplete agreement between the P serotype (based on enzyme immunoassay [EIA] reactivities) and the P genotype (based on sequence similarity). The P genotypes are in brackets, whereas the P serotypes are open numbers. Epidemiological studies have shown that serotypes G1, G2, G3, and G4, associated with P1A (8) or P1B (4), have been the most common serotypes causing severe disease in children globally over the last 20 years (35, 40). Genetic and antigenic variation has been recorded within the G1, G2, G3, and G4 serotypes (38). There is evidence that G9 strains are more susceptible to reassortment, and hence to genetic change, than are these other serotypes (27, 49). The increasing prevalence of G9 strains worldwide makes it important to continue molecular epidemiological studies of their occurrence and genetic and antigenic variability.
The emergence and persistence of serotype G9 has had a major impact on health care services in Australia (33, 34). This report describes the appearance, spread, and prevalence of G9 strains in widely separated areas of Australia (>2,000 km apart) during the 5 years after 1997, when they were first detected. It characterizes serotype G9 strains collected in Australia as part of the National Rotavirus Strain Surveillance Program and describes the distinct antigenic and genetic patterns found in these strains. The results underline the importance of continued detailed epidemiological and virological studies to identify the rotavirus serotypes that cause severe gastroenteritis, including characterization of less common and/or unusual strains. Knowledge of rotavirus strains in circulation in Australia and other countries will aid in assessing the suitability of candidate vaccines to protect against all currently circulating rotavirus serotypes.
MATERIALS AND METHODS
Stool samples.
A total of 2,843 group A rotavirus-positive diarrhea specimens were collected from children (<5 years of age) with severe acute diarrhea who were admitted to hospitals at three widely separated locations in Australia representing eastern (Melbourne), central (Alice Springs), and western (Perth) urban areas from January 1997 to December 2001. Fecal samples were collected within 48 h of hospital admission, and rotavirus detection was undertaken by enzyme immunoassay or latex agglutination. Rotavirus-positive specimens were stored at 4°C in the laboratory of origin and later sent to the Rotavirus Reference Laboratory in Melbourne.
During 1999, additional rotavirus-positive fecal specimens (n = 332) were collected from children admitted to hospitals in other east coast Australian cities (Sydney, Adelaide, and Brisbane) and transported to the Melbourne laboratory. Selected serotype G9 strains identified in these samples were also used for sequence analysis.
Serotyping EIA.
All rotavirus-positive samples were tested in Melbourne for G serotype and VP6 subgroup antigens with a previously described monoclonal antibody (MAb)-based EIA (10). The specific MAbs used for serotype determination were RV4:2 (G1 specific), RV5:3 (G2 specific), RV3:1 (G3 specific), ST3:1 (G4 specific), and F45:1 (G9 specific). Additional serotype G9 MAbs F45:8 and WI61:1 (30) were also used to screen all of the putative serotype G9 strains and nontypeable strains.
RNA electropherotyping.
The electropherotypes of rotavirus-positive samples were determined by polyacrylamide gel electrophoresis as described by Dyall-Smith and Holmes (17).
VP7 and VP4 genotyping.
Specimens that could not be assigned a serotype, or that reacted with more than one MAb by EIA, and were collected from 2000 onward were analyzed by a heminested reverse transcription (RT)-PCR assay to determine the VP7 genotype (22). All specimens identified as non-serotype G1, together with 20% of the G1 samples (selected as representative of different electropherotypes present from 1997 to 2001), were assayed to determine the VP4 genotype. The VP4 genotype was determined by multiplex seminested RT-PCR (21). Classifications of P and G types were designated in accordance with the recommendations of the Rotavirus Nomenclature Working Group (25).
Sequence analysis.
PCR products of the gene encoding the entire VP7 protein of G9 strains were amplified by RT-PCR with the Beg9 and End9 primers (22). The PCR products were gel purified with a Qiagen kit (Qiagen Inc.). The nucleotide sequence of the entire open reading frame of the gene encoding VP7 was determined by the dideoxynucleotide chain terminator method with the BigDye sequencing kit and specific oligonucleotide primers in an automated sequencer.
Sequences were analyzed with the Sequencher program (Gene Codes Corp., Inc., Ann Arbor, Mich.) and subsequently compared with other sequences with E-CLUSTAL W and analyzed by using the DNAdist and Neighbor programs from the PHYLIP software accessed through BioManager, Australian National Genomic Information Service (University of Sydney). The statistical significance of the constructed phylogenies were analyzed by bootstrap analysis with 100 replicates (20). The phylogenetic tree was displayed with the Treeview program.
Nucleotide sequence accession numbers.
The Australian VP7-encoding gene sequences described in this study have been deposited in the GenBank data bank and assigned accession numbers AY307085 to AY307094, inclusive.
RESULTS
Data on the occurrence of the G1, G2, G3, G4, and G9 serotypes from the Australia-wide rotavirus surveillance from January 1997 to December 2001 are presented in Table 1. Results obtained by surveillance prior to 1997 are not included since analysis of all of the strains found, including nontypeable strains, by RT-PCR showed no evidence of G9 occurrence prior to 1997 (39). Table 1 shows that serotype G1 was the dominant type each year, representing 50.7 to 70.8% of the samples. Serotype G2 was identified each year, with a peak in prevalence during 1999, when it was the second most common serotype (12.8%). Serotypes G3 and G4 were present each year but represented less than 2.5% of the strains found. Serotype G9 was first identified in Australia in 1997 (39) and emerged as the second most common type in 2000, with a peak in prevalence during 2001, when it represented 29% of the strains found.
TABLE 1.
Overall serotyping results from three Australian centers, Alice Springs, Perth, and Melbourne, from 1997 to 2001a
| Yr | No. of isolates | % of rotavirus-positive samples by serotype
|
|||||
|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G9 | NTb | ||
| 1997 | 466 | 62.9 | 6.4 | 0 | 1.5 | 0.6 | 28.5 |
| 1998 | 498 | 65.5 | 3.6 | 1.6 | 0.2 | 0 | 29.1 |
| 1999 | 880 | 51.8 | 12.8 | 0.3 | 0.7 | 5.3 | 29 |
| 2000 | 356 | 70.8 | 9 | 0.6 | 2.5 | 10.1 | 7 |
| 2001 | 643 | 50.7 | 1.4 | 0.2 | 1.9 | 29 | 16.8 |
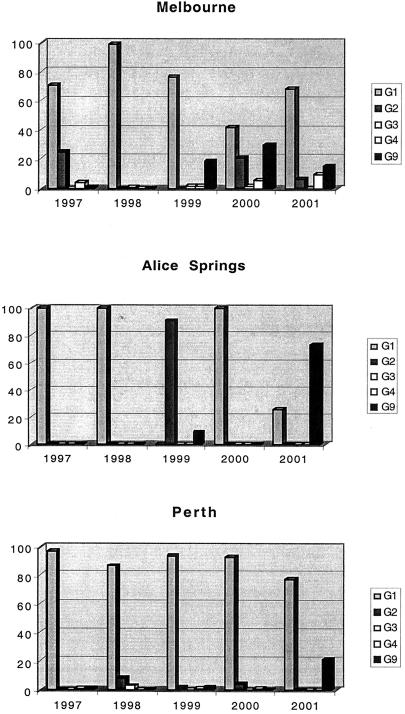
Figure 1 illustrates the prevalence of all of the rotavirus serotypes at the three locations studied. G9 strains were first identified in Melbourne and Perth in 1997 and were the second most prevalent serotype in Melbourne in 1999, 2000, and 2001, representing 19% (n = 30), 29% (n = 35), and 15% (n = 19) of the strains found, respectively. Serotype G9, while present in Perth during 1999 to 2001, represented less than 3% (n = <6) of all of the strains found in 1999 and 2000 but 22% (n = 57) of the strains found in 2001. Serotype G9 strains were identified in Alice Springs in only 2 of the 5 years (1999 and 2001), but this was the dominant serotype, representing 73.5% (n = 111) of all of the strains serotyped in 2001.
FIG. 1.
Relative frequencies of rotavirus serotypes G1, G2, G3, G4, and G9 in hospitalized children in Melbourne, Alice Springs, and Perth, Australia, from 1997 to 2001.
All of the G9 strains identified from all of the sites studied (1997 to 2001) belonged to subgroup II, showed “long” RNA patterns, were VP4 genotype P (8), and reacted with serotype G9-reactive MAbs F45:1 and WI61:1.
Antigenic analysis of the G9 strains identified during 1999 to 2001 showed three distinct antigenic patterns (G9A, G9B, and G9C) on the basis of their EIA reactivities with the additional G9 MAb (F45:8) and G4 serotyping MAb (ST3:1) (Table 2). The G9A group reacted with both serotype G9 MAb F45:8 and G4 MAb ST3:1 and represented 56 to 76% of the strains found. The EIA reactivities of the strains with the ST3:1 MAb ranged from weakly positive (>0.2 above the background) to strongly positive (>1.0 above the background). The G9B group reacted with MAb F45:8 but not with MAb ST3:1 and represented between 2 and 7% of G9 strains identified. The third group, G9C, represented 22 to 37% of the strains found and did not react with either the F45:8 or the ST3:1 MAb. The three antigenic groups were distributed with similar frequencies at each of the geographic sites.
TABLE 2.
Antigenic reactivities of the serotype G9 strains present in Australia from 1999 to 2001
| Subtype | EIA MAb reactivitya
|
Prevalence rate (%)
|
|||||
|---|---|---|---|---|---|---|---|
| F45:1-WI61:1 | F45:8 | ST3:1 | Avg (n = 270) | 1999 | 2000 | 2001 | |
| G9A | + | + | + | 63 | 76 | 61 | 56 |
| G9B | + | + | − | 4 | 2 | 3 | 7 |
| G9C | + | − | − | 33 | 22 | 36 | 37 |
+, positive EIA reactivity; −, negative EIA reactivity. F45:1, WI61:1, and F45:8 are serotype G9-reactive MAbs; ST3:1 is a serotype G4-reactive MAb.
Sequence analysis. (i) Comparison of the VP7-encoding genes of antigenic variants G9A, G9B, and G9C.
Comparison of the complete nucleotide and deduced amino acid sequences of the VP7-encoding genes of G9 strains isolated in Melbourne during 2000 and 2001, representing G9A (four strains), G9B (four strains), and G9C (four strains), revealed greater than 99% nucleotide and amino acid sequence homology among the three antigenic types. In common with other G9 strains sequenced, the gene contained 1,061 nucleotides.
Antigenic regions of VP7 (region A, amino acids [aa] 87 to 101; region B, aa 143 to 152; region C, aa 201 to 221; region D, aa 291; region E, aa 189 to 190; region F, aa 235 to 242) previously identified as neutralization epitopes by sequence analysis of escape variants resistant to neutralization by MAbs were compared among G9A, G9B, and G9C strains. No conserved amino acid substitutions in these major antigenic regions of the VP7 protein of Melbourne G9A, G9B, and G9C strains were identified. A single conserved amino acid difference was identified in strains from group G9B, where a replacement of proline with serine was found outside the major antigenic regions at amino acid position 46 compared with G9A and G9C strains.
The VP7-encoding genes from two additional G9C strains isolated in Sydney and Adelaide in 1999 were also sequenced. A single amino acid difference was identified when these strains were compared with all of the Melbourne G9 strains, involving a change from aspartic acid to asparagine at position 100 in the antigenic A region.
(ii) Phylogenetic relationship between G9 isolates from Australia and other countries.
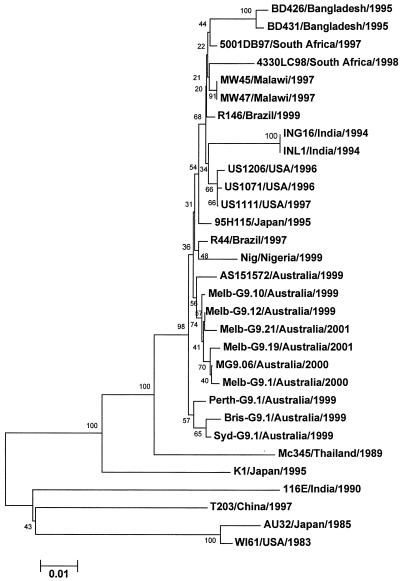
The nucleotide sequences of the VP7-encoding genes from six G9 strains representing Melbourne groups G9A, G9B, and G9C, together with additional single strains from Alice Springs, Perth, Sydney, and Brisbane, were determined and compared with the corresponding sequences available in the GenBank database of representative G9 strains from other worldwide locations. It is evident from the phylogenetic analysis (Fig. 2) that the recently identified serotype G9 strains appear to be distinct from the G9 strains identified in the 1980s (e.g., WI61 and AU32). There is less than 4% nucleotide sequence divergence among the G9 strains that have recently emerged worldwide; however, distinct groups are evident. For example, the Australian G9 strains exhibit greater than 99% nucleotide homology with each other. Compared with Australian prototype G9 strain MG9, the other major groups exhibited slightly lower homologies (US1071, 98.8%; MW47, 98.8%; BD426, 97.6%; India, G16 97.1%).
FIG. 2.
Phylogenetic analysis of the coding region (nucleotides 49 to 1029) of the VP7-encoding gene from serotype G9 strains identified worldwide. The tree was constructed by using the E-CLUSTAL W and neighbor-joining methods, and bootstrap values are indicated. The G9 strains used in the analysis were obtained from the GenBank database. The accession numbers are as follows: AU32, ABO45372; 116E, L14072; T203, AF260959; K1, ABO45374; Mc345, D38055; R44, AF438227; R146, AF27970; Nigeria, AF359358; 95H115, AB260959; 4330LC/98, AF529865; 5001DB/97, AF529864; MW45, HR0250545; MW47, HRO250544; BD426, HRO250541; BD431, HRO250542; 1071, HRO250268; 1111, HRO250269; 1206, HRO250271; ING16, HRO250276; INL1, HRO250277.
DISCUSSION
This study highlights the emergence of serotype G9 as an epidemiologically important rotavirus strain in Australia since 1997. Retrospective studies of previously nontypeable strains showed no evidence of the presence of G9 in Australia before it was identified in 1997 in three children with severe rotavirus-induced gastroenteritis (39). Since then, G9 strains have been identified across the continent and G9 has become the second most prevalent G type identified overall in Australia during 2000 and 2001. This timing reflects that documented worldwide, where G9 strains reemerged in 1995 and 1996 after their initial identification as a cause of acute gastroenteritis in Japan and the United States in 1986 (8). Serotype G9 has been identified in more than 17 countries (2-4, 6, 7, 9, 12-14, 19, 24, 26, 28, 33-37, 41, 42, 47, 49, 50), with incidence rates generally ranging from less than 1 to 8%. Exceptions include countries such as Bangladesh and Nigeria, which had prevalence rates of greater than 34% (47, 49).
Different epidemiological patterns of serotype G9 were identified in Australia among Melbourne, Victoria (representing the temperate eastern seaboard); Perth, Western Australia (representing the temperate western seaboard); and Alice Springs, Northern Territory (representing the hot, arid climate of central Australia). This study shows the persisting importance of G9 in hospitalized Melbourne children in 1999 (19%), 2000 (29%), and 2001 (15%). Serotype G9 exhibited a high prevalence in children admitted to hospitals in Perth in 2001 (22%) after 4 years of low prevalence from 1997 to 2000. Striking yearly differences in prevalence were identified in Alice Springs, ranging from 9% in 1999 to 0% in 1997, 1998, and 2000 and to 73.5% in 2001. The occurrence of rotavirus outbreaks caused by other newly emerged dominant strains, particularly among aboriginal children, has been noted previously in this region, where children from widely separated settlements are transferred to the centralized Alice Springs Hospital (5).
Earlier characterization of G9 strains described variability in the associated P type. The first reports of serotype G9 in Japan and the United States in 1983 showed that these strains contain a P[8] VP4 protein. Since then G9 strains have been reported with a P[11], a P[6], or a P[8] VP4 gene (15, 26, 31, 43). In both the United States and the United Kingdom, the combination of G and P types has switched in the years subsequent to their initial identification. For example, G9 strains were first found in the United Kingdom as G9P[6] in 1995 to 1996 but were displaced by G9P[8] strains in subsequent years (27). This alteration in P type has not been seen in Australia. All of the G9 strains identified in this study from 1997 to 2001 possessed the P[8] VP4 gene, had a long electropherotype, and exhibited VP6 subgroup II antigens. Mixed infections have been rare in Australia, with a frequency of less than 1.7% (33, 34), but have been identified elsewhere at higher frequencies (e.g., 23% in Bangladesh) (28, 49). The occurrence of mixed infections could explain the suggestion that serotype G9 strains undergo reassortment much more easily and rapidly than other G serotypes, as evidenced by the rapid alteration in the VP4 and VP6 gene combinations in Bangladesh (49).
The G9 strains present in Australia from 1999 to 2001 comprised three distinct antigenic groups or monotypes, designated G9A, G9B, and G9C, reflecting three patterns of reaction with neutralizing MAbs (N-MAbs) directed to G4 and G9 VP7. The predominant monotype, G9A, representing the majority of G9 strains (63%), cross-reacted with serotype G4-specific MAb ST3:1, in addition to G9 N-MAbs WI61:1 and F45:8. Previously, Unicomb and coworkers (49) reported that 14 of 19 G9P[6] and 6 of 8 G9P[8] Bangladeshi strains with a short electropherotype exhibited similar cross-reactivity with serotype G4 MAbs. A single strain from India has also been shown to have similar cross-reactivity (11). The reactivity with ST3:1 cannot be explained by changes in the sequences of the MAb binding sites on VP7. It was previously suggested that the binding of ST3:1 may be a result of close homology between G4 and G9 strains in the antigenic A region (11). However, this explanation seems unlikely, at least for these Australian strains, since all three of the antigenic groups identified in this study contained identical A, B, C, and F regions, even though groups G9B and G9C did not react with the ST3:1 MAb. An alternative explanation is that these differing reactivities are related to unidentified changes in the VP4 structure that have been shown previously to influence the expression of VP7 epitopes (16). Clarification of this requires identification of the complete nucleotide and deduced amino acid sequences of these strains.
G9B (4% of the strains found) and G9C (33% of the strains found) reacted only with the G9 N-MAbs. G9B reacted with both MAbs WI61:1 and F45:8, while strains in the G9C group reacted solely with MAb WI61:1, suggesting antigenic drift in the VP7 protein. The monotype G9B differed genetically from G9A and G9C, where a single amino acid substitution was identified at position 42, but this position is unlikely to influence MAb binding. Analysis of the antigenic regions identified on the VP7-encoding gene previously implicated in neutralization by the use of antigenic variants (30) did not identify any genetic basis for the different MAb reactivities of G9A, G9B, and G9C. By using antigenic variants resistant to neutralization by MAb F45:8, we have previously shown that no clear motif is evident for the binding and neutralization of this MAb (30). Variants selected a substitution at position 97. However, mutations in the A, C, and F regions failed to influence MAb binding (30). The present study of Australian G9 strains showed no difference in any antigenic region that may have influenced MAb binding.
The inability of serotype G9-specific MAb F45:8 to bind all of the serotype G9 strains and the observed cross-reactivity with serotype G4-specific MAb ST3:1 have implications for the reliability and accuracy of all surveys to determine the epidemiological importance of serotype G9. For example, reliance on F45:8 alone in a serotyping EIA would have failed to identify as many as one-third of the G9 strains. Failure to use this G9-specific MAb (as occurs in many studies) would also have resulted in almost two-thirds of the G9 strains being falsely identified as serotype G4, while one-third remained untypeable. Conversely, exclusive reliance on RT-PCR to determine the VP7 genotype would have failed to identify the cross-reaction with G4 serotypes that could have immunological importance. Sole reliance on PCR to determine the G genotype of G9 strains has already been shown to have potential problems. A recent publication reported that Brazilian G9 strains could be incorrectly assigned to genotype G3 or G4 or mixed genotype G3-G9 or G4-G9, depending on the primer pool used for typing (45). This is due to the extent of genetic variation identified in the VP7-encoding gene of the G9 strains. Therefore, all typing methods require continued validation and improvements to correctly identify rotavirus serotypes. We are confident that serotype G9 strains were not present in our Australian strain collection prior to 1997 since all of the G3 and G4 strains have been screened with a combination of EIA, RT-PCR, and electropherotyping assays.
Identification of the Australian serotype G9 strains together with other G9 strains from diverse geographic locations, such as the United States, India, Nigeria, China, and Malawi, highlights the global presence of serotype G9, its persistence in locations monitored for several years, and its capacity to undergo antigenic change. Phylogenetic analysis illustrates that the recently identified G9 VP7 protein is divided into several groups, each of which has a limited geographic relationship. Whether these differences are due to differing geographic pressures on the same G9 strain remains to be elucidated. These results, however, suggest that the reemergence of G9 after the initial description from Japan and the United States in 1985 and 1986 was not directly related to evolution of the earlier strains (37).
Since 1997, the increase in reports of G9 from developed and developing countries shows the need for continued surveillance to identify the persistence of G9 strains in the community. Further studies to clarify the basis for the reactivity of certain G9 strains with G4 MAbs are required. In particular, stimulation of cross-neutralizing antibodies to VP7 of both G9 and G4 may be critical to a decision of whether it is necessary to add a G9 reassortant to current candidate vaccines. Monitoring of changes in strain prevalence will provide better understanding of virus evolution and the shifting trends of strain patterns over time. This information is vital for future vaccine strategies, which are predicated on the development of serotype-specific immunity to the globally common G serotypes.
Acknowledgments
This work was partially supported by grants from the Australian Commonwealth Department of Health and Aged Care, Australia, and GlaxoSmithKline.
REFERENCES
- 1.Aijas, S., K. Gowda, H. V. Jagannath, R. R. Reddy, P. P. Maiya, R. L. Wood, H. B. Greenberg, M. Raju, A. Babu, and C. D. Rao. 1996. Epidemiology of symptomatic human rotaviruses in Bangladore and Mysore, India from 1988 to 1994 as determined by electropherotypes, subgroup and serotype analysis. Arch. Virol. 141:715-726. [DOI] [PubMed] [Google Scholar]
- 2.Araújo, I. T., M. S. R. Ferreira, A. M. Fialho, R. M. Assis, C. M. Cruz, M. Rocha, and J. P. G. Leite. 2001. Rotavirus genotypes P[4]G9, P[6]G9, and P[8]G9 in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil. J. Clin. Microbiol. 39:1999-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arista, S., E. Vizzi, D. Ferraro, A. Cascio, and R. di Stefano. 1997. Distribution of VP7 and VP4 genotypes among rotavirus strains recovered from Italian children with diarrhea. Arch. Virol. 142:2065-2071. [DOI] [PubMed] [Google Scholar]
- 4.Armah, G. E., C. T. Pager, R. H. Asmah, F. R. Anto, A. R. Oduro, F. Binka, and A. D. Steele. 2001. Prevalence of unusual human rotavirus strains in Ghanaian children. J. Med. Virol. 63:67-71. [PubMed] [Google Scholar]
- 5.Bishop, R. F., P. J. Masendycz, H. C. Bugg, J. B. Carlin, and G. L. Barnes. 2001. Epidemiology patterns of rotaviruses causing severe gastroenteritis in young children throughout Australia from 1993 to 1996. J. Clin. Microbiol. 39:1085-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bok, K., G. Palacios, K. Sijvarger, D. Matson, and J. Gomez. 2001. Emergence of G9 P[6] human rotaviruses in Argentina: phylogenetic relationships among G9 strains. J. Clin. Microbiol. 39:4020-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bon, F., C. Fromantin, S. Aho, P. Pothier, E. Kohli, and The Azay Group. 2000. G and P genotyping of rotavirus strains circulating in France over a three-year period: detection of G9 and P[6] strains at low frequencies. J. Clin. Microbiol. 38:1681-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, H. F., Y. Hoshino, L. M. Bell, J. Groff, G. Hess, P. Bachman, and P. A. Offit. 1987. Rotavirus isolate W161 representing a presumptive new human serotype. J. Clin. Microbiol. 25:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coluchi, N., V. Munford, J. Manzur, C. Vazquez, M. Escobar, E. Weber, P. Marmol, and M. L. Racz. 2002. Detection, subgroup specificity, and genotype diversity of rotavirus strains in children with acute diarrhea in Paraguay. J. Clin. Microbiol. 40:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coulson, B. S., L. E. Unicomb, G. A. Pitson, and R. F. Bishop. 1987. Simple and specific enzyme immunoassay using monoclonal antibodies for serotyping human rotaviruses. J. Clin. Microbiol. 25:509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulson, B. S., J. R. Gentsch, B. K. Das, M. K. Bhan, and R. I. Glass. 1999. Comparison of enzyme immunoassay and reverse transcriptase PCR for identification of serotype G9 rotaviruses. J. Clin. Microbiol. 37:3187-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cubitt, W. D., A. D. Steele, and M. Iturrize. 2000. Characterisation of rotaviruses from children treated at a London hospital during 1996: emergence of strains G9P2A[6] and G3P2A[6]. J. Med. Virol. 61:150-154. [DOI] [PubMed] [Google Scholar]
- 13.Cunliffe, N. A., J. S. Gondwe, R. L. Broadhead, M. E. Molyneux, P. A. Woods, J. S. Bresee, R. I. Glass, J. R. Gentsch, and C. A. Hart. 1999. Rotavirus G and P types in children with acute diarrhea in Blantyre, Malawi, from 1997 to 1998: predominance of novel P[6]G8 strains. J. Med. Virol. 57:308-312. [PubMed] [Google Scholar]
- 14.Cunliffe, N. A., W. Dove, J. E. G. Bunn, M. B. Ramadam, J. W. O. Nyangao, R. L. Riveron, L. E. Cuevas, and C. A. Hart. 2001. Expanding global distribution of rotavirus serotype G9: detection in Libya, Kenya and Cuba. Emerg. Infect. Dis. 7:890-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, B. K., J. R. Gentsch, H. G. Cicirello, P. A. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 32:1820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn, S. J., J. W. Burns, T. L. Cross, P. T. Vo, R. L. Ward, M. Bremont, and H. B. Greenberg. 1994. Comparison of VP4 and VP7 of five murine rotavirus strains. Virology 203:250-259. [DOI] [PubMed] [Google Scholar]
- 17.Dyall-Smith, M. L., and I. H. Holmes. 1984. Sequence homology between human and animal rotavirus serotype-specific glycoproteins. Nucleic Acids Res. 12:3937-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estes, M. 1996. Rotaviruses and their replication, p. 1625-1655. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 19.Fang, Z.-Y., H. Yang, J. Qi, J. Zhang, L.-W. Sun, J.-Y. Tang, L. Ma, Z.-Q. Du, A. He, J.-P. Xie, Y.-Y. Lu, Z.-Z. Ji, B.-Q. Zhu, H.-Y. Wu, S.-E. Lin, H.-P. Xie, D. D. Griffin, B. Ivanoff, R. I. Glass, and J. R. Gentsch. 2002. Diversity of rotavirus strains among children with acute diarrhea in China: 1998-2001 surveillance study. J. Clin. Microbiol. 40:1875-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 21.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouvea, V., R. I. Glass, P. Woods, K. Taniguichi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouvea, V., and N. Santos. 1999. Rotavirus serotype G5: an emerging cause of epidemic childhood diarrhea. Vaccine 17:1291-1292. [DOI] [PubMed] [Google Scholar]
- 24.Griffin, D. D., C. D. Kirkwood, U. D. Parashar, P. A. Woods, J. S. Bresee, R. I. Glass, J. R. Gentsch, and The National Rotavirus Strain Surveillance System Collaborating Laboratories. 2000. Surveillance of rotavirus strains in the United States: identification of unusual strains. J. Clin. Microbiol. 38:2784-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshino, Y., and A. Z. Kapikian. 1996. Classification of rotavirus VP4 and VP7 serotypes. Arch. Virol. 12(Suppl.):99-111. [DOI] [PubMed] [Google Scholar]
- 26.Iturriza-Gomara, M., J. Green, D. W. G. Brown, M. Ramsay, U. Desselberger, and J. J. Gray. 2000. Molecular epidemiology of human group A rotavirus infections in the United Kingdom between 1995 and 1998. J. Clin. Microbiol. 38:4394-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iturriza-Gomara, B. Isherwood, U. Desselberger, and J. J. Gray. 2001. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J. Clin. Microbiol. 75:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jian, V., B. K. Das, M. K. Bhan, R. I. Glass, J. R. Gentsch, and The Indian Strain Surveillance Collaborating Laboratories. 2001. Great diversity of group A rotavirus strains and high prevalence of mixed rotavirus infections in India. J. Clin. Microbiol. 39:3524-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapikian, A. Z., and R. M. Chanock. 1996. Rotaviruses, p. 1657-1708. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 30.Kirkwood, C., P. J. Masendycz, and B. S. Coulson. 1993. Characteristics and location of cross-reactive and serotype-specific neutralization sites on VP7 of human G type 9 rotaviruses. Virology 196:79-88. [DOI] [PubMed] [Google Scholar]
- 31.Kirkwood, C. D., J. R. Gentsch, Y. Hoshino, H. F. Clark, and R. I. Glass. 1999. Genetic and antigenic characterization of a serotype P[6]G9 human rotavirus strain isolated in the U.S. Virology 256:45-53. [DOI] [PubMed] [Google Scholar]
- 32.Leite, J. P., A. A. Alfieri, P. Woods, R. I. Glass, and J. R. Gentsch. 1996. Rotavirus G and P types circulating in Brazil: characterization by RT-PCR, probe hybridization, and sequence analysis. Arch. Virol. 141:2365-2374. [DOI] [PubMed] [Google Scholar]
- 33.Masendycz, P., N. Bogdanovic-Sakran, C. Kirkwood, R. Bishop, and G. Barnes. 2000. Report of the Australian rotavirus surveillance program, 1999/2000. Commun Dis. Intell. 24:195-198. [PubMed] [Google Scholar]
- 34.Masendycz, P., N. Bogdanovic-Sakran, C. Kirkwood, R. Bishop, and G. Barnes. 2001. Report of the Australian rotavirus surveillance program, 2000/2001. Commun Dis. Intell. 25:144-146. [PubMed] [Google Scholar]
- 35.Nakata, S., Z. Gatheru, S. Ukae, N. Adachi, N. Kobayashi, S. Honma, J. Muli, P. Ogaja, j. Nyango, E. Kiplagat, P. M. Tukei, and S. Chiba. 1999. Epidemiological study of the G serotype distribution of group A rotaviruses in Kenya from 1991 to 1994. J. Med. Virol. 58:296-303. [DOI] [PubMed] [Google Scholar]
- 36.O'Halloran, F., M. Lynch, B. Cryan, H. O'Shea, and S. Fanning. 2000. Molecular characterization of rotavirus in Ireland: detection of novel strains circulating in the population. J. Clin. Microbiol. 38:3370-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oka, T., T. Nakagomi, and O. Nakagomi. 2000. Apparent re-emergence of serotype G9 in 1995 among rotaviruses recovered from Japanese children hospitalized with acute gastroenteritis. Microbiol. Immunol. 44:957-961. [DOI] [PubMed] [Google Scholar]
- 38.Palombo, E. A. 1999. Genetic and antigenic diversity of human rotaviruses: potential impact on the success of candidate vaccines. FEMS Lett. 181:1-8. [DOI] [PubMed] [Google Scholar]
- 39.Palombo, E. A., P. J. Masendycz, H. C. Bugg, N. Bogdanovic-Sakran, G. L. Barnes, and R. F. Bishop. 2000. Emergence of serotype G9 human rotaviruses in Australia. J. Clin. Microbiol. 38:1305-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parasher, U. D., J. S. Bresee, J. R. Gentsch, and R. I. Glass. 1998. Rotavirus. Emerg. Infect. Dis. 4:561-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramachandran, M., B. K. Das, A. Vij, R. Kumar, S. S. Bhambal, N. Kesari, H. Rawat, L. Bahl, S. Thakur, P. A. Woods, R. I. Glass, M. K. Bhan, and J. R. Gentsch. 1996. Unusual diversity of human rotavirus G and P genotypes in India. J. Clin. Microbiol. 34:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramachandran, M., J. R. Gentsch, U. D. Parashar, S. Jin, P. A. Woods, J. L. Holmes, C. D. Kirkwood, R. F. Bishop, H. B. Greenberg, S. Urasawa, G. Gerna, B. S. Coulson, K. Taniguchi, J. S. Bresee, R. I. Glass, and The National Rotavirus Strain Surveillance System Collaborating Laboratories. 1998. Detection and characterization of novel rotavirus strains in the United States. J. Clin. Microbiol. 36:3223-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramachandran, M., C. D. Kirkwood, L. Unicomb, N. A. Cunliffe, R. L. Ward, M. K. Bhan, H. F. Clark, R. L. Glass, and J. R. Gentsch. 2000. Molecular characterization of serotype G9 rotavirus strains from a global collection. Virology 278:436-444. [DOI] [PubMed] [Google Scholar]
- 44.Santos, N., R. C. C. Lima, C. F. A. Pereira, and V. Gouvea. 1998. Detection of rotavirus types G8 and G10 among Brazilian children with diarrhea. J. Clin. Microbiol. 36:2727-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santos, N., E. M. Volotao, C. C. Soares, M. C. Albuquerque, F. M. da Silva, V. Chizhikov, and Y. Hoshino. 2002. VP7 gene polymorphism of serotype G9 rotavirus strains and its impact on G genotype determination by PCR. Virus Res. 90:1-14. [DOI] [PubMed] [Google Scholar]
- 46.Sereno, M. M., and M. I. Gorziglia. 1994. The outer capsid protein VP4 of murine rotavirus strain Eb represents a tentative new P type. Virology 199:500-504. [DOI] [PubMed] [Google Scholar]
- 47.Steele, A. D., L. Nimzing, I. Peenze, M. C. de Beer, A. Geyer, I. Angyo, and N. E. Gomwalk. 2002. Circulation of the novel G9 and G8 rotavirus strains in Nigeria in 1998/1999. J. Med. Virol. 67:608-612. [DOI] [PubMed] [Google Scholar]
- 48.Timenetsky, M. D., V. Gouvea, N. Santos, R. C. Carmona, and Y. Hoshino. 1997. A novel human rotavirus serotype with dual G5-G11 specificity. J. Gen. Virol. 78(Pt. 6):1373-1378. [DOI] [PubMed] [Google Scholar]
- 49.Unicomb, L. E., G. Podder, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. G. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J. Clin. Microbiol. 37:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou, Y., J. Supawadee, C. Khamwan, S. Tonusin, S. Peerakome, B. Kim, K. Kaneshi, Y. Ueda, S. Nakaya, K. Akatani, N. Maneekarn, and H. Ushijima. 2001. Characterization of human rotavirus serotype G9 isolated in Japan and Thailand from 1995 to 1997. J. Med. Virol. 65:619-628. [PubMed] [Google Scholar]