Abstract
We have developed a sensitive in vitro assay for detecting disease-associated prion aggregates by combining an aggregation-specific enzyme-linked immunosorbent assay (AS-ELISA) with the fluorescent amplification catalyzed by T7 RNA polymerase technique (FACTT). The new assay, named aggregation-specific FACTT (AS-FACTT), is much more sensitive than AS-ELISA and could detect prion aggregates in the brain of mice as early as 7 days after an intraperitoneal inoculation of PrPSc. However, AS-FACTT was still unable to detect prion aggregates in blood of infected mice. To further improve the detection limit of AS-FACTT, we added an additional prion amplification step (Am) and developed a third-generation assay, termed Am-A-FACTT. Am-A-FACTT has 100% sensitivity and specificity in detecting disease-associated prion aggregates in blood of infected mice at late but still asymptomatic stages of disease. At a very early stage, Am-A-FACTT had a sensitivity of 50% and a specificity of 100%. Most importantly, Am-A-FACTT also detects prion aggregates in blood of mule deer infected with the agent causing a naturally occurring prion disease, chronic wasting disease. Application of this assay to cattle, sheep, and humans could safeguard food supplies and prevent human contagion.
Transmissible spongiform encephalopathy (TSE), or prion disease, is a group of fatal neurodegenerative diseases (15, 16). For decades, the only diagnostic test for TSEs was the demonstration of the presence of spongiform lesions in the brains of infected animals or humans by histopathology (15, 16). No reliable in vitro diagnostic test was available until the discovery that TSEs are caused by the conversion of a normal cellular prion protein, PrPC, into the pathogenic scrapie PrP isoform, PrPSc (4, 28). An important effect of this conformational change is that while the entire PrPC is protease sensitive, the C-terminal domain of PrPSc becomes relatively protease resistant (12, 25). Consequently, the presence of protease-resistant PrPSc in central nervous system (CNS) tissue has provided the basis for the in vitro diagnosis of all prion diseases (19, 39). However, available tests are mostly postmortem, invasive, not very sensitive, and nonquantitative. These deficiencies have also hampered our ability to follow the migration, replication, and accumulation of PrPSc in infected animals.
The exact chemical composition of an infectious PrPSc particle is not known. In vivo, PrPSc exists as aggregates referred to as scrapie amyloid fibrils (22, 29). PrPSc infectivity in hamster brain has a sedimentation coefficient of 40S (22). In another study, it was estimated that the smallest PrPSc has a molecular mass of about 600 kDa (44). However, ionizing radiation inactivation experiments found that the minimum size of a PrPSc molecule has a molecular mass of 50 kDa, which corresponds to a PrP dimer (3). A recent study found that the smallest infectious PrPSc particle has a molecular mass of between 300 and 600 kDa, which corresponds to between 14 and 28 PrPC molecules (38). The relationships between these smaller PrPSc aggregates and scrapie amyloid fibrils are not known. Whether PrPSc is present in the circulation of infected animals has also been controversial for decades (1, 5, 6, 9, 31, 45). This uncertainty is most likely due to the lack of a more sensitive assay that can detect PrPSc in the circulation. However, a recent study reported that two patients who had received blood from donors who had died from variant Creutzfeldt-Jakob disease (vCJD) had also developed vCJD (21, 27). Therefore, the presence of infectious prion in blood is no longer a theoretical possibility but rather is a tangible threat to public health. An in vitro test that can detect PrPSc in blood would greatly enhance our ability to monitor the occurrence and spread of prion diseases.
To increase the detection limit of currently available immunoblotting assays, Soto and colleagues developed a “protein misfolding cyclic amplification” (PMCA) technique. PMCA allows an undetectable amount of PrPSc to convert exogenously provided PrPC to PrPSc after multiple cycles of sonication (11, 33, 40). Another study suggested that simple incubation without sonication is sufficient (41). PMCA has been used to amplify and detect PrPSc in the blood of terminally sick hamsters with 89% sensitivity and terminally sick mice with 80% sensitivity and specificity (11, 32). Using “seeded polymerization” of PrP, a concept similar to PMCA, it was reported that PrP fibrils could be detected in the plasma of cattle naturally infected with bovine spongiform encephalopathy (43).
Recently, we developed a novel aggregation-specific (AS) enzyme-linked immunosorbent assay (ELISA), termed “AS-ELISA,” for PrPSc aggregates present in the brains of mice infected with the ME7, 139A, or 22L strain of PrPSc (26). The assay can detect disease-associated PrPSc aggregates in the brains of mice 70 days postinfection, a time when protease-resistant PrPSc is undetectable. The fluorescent amplification catalyzed by T7 RNA polymerase technique (FACTT), another newly developed amplification platform, can detect target proteins at femtomolar levels (51). In this paper, we describe the development of an ultrasensitive prion assay. The assay combines an in vitro amplification (Am) step similar to PMCA with AS-ELISA and FACTT. We named the assay Am-A-FACTT. We used Am-A-FACTT to determine whether PrPSc aggregates are present in the brains or blood of infected but asymptomatic mice as well as in deer and elk infected either naturally or experimentally with the agent causing chronic wasting disease (CWD).
MATERIALS AND METHODS
Recombinant PrP protein and monoclonal antibodies.
The generation of recombinant PrPC proteins and anti-PrP monoclonal antibodies (MAbs) was described in detail previously (20, 49, 50). All MAbs were affinity purified. MAbs were biotinylated with the EZ-Link Sulfo-NHS-Biotin kit (Pierce Endogen, Rockford, IL).
Mice.
Mouse-adapted scrapie strain ME7 was injected (0.1 ml of a 10% brain homogenate) intraperitoneally (i.p.) into 7-week-old CD-1 mice as described previously (37). The ME7 inoculant has a titer of about 108 50% infectious doses/ml (26, 37). Sham-infected, age- and sex-matched CD1 mice, normal CD1 mice, and PrPC−/− mice were used as controls. Blood was collected from individual mice by cardiac puncture with a 30-gauge needle. Plasma samples were collected after centrifugation and stored in aliquots at −80°C. All animal experiments were carried out according to institutional regulations and standards. All experiments have been repeated at least twice with comparable results.
Naturally infected mule deer and elk.
Blood (plasma) and brain tissue (5-mm slices of medulla oblongata cut immediately caudal to the obex) were collected from CWD agent-infected and presumably unexposed free-ranging mule deer (Odocoileus hemionus) as described previously (23, 46-48). Five paired sets of blood and brain tissue samples were collected at euthanasia from naturally infected, free-ranging deer previously diagnosed as being CWD agent infected by tonsil biopsy (47, 48). Blood from five unexposed live deer and brain tissue from five different hunter-killed deer were also collected. Three additional CWD brain samples (one mule deer and two elk [Cervus elaphus nelsoni]) and five non-CWD samples (two mule deer and three elk) were kindly provided by the late Elizabeth S. Williams of the Wyoming State Veterinary Laboratory. Plasma and brain tissue samples were stored at −80°C until assayed. All animal handling and sampling were carried out according to institutional policies and standards.
Experimentally infected mule deer.
As part of an ongoing study (M. W. Miller, L. L. Wolfe, and R. V. Lewis, unpublished results), in December 2004, mule deer fawns were inoculated with 1 g of homogenized deer brain tissue by using a small syringe inserted into the diastema of the oral cavity as described previously (36). This deer brain tissue homogenate was prepared from the brains of 26 captive mule deer naturally infected with the CWD agent and was previously demonstrated to contain the agent of infectious CWD (36). All fawns were shown to be negative for CWD infection by tonsil biopsy immunohistochemistry (47, 48) prior to inoculation.
Preparation of mouse brain homogenate.
To prepare 20% (wt/vol) total mouse brain homogenate, individual entire mouse brains were homogenized in ice-cold lysis buffer (phosphate-buffered saline [PBS] with 1% Nonidet P-40, 0.5% sodium deoxycholate, 5 mM EDTA, pH 8.0) in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma, MO). If the homogenate was to be treated with proteases, PMSF was omitted. After centrifugation at 1,000 × g for 10 min, the supernatant was stored in aliquots at −80°C.
AS-ELISA assay.
In a conventional sandwich or capture ELISA, two MAbs with distinct binding epitopes are required: one MAb is immobilized on a solid phase to capture the antigen, and a second MAb that reacts with a distinct epitope is then used to detect the bound antigen. We reasoned that when PrP proteins dimerize or further aggregate, some epitopes would be buried, while others would be present more than once. We further postulated that if the epitope is present more than once, we might be able to use the same MAb as the capture MAb as well as the detecting MAb. Therefore, in our aggregation-specific ELISA, the same MAb is used as the capture MAb and the detecting MAb.
Ninety-six-well plates (Costar, NY) were coated with affinity-purified capture MAbs (0.5 μg/well in 100 μl) at room temperature for 2 to 3 h. The coated plates were blocked with 3% bovine serum albumin (Sigma, MO) in PBS overnight at 4°C. Different amounts of recombinant PrP protein or 100 μl of diluted brain homogenates (containing 60 μg of total brain proteins from a 20% total brain homogenate) was added to the wells. Plates were incubated at room temperature for 2 h and then washed with PBS with 0.05% Tween 20 (PBST) three times before the addition of detecting biotinylated MAbs. After three additional washes with PBST, a horseradish peroxidase streptavidin conjugate (Chemicon, CA) was added to the plates and incubated for 1 h. The plates were washed three times with PBST, and 100 μl of ABTS [2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt] (Roche Diagnostic, IN) was dispensed into each well. After 20 min, the absorbance was read at 405 nm on a Kinetic Micro-Plate reader (Molecular Devices, CA). The results presented are the averages of the duplicates, and all experiments were carried out blind and repeated at least twice.
AS-FACTT assay.
For the AS-FACTT assay, a 384-well plate was coated with capture antibody in carbonate-bicarbonate buffer (pH 9.6) at 5 μg/ml and 20 μl/well overnight at 4°C. The plate was washed three times with PBST (0.1% Tween 20 in PBS) and blocked with 1× casein buffer (PBS-casein, 10× concentrated; BioFX Laboratory) for 1 h. After three washes with PBST, the tested samples (from a 20% total brain homogenate) were diluted in PBS and added to the coated plate, in the amount of 20 μl per well, for a 60-min incubation at room temperature. The plate was washed three times with PBST, and 20 μl of a diluted biotinylated detection antibody (1 μg/ml) was added to each well. Plates were incubated at room temperature for 60 min. Streptavidin (Chemicon) and the biotin-DNA template (the amplification module [AM]) (51) were added sequentially at 5 μg/ml and 250 ng/ml, respectively, with a 30-min incubation at room temperature for each step, followed by three washes with PBST between each incubation. After excess AM and proteins were removed by washing, 20 μl of a reaction mixture containing 60 units of T7 RNA Polymerase Plus (Ambion), 1.25 mM nucleoside triphosphate, and 1× T7 buffer (Ambion) was added to each well. RNA amplification was performed at 37°C for 3 h. The RNA-intercalating dye RiboGreen (Molecular Probes) was added to the reaction mixture (20 μl, diluted 1:200 in the Tris-EDTA buffer supplied by the manufacturer), and the plate was read at an excitation wavelength of 485 nm and an emission wavelength of 530 nm in a TECAN SpectraFluor reader.
In vitro amplification.
For amplification, 10 μl of plasma was mixed with 500 μl of PBS (pH.7.0) and 30 μl of a 20% (wt/vol) brain homogenate from normal mice or mule deer in PBS with 5 μl of a protease inhibitor cocktail, PMSF, in a 15-ml conical tube. Some plasma was also mixed with the same volume of 20% brain homogenate from PrPC−/− mice as controls. The presence of PrPC in the brain homogenate is required for amplification because we did not detect any signal in samples containing infected plasma and brain homogenate from PrPC−/− mice (results not shown). The mixture was incubated in a 37°C shaking (240 rpm) water bath for 1 h. Another 10 μl of a 20% normal brain homogenate with PMSF was then added, and the mixture was again incubated in a 37°C shaking water bath for 1 h. This procedure was repeated four more times. The sample was then placed in a 4°C rotating platform overnight. Tubes were centrifuged, and the supernatants were discarded. The pellet was then dissolved in 120 μl PBS, and 20 μl of the mixture was removed and used for AS-FACTT.
Enzymatic treatment of brain homogenates.
Each brain homogenate was treated with 50 μg/ml of proteinase K (Sigma, MO) at 37°C for 1 h as described previously (26). The protease was inactivated by the addition of PMSF to a final concentration of 3 μM.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting.
To detect PrP species in brain homogenates, brain samples containing 60 μg of total protein were dissolved in 2× sample buffer and heated at 95°C for 5 min before being separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described previously (26). The 12% polyacrylamide gel was then transferred onto a nitrocellulose membrane and probed by MAb 8H4. After incubation with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G Fc (Chemicon, CA), the transferred PrP species were visualized by a chemiluminescence blotting system (Roche Diagnostic, NJ).
Statistical analysis.
A paired Student t test was used to calculate the statistical difference between experimental and control values. Differences were considered statistically significant if the P value was <0.05.
RESULTS
AS-FACTT is more sensitive than AS-ELISA.
AS-ELISA can detect disease-associated PrPSc aggregates in the brains of mice at 70 days but not at 35 days postinfection (23). To improve the sensitivity of AS-ELISA, we combined AS-ELISA with FACTT and developed a new assay, AS-FACTT. Approximately 5% of recombinant PrP (rPrP) occurs as dimers (26). We first demonstrated that AS-FACTT is more sensitive than AS-ELISA for detecting rPrP dimers. With anti-PrP MAb 11G5, which was previously identified as being able to react with the rPrP dimer (26), the lowest detection limit of AS-ELISA is about 20 ng/ml or 2 ng/well of rPrP. On the other hand, AS-FACTT can detect 10 pg/ml or 0.2 pg/well of rPrP (results not shown). We estimate that while AS-ELISA has a detection limit of about 100 pg of rPrP dimers, AS-FACTT has a detection limit of about 10 fg. Thus, AS-FACTT is about 1,000-fold more sensitive than AS-ELISA, an estimation that is in good agreement with recent studies using FACTT (51).
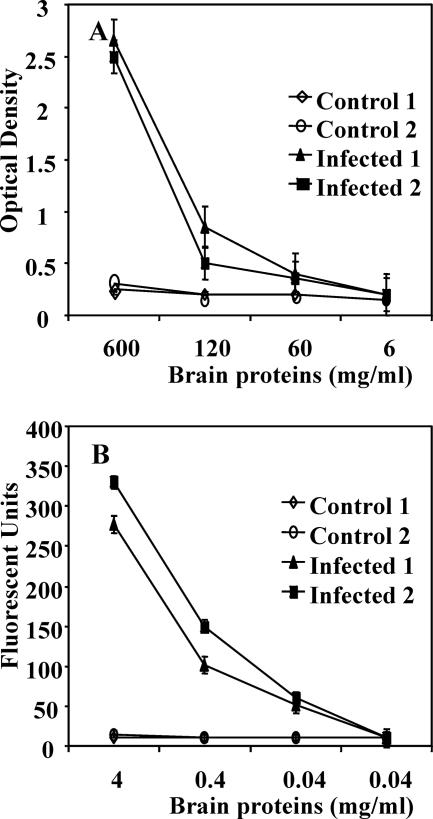
We then compared the sensitivities of AS-FACTT and AS-ELISA for the detection of PrPSc aggregates in the brains of PrPSc-infected mice. Brain homogenates from noninfected or from infected, terminally sick mice were serially diluted and assayed for the presence of PrPSc aggregates by AS-ELISA (Fig. 1A) or AS-FACTT (Fig. 1B). AS-FACTT was more sensitive than AS-ELISA for the detection of PrPSc aggregates. AS-FACTT detected significant reactivity in samples from infected brains, even when diluted to 0.04 μg/ml of total brain protein, which corresponds to ∼0.8 ng/well (P < 0.01). In hamster brains, it was estimated that there were between 100 and 1,000 50% lethal dose units of infectivity per ng of brain protein (13). We estimate that the titer of infectivity in hamster brain is about 20-fold higher than the titer in ME7-infected mouse brains. Hence, AS-FACTT potentially detected between 4 and 40 50% lethal dose units of infectivity.
FIG. 1.
AS-FACTT is more sensitive than AS-ELISA in detecting PrPSc aggregates. (A) Detection of PrPSc aggregates by AS-ELISA. Individual brain homogenates from noninfected, normal, or infected mice were serially diluted and assayed by AS-ELISA (26). AS-ELISA is able to detect signals at 120 μg/ml (P < 0.01) but not at 60 μg/ml of total brain proteins. Results presented are the averages of the duplicates ± standard errors for individual mice. (B) Detection of PrPSc aggregates by AS-FACTT. Individual brain homogenates were assayed by AS-FACTT. Brain homogenates from noninfected, PrPC−/− mice were used to establish backgrounds. Backgrounds were subtracted from all readings from controls as well as from infected samples. AS-FACTT was able to detect small but significant levels of immunoreactivity in samples containing 0.04 μg/ml of total brain protein (P < 0.01).
Detection of PrPSc aggregates during early stages of disease.
Previously, we found that AS-ELISA can first detect PrPSc aggregates in brain tissues at approximately 70 days postinfection (26). We therefore determined whether AS-FACTT could detect PrPSc aggregates earlier. We inoculated mice intraperitoneally rather than intracerebrally to avoid the possibility of detecting the injected PrPSc in the brain tissues. At 35 days after inoculation, brains from some mice individually were assayed for PrPSc aggregates by AS-ELISA and AS-FACTT. Brains from a group of mice at terminal stages of disease were used as positive controls. Both AS-ELISA and AS-FACTT detected aggregates in every animal at terminal stages (Table 1). However, in brains from animals inoculated 35 days earlier, only AS-FACTT was able to detect PrPSc aggregates (P < 0.0001) with 100% sensitivity and specificity.
TABLE 1.
Detection of PrPSc aggregates by AS-FACTT at 35 days after PrPSc inoculation
| Group and assay | No. of animals | Detection of PrpSc aggregatesa |
P valueb | ||
|---|---|---|---|---|---|
| Mean ± SD | Min | Max | |||
| Normal controlsc | |||||
| AS-ELISA | 13 | 0.33 ± 0.07 | 0.200 | 0.500 | |
| AS-FACTT | 17 | 295 ± 21.42 | 243 | 344 | |
| 35 days postinfectiond | |||||
| AS-ELISA | 10 | 0.356 ± 0.05 | 0.290 | 0.450 | 0.352* |
| AS-FACTT | 20 | 487 ± 41.44 | 429 | 585 | <0.0001* |
| Terminally ille | |||||
| AS-ELISA | 10 | 1.95 ± 0.55 | 1.34 | 2.70 | <0.0001* |
| AS-FACTT | 10 | 841 ± 205 | 630 | 1,200 | <0.0001* |
Values for AS-ELISA are ODs. Values for AS-FACTT are numbers of fluorescent units.
The P value was calculated by the paired Student t test.
Individual brain homogenates were assayed.
CD1 mice were inoculated (i.p.) with 0.1 ml of a 10% brain homogenate from terminally sick CD1 mice infected with ME7 PrPSc. At 35 days after inoculation, individual brains were removed, and homogenates were prepared and assayed.
Terminally ill CD-1 mice infected >185 days earlier with ME7 PrPSc and showing clinical signs of prion disease as described previously (37).
Temporal appearance of PrPSc in brain.
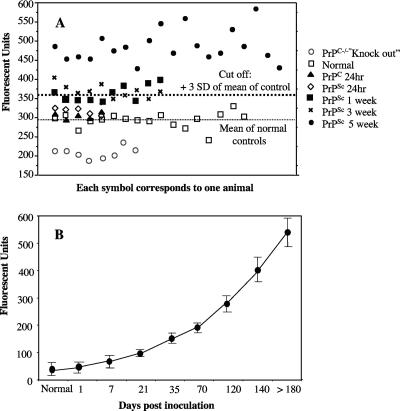
We next investigated the temporal appearance of PrPSc in the brains of mice inoculated intraperitoneally. At 1, 7, 21, or 35 days after inoculation, brain homogenates from individual mice were prepared. A control group of mice was inoculated with identical amounts of normal brain homogenate, and their brains were harvested 1 day later. Brain homogenates from PrPC−/− mice, which did not express PrPC, were also included as controls. Each brain homogenate was assayed for aggregates by AS-FACTT (Fig. 2A). The immunoreactivities detected in normal mice, mice injected with normal homogenates, or mice injected with PrPSc 1 day earlier were comparable. However, significant immunoreactivity was detected in 5 of the 10 mice injected 7 days earlier with PrPSc (a reading that was higher than the mean of controls by 3 standard deviations was considered positive) (Fig. 2A). The results of additional experiments with more time points are summarized in Fig. 2B. The levels of immunoreactivity increased slowly as infection progressed.
FIG. 2.
Temporal appearance of PrPSc in brains of infected mice. (A) PrPSc aggregates can be detected 7 days after injection. Individual brain homogenates from noninfected, normal control mice; mice injected i.p. with normal brain homogenates 24 h earlier; or PrPC−/− mice or mice injected with ME7 PrPSc at various times were assayed for PrPSc by AS-FACTT. Each symbol represents the average of duplicates from an individual mouse. The thick dashed line is the cutoff, which is 3 standard deviations (SD) above the mean of normal controls. The thin dashed line is the mean of the normal controls. There is a small difference between normal (□) and PrPC−/− (○) brain homogenates, suggesting that a small amount of PrP aggregates is present in normal brain. There is no difference among normal mice (□), mice injected with normal homogenates 24 h earlier (⋄), or mice injected with PrPSc 24 h earlier (▴). There are significant differences between controls and mice injected with PrPSc 7 (▪), 21 (×), or 35 (•) days earlier (P < 0.001 in all three groups). (B) Temporal appearance of PrPSc during disease progression. At different times after inoculation (i.p.), individual brain homogenates were assayed by AS-FACTT. The background from PrPC−/− brain was subtracted from each reading. The results presented are the means ± standard errors for each group of animals.
Detection of PrPSc aggregates in the circulation of infected mice.
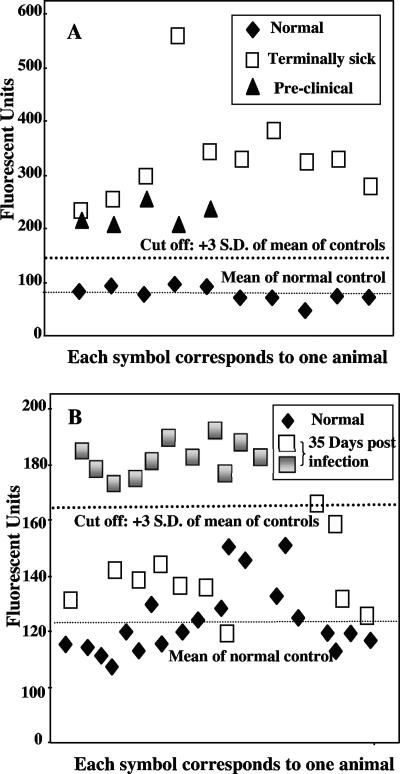
We were unable to detect aggregates in the plasma of infected mice using AS-FACTT. However, Am-A-FACTT detected PrPSc aggregates in the blood of 10 of 10 mice (Fig. 3A) at terminal stages of disease and in 5 of 5 mice (Fig 3A) at late but presymptomatic stages of disease. We failed to detect any protease-resistant PrPSc species in the amplified materials by immunoblotting (not shown). Therefore, the amount of amplified PrPSc aggregates must be below the detection limit of immunoblotting but is detectable by the more sensitive Am-A-FACTT. To validate the utility of Am-A-FACTT, we determined its intra-assay coefficient of variation (CV) and interassay CV. The assay is stable, and its discriminating power is robust. The intra-assay CV was less than 10% and the interassay CV was less than 14% in all groups (Table 2).
FIG. 3.
Detection of PrPSc aggregates in blood by Am-A-FACTT. (A) Detection of PrPSc aggregates in mice at terminal and presymptomatic stages of disease. Individual plasma samples from normal or infected mice were amplified with brain homogenate from either normal mice or PrPC−/− mice. The results presented are the averages of the duplicates from each mouse with the background subtracted. All samples from mice at terminal stages (□) or preclinical stages (▴) of disease have higher immunoreactivity than controls (⧫). (B) Detection of PrPSc aggregates in mice infected 35 days earlier. Experiments were carried out as described above. Am-A-FACTT detected higher levels of immunoreactivity in 11 (░⃞) of the 22 mice inoculated 35 days earlier (░⃞ + □). S.D., standard deviation.
TABLE 2.
Statistical analysis of Am-A-FACTT results
| Variation | Sample size (no. of animals) | Detection of PrPSc aggregates (fluorescent units) |
P valuec | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | SE | Variance | CV | Min | Max | Range | |||
| Interassaya | ||||||||||
| Control animals | 50 | 147.220 | 21.477 | 3.033 | 459.971 | 0.03 | 104 | 192 | 88 | |
| Infected animals | 50 | 579.380 | 33.257 | 4.703 | 1,105.996 | 0.057 | 509 | 641 | 132 | <0.0001 |
| Intra-assayb | ||||||||||
| Control animals | 50 | 145.320 | 18.984 | 2.685 | 360.385 | 0.131 | 113 | 189 | 76 | |
| Infected animals | 50 | 569.200 | 37.908 | 5.361 | 1,437.02 | 0.067 | 504 | 685 | 181 | >0.0001 |
For intra-assay variations, blood samples from five noninfected control mice and blood samples from five infected and terminally sick mice were assayed individually by Am-A-FACTT. Each sample was assayed in 10 tubes (5 × 10 = 50). The background, which was the average of five samples using PrPC−/− mouse brain homogenates as a substrate, was subtracted from the reading.
For interassay variations, 10 normal and 10 infected samples were assayed by Am-A-FACTT on five different days.
P values were calculsted by paired Student t test.
We next investigated if PrPSc aggregates were detectable in the plasma of mice at very early stage of disease, at 35 days postinoculation. Am-A-FACTT detected higher immunoreactivity (3 standard deviations above the mean of controls) in 11 of the 22 infected mice (Fig. 3B). Hence, even at this very early stage of disease, Am-A-FACTT has a sensitivity of 50% and a specificity of 100%.
Detection of PrPSc aggregates in deer and elk infected with the agent causing chronic wasting disease.
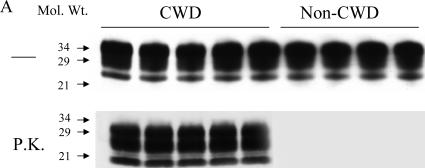
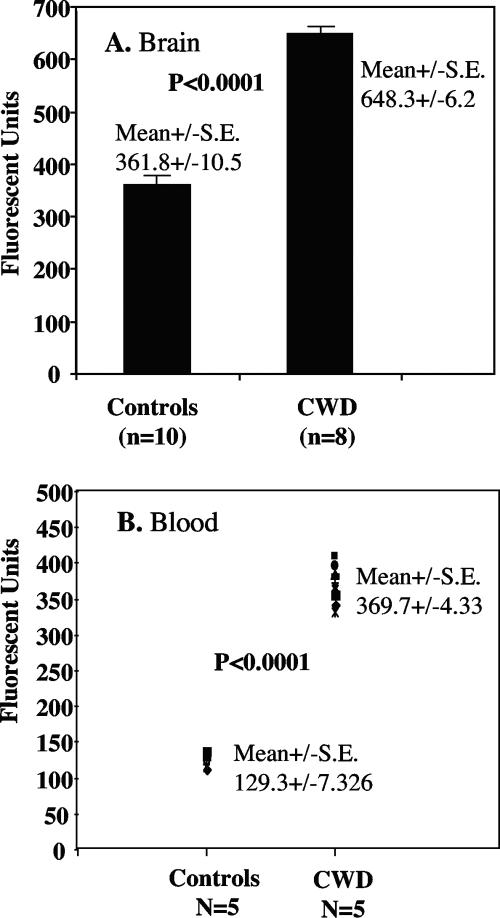
It was important to determine whether the assays can detect PrPSc aggregates in naturally infected animals, such as deer and elk infected with the CWD agent. We first demonstrated that the CWD agent-infected deer have proteinase K (PK)-resistant PrP species. Data from representative experiment using five infected deer and four noninfected, control deer are shown in Fig. 4. It is clear that all CWD agent-infected deer have PK-resistant PrP species. We then determined whether AS-FACTT could detect PrPSc aggregates in CWD agent-infected deer. We found that AS-FACTT detected PrPSc aggregates in the brains of all CWD agent-infected mule deer and elk (Fig. 5A). We next investigated whether Am-A-FACTT could detect PrPSc aggregates in blood. All blood samples from presymptomatic, infected mule deer were positive (Fig. 5B).
FIG. 4.
Detection of PK-resistant PrPSc species in CWD agent-infected deer. Brain homogenates from five CWD agent-infected deer and four noninfected control deer were prepared, treated with PK, and immunoblotted with MAb 8H4 as described in Materials and Methods. PK-resistant PrPSc species were detected in all infected deer but not in control deer. Mol. Wt., molecular weight (in thousands).
FIG. 5.
Detection of PrPSc aggregates in deer and elk with CWD. (A) Detection of PrPSc in brain by AS-FACTT. All CWD agent-infected deer and elk had higher immunoreactivities. Results presented are the means ± standard errors (S.E.) of the two groups. (B) Detection of PrPSc in blood by Am-A-FACTT. Plasma samples from five CWD agent-infected deer and five noninfected deer were assayed by Am-A-FACTT in duplicate. Readings from CWD blood samples amplified with brain homogenates from PrPC−/− mice were used as the background. All CWD deer samples (10 readings) had higher signals.
Detection of PrPSc aggregates in experimentally infected mule deer during disease progression.
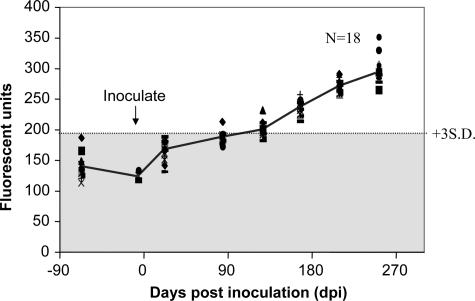
Plasma samples were collected from a group of deer (n = 18) in October or early December 2004, prior to inoculation with PrPSc orally on 21 December 2004. These plasma samples were used as controls. At 22, 84, 127, 167, 209, and 252 days after infection, plasma samples were again collected from individual deer and assayed for PrPSc aggregates by Am-A-FACTT. The results of such an experiment are shown in Fig. 6. Eighty-four days after inoculation, one of the infected deer had detectable PrPSc aggregates (3 standard deviations above the mean of normal controls); by 127 days, eight of the deer had significant PrPSc aggregates; and by 167 days, all inoculated deer had detectable circulating PrPSc aggregates. All inoculated deer had CWD as confirmed by tonsil biopsy immunohistochemistry at 252 days postinoculation. As of August 2006, more than 600 days after inoculation, most of these experimentally infected deer were still alive. Therefore, Am-A-FACTT was able to detect PrPSc aggregates in the plasma of mule deer at very early stages of infection.
FIG. 6.
Detection of PrPSc aggregates in orally inoculated deer. Plasma samples were collected from 18 deer on October or December 2004 prior to oral inoculation with 1 g of brain homogenates from CWD agent-infected deer on 21 December 2004. At various times after inoculation, plasma samples were again collected from individual deer and tested for the presence of PrPSc aggregates by Am-A-FACTT. Am-A-FACTT carried out with brain homogenates from PrPC knockout mice was used as the background and subtracted from each reading. The connecting line from each time point is the mean of results for the 18 deer at each time point. The cutoff line is 3 standard deviations (S.D.) above the mean of the controls. The experiment was carried out blind and was repeated twice. It is clear that by 167 days postinoculation, all deer had PrPSc aggregates in their circulation.
DISCUSSION
A test that can detect asymptomatic carriers infected with PrPSc is needed to prevent accidental transmission of prion diseases from animals to humans and from humans to humans; such a test would also be valuable as a tool in the surveillance and control of animal prion diseases. Preferably, the test should be minimally invasive and able to detect PrPSc in body fluids such as blood. We have taken the first step toward this goal. We improved the detection limits of our original AS-ELISA by incorporating a signal amplification step, FACTT. We then further improved the sensitivity of AS-FACTT by incorporating a PrPC-to-PrPSc amplification step and developed an ultrasensitive assay, termed Am-A-FACTT. Am-A-FACTT detects PrPSc aggregates in the blood of infected but asymptomatic mice with a high degree of sensitivity and specificity. Moreover, Am-A-FACTT was also able to detect disease-associated prion aggregates in mule deer naturally or experimentally infected with the CWD agent.
It is believed that in naturally occurring prion diseases, such as scrapie in sheep and goats, CWD in deer and elk, bovine spongiform encephalopathy in cattle, and vCJD in humans, the infectious agent enters the host via the oral route. However, the mechanisms by which PrPSc enters the CNS are not completely understood (2, 7, 24, 42). It is believed that PrPSc is imported into the CNS either by follicular dendritic cells (10, 18) or by neurons (17, 30). Irrespective of the pathway, disease-associated PrPSc aggregates are detectable in the brains of some mice at 1 week after an intraperitoneal inoculation by AS-FACTT. Therefore, some PrPSc aggregates are able to migrate from the periphery to the CNS within 1 week. It has been reported that peripherally injected hamster PrPSc can reach the CNS of hamsters within 10 days (14). PrPSc could be detected as early as 2 weeks after intracerebral inoculation by PMCA (40). We found that by 35 days after intraperitoneal inoculation, PrPSc aggregates were detected in the brains of all mice by AS-FACTT.
The AS-ELISA is specific for PrP aggregates composed of at least a dimeric complex, because it does not react with either PrP monomer or brain homogenates from PrPC “knockout” mice (26). AS-ELISA can also detect PrPSc aggregates in the brains of mice infected with any one of the three strains of PrPSc, ME7, 139A, or 22L (26). Previous studies revealed that the immunoreactivity detected by AS-ELISA included PK-resistant PrPSc that had a molecular mass of about 2,000 kDa (26). By ultracentrifugation in a sucrose gradient, most of the immunoreactivity detected by AS-ELISA is present in fractions 3, 4, and 5. These fractions are mostly devoid of PrPC, as it partitions into the top two fractions. On the other hand, AS-ELISA reacts poorly with the largest PrPSc aggregates, which tend to partition into the bottom fractions. Recent studies by flow field-flow fractionation found that the most infectious hamster PrPScs have a molecular mass of 300 to 600 kDa, which corresponds to 14 to 28 molecules of PrPC (38). Using flow field-flow fractionation, which is much more refined and precise, we found that the AS-ELISA preferentially reacts with hamster PrPSc aggregates with a molecular mass of between 200 and 420 kDa (B. Chang, J. R. Silveira, B. Caughey, and M.-S. Sy, unpublished results). Therefore, it is likely that the AS-ELISA detects infectious PrPSc.
The level of PrPSc in the blood is much lower than that in the CNS (1, 6, 8). Therefore, an additional in vitro amplification step similar to PMCA is required for the detection of PrPSc in blood. The new assay, Am-A-FACTT, was able to detect PrPSc aggregates in the blood of all mice at terminal stages of disease (total, n > 40), as well as mice at late but presymptomatic stages of disease (total, n > 20). Furthermore, Am-A-FACTT was able to detect higher immunoreactivities in 11 of the 22 mice inoculated 35 days earlier, at very early stages of disease. It should be noted that by 35 days postinoculation, PrPSc aggregates were detected in the brains of all infected animals. On the other hand, at this time, only 50% of mice had detectable PrPSc aggregates in their blood by Am-A-FACTT. This difference most likely reflects the lower levels of PrPSc in the circulation. By using immunoblotting, we did not detect any protease-resistant PrPSc species after the amplification procedures (results not shown). Therefore, the amount of amplified PrPSc aggregates must be below the detection limit of immunoblotting but is detectable by the more sensitive Am-A-FACTT. Am-A-FACTT did not detect any reactivity in blood from animals infected either 1 day or 7 days earlier (results not shown). Hence, the PrPSc aggregates that are detected are most likely host derived rather than the injected PrPSc, and it takes time for PrPSc to appear in the blood. Accordingly, we also failed to detect PrPSc in the blood of PrPC−/− mice inoculated with PrPSc (results not shown).
Perhaps more significantly, Am-A-FACTT was also able to detect disease-associated PrPSc aggregates in mule deer naturally infected with CWD with a high degree of specificity and sensitivity. This is important because experimentally infected animals tend to accumulate much higher levels of PrPSc. In 18 deer orally inoculated with a low dose of CWD PrPSc (1 gram of CWD agent-infected brain), one deer had disease-associated PrPSc aggregates at 84 days postinoculation. By 120 days, all inoculated deer had detectable PrPSc aggregates in their circulation. In a previous CWD pathogenesis study (36), PrPSc could be detected by immunohistochemical staining in lymph nodes at between 42 and 90 days postinoculation; however, these animals were inoculated with concentrations of PrPSc that were 10 times higher than concentrations used to inoculate the deer that we studied here. Therefore, it appears that Am-A-FACTT may be at least as sensitive as immunohistochemical staining in identifying preclinical infected deer.
Currently, all in vitro diagnostic tests for prion diseases require either the demonstration of protease-resistant PrPSc in brain or lymphatic tissue (19, 39) or the uncovering of hidden epitopes after denaturation (34, 35). More recently, PMCA has also been used to detect PrPSc in the blood of terminally sick mice and hamsters with 80 to 89% sensitivity, respectively (11, 32). However, this assay still requires treatment with protease, immunoblotting, and several days to perform. Am-A-FACTT provides distinct advantages: (i) Am-A-FACTT is more sensitive because of the use of a capture antibody and two amplification steps, namely, PrPC-to-PrPSc conversion amplification and signal amplification by FACTT; (ii) the assay detects smaller PrPSc aggregates, which are more likely to be present in the body fluids of infected animals; and (iii) the assay is more quantitative and less time-consuming. Most importantly, Am-A-FACTT detects PrPSc aggregates in the blood of naturally infected mule deer. It is likely that the same approach can be used to establish a blood test for prion diseases in humans and additional animal species, such as cattle, sheep, and elk. Finally, the principle of the aggregation-specific ELISA may be applicable to other diseases caused by abnormal protein aggregation, such as Alzheimer's disease or Parkinson's disease.
Acknowledgments
This work is supported in part by NIH grant NS-045981-02 and an award/contract from the U.S. Department of the Army, DAMD17-03-1-0286, to M.-S.S. and grants from the Abramson Family Cancer Research Institute and the NIH to M.I.G.; mule deer work in Colorado by M.W.M. and L.L.W. was supported by the Colorado Division of Wildlife and an award/contract from the U.S. Department of the Army, DAMD17-03-0542.
We also thank Dacai Liu and M. Lamm for reading of the manuscript and suggestions and T. Sirochman, M. Sirochman, and K. Griffin for animal and sampling assistance.
We declare no competing financial interests.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Aguzzi, A. 2001. Blood simple prion diagnostics. Nat. Med. 7:289-290. [DOI] [PubMed] [Google Scholar]
- 2.Bartz, J. C., C. Dejoia, T. Tucker, A. E. Kincaid, and R. A. Bessen. 2005. Extraneural prion neuroinvasion without lymphoreticular system infection. J. Virol. 79:11858-11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellinger-Kawahara, C. G., E. Kempner, D. Groth, R. Gabizon, and S. B. Prusiner. 1988. Scrapie prion liposomes and rods exhibit target sizes of 55,000 Da. Virology 164:537-541. [DOI] [PubMed] [Google Scholar]
- 4.Bolton, D. C., M. P. McKinley, and S. B. Prusiner. 1982. Identification of a protein that purifies with the scrapie prion. Science 218:1309-1311. [DOI] [PubMed] [Google Scholar]
- 5.Bradley, R. 1999. BSE transmission studies with particular reference to blood. Dev. Biol. Stand. 99:35-40. [PubMed] [Google Scholar]
- 6.Brown, P. 2001. Creutzfeldt-Jakob disease: blood infectivity and screening tests. Semin. Hematol. 38:2-6. [DOI] [PubMed] [Google Scholar]
- 7.Brown, P. 2001. The pathogenesis of transmissible spongiform encephalopathy: routes to the brain and the erection of therapeutic barricades. Cell. Mol. Life Sci. 58:259-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, P., L. Cervenakova, and H. Diringer. 2001. Blood infectivity and the prospects for a diagnostic screening test in Creutzfeldt-Jakob disease. J. Lab. Clin. Med. 137:5-13. [DOI] [PubMed] [Google Scholar]
- 9.Brown, P., C. J. Gibbs, Jr., P. Rodgers-Johnson, D. M. Asher, M. P. Sulima, A. Bacote, L. G. Goldfarb, and D. C. Gajdusek. 1994. Human spongiform encephalopathy: the National Institutes of Health series of 300 cases of experimentally transmitted disease. Ann. Neurol. 35:513-529. [DOI] [PubMed] [Google Scholar]
- 10.Bruce, M. E., K. L. Brown, N. A. Mabbott, C. F. Farquhar, and M. Jeffrey. 2000. Follicular dendritic cells in TSE pathogenesis. Immunol. Today 21:442-446. [DOI] [PubMed] [Google Scholar]
- 11.Castilla, J., P. Saa, and C. Soto. 2005. Detection of prions in blood. Nat. Med. 11:982-985. [DOI] [PubMed] [Google Scholar]
- 12.Cohen, F. E., and S. B. Prusiner. 1998. Pathologic conformations of prion proteins. Annu. Rev. Biochem. 67:793-819. [DOI] [PubMed] [Google Scholar]
- 13.Czub, M., H. R. Braig, and H. Diringer. 1986. Pathogenesis of scrapie: study of the temporal development of clinical symptoms, of infectivity titres and scrapie-associated fibrils in brains of hamsters infected intraperitoneally. J. Gen. Virol. 67:2005-2009. [DOI] [PubMed] [Google Scholar]
- 14.Diringer, H. 1984. Sustained viremia in experimental hamster scrapie. Brief report. Arch. Virol. 82:105-109. [DOI] [PubMed] [Google Scholar]
- 15.Gajdusek, D. C., and V. Zigas. 1957. Degenerative disease of the central nervous system in New Guinea; the endemic occurrence of kuru in the native population. N. Engl. J. Med. 257:974-978. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, R. T., and C. J. Gibbs, Jr. 1998. Creutzfeldt-Jakob disease and related transmissible spongiform encephalopathies. N. Engl. J. Med. 339:1994-2004. [DOI] [PubMed] [Google Scholar]
- 17.Kimberlin, R. H., and C. A. Walker. 1980. Pathogenesis of mouse scrapie: evidence for neural spread of infection to the CNS. J. Gen. Virol. 51:183-187. [DOI] [PubMed] [Google Scholar]
- 18.Kitamoto, T., T. Muramoto, S. Mohri, K. Doh-Ura, and J. Tateishi. 1991. Abnormal isoform of prion protein accumulates in follicular dendritic cells in mice with Creutzfeldt-Jakob disease. J. Virol. 65:6292-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kretzschmar, H. A. 2003. Diagnosis of prion diseases. Clin. Lab. Med. 23:109-128. [DOI] [PubMed] [Google Scholar]
- 20.Li, R., T. Liu, B. S. Wong, T. Pan, M. Morillas, W. Swietnicki, K. O'Rourke, P. Gambetti, W. K. Surewicz, and M. S. Sy. 2000. Identification of an epitope in the C terminus of normal prion protein whose expression is modulated by binding events in the N terminus. J. Mol. Biol. 301:567-573. [DOI] [PubMed] [Google Scholar]
- 21.Llewelyn, C. A., P. E. Hewitt, R. S. Knight, K. Amar, S. Cousens, J. Mackenzie, and R. G. Will. 2004. Possible transmission of variant Creutzfeldt-Jakob disease by blood transfusion. Lancet 363:417-421. [DOI] [PubMed] [Google Scholar]
- 22.McKinley, M. P., M. B. Braunfeld, C. G. Bellinger, and S. B. Prusiner. 1986. Molecular characteristics of prion rods purified from scrapie-infected hamster brains. J. Infect. Dis. 154:110-120. [DOI] [PubMed] [Google Scholar]
- 23.Miller, M. W., and E. S. Williams. 2003. Prion disease: horizontal prion transmission in mule deer. Nature 425:35-36. [DOI] [PubMed] [Google Scholar]
- 24.Nicotera, P. 2001. A route for prion neuroinvasion. Neuron 31:345-348. [DOI] [PubMed] [Google Scholar]
- 25.Pan, K. M., M. Baldwin, J. Nguyen, M. Gasset, A. Serban, D. Groth, I. Mehlhorn, Z. Huang, R. J. Fletterick, F. E. Cohen, et al. 1993. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA 90:10962-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan, T., B. Chang, P. Wong, C. Li, R. Li, S.-C. Kang, J. D. Robinson, A. R. Thompsett, P. Tein, S. Yin, G. Barnard, I. McConnell, D. R. Brown, T. Wisniewski, and M.-S. Sy. 2005. An aggregation-specific enzyme-linked immunosorbent assay: detection of conformational differences between recombinant PrP protein dimers and PrPSc aggregates. J. Virol. 79:12355-12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peden, A. H., M. W. Head, D. L. Ritchie, J. E. Bell, and J. W. Ironside. 2004. Preclinical vCJD after blood transfusion in a PRNP codon 129 heterozygous patient. Lancet 364:527-529. [DOI] [PubMed] [Google Scholar]
- 28.Prusiner, S. B. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216:136-144. [DOI] [PubMed] [Google Scholar]
- 29.Prusiner, S. B., M. P. McKinley, K. A. Bowman, D. C. Bolton, P. E. Bendheim, D. F. Groth, and G. G. Glenner. 1983. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 35:349-358. [DOI] [PubMed] [Google Scholar]
- 30.Race, R., M. Oldstone, and B. Chesebro. 2000. Entry versus blockade of brain infection following oral or intraperitoneal scrapie administration: role of prion protein expression in peripheral nerves and spleen. J. Virol. 74:828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ricketts, M. N., N. R. Cashman, E. E. Stratton, and S. ElSaadany. 1997. Is Creutzfeldt-Jakob disease transmitted in blood? Emerg. Infect. Dis. 3:155-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saa, P., J. Castilla, and C. Soto. 2006. Presymptomatic detection of prions in blood. Science 313:92-94. [DOI] [PubMed] [Google Scholar]
- 33.Saborio, G. P., B. Permanne, and C. Soto. 2001. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411:810-813. [DOI] [PubMed] [Google Scholar]
- 34.Safar, J., H. Wille, V. Itri, D. Groth, H. Serban, M. Torchia, F. E. Cohen, and S. B. Prusiner. 1998. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 4:1157-1165. [DOI] [PubMed] [Google Scholar]
- 35.Safar, J. G., M. D. Geschwind, C. Deering, S. Didorenko, M. Sattavat, H. Sanchez, A. Serban, M. Vey, H. Baron, K. Giles, B. L. Miller, S. J. Dearmond, and S. B. Prusiner. 2005. Diagnosis of human prion disease. Proc. Natl. Acad. Sci. USA 102:3501-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigurdson, C. J., E. S. Williams, M. W. Miller, T. R. Spraker, K. I. O'Rourke, and E. A. Hoover. 1999. Oral transmission and early lymphoid tropism of chronic wasting disease PrPres in mule deer fawns (Odocoileus hemionus). J. Gen. Virol. 80:2757-2764. [DOI] [PubMed] [Google Scholar]
- 37.Sigurdsson, E. M., M. S. Sy, R. Li, H. Scholtzova, R. J. Kascsak, R. Kascsak, R. Carp, H. C. Meeker, B. Frangione, and T. Wisniewski. 2003. Anti-prion antibodies for prophylaxis following prion exposure in mice. Neurosci. Lett. 336:185-187. [DOI] [PubMed] [Google Scholar]
- 38.Silveira, J. R., G. J. Raymond, A. G. Hughson, R. E. Race, V. L. Sim, S. F. Hayes, and B. Caughey. 2005. The most infectious prion protein particles. Nature 437:257-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soto, C. 2004. Diagnosing prion diseases: needs, challenges and hopes. Nat. Rev. Microbiol. 2:809-819. [DOI] [PubMed] [Google Scholar]
- 40.Soto, C., L. Anderes, S. Suardi, F. Cardone, J. Castilla, M. J. Frossard, S. Peano, P. Saa, L. Limido, M. Carbonatto, J. Ironside, J. M. Torres, M. Pocchiari, and F. Tagliavini. 2005. Pre-symptomatic detection of prions by cyclic amplification of protein misfolding. FEBS Lett. 579:638-642. [DOI] [PubMed] [Google Scholar]
- 41.Supattapone, S. 2004. Prion protein conversion in vitro. J. Mol. Med. 82:348-356. [DOI] [PubMed] [Google Scholar]
- 42.Sy, M. S., and P. Gambetti. 1999. Prion replication—once again blaming the dendritic cell. Nat. Med. 5:1235-1237. [DOI] [PubMed] [Google Scholar]
- 43.Trieschmann, L., A. Navarrete Santos, K. Kaschig, S. Torkler, E. Maas, H. Schatzl, and G. Bohm. 2005. Ultra-sensitive detection of prion protein fibrils by flow cytometry in blood from cattle affected with bovine spongiform encephalopathy. BMC Biotechnol. 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tzaban, S., G. Friedlander, O. Schonberger, L. Horonchik, Y. Yedidia, G. Shaked, R. Gabizon, and A. Taraboulos. 2002. Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry 41:12868-12875. [DOI] [PubMed] [Google Scholar]
- 45.Volkel, D., K. Zimmermann, I. Zerr, M. Bodemer, T. Lindner, P. L. Turecek, S. Poser, and H. P. Schwarz. 2001. Immunochemical determination of cellular prion protein in plasma from healthy subjects and patients with sporadic CJD or other neurologic diseases. Transfusion 41:441-448. [DOI] [PubMed] [Google Scholar]
- 46.Williams, E. S. 2003. Scrapie and chronic wasting disease. Clin. Lab. Med. 23:139-159. [DOI] [PubMed] [Google Scholar]
- 47.Wolfe, L. L., M. M. Conner, T. H. Baker, V. J. Dreitz, K. P. Burnham, E. S. Williams, N. T. Hobbs, and M. W. Miller. 2002. Evaluation of antemortem sampling to estimate chronic wasting disease prevalence in free-ranging mule deer. J. Wildlife Manag. 66:564-573. [Google Scholar]
- 48.Wolfe, L. L., M. W. Miller, and E. S. Williams. 2004. Feasibility of “test-and-cull” for managing chronic wasting disease in urban mule deer. Wildl. Soc. Bull. 32:500-505. [Google Scholar]
- 49.Yin, S. M., Y. Zheng, and P. Tien. 2003. On-column purification and refolding of recombinant bovine prion protein: using its octarepeat sequences as a natural affinity tag. Protein Expr. Purif. 32:104-109. [DOI] [PubMed] [Google Scholar]
- 50.Zanusso, G., D. Liu, S. Ferrari, I. Hegyi, X. Yin, A. Aguzzi, S. Hornemann, S. Liemann, R. Glockshuber, J. C. Manson, P. Brown, R. B. Petersen, P. Gambetti, and M. S. Sy. 1998. Prion protein expression in different species: analysis with a panel of new mAbs. Proc. Natl. Acad. Sci. USA 95:8812-8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, H., X. Cheng, M. Richter, and M. I. Greene. 2006. A sensitive and high-throughput assay to detect low-abundance proteins in serum. Nat. Med. 12:473-477. [DOI] [PubMed] [Google Scholar]