Abstract
The culture of Tropheryma whipplei, the bacterium responsible for Whipple's disease, has been established only recently. Our objective is to describe, based on our experience, the culture of T. whipplei in HEL cells detected by immunofluorescence staining. Over 3 years, we received 18 samples for T. whipplei culture from 15 patients with Whipple's disease. Ten duodenal biopsy specimens from 10 patients with digestive symptoms were available. Five cardiac valves and three blood samples from five patients with endocarditis were also available. We correlated the results of culture with the type of sample and the culture procedure. Seven isolates were obtained, and three were subsequently established for more than 4 passages. The mean delay for the primary detection was 30 days. The bacterium was isolated more frequently from sterile specimens (5 of 8) than from duodenal biopsy specimens (2 of 10), but the difference (P = 0.14) was not significant. Decontamination of digestive samples containing colistin, amphotericin B, and cephalotin or ciprofloxacin did not impair the isolation of T. whipplei. The use of vancomycin precludes the primary isolation (7 of 12 versus 0 of 6; P = 0.08) and the establishment of T. whipplei (3 of 12 versus 0 of 6; P = 0.5). Omitting samples cultured with vancomycin, the establishment of the strain was significantly higher when antibiotics were prescribed for no more than 7 days (3 of 4 versus 0 of 8; P = 0.03). Our results demonstrate that samples must be collected within 1 week of an antibiotic regimen's initiation for the successful establishment of the bacterium.
Whipple's disease is a systemic disease caused by a bacterium, Tropheryma whipplei (17). Described by Whipple in 1907, this disease is known mainly as a chronic pathology involving the intestine. Malabsorption, diarrhea, and weight loss eventually associated with adenopathies and polyarthritis correspond to the classical symptoms (5, 8, 24, 28). In the absence of specific antibiotic therapy, the disease is always fatal (8, 24, 28). The reference method for diagnosis of the disease is the histological examination of small-bowel biopsy specimens where periodic acid-Schiff (PAS)-positive, diastase-resistant organisms are observed in macrophages (5, 8, 24, 28). However, gastrointestinal symptoms are minimal or absent for approximately 15% of patients (5, 7, 8, 24, 28). Neurologic Whipple's disease without digestive symptoms has been reported (12, 23) as well as cardiac involvement (4, 7, 11, 13, 30). In 1991, Wilson et al. used broad-range primers to amplify bacterial 16S rDNA directly from infected tissue and then determined that this bacterium belongs to the high-G+C-content, gram-positive bacteria among the class Actinomycetes (36). To date, six genotypes (numbered 1 to 6) of T. whipplei have been described based on sequence variation within the 16 to 23S ribosomal DNA (rDNA) intergenic spacer, and two genotypes (named A and B) have been described based on the 23S rRNA gene (14-16, 25).
For a long time, the culture of the bacterium responsible for Whipple's disease has been an elusive goal. The first isolation of T. whipplei, using human macrophages inactivated with interleukin-4, was reported in 1997 (35). Unfortunately, this work has not been pursued or reproduced. For several years, our laboratory has specialized in the growth of fastidious microorganisms by using a centrifugation-shell vial technique called JNSP, for “je ne sais pas,” or “I do not know” (what I am growing) (9, 3, 10, 18-20, 27, 29). From 1999 to 2002, we attempted to achieve isolation of microorganisms by using this protocol on 5,690 samples (unpublished data). In 2000, Raoult et al. described the stable isolation of T. whipplei from a cardiac valve of a patient with Whipple's disease endocarditis by using an adapted JNSP protocol (31). This first strain has been distributed to 14 laboratories, and we have received follow-up from 9 laboratories. Among these, culture has been attempted in seven cases and was successful in five cases (unpublished data). In 2001, Raoult et al. reported the isolation of T. whipplei from a duodenal biopsy sample from a patient with typical relapsing Whipple's disease who had received cotrimoxazole treatment (32). The establishment of the first strain of the Whipple's disease bacterium in our laboratory has promised new perspectives on the diagnosis and treatment of Whipple's disease. In this work, we summarize our 3-year experience of attempted isolation of T. whipplei from patients with digestive Whipple's disease and from patients with Whipple's disease endocarditis.
MATERIALS AND METHODS
In our laboratory, we routinely used a JNSP protocol on various clinical samples, and more than 5,000 JNSP protocols have been performed to date. To detect the growth of bacteria, we used 16S rDNA amplification followed by sequencing, and we also stained the cells with either the Gimenez stain or an immunofluorescence stain using the patient serum diluted to 1/100 or specific antibodies. PAS staining is not useful, because false positives may be observed. We have adapted this JNSP protocol for the growth of T. whipplei. Since 1999, we have tried to culture the Whipple's disease bacterium from clinical samples. We have tested 18 samples from 15 patients with Whipple's disease diagnosed on the basis of clinical features, histological findings, and PCR studies for whom fresh samples were available for culture (Table 1). For each patient, a standardized questionnaire, including epidemiological and clinical features, was completed by the physician in charge. Ten fresh duodenal biopsy specimens, which were naturally contaminated by intestinal flora, were obtained from 10 patients with digestive Whipple's disease. Eight noncontaminated samples consisting of five cardiac valves and three blood samples were also obtained from five patients with Whipple's disease endocarditis.
TABLE 1.
Data for 18 inoculated samples from 15 patients with diagnosed Whipple's diseasea
| Patient | Sexa/age (yr) | Localization | Sample | PAS resultd | Immuno- histology resultd | PCR result/ type | Antibiotic therapy before biopsy/length | Medium with van- comycin | Culture/day of primary detection | Passage no./strain name |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F/33 | Digestive | Duodenal biopsy | + | + | +/1A | No | No | Yes/30 days | 3/Slow 1 |
| 2 | M/70 | Digestive | Duodenal biopsy | + | + | +/1A | No | Yes | No | |
| 3 | M/63 | Digestive | Duodenal biopsy | + | + | +/1A | Ofloxacin and cotrimoxazole/2 wk | No | No | |
| 4 | M/47 | Digestive | Duodenal biopsy | + | + | +/1A | No | Yes | No | |
| 5 | M/58 | Digestive | Duodenal biopsy | + | + | +/2A | No | Yes | No | |
| 6 | F/70 | Digestive, arthralgia | Duodenal biopsy | − | + | +/1A | No | Yes | No | |
| 7 | M/60 | Digestive | Duodenal biopsy | + | + | +/2A | Cotrimoxazole/3 wk | Yes | No | |
| 8 | F/74 | Digestive | Duodenal biopsy | + | NAb | +/1A | No | No | Yes/30 days | 10/Slow 2 |
| 9 | M/52 | Digestive | Duodenal biopsy | + | NA | +/1A | Cotrimoxazole/3 wk | No | No | |
| 10 | M/66 | Digestive, uveitis | Duodenal biopsy | + | + | +/1A | No | Yes | No | |
| 11 | M/42 | Endocarditis | Cardiac valve | + | + | +/2A | Penicillin G and gentamicin/1 wk | No | Yes/72 days | 36/Twist |
| 12 | M/63 | Endocarditis | Cardiac valve | + | + | +/2A | Amoxicillin and gentamicin/3 wk | No | Yes/15 days | 3/Endo 2 |
| Blood | NPc | − | − | Amoxicillin/7 wk | No | No | ||||
| 13 | M/61 | Endocarditis | Cardiac valve | + | + | +/2A | Ofloxacin and vibramycin/3 wk | No | Yes/15 days | 3/Endo 3 |
| Blood | NP | − | − | Ofloxacin and vibramycin/5 wk | No | |||||
| 14 | M/59 | Endocarditis | Cardiac valve | + | + | +/1A | Amoxicillin/6 wk | No | Yes/30 days | 2/Endo 4 |
| 15 | M/61 | Endocarditis | Cardiac valve | + | + | +/1A | Amoxicillin and gentamicin/5 wk | No | No | |
| Blood | NP | NP | − | Amoxicillin and gentamicin/1 wk | No | Yes | 6/Endo 5 |
M, male; F, female.
NA, not available.
NP, not performed.
+, positive; −, negative.
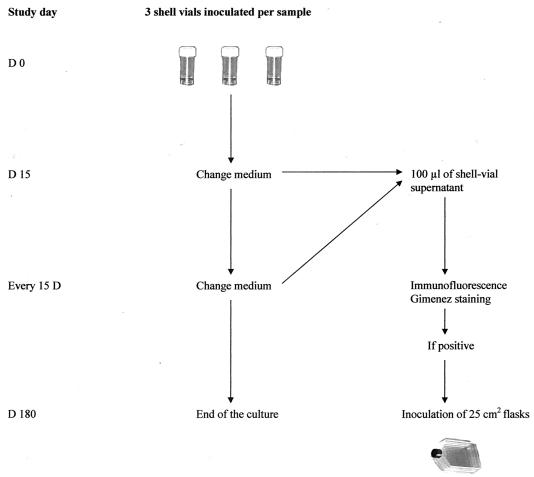
The several steps of T. whipplei cell culture are summarized in Fig. 1. Culture was performed by the centrifugation-shell vial technique with a human fibroblast cell line (HEL). These cells are routinely used, and more than 6,000 attempts to culture intracellular organisms have been performed in our laboratory using this cell line (unpublished data). For cardiac valves, frozen tissues were placed in minimal essential medium (MEM) and crushed. Duodenal biopsy specimens were incubated for 30 min in 2 ml of Rinaldini medium (6.8 g of NaCl, 0.4 g of KCl, 0.15 g of NaH2PO4, 1.0 g of glucose, 2.2 g of NaHCO3, and 0.002 g of phenol red in 1.0 liter of distilled water) containing a mix of antibiotics. This mix was composed of colistin sulfate (10 μg/ml), amphotericin B (1 μg/ml), and ciprofloxacin (1 μg/ml) or cephalotin (2 μg/ml). This protocol was established using our preliminary in vitro data. If the culture was contaminated with a Staphylococcus sp., vancomycin (2 μg/ml) was added. After incubation in the antibiotic solution, the specimen was rinsed by immersion for 5 min in 5 ml of MEM (Gibco, Gaithersburg, Md.). Rinsing was repeated twice, and the biopsy specimen was crushed in MEM. The suspension was used to inoculate three shell vials with a centrifugation step of 4,000 × g for 1 h in MEM. The cells were then incubated in MEM with 10% fetal calf serum and 2 mM glutamine at 37°C under a 5% CO2 atmosphere. All cell lines and culture reagents were checked weekly for bacterial contamination. For decontamination control, the biopsy specimen was inoculated onto Columbia sheep blood agar and Polyvitex chocolate agar plates (Biomérieux, Marcy L'Etoile, France) and was incubated at 37°C for 10 days under a 5% CO2 atmosphere.
FIG. 1.
Schematic representation of the procedure for isolation of T. whipplei.
Based on our previous experience, inoculated vials were processed as follows. Every 15 days for 180 days, the culture medium was changed. The medium was replaced on days 15 and 30 with a medium supplemented with the same mix of antibiotics used the first day for the duodenal biopsy specimens and with a medium free of antibiotics for all the other samples. On day 45, the culture medium was replaced with a medium free of antibiotics for all the specimens. Every 15 days, before the medium was replaced, 100 μl of the supernatant was obtained for centrifugation and stained with Gimenez and immunofluorescence stains. For immunofluorescence staining, after centrifugation and fixation with methanol, 100 μl of a homemade rabbit anti-T. whipplei antiserum diluted 1:200 in phosphate-buffered saline (PBS) with 3% nonfat dry milk was added, and the slides were incubated at 37°C for 30 min (1). The specificity of this antibody was established against 34 different bacterial strains (22). After three washes with PBS, 100 μl of fluorescein isothiocanyate-conjugated goat-anti rabbit immunoglobulin G (Jackson Immunoresearch Laboratories, West Grove, Pa.) diluted 1:200 in PBS with 3% nonfat dry milk was added, and the slides were incubated at 37°C for 30 min. After three washes with PBS, a coverslip was mounted in phosphate-buffered glycerol medium (pH 8.0) and examined at a magnification of ×400 with an epifluorescence microscope.
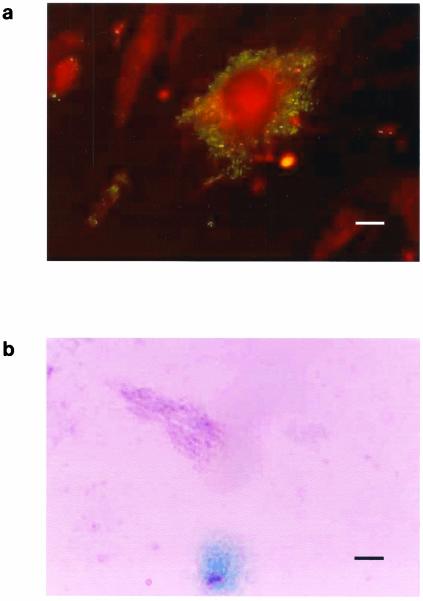
If the Gimenez staining and the immunofluorescence staining were positive (Fig. 2), the shell vial supernatant and inoculated cells were harvested, inoculated into a 25-cm2 (internal surface) cell culture flask with 5 ml of medium, and incubated at 37°C under a 5% CO2 atmosphere to establish the isolate. When cultures were positive, small, pale-pink, poorly staining bacilli were observed with Gimenez staining and small, brightly fluorescent bacilli were observed with immunofluorescent staining. A primary isolation of T. whipplei was defined as a positive specific immunofluorescence or Gimenez staining in the supernatant of the inoculated shell vials at the 1st passage. A strain of T. whipplei was considered to be established when at least four subcultures were obtained. Histological and immunochemistry analysis were performed on 5-μm-thick formalin-fixed and paraffin-embedded biopsy sections, as previously described (1, 21, 32, 33).
FIG. 2.
Whipple's disease bacterium. (a) Immunofluorescence staining of the bacteria in infected HEL cells. Bar, 6 μm. (b) Gimenez staining of the bacteria in infected HEL cells. Bar, 15 μm.
PCR and sequencing were performed on both the fresh biopsy specimen and the isolate. The biopsy sample was ground mechanically and resuspended in 500 μl of sterile deionized water in a sterile tube. Five hundred microliters of cell culture supernatant was centrifuged at 12,000 × g for 5 min. The resulting pellet was resuspended in 90 μl of Tris-EDTA buffer for PCR studies. The DNA was extracted using Qiagen (Hilden, Germany) columns (QIAamp tissue kit) as described by the manufacturer. To perform PCR, the 16 to 23S rDNA intergenic spacer region was amplified and sequenced using primers tw3f and tw4f, domain III of the 23S rRNA gene was amplified and sequenced using primers TW-23InsF and TW-InsR2, and the rpoB gene was amplified and sequenced using primers TWRPOB.F and TWRPOB.R, as previously described (6, 14, 16). PCR products were detected by 1% agarose gel electrophoresis analysis and ethidium bromide staining. Amplicons were purified using Qiagen columns (QIAquick Spin PCR purification kit).
For sequencing, a commercially available sequencing kit (dRhodamine Terminator Cycle sequencing kit; Perkin-Elmer Applied Biosystems, Warrington, England) was used according to the manufacturer's recommendations, as previously reported. All the sequences from the isolated strains were aligned with the sequences in the GenBank DNA sequence database with the BLAST program (version 2.0; National Center for Biotechnology Information).
We compared the number of strains isolated from the valves with the number of strains isolated from the duodenal biopsy specimens by using Fisher's exact test with Epi Info (version 6.04a; Centers for Disease Control and Prevention, Atlanta, Ga.). A P value of <0.05 was considered statistically significantly different.
RESULTS
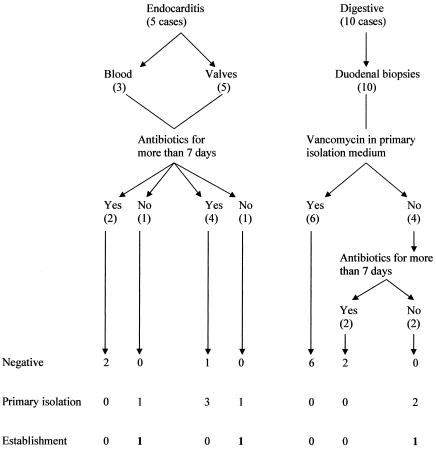
Since 1999, after the first successful isolation of T. whipplei in our laboratory, we have never observed the growth of this bacterium when we did not expect it. Each time a T. whipplei strain was isolated, a diagnosis of Whipple's disease had been established previously by histological or PCR analysis. All together, we obtained primary isolation of seven isolates and establishment of three strains (Table 1 and Fig. 3). The mean time for the primary detection was 30 days (standard deviation, ±20 days). T. whipplei was isolated more frequently from sterile specimens (5 of 8) than from contaminated samples (2 of 10), but this difference was not statistically significant (P = 0.14). However, the statistical results should be regarded with caution because the number of isolates is still small.
FIG. 3.
Schematic of culture of T. whipplei in cases of Whipple's disease. Established strains that were obtained from patients receiving either no antibiotics or short antibiotic regimens and with no vancomycin in the isolation medium are boldfaced.
Our first isolate of T. whipplei, the Twist-Marseille strain, was obtained from a cardiac valve (31). The strain is now in its 36th passage, and its genome has been entirely sequenced (GenBank accession number AE0116852). Three new strains (Endo 2, Endo 3, and Endo 4) were isolated from three of the four other infected cardiac valves from patients with negative-blood-culture endocarditis. Of these, two strains were subcultured three times and one was subcultured only once. All the valves presented positive PCR amplification and immunochemistry. Three isolates were identified as genotype 2A and two as genotype 1A.
Another endocarditis strain (Endo 5) was also isolated, from a blood culture obtained from a patient (patient 15) for whom conventional blood culture was negative. This blood culture was performed at the beginning of antibiotic therapy. T. whipplei growth was observed 15 days after inoculation. Cardiac valve removal was performed for this patient approximately 5 weeks after the beginning of antibiotics. No bacterial growth was observed for this cardiac valve, but PCR and immunochemistry yielded positive results. The two other blood samples cultured were taken 5 and 7 weeks after the beginning of antibiotic therapy and were both sterile. PCR was performed only for the two last blood samples, and both were negative.
Before the cardiac surgery or the blood sampling, all patients received antibiotic therapy. Previous antibiotic therapy did not prevent the isolation of T. whipplei from sterile specimens but may have prevented the establishment of strains. There was an apparent trend for isolates obtained from patients treated with antibiotics for more than 7 days not to become established in culture. Both strains obtained from patients treated for fewer than 7 days could be propagated beyond initial isolation, while none of the six strains obtained from patients treated longer (P = 0.03) could be continued in long-term culture. None of the patients with negative-blood-culture endocarditis had intestinal symptoms, and their duodenal biopsy specimens were negative by PAS staining, immunochemistry, and PCR.
The second strain of T. whipplei isolated in our laboratory was obtained from a duodenal biopsy specimen (Slow 1) of a patient with a relapsing digestive Whipple's disease (32). Because the duodenal biopsy specimens were naturally contaminated, we used colistin, cephalotin, and amphotericin B as antimicrobial agents to control contamination. Unfortunately, the strain could not be continued after 3 passages. Another strain (Slow 2) was isolated from a duodenal biopsy specimen. This isolate, for which colistin, amphotericin B, and ciprofloxacin were used as antimicrobial agents, is currently in its 10th passage. The antibiotic solution, when used without vancomycin, did not affect the culture, as two primary isolations and one strain establishment were obtained for two duodenal biopsy specimens, results comparable to those for sterile samples treated without antibiotics (2 of 8 versus 5 of 8, respectively [P = 1]). Addition of vancomycin to the antibiotic solution had a profound effect on the isolation of T. whipplei: none of the six samples exposed to vancomycin-containing solutions grew. In contrast, 7 of 12 samples in solutions without vancomycin grew (P = 0.03), and 3 of these became established (P = 0.05).
The prescription of antibiotics for more than 7 days before sampling (excluding samples cultured with vancomycin) prevented culture of T. whipplei from any samples. Four of four strains from patients without antibiotic therapy were primary isolates, and three of four became established, versus three of eight (P = 0.3) and none of eight (P = 0.07), respectively, for patients treated with antibiotics for 7 days. Four strains were isolated from the 10 genotype-1A samples, and 3 strains were isolated from the 5 genotype-2A samples. Two of the genotype-1A isolates and one genotype-2A isolate were successfully established.
DISCUSSION
The epidemiology and pathogenesis of Whipple's disease are still poorly understood. The isolation of infecting bacteria can serve, therefore, as a basis for the evaluation of much-needed improved diagnostic assays and as a way to enhance our understanding of the diversity and epidemiology of T. whipplei and the infections that it causes. In 1997, the first attempt at isolation of the bacterium by using intestinal biopsy samples from a patient with Whipple's disease failed because of the inevitable bacterial overgrowth (35). The isolation of T. whipplei from tissues of two heart valves, obtained from two patients with Whipple's disease endocarditis, was reported after inoculation in interleukin-4-deactivated macrophages (35). However, the two strains could not be established and propagated, and the work was not reproduced. Since 1999, two isolates of T. whipplei have been recovered after inoculation of HEL cells in shell vials by our team (31, 32). In vitro, growth has been observed not only in HEL or MRC5 cells (17) but also in HeLa cells in an acidic vacuole at pH 5 (35).
The centrifugation-shell vial system is a cell culture technique for the culture of viruses and facultatively or strictly intracellular bacteria. We have used this technique routinely in our laboratory for several years, and it has led to the recovery of fastidious bacteria including Rickettsia spp. (3, 19), Coxiella burnetii (29), Bartonella spp. (20), Francisella tularensis (9), Chlamydia trachomatis (27), and mycobacteria (10) from various clinical specimens. The data reported here represent the first collection of established T. whipplei isolates obtained from clinical specimens and the first isolation of the bacterium from blood. In addition, another strain of T. whipplei was isolated from cerebrospinal fluid by an American-English team using our protocol (2).
Culture of T. whipplei is currently achievable, but it is fastidious work. A difference exists between primary isolation and establishment of a strain. Successful isolation has not always led to propagation of the isolate. Only three of seven T. whipplei isolates were successfully established for more than 4 passages. This is in line with previous studies that have shown the potential of the shell vial technique for isolation of fastidious organisms but not necessarily for further propagation (19, 20). In these studies, we could establish only 15 of 34 (44%) Rickettsia strains and 65 of 81 (80%) Bartonella strains isolated by the shell vial method. The inability of these isolates to grow may be explained by specimens not being inoculated immediately after isolation, small numbers of organisms, and/or antibiotic therapy given prior to biopsy. Our experience with C. burnetii (29), Bartonella spp. (20), and Rickettsia conorii (19) demonstrated that samples must be collected prior to the initiation of an antibiotic regimen if the bacterium is to be successfully isolated. However, in our present study, most of our patients had antibiotic therapy before the sampling. Five of our seven isolates were from patients with previous antibiotic therapy. Our data show that the critical factor is the length of the antibiotic therapy before the sampling. Antibiotic therapy for more than 7 days before sampling significantly affects the establishment of the bacterium in culture, as evidenced by the fact that none of the bacteria isolated from patients on therapy for more than 7 days could be grown.
In addition, due to the presence of intestinal flora in duodenal biopsies, antibiotics must always be used in the culture medium to decontaminate the samples. It is therefore necessary to design specific antibiotic protocols. Our first antibiotic protocols were based on preliminary results derived from studying the in vitro antibiotic susceptibility of our first isolate (unpublished data). Ciprofloxacin, cephalotin, and colistin were used at concentrations which appeared not to inhibit the growth of T. whipplei. When these protocols were used, T. whipplei could be isolated and established. However, the principal limitation of these antibiotic mixes, directed against the intestinal flora, is that they could not always inhibit the growth of Staphylococcus. Problems with staphylococcal contamination led us to use vancomycin, with the result that cultures of duodenal biopsy specimens exposed to this drug were negative. This finding is in keeping with 16S rDNA sequence results that suggest that T. whipplei might be classified among gram-positive organisms (34), and it also suggests that vancomycin might be active in vivo against Whipple's disease. Since vancomycin must be avoided in the culture medium, we have no antibiotic regimen to propose to control staphylococcal overgrowth. Thus, more information about T. whipplei's antibiotic susceptibility profile is necessary to improve the culture of duodenal biopsy specimens.
In our study, T. whipplei strains of the two most common genotypes, 1A and 2A, were found and were successfully cultivated. In addition, there is no difference in primary isolation and establishment between these two different genotypes of T. whipplei.
Several factors have allowed the successful culture of T. whipplei. The first is patience, as the growth of T. whipplei is observed after an average of 30 days of incubation and sometimes more. The second factor is the treatment of patients, as our data clearly demonstrate that samples must be collected before the completion of 1 week of antibiotic treatment in order to isolate and establish the bacterium. Third, for contaminated samples, antimicrobial protocols consisting of colistin (10 μg/ml), amphotericin B (1 μg/ml), and ciprofloxacin (1 μg/ml) or cephalotin (2 μg/ml) can be used but vancomycin should absolutely be avoided. Currently, culture cannot be considered a practical tool for the diagnosis of Whipple's disease, but obtaining several more strains may be useful for better understanding of the pathology of Whipple's disease. The one great disadvantage of this method of culturing T. whipplei is that culture is currently performed only in specialized laboratories with the capacity for cell culture (26), making it necessary to improve the culturing technique in order to more efficiently isolate and propagate this organism. Finally, we conclude that the isolation and establishment of T. whipplei in culture was not an epiphenomenon and that we have defined a strategy for accomplishing this on a routine basis. Our success will no doubt improve as we gain more experience in isolating this very fastidious organism.
Acknowledgments
We are indebted to Kelly Johnston for reviewing the manuscript and to the following colleagues who provided biopsy and clinical data: A. Armengan (Barcelona, Spain), A. Bammert (Nantes, France), J. M. Devaster (Brussels, Belgium), N. Costedoat-Chalumeau (Paris, France), J. M. Favriel (Charleville-Mézières, France), P. Lecoq (Denain, France), H. Jeanmaire (Epinal, France) C. Poyart (Paris, France), and Y. Van Laethem (Brussels, Belgium).
REFERENCES
- 1.Baisden, B., H. Lepidi, D. Raoult, P. Argani, J. Yardley, and J. Dumler. 2002. Diagnosis of Whipple disease by immunohistochemical analysis. A sensitive and specific method for the detection of Tropheryma whipplei (the Whipple bacillus) in paraffin-embedded tissue. Am. J. Clin. Pathol. 118:742-748. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S., M. Maiwald, L. Murphy, M. Pallen, C. Yeats, L. Dover, H. T. Nobertczak, G. Besra, M. Quail, D. Harris, A. von Herbay, A. Goble, S. Rutter, R. Squares, S. Squares, B. Barrell, J. Parkhill, and D. Relman. 2003. Sequencing and analysis of the genome of the Whipple's disease bacterium Tropheryma whipplei. Lancet 361:637-644. [DOI] [PubMed] [Google Scholar]
- 3.Birg, M., B. La Scola, V. Roux, P. Brouqui, and D. Raoult. 1999. Isolation of Rickettsia prowazekii from blood by shell vial cell culture. J. Clin. Microbiol. 37:3722-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Célard, M., G. de Gevigney, S. Mosnier, P. Buttard, Y. Benito, J. Etienne, and F. Vandenesch. 1999. Polymerase chain reaction analysis for diagnosis of Tropheryma whippelii infective endocarditis in two patients with no previous evidence of Whipple's disease. Clin. Infect. Dis. 29:1348-1349. [DOI] [PubMed] [Google Scholar]
- 5.Dutly, F., and M. Altwegg. 2001. Whipple's disease and “Tropheryma whippelii.” Clin. Microbiol. Rev. 14:561-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenollar, F., P. Fournier, R. Gerolami, H. Lepidi, C. Poyart, and D. Raoult. 2002. Quantitative detection of Tropheryma whipplei DNA by real-time PCR. J. Clin. Microbiol. 40:1119-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenollar, F., H. Lepidi, and D. Raoult. 2001. Whipple's endocarditis: review of the literature and comparisons with Q fever, Bartonella infection, and blood culture-positive endocarditis. Clin. Infect. Dis. 33:1309-1316. [DOI] [PubMed] [Google Scholar]
- 8.Fenollar, F., and D. Raoult. 2001. Whipple's disease. Clin. Diagn. Lab. Immunol. 8:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fournier, P., L. Bernabeu, B. Schubert, M. Mutillod, V. Roux, and D. Raoult. 1998. Isolation of Francisella tularensis by centrifugation of shell vial cell culture from an inoculation eschar. J. Clin. Microbiol. 36:2782-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fournier, P., M. Drancourt, H. Lepidi, M. Gevaudan, and D. Raoult. 2000. Isolation of mycobacteria from clinical samples using the centrifugation-shell vial technique. Eur. J. Clin. Microbiol. Infect. Dis. 19:69-70. [DOI] [PubMed] [Google Scholar]
- 11.Geissdorfer, W., I. Wittmann, G. Seitz, R. Cesnjevar, M. Rollinghoff, C. Schoerner, and C. Bogdan. 2001. A case of aortic valve disease associated with Tropheryma whippelii infection in the absence of other signs of Whipple's disease. Infection 29:44-47. [DOI] [PubMed] [Google Scholar]
- 12.Gerard, A., F. Sarrot-Reynauld, E. Liozon, P. Cathebras, G. Besson, C. Robin, A. Vighetto, J. Mosnier, I. Durieu, D. Vital-Durand, and H. Rousset. 2002. Neurologic presentation of Whipple disease: report of 12 cases and review of the literature. Medicine 81:443-457. [DOI] [PubMed] [Google Scholar]
- 13.Gubler, J., M. Kuster, F. Dutly, F. Bannwart, M. Krause, H. Vögelin, G. Garzoli, and M. Altwegg. 1999. Whipple endocarditis without overt gastrointestinal disease: report of four cases. Ann. Intern. Med. 131:112-116. [DOI] [PubMed] [Google Scholar]
- 14.Hinrikson, H., F. Dutly, and M. Altwegg. 2000. Analysis of the actinobacterial insertion in domain III of the 23S rRNA gene of uncultured variants of the bacterium associated with Whipple's disease using broad-range and “Tropheryma whippelii”-specific PCR. Int. J. Syst. E vol. Microbiol. 50:1007-1011. [DOI] [PubMed] [Google Scholar]
- 15.Hinrikson, H., F. Dutly, and M. Altwegg. 2000. Evaluation of a specific nested PCR targeting domain III of the 23S rRNA gene of “Tropheryma whippelii” and proposal of a classification system for its molecular variants. J. Clin. Microbiol. 38:595-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinrikson, H., F. Dutly, S. Nair, and M. Altwegg. 1999. Detection of three different types of “Tropheryma whippelii” directly from clinical specimens by sequencing, single-strand conformation polymorphism (SSCP) analysis and type-specific type PCR of their 16S-23S ribosomal intergenic spacer region. Int. J. Syst. Bacteriol. 49:1701-1706. [DOI] [PubMed] [Google Scholar]
- 17.La Scola, B., F. Fenollar, P. Fournier, M. Altwegg, M. Mallet, and D. Raoult. 2001. Description of Tropheryma whipplei gen. nov., sp. nov., the Whipple's disease bacillus. Int. J. Syst. E vol. Microbiol. 51:1471-1479. [DOI] [PubMed] [Google Scholar]
- 18.La Scola, B., G. Michel, and D. Raoult. 1999. Isolation of Legionella pneumophila by centrifugation of shell vial cell cultures from multiple liver and lung abscesses. J. Clin. Microbiol. 37:785-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.La Scola, B., and D. Raoult. 1996. Diagnosis of Mediterranean spotted fever by cultivation of Rickettsia conorii from blood and skin samples using the centrifugation-shell vial technique and by detection of R. conorii in circulating endothelial cells: a 6-year follow-up. J. Clin. Microbiol. 34:2722-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.La Scola, B., and D. Raoult. 1999. Culture of Bartonella quintana and Bartonella henselae from human samples: a 5-year experience (1993 to 1998). J. Clin. Microbiol. 37:1899-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lepidi, H., N. Costedoat, J. Piette, J. Harlé, and D. Raoult. 2002. Immunohistological detection of Tropheryma whipplei (Whipple bacillus) in lymph nodes. Am. J. Med. 113:334-336. [DOI] [PubMed] [Google Scholar]
- 22.Lepidi, H., F. Fenollar, R. Gerolami, J. L. Mege, M. F. Bonzi, M. Chappuis, J. Sahel, and D. Raoult. Whipple's disease: immunospecific and quantitative immunohistochemical study of intestinal biopsies. Hum. Pathol., in press. [DOI] [PubMed]
- 23.Louis, E., T. Lynch, P. Kaufmann, S. Fahn, and J. Odel. 1996. Diagnostic guidelines in central nervous system Whipple's disease. Ann. Neurol. 40:561-568. [DOI] [PubMed] [Google Scholar]
- 24.Maiwald, M., and D. Relman. 2001. Whipple's disease and Tropheryma whippelii: secrets slowly revealed. Clin. Infect. Dis. 32:457-463. [DOI] [PubMed] [Google Scholar]
- 25.Maiwald, M., A. von Herbay, P. Lepp, and D. Relman. 2000. Organization, structure, and variability of the rRNA operon of the Whipple's disease's bacterium (Tropheryma whippelii). J. Bacteriol. 182:3292-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marth, T., and D. Raoult. 2003. Whipple's disease. Lancet 361:239-246. [DOI] [PubMed] [Google Scholar]
- 27.Maurin, M., and D. Raoult. 2000. Isolation of endothelial cell cultures of Chlamydia trachomatis LGV (serovar L2) from a lymph node of a patient with suspected cat scratch disease. J. Clin. Microbiol. 38:2062-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misbah, S., and N. Mapstone. 2000. Whipple's disease revisited. J. Clin. Pathol. 53:750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musso, D., and D. Raoult. 1995. Coxiella burnetii blood cultures from acute and chronic Q-fever patients. J. Clin. Microbiol. 33:3129-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naegeli, B., F. Bannwart, and O. Bertel. 2000. An uncommon cause of recurrent strokes: Tropheryma whippelii endocarditis. Stroke 31:2002-2003. [DOI] [PubMed] [Google Scholar]
- 31.Raoult, D., M. Birg, B. La Scola, P. Fournier, M. Enea, H. Lepidi, V. Roux, J. Piette, F. Vandenesch, D. Vital-Durand, and T. Marrie. 2000. Cultivation of the bacillus of Whipple's disease. N. Engl. J. Med. 342:620-625. [DOI] [PubMed] [Google Scholar]
- 32.Raoult, D., B. La Scola, P. Lecocq, H. Lepidi, and P. Fournier. 2001. Culture and immunological detection of Tropheryma whippelii from the duodenum of a patient with Whipple disease. JAMA 285:1039-1043. [DOI] [PubMed] [Google Scholar]
- 33.Raoult, D., H. Lepidi, and J. Harlé. 2001. Tropheryma whipplei circulating in blood monocytes. N. Engl. J. Med. 345:548. [DOI] [PubMed] [Google Scholar]
- 34.Relman, D., T. Schmidt, R. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 35.Schoedon, G., D. Goldenberger, R. Forrer, A. Gunz, F. Dutly, M. Höchli, M. Altwegg, and F. Schaeffer III. 1997. Deactivation of macrophages with interleukin-4 is the key to the isolation of Tropheryma whippelii. J. Infect. Dis. 176:672-677. [DOI] [PubMed] [Google Scholar]
- 36.Wilson, K., R. Blitchington, R. Frothingham, and J. Wilson. 1991. Phylogeny of the Whipple's-disease-associated bacterium. Lancet 338:474-475. [DOI] [PubMed] [Google Scholar]