Abstract
Most human immunodeficiency virus type 1 (HIV-1) strains require either the CXCR4 or CCR5 chemokine receptor to efficiently enter cells. Blocking viral binding to these coreceptors is an attractive therapeutic target. Currently, several coreceptor antagonists are being evaluated in clinical trials that require characterization of coreceptor tropism for enrollment. In this report, we describe the development of an automated and accurate procedure for determining HIV-1 coreceptor tropism (Trofile) and its validation for routine laboratory testing. HIV-1 pseudoviruses are generated using full-length env genes derived from patient virus populations. Coreceptor tropism is determined by measuring the abilities of these pseudovirus populations to efficiently infect CD4+/U87 cells expressing either the CXCR4 or CCR5 coreceptor. Viruses exclusively and efficiently infecting CXCR4+/CD4+/U87 cells are designated X4-tropic. Conversely, viruses exclusively and efficiently infecting CCR5+/CD4+/U87 cells are designated R5-tropic. Viruses capable of infecting both CXCR4+/CD4+/U87 and CCR5+/CD4+/U87 cells are designated dual/mixed-tropic. Assay accuracy and reproducibility were established by evaluating the tropisms of well-characterized viruses and the variability among replicate results from samples tested repeatedly. The viral subtype, hepatitis B virus or hepatitis C virus coinfection, and the plasma viral load did not affect assay performance. Minority subpopulations with alternate tropisms were reliably detected when present at 5 to 10%. The plasma viral load above which samples can be amplified efficiently in the Trofile assay is 1,000 copies per ml of plasma. Trofile has been automated for high-throughput use; it can be used to identify patients most likely to benefit from treatment regimens that include a coreceptor inhibitor and to monitor patients on treatment for the emergence of resistant virus populations that switch coreceptor tropism.
Infection of cells by human immunodeficiency virus type 1 (HIV-1) results from complex interactions between two viral and two cellular proteins. Both viral proteins necessary for HIV-1 entry, the surface protein (gp120) and the transmembrane protein (gp41), are encoded by the envelope gene (env), which is translated as a single polyprotein (gp160) and subsequently processed via proteolytic cleavage (2, 32). The gp41 and gp120 proteins associate with each other to form heterotrimeric structures that are integrated within the viral membrane upon virus assembly (15).
The cellular proteins necessary for HIV-1 entry include the CD4 receptor (10) and one of two chemokine receptors, commonly referred to as coreceptors (3). Of the repertoire of potential chemokine receptors, only CXCR4 (14) and CCR5 (33) are clinically and pharmacologically relevant. No other coreceptors are known to play roles in pathogenesis (24), and these two coreceptors account for almost 100% of the viral load in vivo (9).
HIV-1 entry begins with the attachment of gp120 to CD4 molecules on the surfaces of target cells. Attachment activates the viral protein by exposing its coreceptor binding site (36, 43) and initiating interaction of gp120 with the coreceptor (CXCR4 or CCR5). Coreceptor binding triggers the insertion of the gp41 N-terminal fusion peptide into the host membrane and the formation of a “coiled-coil” structure. Multiple coiled-coil structures permeating the cell membrane allow the release of the HIV-1 core into the cell.
Novel antiretroviral agents in clinical development target all aspects of this HIV-1 entry process and range from orally available small-molecule inhibitors to injectable monoclonal antibodies (5). There are three broad classes of entry inhibitors: attachment inhibitors, coreceptor antagonists (CRAs), and fusion inhibitors. Attachment inhibitors represent a diverse class of compounds that disrupt the initial binding of the HIV-1 gp120 surface protein to the CD4 cell surface receptor by one of several mechanisms (44). CRAs block HIV entry by preventing the virus from interacting with either of its two principal coreceptors, CCR5 and CXCR4. Fusion inhibitors inhibit the final conformational change of gp41 required for membrane fusion and virus entry.
The abilities of coreceptor antagonists to inhibit HIV infection have been demonstrated in cell culture systems and animal models (35), and several compounds are advancing in clinical trials. Lead candidates targeting the CCR5 coreceptor include the monoclonal antibody Pro140 (Progenics, Tarrytown, NY), which blocks HIV-1 infection by occupying the gp120 binding site on CCR5 (41). Two orally available small-molecule CCR5 antagonists are also currently in phase IIb/III clinical trials: vicriviroc (formerly SCH-D; Schering-Plough, Kenilworth, NJ) (38) and maraviroc (formerly UK427857; Pfizer, New York, NY) (11, 13). Those targeting the CXCR4 coreceptor include AMD11070 (AnorMED; Langley, Canada), which is in phase II trials (N. Stone, S. Dunaway, C. Flexner, G. Calandra, I. Wiggins, J. Conley, S. Synder, C. Tierney, C. Hendrix, et al., presented at the 15th International AIDS Conference, Bangkok, Thailand, 2004; D. Schols, S. Claes, S. Hatse, K. Princen, K. Vermeire, E. DeClercq, R. T. Skerlj, G. Bridger, and G. Clandra, presented at the 10th Conference on Retroviruses and Opportunistic Infections, Boston, MA, 2003).
Essentially, all HIV-1 strains use the CD4 receptor. However, coreceptor usage may vary among strains. HIV-1 strains using CXCR4 exclusively are classified as X4-tropic, those using CCR5 exclusively are classified as R5-tropic, and strains capable of using both receptors are deemed R5X4-tropic (or dual-tropic). The coreceptor specificity and binding are determined in part by sequences in both the coreceptor binding site and the third variable loop (V3) of gp120 (6, 7, 21, 30, 31, 37, 46). As HIV-1 evolves during the course of infection, coreceptor usage may also change. HIV-1 strains present in early infection preferentially use the CCR5 coreceptor, while CXCR4-using strains emerge in about 50% of patients over time and are associated with accelerated immunologic decline and progression to AIDS (16, 18, 19, 39, 40). It is unclear what factors suppress the emergence of CXCR4-using viruses during early-stage infection or contribute to their appearance in advanced disease. Moreover, it is unknown whether the emergence of CXCR4-using strains is a consequence or a cause of accelerated immunologic decline and progression to AIDS. Until the causes of coreceptor switching during infection are fully understood, it is prudent to consider the coreceptor tropism of the patient's virus when initiating treatment with entry inhibitors designed to interfere with coreceptor binding. Periodic monitoring for escape variants capable of using the alternate coreceptor should also be performed during treatment, particularly during failure of a CRA-containing treatment regimen (45).
To accelerate the development of entry inhibitors and to assist clinicians in the selection of appropriate treatment regimens, we developed and automated a novel recombinant-virus coreceptor tropism assay, Trofile, that can accurately assess the HIV-1 coreceptor tropisms of patient viruses from plasma samples. Trofile is capable of detecting both CXCR4 and CCR5 usage during a single round of virus replication.
The performance of the Trofile assay was formally validated according to regulations specified by the Clinical Laboratory Improvement Amendments and included characterizations of the assay's accuracy, reproducibility, specificity, and sensitivity. The formal validation was conducted in order to provide the assay to clinicians for therapy guidance during clinical trials and to provide widespread access once CRAs become part of standard antiretroviral therapy. The assay is currently used in clinical trials to select patients based on their coreceptor usage prior to treatment (i.e., at trial enrollment), as well as to detect coreceptor switching in the setting of viral rebound after initiation of therapy.
MATERIALS AND METHODS
Vectors. (i) pCXAS-PXMX, an env expression vector.
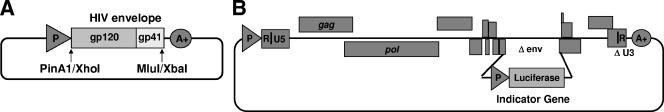
An expression vector (pCXAS-PXMX) containing a cytomegalovirus promoter/enhancer, a multiple cloning site polylinker (PinAI, XhoI, MluI, and XbaI), and a simian virus 40 polyadenylation signal sequence was constructed. The vector was designed to accept reverse transcription-PCR-amplified cDNA fragments derived from the full-length HIV-1 env genes of patient samples. The expression of patient env libraries in the resulting env expression test vectors (eETVs) was driven by the cytomegalovirus promoter (Fig. 1A).
FIG. 1.
Structures of vectors used in the Trofile assay. (A) Patient eETVs were constructed by cloning the amplified env genes from patient plasma samples into pCXAS-PXMX (see the text for details). The amplified fragment comprised the entire open reading frame of HIV-1 gp160. (B) A replication-defective genomic vector, RTV1.F-lucP.CNDOΔU3, was constructed with a luciferase cassette inserted into a deleted region of the env gene of the NL4-3 strain of HIV-1.
(ii) RTV1.F-lucP.CNDOΔU3, a HIV-1 genomic vector.
A retroviral vector (RTV1.F-lucP.CNDOΔU3) was modified from a previously designed vector (RTV1.F-lucP.CNDO) based on an infectious molecular clone of HIV-1 (NL4-3) (1, 27). The vector is replication defective and contains a luciferase expression cassette inserted within a deleted region of the env gene. To minimize the potential for generation of replication-competent virus upon cotransfection of target cells with eETV libraries, a self-inactivating deletion in the U3 region of the 3′ long terminal repeat (LTR) (ΔU3) was introduced that substantially reduces viral-gene transcription from the 5′ LTR of an integrated provirus in infected cells (23) (Fig. 1B).
Viruses. (i) Reference viruses.
Four viruses with well-documented coreceptor tropism were used as assay controls, or references, in all experiments. NL4-3 and HXB2 are laboratory-adapted X4-tropic strains of HIV-1, JRCSF is an R5-tropic primary isolate, and 92HT594 is a low-passage dual-tropic strain. All of the viruses can be obtained from the AIDS Research Reagent and Reference Program (ARRRP), NIH.
(ii) Validation samples.
Envelopes from 287 viruses were included in validation studies. Viruses included 38 well-defined, patient-derived primary isolates (ARRRP, NIH), 12 viruses isolated from HIV+ plasma samples purchased from a commercial source (Teragenix, Ft. Lauderdale, FL), 207 viruses isolated from plasma samples from the SCOPE (Study on the Consequences of the Protease Inhibitor Era) cohort at San Francisco General Hospital, and 40 viruses isolated from plasma aliquots archived from samples submitted to the Monogram Biosciences Clinical Reference Laboratory for routine HIV-1 resistance testing.
Methods. (i) Reverse transcription-PCR amplification of env genes from patient plasma samples.
HIV-1 virus was pelleted by centrifugation (20,400 × g; 60 min) from 1 ml of plasma. Virus particles were disrupted by resuspending pellets in 200 μl of lysis buffer (4 M guanidine thiocyanate, 0.1 M Tris HCl [pH 8.0], 0.5% sodium lauryl sarcosine, 1% dithiothreitol). RNA was isolated and purified from viral lysates using oligo(dT) linked to magnetic beads (Dynal, Oslo, Norway). Reverse transcription was performed with Thermoscript (Invitrogen, Gaithersburg, MD); env cDNA sequences were amplified with the Advantage 2 PCR kit (Clontech, Mountain View, CA), using a forward primer containing PinAI and XhoI sites and a reverse primer containing MluI and XbaI sites. The amplification products were libraries of env genes that represented the diversity of the viral env sequences present in the patient population. Each fragment was approximately 2.5 kb in length, spanning the entire open reading frame of the HIV-1 gp160 polyprotein.
(ii) eETV construction.
PCR amplification products were digested with PinAI and MluI restriction enzymes (Gibco/BRL), purified by agarose gel electrophoresis, and ligated into the PinAI- and MluI-digested expression vectors (pCXAS-PXMX). The resulting eETVs representing the amplified patient virus populations were propagated by transformation of Multishot TOP10 chemically competent Escherichia coli (Invitrogen, Carlsbad, CA). Libraries of eETV plasmids were isolated and purified from transformed cultures using silica column chromatography (QIAGEN, Valencia, CA).
(iii) Preparation of viral stocks from eETVs.
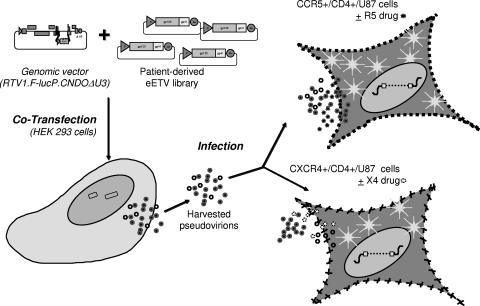
Replication-defective recombinant-virus stocks were produced by cotransfecting human embryonic kidney 293 (HEK293) cell cultures (ARRRP, NIH) with eETV libraries and RTV1.F-lucP.CNDOΔU3, resulting in virus particles pseudotyped by the patient-derived env proteins (Fig. 2). Viral stocks were harvested approximately 48 h after transfection.
FIG. 2.
Schematic diagram of the assay. Replication-defective virus particles were produced by cotransfecting HEK 293 cells with eETV and RTV1.F-lucP.CNDOΔU3. Following transfection, the virus particles were harvested and used to infect target cells (U87 cells expressing CD4 and either the CCR5 or CXCR4 coreceptor). The abilities of virus particles to complete a single round of replication in the presence or absence of CRAs was assessed by measuring the luciferase activities in target cells.
(iv) Determination of coreceptor tropism.
CD4+/U87 cells expressing the CXCR4 or the CCR5 coreceptor were inoculated with harvested pseudovirus in the presence and absence of increasing concentrations of either CXCR4- or CCR5-specific CRAs in a 96-well format. Successful infection led to the production of luciferase in target cells, and coreceptor tropism was assessed by evaluating the luciferase activity in lysed cells 72 h postinoculation. Coreceptor tropism (X4, R5, or dual/mixed [DM]) was determined by assessing the ability of the recombinant viruses to infect CXCR4+/CD4+/U87 or CCR5+/CD4+/U87 cells in the presence and absence of CXCR4 and CCR5 CRA, respectively (Fig. 2). The bicyclam AMD-3100 (AnorMED, Langley, Canada) was used to block CXCR4-mediated entry (12), and a member of the 4-(piperidin-1-yl) butane family of CCR5 antagonists (Merck, West Point, PA) was used to block CCR5-mediated entry (C. G. C. P. Caldwell, K. F. Donnelly, P. E. Finke, K. Shankaran, L. C. Meurer, B. Oates, C. G. Caldwell, M. MacCoss, S. G. Mills, L. Malkowitz, M. S. Springer, W. A. Schlief, A. Carella, G. Carver, K. Holmes, and E. A. Emini, presented at the American Chemical Society 220th National Meeting, Washington, DC, 2000). For inhibition studies, three concentrations for each compound were used in serial 1:4 dilutions, starting at 0.67 μg/ml for AMD3100 and 0.45 μg/ml for the CCR5 antagonist.
(v) Data analyses.
Luciferase activity was measured in lysed cells as relative light units (RLU). RLU were normalized across CXCR4+/CD4+/U87 or CCR5+/CD4+/U87 cell lines using the RLU results from inoculations of both cell types with pseudoviruses of a well-characterized dual-tropic reference strain (i.e., 92HT594). Specifically, RLUs from 92HT594-infected CXCR4+/CD4+/U87 or CCR5+/CD4+/U87 cells were adjusted to 1,000,000 counts to give two independent scaling factors (i.e., to normalize counts on CXCR4+ and CCR5+ cells, respectively). These scaling factors were applied to the RLU from CXCR4+/CD4+/U87 or CCR5+/CD4+/U87 cells infected with viral stocks generated from sample-derived eETV libraries, and the results were reported as “balanced” RLU (bRLU).
In order for a viral stock to be scored as “positive” for coreceptor (i.e., CXCR4 or CCR5) usage, two discreet logical tests were applied to the RLU data. (i) Was luciferase activity in the absence of drug above background? (Note: the run-to-run background typically ranged from 50 to 200 RLU). (ii) Was there clear evidence of inhibition of RLU by the specific coreceptor inhibitor? Inhibition was defined as a >50% decrease in RLU in the presence of drug relative to the RLU measured in the absence of drug. A positive score on both cell lines was designated dual-mixed (DM), as we could not distinguish whether the population was a heterogeneous mixture of exclusively X4- and R5-tropic viruses or a homogeneous population of viruses that were dual-tropic (i.e., capable of using either receptor).
RESULTS
Reference strains. (i) CXCR4 coreceptor tropism.
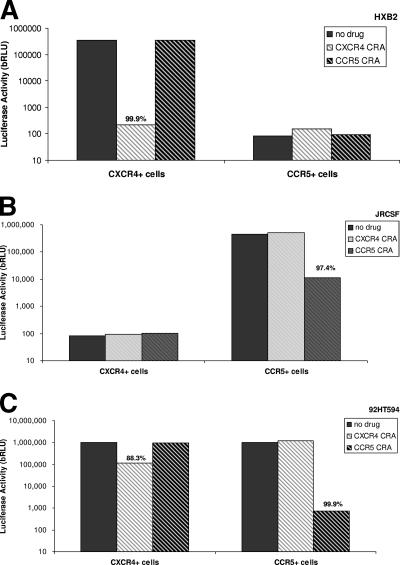
Replication-defective recombinant-virus stocks derived from the X4-tropic strain HXB2 were used to inoculate both CXCR4+/CD4+/U87 and CCR5+/CD4+/U87 cells. In the absence of CRAs, high luciferase activity was observed only in CXCR4+ cells; luciferase activity in CCR5+ cells was below background levels (Fig. 3A). The addition of AMD3100, a CXCR4-specific CRA, effectively abolished luciferase production in CXCR4+ cells, while addition of a CCR5-specific CRA [a member of the 4-(piperidin-1-yl) butane family] had no effect (Fig. 3A). As HXB2 was able to infect CXCR4+/CD4+/U87 cells, but not CCR5+/CD4+/U87 cells, and as infection was blocked by the CXCR4-specific CRA (AMD3100), we confirmed that HXB2 is an exclusively X4-tropic virus.
FIG. 3.
Trofile coreceptor tropism determinations of well-characterized reference strains. CXCR4+ and CCR5+ cells were infected with (A) the X4-tropic strain HXB2, (B) the R5-tropic strain JRCSF, and (C) the dual-tropic strain 92TH594. Infectivity was analyzed in the presence and absence of either CXCR4 or CCR5 antagonists. The percent inhibition by these compounds is given above the appropriate columns. (A) bRLU from inoculation of CXCR4+ and CCR5+ cell lines with strain HXB2. (B) bRLU from inoculation of CXCR4+ and CCR5+ cell lines with strain JRCSF. (C) bRLU from inoculation of CXCR4+ and CCR5+ cell lines with strain 92TH594.
(ii) CCR5 coreceptor tropism.
Replication-defective recombinant-virus stocks derived from the R5-tropic strain JRCSF were used to inoculate both CXCR4+/CD4+/U87 and CCR5+/CD4+/U87 cells. In the absence of CRAs, high luciferase activity was observed only in CCR5+ cells. Luciferase activity in CXCR4+ cells was below background levels (Fig. 3B). Furthermore, the addition of a CCR5-specific CRA dramatically reduced luciferase production in CCR5+ cells, while the addition of AMD3100 had no inhibitory effect. As JRCSF was able to infect CCR5+/CD4+/U87 cells, but not CXCR4+/CD4+/U87 cells, and as infection was blocked by the CCR5-specific CRA, we confirmed that JRCSF is an R5-tropic virus.
(i) DM coreceptor tropism.
Replication-defective recombinant-virus stocks derived from the R5X4-tropic strain 92HT594 were used to inoculate both CXCR4+/CD4+/U87 and CCR5+/CD4+/U87 cells. In the absence of CRAs, high luciferase activity was observed in CXCR4+ and CCR5+ cells (Fig. 3C). In CXCR4+ cells, luciferase activity was inhibited in the presence of AMD3100. However, the addition of an R5-specific CRA had no inhibitory effect. Conversely, in CCR5 cells, the addition of the CCR5-specific CRA strongly inhibited luciferase activity, while the addition of AMD3100 had no inhibitory effect (Fig. 3C). As 92HT594 is able to infect both CXCR4+/CD4+/U87 and CCR5+/CD4+/U87 cells, and as infection was inhibited by specific CRAs in the appropriate cell types, we confirmed that 92HT594 is DM tropic.
Assay performance. (i) Accuracy.
The ability of the assay to accurately assess HIV-1 coreceptor tropism was demonstrated by testing 38 HIV-1 isolates representing six different HIV-1 Env subtypes (i.e., subtypes A, B, C, D, E, and G). All 38 isolates had been previously characterized and documented as R5, X4, or dual tropic by various independent laboratories using conventional assays. In this assessment, 100% of the test results matched the expected tropism assessments. The tropism determinations and bRLUs are listed in Table 1.
TABLE 1.
Trofile results from 38 viral isolates with well-documented coreceptor tropisms consistent with published data
| Virus identifiera | env subtypeb | Tropismc | Median bRLU
|
|
|---|---|---|---|---|
| CCR5 cells | CXCR4 cells | |||
| 92RW016 | A | R5 | 566,433 | 97 |
| 92RW023 | A | R5 | 198,966 | 88 |
| 92RW025 | A | R5 | 1,548,099 | 115 |
| 92TH001 | E | R5 | 188,447 | 68 |
| 92TH003 | E | R5 | 95,968 | 55 |
| 92TH005 | E | R5 | 20,318 | 67 |
| 92TH006 | EA | R5 | 385,363 | 91 |
| 92TH007 | E | R5 | 23,192 | 62 |
| 92US660 | B | R5 | 650,755 | 87 |
| 92US712 | B | R5 | 295,317 | 77 |
| 92US727 | B | R5 | 717,949 | 98 |
| 93IN905 | C | R5 | 221,839 | 71 |
| 93MW965 | C | R5 | 65,191 | 56 |
| 93RW002 | A | R5 | 265,663 | 79 |
| 93RW005 | A | R5 | 433,907 | 75 |
| 93RW018 | A | R5 | 449,778 | 70 |
| 93RW022 | A | R5 | 36,018 | 55 |
| 93UG082 | D | R5 | 257,959 | 89 |
| 94KE105 | C | R5 | 633,645 | 81 |
| 94UG108 | A | R5 | 913,568 | 92 |
| 98CN006 | A | R5 | 2,223,534 | 82 |
| 98TZ013 | A | R5 | 664,761 | 87 |
| ASM 3 | B | R5 | 666,022 | 85 |
| ASM 80 | B | R5 | 208,830 | 73 |
| ASM 93765 | B | R5 | 646,804 | 91 |
| G3 | G | R5 | 96,450 | 54 |
| 92UG038 | D | X4 | 121 | 2,353,159 |
| 92UG046 | D | X4 | 56 | 47,063 |
| 93UG053 | D | X4 | 95 | 2,978,928 |
| 93UG065 | D | X4 | 73 | 881,081 |
| 93UG070 | D | X4 | 57 | 221,358 |
| CMU02 | EA | X4 | 93 | 204,312 |
| NL43 | B | X4 | 62 | 463,980 |
| 92HT593 | B | DM | 87,086 | 671,438 |
| 92HT596 | E | DM | 416,785 | 1,108,280 |
| 93RW024 | A | DM | 607,101 | 1,434,093 |
| 93TH051 | E | DM | 20,772 | 494,291 |
| ASM 54 | B | DM | 37,408 | 2,168 |
All viruses except NL4-3 were obtained from ARRRP, NIH; NL4-3 was obtained from David Ho, Aaron Diamond AIDS Research Center, NY.
env subtype determined by in-house sequencing; concordant with published (ARRRP) results.
By Trofile assay.
(ii) Reproducibility.
In one set of experiments, three well-characterized virus stocks (NL4-3, 92US727, and 92HT596) were repeatedly tested at two different inputs to mimic different viral loads (Table 2). Ten replicates at each dilution (20 replicates per sample) were tested by multiple operators using different lots of critical reagents over a 4-week period. Reproducibility was evaluated using the standard practice of pairwise comparison among multiple replicates. The total number of comparisons for a given sample on each cell line (i.e., either CXCR4 or CCR5 expressing) was 190. Therefore, 380 comparisons were made for each sample on both cell lines, with a total of 1,140 possible pairwise comparisons for all three samples. In this analysis, 100% of the 1,140 possible pairwise coreceptor tropism determinations were concordant.
TABLE 2.
Characteristics of three virus samples used for reproducibility studies
| Virus identifier | Tropism | Virus input (c/ml)a
|
|
|---|---|---|---|
| Neat | Diluted | ||
| NL4-3 | X4 | 3,800 | 1,617 |
| 92US727 | R5 | ∼75,000 | 3,100 |
| 92HT596 | Dual | ∼75,000 | 3,340 |
Multiple replicates were prepared at two concentrations (“neat” and “diluted). See the text for details.
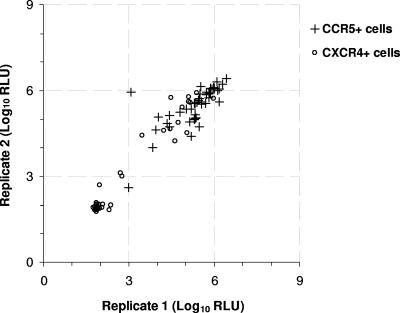
In a second set of experiments, separate aliquots of 46 plasma samples were tested in two assay runs on both CCR5+/CD4+/U87 and CXCR4+/CD4+/U87 cells. The testing of each aliquot involved multiple operators and instrumentations and different lots of critical reagents and was conducted over at least a 14-day period. All 46 samples were able to infect CCR5+/CD4+/U87 cells, and 18 were also able to infect CXCR4+/CD4+/U87 cells and were designated DM tropic. The designations were confirmed by CRA-specific inhibition of infection; 100% of the 46 paired tropism determinations made on each cell line were concordant. Additionally, RLU measurements were concordant for all 46 pairs on both cell lines (Fig. 4).
FIG. 4.
Concordance of duplicate tropism determinations. Two replicates of 46 plasma samples were evaluated in the Trofile assay. The infectivities (luciferase activities) of paired replicates on CCR5+/CD4+/U87 cells (crosses) and CXCR4+/CD4+/U87 cells (circles) were graphed pairwise to demonstrate the concordance between measurements.
(iii) Sensitivity of amplification.
A detailed study of amplification sensitivity was conducted using 207 patient plasma samples with well-defined viral loads from the SCOPE cohort. SCOPE is a prospective cohort study designed to characterize the natural history of both antiretroviral-treated and untreated HIV disease, with a focus on the pathogenesis of drug-resistant viremia. We considered SCOPE samples to be representative of clinical samples likely to be submitted to Monogram Biosciences for coreceptor tropism testing. Viral loads were determined by the branched-DNA assay (Quantiplex Assay, version 3.0; Bayer Nucleic Acid Diagnostics, Emeryville, CA). Seven viral-load bins were established to distribute the sample set, with viral loads ranging from undetectable (<50) to 5,000 copies/ml (c/ml) (Table 3). Successfully amplified samples were coded as 1, while unsuccessful amplifications were coded as 0. Statistical analyses were carried out with the SAS/STAT package (SAS Institute Inc., Cary, NC). The relationship between the log10-transformed viral load and the probability of successful amplification was fitted to a logistic function (PROC LOGISTIC). Asymptotic 95% calibration intervals corresponding to 0.70 to 0.95 (0.05 increments) probabilities were calculated by inverting the asymptotic 95% confidence intervals for these predicted probabilities. The Hosmer-Lemeshow goodness-of-fit statistic was used to assess the appropriateness of the linear model (22). The overall model was significant by chi-square statistics. An amplification success rate of 90% was obtained at an average viral load of 680 c/ml, with a 95% confidence interval of 400 to 1,320 c/ml. Ninety-five percent positive amplification was obtained at an average viral load of 1,430 c/ml, with a 95% confidence interval of 790 to 3,630 c/ml. Among all samples with viral loads of >1,000 c/ml, 95% (104/109) were successfully amplified. Based on these results, it is recommended that 1,000 c/ml be specified as the viral-load threshold at which samples can be amplified efficiently in the Trofile assay.
TABLE 3.
Amplification sensitivitya
| Viral load (c/ml) | No. of samples | No. amplified | % Success |
|---|---|---|---|
| <50b | 28 | 5 | 17.9 |
| 50-100 | 13 | 6 | 46.2 |
| 100-500 | 9 | 7 | 77.8 |
| 500-1,000 | 48 | 45 | 93.8 |
| 1,000-2,500 | 52 | 49 | 94.2 |
| 2,500-5,000 | 36 | 34 | 94.4 |
| 5,000-10,000 | 21 | 21 | 100.0 |
| ≥1,000 | 109 | 104 | 95.4 |
| 50-1,000 | 122 | 107 | 87.7 |
Evaluation of the amplification sensitivity involved the testing of 207 samples from the SCOPE cohort (see the text for details).
The plasma viral load of some samples in this bin may have been zero.
(iv) Linearity of results.
The linearity of the assay was demonstrated across a wide range of virus concentrations (3 to 4 log10 units) using HIV-positive plasma samples. Specifically, three high-viral-load samples (>200,000 c/ml) were tested at their original concentrations and at 10-fold and 100-fold dilutions in HIV-1-negative human plasma. Actual concentrations were determined by Amplicor Monitor version 1.5. Eight replicates at each of the three dilutions for each sample were used to infect CXCR4+/CD4+/U87 and CCR5+/CD4+/U87 cells. The virus input did not affect tropism calls; 100% of the 1,104 possible pairwise tropism determinations were concordant.
(v) Sensitivity of detection of minor variants.
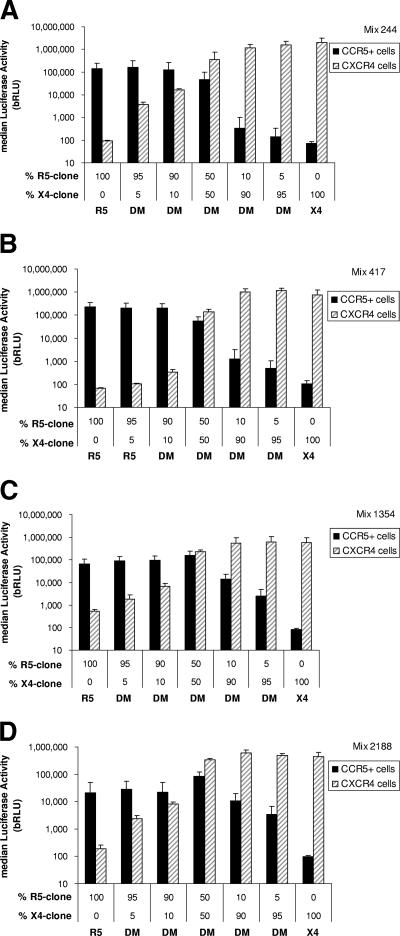
The ability of the assay to detect minor subpopulations of viruses was assessed by mixing molecular clones of X4- and R5-tropic env variants isolated from patient plasma with naturally occurring mixed-tropic HIV-1 populations. One X4- and one R5-tropic env clone were selected from each of four subjects. Clone pairs were selected based on similar infectivities (i.e., RLU) of CXCR4+/CD4+/U87 and CCR5+/CD4+/U87 cells. For each subject, the paired clones were mixed at seven R5/X4 DNA ratios of 100:0, 95:5, 90:10, 50:50, 10:90, 5:95, and 0:100. Figure 5 shows the RLU produced using each of these mixture ratios. For each patient, each mixture was tested in quadruplicate. In this investigation of four patient viral populations, the minor variant was always detectable (40/40) when present at a frequency of 10% and was detectable in 85% of the mixtures (34/40) when present at a frequency of 5% (Fig. 5).
FIG. 5.
Sensitivity to detect minor variants within mixed populations of clones derived from four patients (244, 417, 1354, and 2188). The results are displayed as median RLU from replicates (see the text for details); the error bars are 1 standard deviation from the median (see the text for details). (A) Mixture 244. Minority species of X4- and R5-tropic viruses were both detected in mixed viral stocks at the lowest frequency tested (5% of the population). (B) Mixture 417. Minority species of R5-tropic viruses were detected in mixed viral stocks at the lowest frequencies tested (5% of the population). The minority X4-tropic variant was detectable only when its frequency was greater than 10% of the total population. (C) Mixture 1354. The minority species of X4- and R5-tropic viruses were both detected in mixed viral stocks at the lowest frequency tested (5% of the population). (D) Mixture 2188. The minority species of X4- and R5-tropic viruses were both detected in mixed viral stocks at the lowest frequency tested (5% of the population).
(vi) Specificity.
Assay specificity was evaluated using two lots of HBV+ HIV− plasma and HCV+ HIV− plasma. No false-positive amplification results were observed in three replicate evaluations of each lot. Assay interference was evaluated by spiking an HIV+ plasma sample with HBV+ plasma or HCV+ plasma. No false-negative amplification results were observed in three replicate evaluations of each lot.
DISCUSSION
Assay performance.
The performance of the Trofile assay was formally validated according to regulations specified by the Clinical Laboratory Improvement Amendments under the jurisdiction of the Centers for Medicare and Medicaid Services. In addition, certifications by the College of American Pathologists and state regulatory agencies were obtained. All testing was conducted in the Monogram Biosciences Clinical Reference Laboratory by trained and certified laboratory personnel. The results of this formal validation were presented to the Food and Drug Administration (Center for Drug Evaluation and Research; Division of Antiviral Drug Products) on 1 July 2004 (drug master file 18047).
Our experiments demonstrated that Trofile is a sensitive and reproducible assay capable of measuring the coreceptor tropism of HIV-1 (i.e., R5, X4, or DM) from the vast majority of patient plasma samples with viral loads of ≥1,000 c/ml. Viral loads of <1,000 c/ml are unlikely to invoke dramatic changes in antiretroviral treatment but may warrant intensification of the existing treatment regimen. The assay can be used to assign tropisms to viral populations across diverse viral subtypes. In preliminary experiments, Trofile was capable of detecting minority species in viral populations at frequencies as low as 5 to 10%. In reality, sensitivity to detect minor species varies with relative infectivity. Specifically, detecting minority species with lower infectivities (RLU) might be more difficult in a heterogeneous population dominated by many strains of higher infectivities. Conversely, such detection may be easier if the minority species has high infectivity relative to the other strains in the population. Detection of minority variants is a limitation of all population-based assays, including all commercial and research use HIV drug resistance assays (both phenotyping and genotyping); however, this limitation has not precluded their usefulness in the clinic. For research applications, our approach does allow us to perform clonal analyses and env gene sequencing to identify low levels of particular mutants (e.g., X4 variants). However, these types of analyses are too labor-intensive and costly for routine clinical use.
Since the assay was validated, we have generated results from more than 30,000 virus stocks that are capable of infecting target cells with high efficiency. Although neither comprehensive nor random, our sampling of more than 100 DM-tropic patient virus populations to date using clonal analyses indicates that mixed virus populations comprised of R5 plus dual, R5 plus dual plus X4, or R5 plus X4 variants are relatively common (W. Huang and J. Whitcomb, unpublished observations). The assay has been used in the evaluation and clinical development of several entry inhibitors, including four that are currently in clinical trials (Table 4).
TABLE 4.
CRA trials that used the Trofile assay at patient screening or baseline and/or during treatment
| Company | Agent | Status | Trial name | Antiretroviral experience | Baseline tropism | Anti-HIV regimenb |
|---|---|---|---|---|---|---|
| GSK | Aplaviroc (APV) | Phase II complete | CCR100136 | No | R5 | LPV/r + APV |
| Phase II complete | CCR102881 | Yes | R5 | ZDV-3TC + EFV vs. ZDV-3TC + APV | ||
| Phase III on hold | CCR104627a | Yes | R5 and DM | OB vs. OB + APV | ||
| Pfizer | Maraviroc (MVC) | Phase III ongoing | A4001026 | No | R5 | 3TC-ZDV + EFV vs. 3TC-ZDV + MVC |
| A4001027c | Yes | R5 | OB vs. OB + MVC | |||
| A4001028d | Yes | R5 | OB vs. OB + MVC | |||
| A4001029 | Yes | DM | OB vs. OB + MVC | |||
| Schering-Plough | Vicriviroc (VCV) | Phase II ongoing | P03802 | No | R5 | 3TC-ZDV + EFV vs. 3TC-ZDV + VCV |
| ACTG5211 | Yes | R5 | OB + VCV | |||
| Progenics | Pro140 | Phase Ib ongoing | Pro140 1302 | No | R5 | Pro140 (monotherapy) |
| AnorMED | AMD11070 | Phase I complete | XACT | No | X4 | AMD11070 (monotherapy) |
| Phase II ongoing | ACTG5210 | No | X4 | AMD11070 (monotherapy) |
Screening study. After screening, the subjects were enrolled in phase III trials: CCR104458-R5/X4, CCR104456-R5, and CCR102709-R5.
OB, optimized background therapy; ZDV, zidovudine; 3TC, lamivudine; LPV/r, ritonavir-boosted lopinavir; EFV, efavirenz; APV, amprenavir.
United States and Canada.
Europe, Australia, and Brazil.
Trofile has several distinct advantages over coreceptor assays that rely on culturing virus with either uninfected peripheral blood mononuclear cells (PBMCs) or transformed T-cell lines. First, as Trofile determines coreceptor tropism by the expression of the complete env genes of multiple viruses isolated from patient plasma, it best approximates the viral population replicating in the patient. In contrast, traditional tropism assays use viruses derived from patient PBMCs stimulated by phytohemagglutinin or anti-CD3 antibodies in the presence of interleukin 2 (43). Such stimulation can reactivate HIV-1 genomes integrated into host DNA, some of which may not be currently replicating in the patient. Moreover, in these methods, patient samples must be passaged for prolonged periods in donor PBMCs or MT2 cells to generate sufficient virus for testing. Passaging potentially selects for viruses adapted to conditions unique to the in vitro culture systems (4, 17). Both can result in a skewed representation of an already unrepresentative viral population within a few passages. In addition to increased accuracy of the assay in representing the coreceptor tropism of the patient's current viremia, Trofile can distinguish between X4, R5, and DM viral populations. Standard coculture techniques using CXCR4+ indicator cells (e.g., MT2 cells) can only determine whether the virus is capable of infecting CXCR4-expressing cells. Consequently, such assays cannot distinguish between exclusively CXCR4-using strains and those capable of using both CXCR4 and CCR5 coreceptors (i.e., dual-tropic strains or mixed cultures).
Further, there are safety features engineered into the Trofile vector system that ensure that our recombinant-virus vectors remain replication incompetent. A self-inactivating deletion in the U3 region of the 3′ LTR (ΔU3) was introduced that substantially reduces viral-gene transcription from the 5′ LTR of an integrated provirus in infected target cells (23). In contrast, other assays do not have such features engineered into their vector systems, increasing the potential for generating replication-competent virus during the assay (17, 34, 43).
A further advantage of Trofile over other recombinant assays stems from the inclusion of entire 2.5-kb env genes from multiple plasma viruses. Previously described recombinant methods have included only ∼600 bp of the V1 to V3 regions of env (43). The V3 region is a major determinant of coreceptor specificity (8). However, the coreceptor binding site also includes discontinuous regions of gp120, collectively referred to as the “bridging sheet domain” (28-30), which is formed from conserved regions of the V1/V2 stem, as well as two strands derived from the C4 region (21). Further, regions outside of V1 to V3, and even outside gp120, can determine or contribute to coreceptor tropism, as well as entry inhibitor susceptibility (W. Huang, J. Toma, S. Fransen, N. Parkin, J. Whitcomb, and C. Petropoulos, presented at the Drug Resistance Workshop, Antiretroviral Therapy, Sitges, Spain, 2006). As the majority of genetic determinants of the HIV-1 envelope phenotype remain largely intractable due to the extreme genetic variability of the env gene, it is essential that the entire gene be included in any assays of coreceptor tropism. Such inclusion allows the elucidation of the complex molecular determinants and mechanisms of inhibition and neutralization for many CRAs, which may map to other regions of the env gene.
Trofile results can be obtained in 2 to 3 weeks, comparable with commercially available phenotypic drug resistance assays relying on similar recombinant-virus vector technologies (1, 27).
Clinical utility.
Trofile provides a quick and accurate measure of HIV-1 coreceptor tropism that can be easily applied to clinical samples. Consequently, Trofile has been used to determine patient eligibility for enrollment in several clinical trials, which include all CRAs in clinical development and several previous candidates (e.g., aplaviroc; GSK). It has also been used to monitor coreceptor switching as a consequence of treatment with these CRAs (Table 4).
Trofile represents a “pharmacogenomic” technology applied to the HIV-1 genome. Pharmacogenomics is a scientific discipline that focuses on how the genetic differences among patients can determine or predict responsiveness or adverse reactions to particular drugs. The application of pharmacogenomics increases the likelihood of success in the clinic and the marketplace. By excluding patients who are unlikely to respond to CRAs based on the coreceptor tropisms of their current viremias, the assay limits exposure to those study participants most likely to benefit from treatment. Such prescreening also minimizes the size of the trials required to achieve the overall objective of obtaining initial proof of concept or efficacy for a particular compound.
Monitoring for coreceptor switching during trials is also important, as we do not yet understand the risks of selective inhibition of coreceptor usage. Concerns have been raised that treatments targeting CCR5-mediated virus entry might induce a switch to CXCR4 coreceptor usage. As CXCR4-using viruses are associated with increased pathogenicity, drug-mediated coreceptor switching may result in accelerated disease progression. It is unclear whether the emergence of CXCR4-using viruses is a cause or a consequence of accelerated disease. Until more information is available, it will be important to closely monitor coreceptor usage during trials of new CCR5 antagonists in the setting of viral rebound. For example, R5X4- and X4-tropic viruses were detected following short-course monotherapy with maraviroc in 2/62 patients (45). Closer analyses suggested these viruses emerged from preexisting reservoirs that were not detected at enrollment. It is important to note that in some cases a “switch” in tropism may be caused by a selective suppression of the majority species by a specific CRA and subsequent “unmasking” of a minor variant. For example, for a patient with a viral load of 100,000 c/ml and a mixed viral infection in which 99.9% of the virus is R5 tropic and 0.1% is X4 tropic, successful treatment with a combination regimen that included a CCR5 antagonist would reasonably be expected to result in a 2- to 3-log-unit reduction in the viral load (to 100 to 1,000 c/ml) and a subsequent ability to detect the minor X4 variant in circulation. While this would be classified as a “switch,” we do not yet understand the clinical consequences of this type of event. Clinical trials of CCR5 inhibitors that are now being conducted should provide some insight into this question. Finally, R5-tropic viruses can become resistant to CCR5 antagonists in the absence of a switch in coreceptor use. A recent study demonstrated that an R5-tropic HIV-1 virus strain developed resistance to the CCR5 antagonists AD101 and SCH-C by acquiring the ability to use drug-bound CCR5 rather than switching to use the CXCR4 coreceptor (42). The reasons for this are unknown, although coreceptor switching does come at a fitness cost and such alternate mechanisms may have lower relative costs (25, 26). However, the resistant virus exhibited reduced infectivity and a diminished affinity for CCR5 (20, 42), also indicating a reduction in fitness. In such instances, changing therapy to include a CXCR4 inhibitor upon virologic failure of CCR5 antagonist therapy in the absence of a coreceptor switch will likely be of no therapeutic benefit, while switching to a CCR5 antagonist that retains activity likely will.
The efficacies of several coreceptor antagonists are being evaluated in ongoing clinical trials (Table 4). In each case, appropriate coreceptor tropism as specified by the Trofile assay was required for enrollment. The preliminary outcomes of two studies were recently reported at the XVIth International AIDS Conference (13 to 18 August 2006, Toronto, Canada). The interim (24-week) analysis of a phase IIb/III study evaluating maraviroc (Pfizer), a CCR5 antagonist, in subjects with dual-tropic (R5X4) or mixed-tropic (R5 plus X4) virus demonstrated that viral-load responses in subjects receiving optimized background plus maraviroc were not significantly different from those in subjects receiving optimized background plus placebo (H. Mayer, E. van der Ryst, M. S. Saag, B. Clotet, G. Fatkenheuer, N. Clumeck, K. Turner, and J. M. Goodrich, presented at the XVIth International AIDS Conference, Toronto, Canada, 2006). Conversely, the interim (24-week) analysis of a phase II study evaluating vicriviroc (Schering-Plough), a CCR5 antagonist, in subjects with R5-tropic virus infections demonstrated that subjects receiving vicriviroc plus optimized background showed significantly greater reductions in viral load than subjects receiving optimized background plus placebo (R. Gulick, Z. Su, C. Flexner, M. Hughes, P. Skolnik, C. Godfrey, W. Greaves, T. Wilkin, R. Gross, E. Coakley, A. Zolopa, M. Hirsch, and D. Kuritzkes, presented at the XVIth International AIDS Conference, Toronto, Canada, 2006). Although not definitive, these recent findings suggest that tropism determinations are likely to aid in the selection of patients most likely to respond to treatment with coreceptor antagonists. More effective treatment regimens can be designed with knowledge of coreceptor tropism in addition to susceptibility information.
Since coreceptor tropism is an important consideration for evaluating novel drugs, as well as for evaluating disease progression in patients enrolled in clinical trials, the addition of Trofile to other clinical tools available for management of HIV-1 infection permits clinicians to select more effective antiretroviral treatment regimens.
Acknowledgments
This work was funded in part by a Small Business Innovation Research Program Grant (SBIR 2 R44 AI048990) from the NIH/NIAID.
Cell lines used for infection experiments (i.e., CXCR4+/CD4+/U87 and CCR5+/CD4+/U87) were generated by Nathaniel Landau, Salk Institute, La Jolla, California. We thank Steven Deeks and Jeffrey Martin from the SCOPES study for samples and are grateful to both Merck and AnorMED for supplying compounds to complete this work. We gratefully acknowledge the help and support of both the Research and Development Department and the Clinical Reference Laboratory at Monogram Biosciences. Eoin Coakley, Neil Parkin, and Jacqueline Reeves provided thoughtful discussion and critical review of the manuscript.
Footnotes
Published ahead of print on 20 November 2006.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan, J. S., J. E. Coligan, F. Barin, M. F. McLane, J. G. Sodroski, C. A. Rosen, W. A. Haseltine, T. H. Lee, and M. Essex. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228:1091-1094. [DOI] [PubMed] [Google Scholar]
- 3.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 4.Bleul, C. C., L. Wu, J. A. Hoxie, T. A. Springer, and C. R. Mackay. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA. 94:1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briz, V., E. Poveda, and V. Soriano. 2006. HIV entry inhibitors: mechanisms of action and resistance pathways. J. Antimicrob. Chemother. 57:619-627. [DOI] [PubMed] [Google Scholar]
- 6.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 7.Cocchi, F., A. L. DeVico, A. Garzino-Demo, A. Cara, R. C. Gallo, and P. Lusso. 1996. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med. 2:1244-1247. [DOI] [PubMed] [Google Scholar]
- 8.Cormier, E. G., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cormier, E. G., and T. Dragic. 2000. An overview of HIV-1 coreceptor function and its inhibitors, p. 19-34. In C. Kuiken, F. McCutchan, B. Foley, J. W. Mellors, B. Hahn, J. Mullins, P. Marx, S. Wolinsky, and B. Korber (ed.), HIV sequence compendium 2000. Los Alamos National Laboratory, Los Alamos, NM.
- 10.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 11.Dorr, P., M. Westby, S. Dobbs, P. Griffin, B. Irvine, M. Macartney, J. Mori, G. Rickett, C. Smith-Burchnell, C. Napier, R. Webster, D. Armour, D. Price, B. Stammen, A. Wood, and M. Perros. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49:4721-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Este, J. A., C. Cabrera, E. De Clercq, S. Struyf, J. Van Damme, G. Bridger, R. T. Skerlj, M. J. Abrams, G. Henson, A. Gutierrez, B. Clotet, and D. Schols. 1999. Activity of different bicyclam derivatives against human immunodeficiency virus depends on their interaction with the CXCR4 chemokine receptor. Mol. Pharmacol. 55:67-73. [DOI] [PubMed] [Google Scholar]
- 13.Fatkenheuer, G., A. L. Pozniak, M. A. Johnson, A. Plettenberg, S. Staszewski, A. I. Hoepelman, M. S. Saag, F. D. Goebel, J. K. Rockstroh, B. J. Dezube, T. M. Jenkins, C. Medhurst, J. F. Sullivan, C. Ridgway, S. Abel, I. T. James, M. Youle, and E. van der Ryst. 2005. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat. Med. 11:1170-1172. [DOI] [PubMed] [Google Scholar]
- 14.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 15.Gelderblom, H. R., E. H. Hausmann, M. Ozel, G. Pauli, and M. A. Koch. 1987. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology 156:171-176. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman, T. L., and R. W. Doms. 1998. Chemokines and coreceptors in HIV/SIV-host interactions. AIDS 12(Suppl. A):S17-S26. [PubMed] [Google Scholar]
- 17.Japour, A. J., S. A. Fiscus, J. M. Arduino, D. L. Mayers, P. S. Reichelderfer, and D. R. Kuritzkes. 1994. Standardized microtiter assay for determination of syncytium-inducing phenotypes of clinical human immunodeficiency virus type 1 isolates. J. Clin. Microbiol. 32:2291-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jekle, A., O. T. Keppler, E. De Clercq, D. Schols, M. Weinstein, and M. A. Goldsmith. 2003. In vivo evolution of human immunodeficiency virus type 1 toward increased pathogenicity through CXCR4-mediated killing of uninfected CD4 T cells. J. Virol. 77:5846-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koot, M., I. P. Keet, A. H. Vos, R. E. de Goede, M. T. Roos, R. A. Coutinho, F. Miedema, P. T. Schellekens, and M. Tersmette. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118:681-688. [DOI] [PubMed] [Google Scholar]
- 20.Kuhmann, S. E., P. Pugach, K. J. Kunstman, J. Taylor, R. L. Stanfield, A. Snyder, J. M. Strizki, J. Riley, B. M. Baroudy, I. A. Wilson, B. T. Korber, S. M. Wolinsky, and J. P. Moore. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 78:2790-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemeshow, S., and D. W. Hosmer, Jr. 1982. A review of goodness of fit statistics for use in the development of logistic regression models. Am. J. Epidemiol. 115:92-106. [DOI] [PubMed] [Google Scholar]
- 23.Miyoshi, H., U. Blomer, M. Takahashi, F. H. Gage, and I. M. Verma. 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72:8150-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moyle, G. J., A. Wildfire, S. Mandalia, H. Mayer, J. Goodrich, J. Whitcomb, and B. G. Gazzard. 2005. Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J. Infect. Dis. 191:866-872. [DOI] [PubMed] [Google Scholar]
- 25.Pastore, C., R. Nedellec, A. Ramos, S. Pontow, L. Ratner, and D. E. Mosier. 2006. Human immunodeficiency virus type 1 coreceptor switching: V1/V2 gain-of-fitness mutations compensate for V3 loss-of-fitness mutations. J. Virol. 80:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pastore, C., A. Ramos, and D. E. Mosier. 2004. Intrinsic obstacles to human immunodeficiency virus type 1 coreceptor switching. J. Virol. 78:7565-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeves, J. D., J. L. Miamidian, M. J. Biscone, F. H. Lee, N. Ahmad, T. C. Pierson, and R. W. Doms. 2004. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J. Virol. 78:5476-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 31.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 32.Robey, W. G., B. Safai, S. Oroszlan, L. O. Arthur, M. A. Gonda, R. C. Gallo, and P. J. Fischinger. 1985. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science 228:593-595. [DOI] [PubMed] [Google Scholar]
- 33.Samson, M., O. Labbe, C. Mollereau, G. Vassart, and M. Parmentier. 1996. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry 35:3362-3367. [DOI] [PubMed] [Google Scholar]
- 34.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaheen, F., and R. G. Collman. 2004. Co-receptor antagonists as HIV-1 entry inhibitors. Curr. Opin. Infect. Dis. 17:7-16. [DOI] [PubMed] [Google Scholar]
- 36.Si, Z., N. Madani, J. M. Cox, J. J. Chruma, J. C. Klein, A. Schon, N. Phan, L. Wang, A. C. Biorn, S. Cocklin, I. Chaiken, E. Freire, A. B. Smith III, and J. G. Sodroski. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc. Natl. Acad. Sci. USA 101:5036-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speck, R. F., K. Wehrly, E. J. Platt, R. E. Atchison, I. F. Charo, D. Kabat, B. Chesebro, and M. A. Goldsmith. 1997. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J. Virol. 71:7136-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strizki, J. M., C. Tremblay, S. Xu, L. Wojcik, N. Wagner, W. Gonsiorek, R. W. Hipkin, C. C. Chou, C. Pugliese-Sivo, Y. Xiao, J. R. Tagat, K. Cox, T. Priestley, S. Sorota, W. Huang, M. Hirsch, G. R. Reyes, and B. M. Baroudy. 2005. Discovery and characterization of vicriviroc (SCH 417690), a CCR5 antagonist with potent activity against human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 49:4911-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tersmette, M., R. A. Gruters, F. de Wolf, R. E. de Goede, J. M. Lange, P. T. Schellekens, J. Goudsmit, H. G. Huisman, and F. Miedema. 1989. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J. Virol. 63:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tersmette, M., J. M. Lange, R. E. de Goede, F. de Wolf, J. K. Eeftink-Schattenkerk, P. T. Schellekens, R. A. Coutinho, J. G. Huisman, J. Goudsmit, and F. Miedema. 1989. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet i:983-985. [DOI] [PubMed] [Google Scholar]
- 41.Trkola, A., T. J. Ketas, K. A. Nagashima, L. Zhao, T. Cilliers, L. Morris, J. P. Moore, P. J. Maddon, and W. C. Olson. 2001. Potent, broad-spectrum inhibition of human immunodeficiency virus type 1 by the CCR5 monoclonal antibody PRO 140. J. Virol. 75:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trouplin, V., F. Salvatori, F. Cappello, V. Obry, A. Brelot, N. Heveker, M. Alizon, G. Scarlatti, F. Clavel, and F. Mammano. 2001. Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J. Virol. 75:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vermeire, K., D. Schols, and T. W. Bell. 2006. Inhibitors of HIV infection via the cellular CD4 receptor. Curr. Med. Chem. 13:731-743. [DOI] [PubMed] [Google Scholar]
- 45.Westby, M., M. Lewis, J. Whitcomb, M. Youle, A. L. Pozniak, I. T. James, T. M. Jenkins, M. Perros, and E. van der Ryst. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80:4909-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]